Abstract
Tetracycline-resistant (Tetr) bacteria were isolated from fishes collected at three different fish farms in the southern part of Japan in August and September 2000. Of the 66 Tetr gram-negative strains, 29 were identified as carrying tetB only. Four carried tetY, and another four carried tetD. Three strains carried tetC, two strains carried tetB and tetY, and one strain carried tetC and tetG. Sequence analyses indicated the identity in Tetr genes between the fish farm bacteria and clinical bacteria: 99.3 to 99.9% for tetB, 98.2 to 100% for tetC, 99.7 to 100% for tetD, 92.0 to 96.2% for tetG, and 97.1 to 100% for tetY. Eleven of the Tetr strains transferred Tetr genes by conjugation to Escherichia coli HB-101. All transconjugants were resistant to tetracycline, oxycycline, doxycycline, and minocycline. The donors included strains of Photobacterium, Vibrio, Pseudomonas, Alteromonas, Citrobacter, and Salmonella spp., and they transferred tetB, tetY, or tetD to the recipients. Because NaCl enhanced their growth, these Tetr strains, except for the Pseudomonas, Citrobacter, and Salmonella strains, were recognized as marine bacteria. Our results suggest that tet genes from fish farm bacteria have the same origins as those from clinical strains.
Many different kinds of antibiotics have been used as therapeutic agents in aquaculture in Japan. Intensive work was done until the 1980s to develop guidelines for antibiotic usage in fish farms. The guidelines regulated doses and required a period of drug-free rearing before sale of fish and succeeded in keeping the residual antibiotics in cultured fish to nondetectable levels. However, Samuelsen et al. (35) found that antibiotic-resistant bacteria persisted in fish farm sediments for at least 18 months after chemotherapy. Since the products of aquaculture are consumed by humans and since many antibiotic resistance determinants are encoded by transferable plasmids, cultured fish may serve as a vehicle for transmission of antibiotic resistance to bacteria that are commensal or pathogenic to humans (34).
Tetracyclines are among the therapeutic agents most commonly used in human and veterinary treatment. Oxytetracycline is permitted to be mixed with feed for fish, and food sanitation law in Japan permits certain residual levels in fish. Because of the widespread use of tetracycline, resistance to it has been disseminated to many species of marine bacteria (4, 6, 18, 42, 46).
More than 30 different kinds of tetracycline resistance determinants have been published. Resistance genes have been mainly categorized into two major groups, those responsible for proton-dependent efflux of tetracycline (24) and those conferring ribosomal protection by cytoplasmic proteins (9). Dissemination of the proton-dependent tetracycline efflux protein in aquaculture environments has been reported (5, 12, 13, 14, 22, 34, 37). Previous work has identified the relevant genes by using DNA hybridization or PCR methods (4, 12, 13, 14, 15, 16, 27, 28, 34, 37), but the nucleotide sequences of these determinants remain unknown. In the present study, we isolated numerous Tetr gram-negative fish farm bacteria and determined the DNA sequences of the Tetr genes. We found that these genes were identical to the Tetr genes identified in clinical isolates and that some were transferable to a laboratory Escherichia coli strain.
MATERIALS AND METHODS
Isolation of bacteria resistant to tetracycline.
Bacteria resistant to tetracycline were isolated from fishes collected at three different fish farms (A, B, and C) in August and September 2000. Fish farm A cultured yellowtail amberjack (Seriola lalandi) and northern bluefin tuna (Thunnus thynnus) and was located on a coastline facing the Sea of Japan. The culture area is encircled by a breakwater, and no river is present. Fish farm B cultured 1- and 2-year-old yellowtails (Seriola quinqueradiata) in a bay with a depth of 100 m. Fish farm C cultured greater amberjack (Seriola dumerili) and yellowtail amberjack (Seriola lalandi) in a coastal area of the Seto Inland Sea.
Fishes ranging from 4 to 6 kg were collected and were immediately processed in coastal facilities. About 1 cm2 of fish skin, 1 g of flesh, and the contents of the rectum were homogenized in 9 ml of sterilized artificial seawater (ASW; 0.3 M NaCl, 0.02 M MgSO4, 0.01 M CaCl2, 0.01 M KCl), and then 0.1 ml of a decimal dilution was spread on modified ZoBell 2216E agar plate medium (47) in which 0.5% polypeptone and 0.1% yeast extract were dissolved in 1,000 ml of ASW. After incubation for 3 days at 25°C, the colonies were counted, and randomly selected colonies were purified with the modified ZoBell 2216E plate. These purified colonies were examined for Gram reaction by the KOH method (33). The gram-negative strains were transferred with a platinum needle to Mueller-Hinton agar plates (Becton Dickinson Microbiology Systems, Sparks, Md.) prepared with a solution containing 1% NaCl and 32 μg of tetracycline ml−1. The bacterial colonies grown on the Mueller-Hinton plates after incubation for 3 days at 25°C were considered to be tetracycline resistant.
Seawater was also collected by using a sterilized Hyroth water sampler (Shibata, Tokyo, Japan) to determine viable counts of heterotrophic bacteria.
Antibacterial susceptibility test.
Antibacterial susceptibility was examined according to a standard method recommended by the National Committee for Clinical Laboratory Standards (30), with Mueller-Hinton II agar (Becton Dickinson Microbiology Systems) and susceptibility test disks (Becton Dickinson Microbiology Systems) that contained ampicillin (10 μg), kanamycin (30 μg), tetracycline (30 μg), chloramphenicol (30 μg), oxytetracycline (30 μg), doxycycline (30 μg), and minocycline (30 μg).
Identification of Tetr strains.
Tetr bacteria were identified on the basis of the 16S rRNA gene sequence. Isolated bacterial colonies were suspended in 100 μl of sterilized distilled water. The suspensions were boiled and centrifuged, and the supernatants were used as template DNA for PCR. The 16S rRNA gene was amplified by PCR with forward primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and reverse primer 1492R (5′-TACGGYTACCTTGTTACGACTT-3′), which are located in E. coli at nucleotide positions 8 to 27 and 1492 to 1512, respectively (8). A 100-μl portion of the reaction mixture contained 1.5 U of Taq polymerase (Takara, Kyoto, Japan), 20 nmol of each deoxynucleoside triphosphate, 0.1 μl of the template DNA, 100 pmol of each primer, and 10 μl of the reaction buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2). The PCR amplification included 30 thermal cycles of 60 s at 94°C, 45 s at 60°C, and 90 s at 72°C, with an additional extension of 300 s at 72°C.
The amplified 16S rRNA gene was purified by agarose gel electrophoresis. The partial 479-bp nucleotide sequences of the PCR products were determined with forward primer 27F and reverse primer 517R (5′-GTATTACCGCGGCTGCTGGC-3′). The Tetr bacteria were identified by referring to the most similar sequence found in the Ribosomal Database Project database (26).
The ability of an isolated strain to produce acid was examined on glucose with Hugh-Leifson medium (20). Oxidase production was examined by using a cytochrome oxidase test strip (Nissui, Tokyo, Japan). Motility was examined with hanging-drop preparations under a phase-contrast microscope. Catalase production was examined with 3% H2O2 solution by the method of Taylor and Achanzar (44).
Identification of Tetr genes.
Tetracycline efflux genes were amplified by PCR with the primers listed in Table 1. According to structural identities, the genes were clustered into four groups: (i) tetA and tetC, (ii) tetB and tetD, (iii) tetG and tetY, and (iv) tetE, tetH, and tetJ. Four different degenerate primer pairs, tetAC-F and tetAC-R for tetA and tetC, tetBDEHJ-F and tetBD-R for tetB and tetD, tetGY-F and tetGY-R for tetG and tetY, and tetBDEHJ-F and tetEHJ-R for tetE, tetH, and tetI, were designed based on regions showing high homology in each group. The predicted sizes of the PCR-amplified products were 417 bp for the tetA and tetC product, 967 or 964 bp for the tetB and tetD product, 917 or 911 bp for the tetG and tetY product, and 650 bp for tetE, tetH, and tetJ product (Table 1). GenBank accession numbers of the tet genes referred to in this experiment are summarized in Table 2.
TABLE 1.
PCR primer sets and restriction enzymes used for the detection of tetracycline efflux genes
| Target gene | Primersa (sequences) | Predicted amplified fragment size (bp) | Restriction enzyme | Restriction fragment sizes (bp) |
|---|---|---|---|---|
| tetA | tetAC-F (5′-CGCYTATATYGCCGAYATCAC-3′), tetAC-R (5′CCRAAWKCGGCWAGCGA-3′) | 417 | SmaI | 267, 150 |
| tetC | 417 | SalI | 216, 201 | |
| tetB | tetBDEFHJ-F (5′-GGDATTGGBCTTATYATGCC-3′), tetBD-R (5′-ATMACKCCCTGYAATGCA-3′) | 967 | SphI | 173, 302, 492 |
| tetD | 964 | SphI | 324, 640 | |
| tetG | tetGY-F (3′-TATGCRTTKATGCAGGTC-5′), tetGY-R (5′-GACRAKCCAAACCCAACC-3′) | 917 | EcoRI | 368, 549 |
| tetY | 911 | SphI | 197, 714 | |
| tetE | tetBDEFHJ-F, tetEHJ-R (5′-AWDGTGGCDGGAATTTG-3′) | 650 | NdeII | 77, 148, 425 |
| tetH | 650 | NdeII | 267, 383 | |
| tetJ | 650 | NdeII | 117, 297, 236 |
F, forward; R, reverse.
TABLE 2.
Sequence data used in this study and sequence identity of tet genes
| Gene | Element | Source | Accession no. | Reference(s)a | Identityb (%) |
|---|---|---|---|---|---|
| tetB | Tn10 | E. coli | J01830 | 10, 19 | 99.3-99.6 |
| pGBG1 | E. coli | AJ277653 | 39 | 99.3-99.6 | |
| pHCM1 | Salmonella serovar Typhi CT18 | AL513383 | 32 | 99.5-99.8 | |
| Shigella flexneri 2a | AF326777 | 25 | 99.5-99.8 | ||
| R100 | Shigella flexneri 2b | AP000342 | UP (SM) | 99.6-99.9 | |
| pHA162 | Vector | AY048738 | 17 | 99.3-98.9 | |
| tetC | pRAS3.2 | Aeromonas salmonicida | AY043299 | 23 | 100 |
| pSC101 | Salmonella serovar Typhimurium | X01654 | 7 | 99.9 | |
| pBR329 | Cloning vector | L08859 | UP (G) | 99.8 | |
| Lambda TXF97 | Cloning vector | U37692 | 43 | 99.8 | |
| pMTet1 | Positive selection vector | AF438204 | 29 | 99.7 | |
| pUCP26 | Cloning vector | U07168 | 40 | 99.7 | |
| Mini-CTX-lacZ | Integration vector | AF140579 | UP (HKBS) | 99.6 | |
| pBSL193 | Cloning vector | U35135 | 2 | 99.6 | |
| pALTER<R>-Ex2 | Cloning vector | U47103 | UP (M) | 99.7 | |
| pRAS3.1 | Aeromonas salmonicida | AY043298 | 23 | 98.2 | |
| tetD | pRA1 | Aeromonas sp. | L06798 | 45 | 99.9-100 |
| Shigella sonnei 119 | AF467074 | UP (HEIL) | 99.9-100 | ||
| Shigella flexneri Gibbon32055 | AF467077 | UP (HEIL) | 99.9-100 | ||
| Salmonella Ordnez | X65876 | 3 | 99.7-99.8 | ||
| tetG | Salmonella serovar Typhimurium DT104 | AF071555 | 31 | 96.2 | |
| pSTG2 | Pseudomonas sp. | AF133140 | 38 | 96.2 | |
| pJA8122 | Vibrio anguillarum | S52437 | 46 | 96.0 | |
| pSTG1 | Pseudomonas sp. | AF133139 | 38 | 92.0 | |
| tetY | pIE1120 | IncQ-like plasmid | AF070999 | 41 | 97.1-100 |
UP, unpublished data; SM, G. Sampei and K. Mizobuchi; G, W. Gilbert; HKBS, T. T. Hoang, A. J. Kutchma, A. Becher, and H. P. Schweizer; M. J. Miles; HEIL, A. B. Hartman, I. I. Essiet, D. W. Isenbarger, and L. E. Lindler.
Sequence identity to the tet genes isolated from fish farm bacteria.
The PCR protocol was basically the same as that used for the 16S rRNA gene, but the annealing temperature of the PCR was set at 50°C, except for the tetAC primer pair, where it was set at 55°C. The PCR-amplified products were analyzed by restriction fragment length polymorphism (RFLP) with the restriction enzymes listed in Table 1.
Sequence analysis of Tetr genes.
For determination of entire nucleotide sequences, tetBCDGY were amplified by PCR with the primers listed in Table 3.
TABLE 3.
PCR primer sets for determination of nucleotide sequences of tetracycline efflux genes
| Target gene | Primersa (sequences [5′-3′]) | Amplified fragment size (bp) | Reported size of tet gene (bp) |
|---|---|---|---|
| tetB | tetB-F (CTAATCTAGACATCATTAATTCC), tetB-R (TTTGAAGCTAAATCTTCTTTAT) | 1,397 | 1,206 |
| tetC | tetC-F (ATGAAATCTAACAATGCGC), tetC-R (TCAGGTCGAGGTGGCCCGG) | 1,191 | 1,191 |
| tetD | tetD-F (ATGAATAAACCCGCTGTCATCGC), tetD-R (AATGCCGTCCACCATACC) | 1,232 | 1,185 |
| tetG | tetG-F (AGTTTCAGGTGCGCAGC), tetG-R (CCAAWTCGCCATGACTMAAT) | 1,232 | 1,176 |
| tetY | tetY-F (ATGTCAAAATCACTTATAACCGC), tetY-R (TCTCCCGCCAAGATTTTA) | 1,191 | 1,176 |
F, forward; R, reverse. Amplified fragments included the full lengths of tet genes, and some also included up- and/or downstream regions.
The PCR protocol was basically the same as that for the 16S rRNA gene, but the annealing temperature was set at 50°C for tetB and tetC and at 55°C for tetD, tetG, and tetY. Nucleotide sequences of the PCR products were determined with a dye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.).
Conjugation.
The ability of bacteria to transfer the Tetr gene to E. coli HB-101 was examined by a conjugation experiment using a filter mating method (36). The Tetr strains were grown on tryptic soy broth (Difco, Detroit, Mich.) supplemented with 1% NaCl-32 μg of tetracycline ml−1 and incubated at 25°C. E. coli HB-101 was grown on Luria-Bertani (LB) medium and incubated at 37°C. Exponential-growth phase cultures of the Tetr bacteria and the recipient were mixed in 2 ml of tryptic soy broth supplemented with 1% NaCl. Cell densities of both the Tetr bacteria and the recipient were on the order of 108 cells ml−1. The mixed cell suspension was collected on a 0.45-μm-pore-size HA membrane filter (Millipore). The filter was then transferred onto a tryptic soy agar (Difco) plate supplemented with 1% NaCl. After incubation for 24 h at 25°C, the filter was transferred into 2 ml of A-14 buffer (39.3 mM Na2HPO4, 22.0 mM KH2PO4, 68.5 mM NaCl, 0.8 mM MgSO4, pH 7.2) and stirred. The 100 μl of the appropriate dilution was spread on an LB plate supplemented with 200 μg of streptomycin ml−1, 20 μg of tetracycline ml−1, 25 μl of 4% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and 0.1 M IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated at 40°C, at which the growth of the donor bacteria was suppressed.
Isolation of plasmids.
The extraction of large plasmids was performed according to the method of Kado and Liu (21). The large plasmids were confirmed by an agarose gel electrophoresis.
Growth in the presence of Na+.
The Tetr strains were grown on tryptic soy agar supplemented with 1% NaCl. Bacterial colonies were suspended in saline solution, and the turbidity of each cell suspension was adjusted to that of the McFarland 0.5 barium sulfate standard. One loopful of the cell suspension was transferred to Mueller-Hinton agar with or without NaCl (1%). After incubation for 10 to 18 h at 25°C, bacteria that showed faster growth in the presence of NaCl were considered to be marine bacteria.
Nucleotide sequence accession numbers.
The tetracycline resistance gene sequences determined in this study were submitted to the GenBank and have been given accession numbers AB089585 to AB089608.
RESULTS
Distribution of Tetr genes.
Bacteria resistant to tetracycline were isolated from fishes collected at three different fish farms (A, B, and C) located in the southern part of Japan. The viable counts of heterotrophic bacteria in seawater of the culture area were on the order of 102 ml−1 for fish farms A and B and 103 ml−1 for fish farm C.
We randomly isolated 57 gram-negative strains from the fishes at fish farm A, 90 from farm B fishes, and 218 from farm C fishes and examined susceptibility to tetracycline. Of these strains, 21 in fish farm A, 6 in farm B, and 39 in farm C were resistant to tetracycline. The total number of Tetr bacterial strains was 66.
Tetr genes of the strains were amplified by PCR with four different sets of primers targeting tetracycline efflux genes (Table 1). PCR products were obtained for 51 Tetr strains. Restriction fragment patterns of the PCR products of 43 Tetr strains were identical to the patterns of the tetracycline efflux genes tetB, tetC, tetD, tetG, and tetY, while the patterns of the other 8 Tetr strains were not identical to those of any of the genes. No PCR product was obtained for the other 15 strains.
Table 4 summarizes the characteristics of the 43 Tetr strains. The tetB gene was found in 31 strains of the Tetr bacteria. All of the tetB strains were motile rods and oxidase positive. These strains showed fermentative metabolism of glucose in Hugh-Leifson medium, while only TC69 showed oxidative metabolism. The tetC gene was found in four strains, which all showed oxidative reactions in the Hugh-Leifson medium, except for TA80, which was unable to grow in the medium. The tetD gene was found in three fermentative strains and one oxidative strain. The tetY gene was found in six strains, four fermentative strains and two strains that showed no color change in Hugh-Leifson medium. Strain TA57 was identified as carrying two different genes, tetC and tetG, while strains TC33 and TC34 carried both tetB and tetY.
TABLE 4.
Tetracycline resistance gene and general characteristicsa of the tetracycline-resistant strains
| Strain(s) | Fish farm | Origin | Gene type(s)b | Metabolism | Oxidase | Motility | Catalase |
|---|---|---|---|---|---|---|---|
| TA5, TA45 | A | Flesh | B | F | + | + | + |
| TA49, TA51 | A | Rectum | B | F | + | + | + |
| TB61 | B | Rectum | B | F | + | + | + |
| TC16-23, TC65, TC74 | C | Rectum | B | F | + | + | + |
| TC26-31, TC35-38 | C | Flesh | B | F | + | + | + |
| TC68, TC71, TC76 | C | Skin | B | F | + | + | + |
| TC69 | C | Flesh | B | O | + | + | + |
| TC33, TC34 | C | Flesh | B, Y | F | + | + | + |
| TC32 | C | Flesh | Y | F | + | + | + |
| TC75 | C | Skin | Y | F | + | + | + |
| TC72, TC73 | C | Flesh | Y | + | − | + | |
| TA3 | A | Skin | D | F | − | + | + |
| TA6 | A | Flesh | D | F | − | + | + |
| TA55 | A | Skin | D | O | + | + | + |
| TC67 | C | Rectum | D | F | − | + | + |
| TA58 | A | Flesh | C | O | + | − | + |
| TA59 | A | Flesh | C | O | − | + | + |
| TA80 | A | Flesh | C | NO | + | + | + |
| TA57 | A | Flesh | C, G | O | − | + | + |
+, positive; −, negative; F, fermentative; O, oxidative; NO, no growth.
B, tetB; Y, tetY; D, tetD; C, tetC; G, tetG.
Sequences of Tetr genes.
To examine the identity of genes, entire DNA sequences of 12 tetB genes (including those of oxidative strains TC33 and TC34), three tetC genes (those of TA57 and two others), four tetD genes, one tetG gene, and five tetY genes were examined. The identity between sequences of tet genes isolated from fish farm bacteria and published sequences is summarized in Table 2. The sequences of tetB genes from the fish farm bacteria were 99.3 to 99.9% identical to the sequences reported for clinical bacteria (10, 19). There was 100% identity in tetB between one fish farm B strain of bacteria (TB61) and four strains of fermentative fish farm C bacteria. The tetC nucleotide sequences of the three fish farm strains were 98.2 to 100% identical to the sequences found in Aeromonas salmonicida (23) and Salmonella enterica serovar Typhimurium (7). The tetD sequences were 99.7 to 100% identical to those of Shigella sonnei (AF467074), Shigella flexneri (AF467077), and pRA1 harbored by Aeromonas (45). The tetG nucleotide sequence of TA57 was 92.0 to 96.2% identical to the sequences reported for Salmonella serovar Typhimurium DT104 (31), pSTG2 and pSTG1 of Pseudomonas spp. (38), and pJA8122 of Vibrio anguillarum (46). Identity of the tetY genes to the sequence of IncQ-like plasmid pIE1120 (41) ranged from 97.1 to 100%.
Sequences for the PCR products of the eight strains whose tet gene restriction fragment patterns showed no identity to those of known tet genes by PCR-RFLP were also determined; the sequences did not show any identity to those of the known tet genes.
Conjugation.
The transferability of Tetr determinants was examined for TC43. As summarized in Table 5, 11 strains were able to transfer their tetracycline resistance to E. coli HB-101 in conjugation experiments. According to analyses of the 16S rRNA gene, these 11 included 2 strains of a Photobacterium sp. that carried tetB, 2 strains of a Photobacterium sp. that carried both tetB and tetY, and 2 strains of a Citrobacter sp. that carried tetD. The other strains included one each of a Vibrio sp. (tetB), a Pseudomonas sp. (tetB), a Photobacterium sp. (tetY), an Alteromonas sp. (tetD), and a Salmonella sp. (tetD). There was no difference in the PCR-RFLPs of Tetr genes between the nine transconjugants and their donor strains. Although Photobacterium sp. strains TC33 and TC34 were identified as carrying both tetB and tetY, only the tetY gene was transferred to the recipient.
TABLE 5.
Results of conjugation experiment
| Donor strain | Genus | Gene(s)a | Transconjugant strain | Transferred genea |
|---|---|---|---|---|
| TA51 | Photobacterium | B | E-TA51 | B |
| TC21 | Photobacterium | B | E-TC21 | B |
| TC68 | Vibrio | B | E-TC68 | B |
| TC69 | Pseudomonas | B | E-TC69 | B |
| TC33 | Photobacterium | B, Y | E-TC33 | Y |
| TC34 | Photobacterium | B, Y | E-TC34 | Y |
| TC32 | Photobacterium | Y | E-TC32 | Y |
| TA3 | Citrobacter | D | E-TA3 | D |
| TA6 | Citrobacter | D | E-TA6 | D |
| TA55 | Alteromonas | D | E-TA55 | D |
| TC67 | Salmonella | D | E-TC67 | D |
B, tetB; Y, tetY; D, tetD.
The antibacterial susceptibilities of the donor and the transconjugant strains were determined by a disk diffusion method. As shown in Table 6, all the donor strains were resistant to tetracycline and oxytetracycline but susceptible to minocycline except for Vibrio (TC68) and Citrobacter (TA3 and TA6) strains. Strains of Photobacterium (TA51, TC21, TC32, TC33, and TC34), Vibrio (TC68), Citrobacter (TA3 and TA6), and Salmonella (TC67) showed resistance to doxycycline, while an Alteromonas strain (TA55) was susceptible.
TABLE 6.
Antimicrobial susceptibilitya of donor strains
| Strain | Genus | Gene(s)b | Susceptibility to:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Tetracycline
|
ABPC | KM | CM | ||||||
| TC | OXY | DOXY | MINO | ||||||
| TA51 | Photobacterium | B | R | R | R | I | R | S | R |
| TC21 | Photobacterium | B | R | R | R | S | R | S | R |
| TC68 | Vibrio | B | R | R | R | R | R | S | R |
| TC69 | Pseudomonas | B | R | R | I | S | S | S | R |
| TC33 | Photobacterium | B, Y | R | R | R | S | R | S | S |
| TC34 | Photobacterium | B, Y | R | R | R | S | R | S | S |
| TC32 | Photobacterium | Y | R | R | R | S | R | S | S |
| TA3 | Citrobacter | D | R | R | R | R | S | R | R |
| TA6 | Citrobacter | D | R | R | R | R | S | R | R |
| TA55 | Alteromonas | D | R | R | S | S | S | R | I |
| TC67 | Salmonella | D | R | R | R | I | S | S | R |
Susceptibility to antibiotics was identified as resistant (R), intermediate (I), or sensitive (S), according to guidelines of the National Committee for Clinical Laboratory Standards (30). Abbreviations for antibiotics: TC, tetracycline; OXY, oxytetracycline; DOXY, doxycycline; MINO, minocycline; ABPC, ampicillin; KM, kanamycin; CM, chloramphenicol.
B, tetB; Y, tetY; D, tetD.
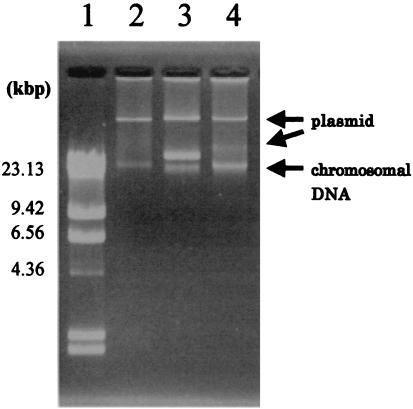
All of the transconjugants were resistant to tetracycline and oxytetracycline (Table 7). Although the Alteromonas strain (TA55) was susceptible to doxycycline and minocycline, the recipient (E-TA55) became resistant to these antibiotics after conjugation. The recipient of tetY also became resistant to chloramphenicol, whereas the donor was susceptible. Interestingly, some of the transconjugants showed different susceptibilities to ampicillin from those of the donor strains. All transconjugants acquired one or two large plasmids (Fig. 1).
TABLE 7.
Antimicrobial susceptibility of transconjugant strains
| Strain | Geneb | Susceptibilitya to:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Tetracycline
|
ABPC | KM | CM | |||||
| TC | OXY | DOXY | MINO | |||||
| E-TA51 | B | R | R | R | R | S* | S | R |
| E-TC21 | B | R | R | I | I | R | S | R |
| E-TC68 | B | R | R | R | R | S* | S | R |
| E-TC69 | B | R | R | R | R* | S | S | R |
| E-TC33 | Y | R | R | R | I | R | S | R* |
| E-TC34 | Y | R | R | R | I | R | S | R* |
| E-TC32 | Y | R | R | R | I | R | S | R* |
| E-TA3 | D | R | R | R | R | S | R | R |
| E-TA6 | D | R | R | R | R | S | R | R |
| E-TA55 | D | R | R | R* | R* | S | R | R |
| E-TC67 | D | R | R | R | R | S | S | R |
| HB-101 | S | S | S | S | S | S | S | |
All abbreviations are as defined for Table 6. *, susceptibility different from that of the donor strain.
B, tetB; Y, tetY; D, tetD.
FIG. 1.
Gel electrophoresis of large plasmids of a transconjugant strain. Lane 1, λHindIII, DNA size marker; lane 2, E-TC68; lane 3, E-TA3; lane 4, E-TC33.
DISCUSSION
From the fishes collected at the three fish farms, 66 gram-negative Tetr bacterial strains were isolated. The percentages of resistant strains were 36.8% for fish farm A, 6.7% for fish farm B, and 17.9% for fish farm C. These values may be affected by the history of antibiotic usage at each fish farm, but no reliable data on antibiotic use were obtained from the fish farmers. However, we note that fish farm B was located in far deeper water (100 m) than fish farms A and C.
The tet genes of 43 Tetr gram-negative strains of bacteria were identified. Thirty-one contained tetB, and the other 12 contained tetC, tetD, tetG, or tetY. The genes of 23 Tetr strains were not identified. The prevalence of tetB in the fish farms was higher than that previously reported for tetB in clinical strains (15, 27, 28) or in phylloplane bacteria in an apple orchard (38). In place of tetB, tetA and tetE have been reported as prevalent in fish farms and marine environments (1, 4, 12, 13, 14, 34, 37). Although neither tetA nor tetE was amplified by PCR in this experiment, the number of the strains that produced no PCR product was only 15 and did not exceed the number of tetB strains.
The nucleotide sequences of tetB from fish farm bacteria were very similar to those reported for clinical strains, but the identity to the tetB sequence of Shigella flexneri was even higher. High identity to clinical strains was also found for tetC, tetD, and tetY from fish farm bacteria. The sequences of tetC were 100% identical to those of pRAS3.2 of clinical strains. The sequences of tetD of TC67 and tetY of both TC72 and TC73 were 100% identical to the sequences from Shigella spp. and pIE1120, respectively. These high identities suggest that tet genes from fish farm bacteria have the same origins as those from clinical strains.
tetG has been found previously in Vibrio spp. (46), Pseudomonas spp. (38), and Salmonella spp. (31). Since the tetG identified here has only 92.0 to 96.2% identity to the previously identified tetG, the gene may be a new tetG variant.
In the present study, the tet genes of 11 strains of fish farm bacteria were transferred to E. coli in conjugation experiments. The 11 strains of fish farm bacteria included strains of Photobacterium, Vibrio, Alteromonas, and Pseudomonas. The transferred genes were tetB, tetD, and tetY. Higher growth rates in the presence of Na+ suggested that the strains of Photobacterium, Vibrio, and Alteromonas are marine bacteria. Hence we suggest that the Tetr gene of marine bacteria can be transferred to human intestinal bacteria, if the marine Tetr bacteria are ingested together with fish flesh. Since many human commensal bacteria carry the same tet genes, plasmids, transposons, and integrons as disease-causing bacterial species (11), our results suggest that marine Tetr bacteria can also transfer tet genes to disease-causing bacteria.
In the conjugation experiments, we observed interesting phenomena. Although Photobacterium sp. strains TC33 and TC34 carried both tetB and tetY, only tetY was transferred to the recipient. The difference in genotype is possible if each determinant was loaded on a different vehicle. Although no published work provides evidence that this process occurs, Smalla et al. (41) reported that the IncQ-like plasmid pIE1120 carries tetY but not tetB. Another interesting phenomenon is the difference in resistance between Alteromonas sp. strain TA55 and its transconjugant, although no difference in the type of tet gene was found. An alternate explanation may be that expression of antibiotic resistance may vary among different host strains. Phenotypic differences reported by Sandaa et al. (36) include the appearance of novobiocin resistance after conjugation between fish farm bacteria and E. coli HB-101.
In this study, we found that each of three Photobacterium strains carried two different Tetr genes, both tetB and tetY or both tetC and tetG (Table 4). These bacteria accounted for about 4.5% of all Tetr strains. This value was higher than those previously reported for lactose-fermenting coliforms (27), Salmonella serovar Typhimurium (28), Salmonella serovar Hadar (16), Aeromonas, and Enterobacteriaceae isolated from catfish, catfish ponds, and rainbow trout farms (12, 13, 14, 37). Although environmental factors have been reported to affect the number of resistance genes carried by a single cell (11), the difference is more likely to be caused by a difference in the number of PCR primers utilized. Previous studies have examined only two to five resistance determinants, while nine different resistance genes were examined in this study.
Although the resistance caused by the tetY gene has not yet been well characterized, we found that transconjugants with the tetY gene became resistant to all tetracyclines. Hence, tetY may cause problems in future clinical treatment.
Acknowledgments
We express sincere thanks to H. Ishizuka, A. Okamoto, and M. Ozawa for helpful technical assistance.
REFERENCES
- 1.Adams, C. A., B. Austin, P. G. Meaden, and D. McIntosh. 1998. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl. Environ. Microbiol. 64:4194-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160:63-67. [DOI] [PubMed] [Google Scholar]
- 3.Allard, J. D., M. L. Gibson, L. H. Vu, T. T. Nguyen, and K. P. Bertrand. 1993. Nucleotide sequence of class D tetracycline resistance genes from Salmonella ordonez. Mol. Gen. Genet. 237:301-305. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, S., and R. Sandaa. 1994. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl. Environ. Microbiol. 60:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoki, T., T. Satoh, and T. Kitano. 1987. New tetracycline resistance determinant on R plasmids from Vibrio anguillarum. Antimicrob. Agents Chemother. 31:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai, T., H. Hamashima, and H. Hasegawa. 1985. Isolation of a new drug-resistance plasmid from a strain of Vibrio parahaemolyticus. Microbiol. Immunol. 29:103-112. [DOI] [PubMed] [Google Scholar]
- 7.Bernardi, A., and F. Bernardi. 1984. Complete sequence of pSC101. Nucleic Acids Res. 12:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius, J., M. L. Poindexter, J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdett, V. 1986. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J. Bacteriol. 165:564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalmers, R., S. Sewitz, K. Lipkow, and P. Crellin. 2000. Complete nucleotide sequence of Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePaola, A., W. E. Hill, and F. M. Harrell. 1993. Oligonucleotide probe determination of tetracycline-resistant bacteria isolated from catfish ponds. Mol. Cell. Probes 7:345-348. [DOI] [PubMed] [Google Scholar]
- 13.DePaola, A., and M. C. Roberts. 1995. Class D and E tetracycline resistance determinants in gram-negative bacteria from catfish ponds. Mol. Cell. Probes 9:311-313. [DOI] [PubMed] [Google Scholar]
- 14.DePaola, A., P. F. Flynn, R. M. McPhearson, and S. B. Levy. 1988. Phenotypic and genotypic characterization of tetracycline- and oxytetracycline-resistant Aeromonas hydrophila from cultured channel catfish (Ictalurus punctatus) and their environments. Appl. Environ. Microbiol. 54:112-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guardabassi, L., L. Dijkshoorn, J. M. Collard, J. E. Olsen, and A. Dalsgaard. 2000. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49:929-936. [DOI] [PubMed] [Google Scholar]
- 16.Guillaume, G., D. Verbrugge, M. Chasseur-Libotte, W. Moens, and J. Collard. 2000. PCR typing of tetracycline resistance determinants (Tet A-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 32:77-85. [DOI] [PubMed] [Google Scholar]
- 17.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa, H., H. Hamashima, and T. Arai. 1986. Mechanisms of plasmid-mediated antibiotic resistances in Vibrio parahaemolyticus. Microbiol. Immunol. 30:437-444. [DOI] [PubMed] [Google Scholar]
- 19.Hillen, W., and K. Schollmeier. 1983. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 11:525-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugh, R., and E. Leifson. 1953. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram-negative bacteria. J. Bacteriol. 66:24-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, E. H., and T. Aoki. 1994. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microbiol. Immunol. 38:31-38. [DOI] [PubMed] [Google Scholar]
- 23.L'Abee-Lund, T. M., and H. Sorum. 2002. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172-181. [DOI] [PubMed] [Google Scholar]
- 24.Levy, S. B., and L. McMurry. 1978. Plasmid-determined tetracycline resistance involves new transport systems for tetracycline. Nature 276:90-92. [DOI] [PubMed] [Google Scholar]
- 25.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, B., C. Tachibana, and S. B. Levy. 1983. Frequency of tetracycline-resistant determinant classes among lactose-fermenting coliforms. Antimicrob. Agents Chemother. 24:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Salazar, J. M., G. Alvarez, and M. C. Gómez-Eichelmann. 1986. Frequency of four classes of tetracycline resistance determinants in Salmonella and Shigella spp. clinical isolates. Antimicrob. Agents Chemother. 30:630-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matin, M. M., and D. P. Hornby. 2000. A positive selection vector combining tetracycline resistance that eliminates the need for bacterial plating. Anal. Biochem. 278:46-51. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk susceptibility tests. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 31.Ng, L. K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 33.Powers, E. M. 1995. Efficacy of the Ryu nonstaining KOH technique for rapidly determining Gram reactions of food-borne and waterborne bacteria and yeasts. Appl. Environ. Microbiol. 61:3756-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes, G., G. Huys, J. Swings, P. McGann, M. Hiney, P. Smith, and R. W. Pickup. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tetA. Appl. Environ. Microbiol. 66:3883-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuelsen, O. B., V. Torsvik, and A. Ervik. 1992. Long-range changes in oxytetracycline concentration and bacterial resistance toward oxytetracycline in a fish farm sediment after medication. Sci. Total Environ. 114:25-36. [DOI] [PubMed] [Google Scholar]
- 36.Sandaa, R. A., V. L. Torsvik, and J. Goskoyr. 1992. Transferable drug resistance in bacteria from fish-farm sediment. J. Can. Microbiol. 38:1061-1065. [Google Scholar]
- 37.Schmidt, A. S., M. S. Bruun, I. Dalsgaard, and J. L. Larsen. 2001. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67:5675-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, D., D. Faure, M. Noirclerc-Savoye, A. C. Barriere, E. Coursange, and M. Blot. 2000. A broad-host-range plasmid for isolating mobile genetic elements in gram-negative bacteria. Plasmid 44:201-207. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 41.Smalla, K., H. Heuer, A. Götz, D. Niemeyer, E. Krögerrecklenfort, and E. Tietze. 2000. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorum, H., M. C. Roberts, and J. H. Crosa. 1992. Identification and cloning of a tetracycline resistance gene from the fish pathogen Vibrio salmonicida. Antimicrob. Agents Chemother. 36:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St Pierre, R., and T. Linn. 1996. A refined vector system for the in vitro construction of single-copy transcriptional or translational fusions to lacZ. Gene 169:65-68. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, W. I., and D. Achanzar. 1972. Catalase test as an aid in the identification of Enterobacteriaceae. Appl. Microbiol. 24:58-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varela, M. F., and J. K. Griffith. 1993. Nucleotide and deduced protein sequences of the class D tetracycline resistance determinant: relationship to other antimicrobial transport proteins. Antimicrob. Agents Chemother. 37:1253-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao, J., and T. Aoki. 1992. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol. Immunol. 36:1051-1060. [DOI] [PubMed] [Google Scholar]
- 47.ZoBell, C. E. 1941. Studies on marine bacteria. Int. J. Mar. Res. 4:42-75. [Google Scholar]