Abstract
Sulfate-reducing bacteria (SRB) in anoxic waters and sediments are the major producers of methylmercury in aquatic systems. Although a considerable amount of work has addressed the environmental factors that control methylmercury formation and the conditions that control bioavailability of inorganic mercury to SRB, little work has been undertaken analyzing the biochemical mechanism of methylmercury production. The acetyl-coenzyme A (CoA) pathway has been implicated as being key to mercury methylation in one SRB strain, Desulfovibrio desulfuricans LS, but this result has not been extended to other SRB species. To probe whether the acetyl-CoA pathway is the controlling biochemical process for methylmercury production in SRB, five incomplete-oxidizing SRB strains and two Desulfobacter strains that do not use the acetyl-CoA pathway for major carbon metabolism were assayed for methylmercury formation and acetyl-CoA pathway enzyme activities. Three of the SRB strains were also incubated with chloroform to inhibit the acetyl-CoA pathway. So far, all species that have been found to have acetyl-CoA activity are complete oxidizers that require the acetyl-CoA pathway for basic metabolism, as well as methylate mercury. Chloroform inhibits Hg methylation in these species either by blocking the methylating enzyme or by indirect effects on metabolism and growth. However, we have identified four incomplete-oxidizing strains that clearly do not utilize the acetyl-CoA pathway either for metabolism or mercury methylation (as confirmed by the absence of chloroform inhibition). Hg methylation is thus independent of the acetyl-CoA pathway and may not require vitamin B12 in some and perhaps many incomplete-oxidizing SRB strains.
High levels of methylmercury found in aquatic food webs are of paramount public health concern (37). In most aquatic ecosystems, the external supply of methylmercury is insufficient to account for methylmercury accumulation in biota and sediments (21) and in situ methylmercury formation plays a key role in determining the amount of methylmercury reaching higher trophic levels. Inorganic mercury, primarily deposited from the atmosphere, is methylated by microorganisms in the anoxic layer of the water column or sediments (for review, see reference 36). Multiple field studies have showed that sulfate-reducing bacteria (SRB) are the key mercury-methylating organisms in nature (10, 15-17, 19, 22, 28). Considerable work on the environmental factors controlling methylmercury production has been conducted (for review, see references 7 and 21). Recent research has focused on the conditions affecting the bioavailability of inorganic mercury to SRB (2-6, 27). However, little is known about the biochemical mechanism of methylmercury formation in SRB. Despite a few previous studies (7, 29, 34; C. C. Gilmour, R. Devereux, M. R. Winfrey, and D. A. Stahl, unpublished data.), we do not understand why some SRB can methylate mercury and others cannot or what effects the availability of various organic substrates may have on the enzymatic pathway of mercury methylation.
Wood (51) originally proposed that a methylcorrinoid derivative is the methyating agent for inorganic mercury, since it was the only major coenzyme capable of donating a methyl group in the carbanion form. Later work identified methylcorrinoid compounds capable of abiotic methylmercury synthesis (41). Research on the biological mechanism of mercury methylation has been conducted on one strain of SRB, Desulfovibrio desulfuricans LS (12-14). A corrinoid-containing protein was identified as key to mercury methylation capacity in inhibition experiments (13). From 14C-labeling studies and enzyme activity experiments, Choi et al. (14) concluded that the corrinoid-containing protein responsible for mercury methylation in D. desulfuricans LS is involved in the acetyl-coenzyme A (CoA) pathway.
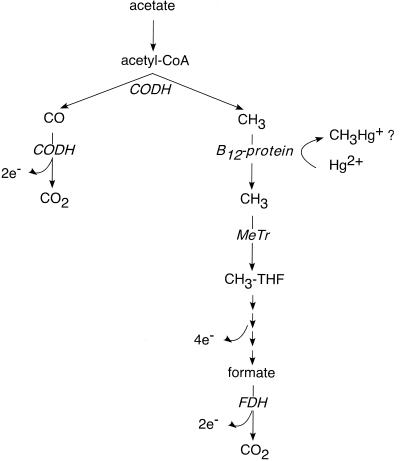
The acetyl-CoA pathway (Fig. 1) is a carbon metabolism pathway that converts acetate into carbon dioxide (and vice versa), through the breakdown of acetate into CO and a methyl moiety by carbon monoxide dehydrogenase (CODH), and subsequent oxidation of both to CO2. Prior research with Moorella thermoacetica, a non-Hg-methylating acetogen, identified a corrinoid-containing protein in the pathway that donated a methyl group to CODH (40). Presumably, a similar corrinoid protein is involved in the acetyl-CoA pathway in SRB. The existence of the acetyl-CoA pathway in D. desulfuricans LS is surprising, given that the strain is an incomplete-oxidizing SRB (converting carbon substrates only to acetate rather than to CO2), which does not use the acetyl-CoA pathway in major carbon metabolism. Biochemical studies using this organism suggest that the pathway is used for minor biosynthetic purposes and/or may also be available for the metabolism of atypical carbon substrates (14). The only other biochemical pathway that has been suggested for Hg methylation is the methionine synthase pathway in Neurospora crassa (33), an aerobic fungus not considered to be an important producer of methylmercury in nature.
FIG. 1.
The acetyl-CoA pathway as used for acetate oxidation. MeTr, methyltransferase.
On the basis of the studies of Bartha, Choi, and coworkers with D. desulfuricans LS, the acetyl-CoA pathway is now generally accepted as being responsible for Hg methylation in SRB. However, there has been no systematic study to verify that this is indeed the case in any other species. Here we report a study of mercury methylation and acetyl-CoA activity in several SRB strains that, like D. desulfuricans LS, do not utilize the acetyl-CoA pathway as a major metabolic pathway.
MATERIALS AND METHODS
Microorganisms and culture conditions.
To probe the hypothesized correlation between the acetyl-CoA pathway and mercury methylation, eight strains of SRB were obtained and assayed for enzyme activity and/or methylmercury production. Pure cultures of Desulfovibrio africanus (DSMZ 2603), Desulfovibrio desulfuricans subsp. desulfuricans (DSMZ 6949), Desulfovibrio vulgaris subsp. vulgaris Marberg (DSMZ 2119), Desulfobulbus propionicus 1pr3 (DSMZ 2302), Desulfobulbus propionicus MUD (DSMZ 6523), Desulfococcus multivorans 1be1 (DSMZ 2059), and Desulfobacter curvatus (DSMZ 3379) were obtained from the German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH [DSMZ]). Environmental isolate BG-8, which belongs to the genus Desulfobacter and was identified as having 96.6% 16S rRNA sequence homology to D. curvatus (42), was kindly provided by Richard Devereux.
All strains were cultured by strict Hungate and asceptic techniques and were grown on variations of Postgate's lactate-sulfate medium (39). The base medium contained sulfate (28 mM Na2SO4), salts (1.7 mM NaH2PO4, 4.7 mM NH4Cl, 6.7 mM MgCl, 6.7 mM KCl, 1.4 mM CaCl2), trace metal (0.5 ml liter−1) and vitamin (5 ml liter−1) stock solutions (24), 0.5 g of yeast extract liter−1, 23 nM selenite, 24 nM tungstate, and 3.6 μM FeCl2 and was buffered at pH 7.5 with 10 mM MOPS (3-[N-morpholino]propanesulfonic acid). D. africanus, D. vulgaris, D. propionicus 1pr3, D. multivorans, and D. desulfuricans subsp. desulfuricans were grown on 35 mM lactate, D. propionicus MUD and strain BG-8 were grown on 20 mM propionate, and D. curvatus was grown on 15 mM acetate. Estuarine SRB strains D. multivorans and D. desulfuricans subsp. desulfuricans were cultured at 10‰ salinity established by adding 0.17 M NaCl; marine D. curvatus and BG-8 were grown at 25‰ salinity (0.43 M NaCl). To maintain reducing conditions, l mM thioglycolate and 1.1 mM ascorbic acid were added to media. Resazurin (1 mg liter−1) was included as a redox indicator. All media were boiled under N2 gas, dispensed into acid-washed serum bottles or Hungate tubes, sealed with rubber stoppers, and autoclaved.
Enzyme assays.
To assay for the presence of the acetyl-CoA pathway in the various SRB strains, spectrophotometric enzyme assays for formate dehydrogenase (FDH) and CODH were conducted on Triton X-100-permeabilized cells (45, 53). Five hundred milliliters of cell cultures was grown to the late-exponential phase as determined by optical density at 660 nm (OD660) recorded on a Beckman DU-7 spectrophotometer. All steps were either performed in a Coy anaerobic chamber filled with N2 (with oxygen levels monitored by methylene blue anaerobic indicator strips) or conducted in degassed, sealed containers. On the same day the enzyme assays were performed, the cells were concentrated by centrifuging the cultures in 50-ml centrifuge tubes for 15 min at 3,900 rpm in a Sorvall RT7 benchtop centrifuge, removing the supernatant, and resuspending the cell pellet in a 5 mM MgCl2 and 2.5 mM dithioerythritol (DTE) solution. To permeabilize the cells, the biological detergent Triton X-100 was added to a final concentration of 0.2% (wt/vol), and the suspension was shaken vigorously.
The activities of FDH and CODH were determined by the reduction of methyl viologen (MeV). Pyruvate dehydrogenase (PDH), a relatively ubiquitous enzyme in sulfate-reducing species, was assayed to confirm the overall enzymatic integrity of the permeabilized cells. α-Ketoglutarate dehydrogenase (α-KETO), a key enzyme of the citric acid cycle, was assayed as an alternative positive control for Desulfobacter strains.
All enzyme assays were performed in anaerobic glass cuvettes (Starna, Atascadero, Calif.) and contained 3 mM MeV, 1 mM DTE, 1 mM MgCl2, 0.01 mM dithionite, 93 to 96 mM Tris buffer (pH 7.8), and 10 to 50 μl of concentrated cells. To determine CODH activity, the cuvettes were saturated with pure CO gas by flushing the vials for 5 min. FDH assays were conducted by adding 2 mM formate, while PDH assays were performed by including 4 mM pyruvate and 1 mM CoA to the reaction mixture. α-KETO assays contained 3 mM α-ketoglutarate and 1 mM CoA. Blank assays including Triton X-100-permeabilized cells but excluding the substrate (CODH blanks were bubbled with N2 gas) were performed to determine the background reducing capacity of the cellular assays. In the absence of cells, no reduction of MeV was observed, which indicated that assay conditions precluded abiotic reduction. The reduction of MeV was recorded at a wavelength of 578 nm by a Cary 100 Bio UV-Vis spectrophotometer with a 6-by-6 multiblock Peltier device set at 37°C. The initial slopes of the kinetic curves, the absorptivity of MeV (9.7 mM−1 cm−1), and the substrate/MeV reduction ratio (1 μmol of substrate oxidized ≅ 2 μmol of MeV reduced) were utilized to determine the enzymatic rates. Rates were normalized based on permeabilized-cell protein concentration (9).
Mercury methylation assays.
SRB strains were tested for their ability to methylate mercury (5). Cultures were grown to early exponential growth (to reduce sulfide inhibition) as determined by OD measurements taken just prior to the methylation assay. The 10-ml cultures were spiked with Hg(II) standard (0.5% HCl) to a final concentration of 5 nM HgCl2 (1 μg liter−1). For controls, cultures were autoclaved and spiked with mercury. To determine the background level of methylmercury in the cultures, blank assays were performed that contained the bacterial strains but were not spiked with mercury. All assays were incubated for 5 h at 28°C. Positive control assays consisting of a known methylating strain spiked with inorganic mercury were included in each assay batch. After 5 h, the assays were stopped by putting the test tubes in an ice bath, and the OD was recorded to track the growth rate of the cultures. The assays were frozen at −20°C awaiting methylmercury distillation and measurement.
Methylmercury concentrations in the assays were determined by distillation (25, 26), followed by aqueous-phase derivitization and cold vapor atomic fluorescence detection (8). Distillation blanks were routinely analyzed and found to contain no detectable methylmercury. To determine the recovery of distillation, assay blanks spiked with 200 pg of methylmercury were included in each distillation round. The average recovery for distilled samples was 0.92 ± 0.23 (n = 15).
Dimethylmercury was not measured in methylation assays. Previous research on SRB has identified mono-methylmercury as the primary biologically produced organomercury species, with dissolved gaseous mercury (either Hg0 or dimethylmercury) accounting for less than 0.1% of the mercury pool in methylation assays (5).
Inhibition assays.
Based on the work of Scholten et al. (44), who tested inhibition of the acetyl-CoA pathway in methanogens, acetogens, and SRB, we chose a concentration of 50 μM for our chloroform inhibition assays. To evaluate the specificity of chloroform as an inhibitor of the acetyl-CoA pathway, chloroform was added to three SRB strains with known biochemistry: D. multivorans 1be1, which obligately utilizes the acetyl-CoA pathway for complete carbon oxidization (45); D. curvatus, which uses the citric acid cycle for complete oxidization (45); and D. africanus, which is an incomplete oxidizer. Fluoroacetate (500 μM), a known inhibitor of the citric acid cycle (11), was used to compare the effect of the inhibitors. Anaerobic chloroform and fluoroacetate stock solutions were added to bacterial cultures after 6.5 h of growth. The cultures were regularly subsampled, and the OD was recorded.
To test the effectiveness of chloroform in inhibiting Hg methylation and the acetyl-CoA pathway, D. multivorans cultures were incubated with and without 50 μM chloroform for 7 h, after which mercury methylation and CODH enzyme activity assays were conducted as described above. To compare the effect of a more commonly used inhibitor on methylation rates, 28 mM Na2MoO4, a specific inhibitor of sulfate reduction (38), was added to D. multivorans pure cultures. OD readings were taken before the inhibitor was added, before the methylation assay, and at the end of the methylation assay.
D. propionicus 1pr3 and D. africanus strains were also incubated with chloroform for 7 h, with 5-h Hg methylation assays. To ensure that the acetyl-CoA pathway was inhibited, another experiment was set up in which D. africanus cultures were incubated with 50 μM chloroform for 3 days prior to the experiment. Two culture stocks were inoculated from the same vial, with chloroform added to one, and both were allowed to grow for 3 days until late exponential phase. These inhibited and noninhibited stocks were each used to inoculate triplicate blank, control, and assay vials (18 vials total) either containing or not containing chloroform, respectively. Cells were grown to early log growth for the Hg methylation assay. Duplicate cultures were run in parallel for determination of CODH and PDH enzyme activities.
RESULTS
Mercury methylation assays.
To investigate the connection between the acetyl-CoA pathway and Hg methylation, seven SRB strains that do not utilize the acetyl-CoA pathway for major carbon metabolism and one control strain were investigated. Three known Hg-methylating SRB strains (D. africanus, D. propionicus 1pr3, Desulfobacter strain BG-8) and one non-Hg-methylating strain (D. desulfuricans subsp. desulfuricans) (29; Gilmour et al., unpublished) were tested for acetyl-CoA pathway activities. Two strains (D. vulgaris subsp. vulgaris Marberg and D. propionicus MUD) previously shown not to have measurable acetyl-CoA pathway activities (45) were analyzed for the capacity to methylate Hg. D. curvatus, a citric acid cycle-utilizing SRB closely related to methylating strain BG-8 (42), was assayed for acetyl-CoA pathway activity and Hg methylation. D. multivorans, a known Hg methylator (Gilmour et al., unpublished) with substantial acetyl-CoA pathway enzyme activities (45) was used as a positive control. D. desulfuricans LS, which was investigated by Bartha and coworkers (12-14), is no longer available and could not be tested. The strains chosen are representatives of four of seven distinct phylogenetic groups (18) of SRB found in marine, estuarine, and freshwater sediments based on 16S rRNA probes (19, 29, 30, 43).
Of the six strains tested for the capacity to methylate Hg, four—D. africanus, D. propionicus 1pr3, D. propionicus MUD, and D. multivorans—methylated Hg. Only D. vulgaris subsp. vulgaris Marberg and D. curvatus had methylation rates that were less than the average method detection limit (Table 1). For all methylation assays, there were no significant differences in growth between blank and Hg assays (P > 0.05). Blank corrected methylation rates varied from 44.5 to 141 pM MeHg h−1. Mercury methylation rates normalized by cell number varied by an order of magnitude (Table 3). Care was taken to ensure that all strains were tested in early exponential growth to minimize the problem of Hg methylation inhibition by sulfide (5, 6). Nonetheless, differences in sulfide levels could lead to variations in methylation rates. In previous experiments with the same D. propionicus 1pr3 and D. multivorans strains, King et al. (28) measured methylation rates that were 230 and 3 times lower, respectively (on a picomole hour−1 cell−1 basis), and greater variability of replicate assays, presumably as a result of the presence of sulfide (1 to 2 mM used as reductant) in their growth media. The normalized Hg methylation rate reported by Benoit et al. (5) for D. propionicus 1pr3 under low-sulfide conditions (0.55 mM) was 22 times lower than the rate measured in this study, likely due to differences in growth stage in the 5-day incubation (5).
TABLE 1.
Specific Hg methylation rates by pure culture SRB
| Species | Hg methylation rate (pM MeHg h−1)a
|
|
|---|---|---|
| Assayb | Controlc | |
| Incomplete oxidizers | ||
| D. africanus | 64.4 ± 28.1 | <DL |
| D. vulgaris subsp. vulgaris Marberg | <DL | <DL |
| D. propionicus 1pr3 | 53.1 ± 7.32 | <DL |
| D. propionicus MUD | 141 ± 31.2 | <DL |
| Complete oxidizers | ||
| D. multivorans 1be1 | 44.5 ± 16.9 | <DL |
| D. curvatus | <DL | <DL |
Values are averages ± standard deviation of three replicate assays. The average method detection limit (DL) = 6.2 pM MeHg h−1 (n = 10).
Live cultures were incubated with 5 nM HgCl2 for 5 h. Rates were corrected for the average blank MeHg concentration.
Autoclaved cultures were incubated with 5 nM HgCl2 for 5 h.
TABLE 3.
Methylmercury concentration and production rates from inhibition assays
| Strain | Absolute concn (pM MeHg)a
|
Specific rate (pM MeHg h−1)a,b
|
Specific normalized rate (pmol of MeHg h−1 cell−1)a,c
|
|||
|---|---|---|---|---|---|---|
| Assay | +CHCl3 | Assay | + CHCl3 | Assay | + CHCl3 | |
| D. multivorans 1be1 | 340.0 ± 84.3 | 115.5 ± 32.9 | 44.5 ± 16.9 | <DLd | 4.2 × 10−9 ± 1.6 × 10−9 | NGe |
| D. propionicus 1pr3 | 332.2 ± 36.6 | 274.3 ± 62.0 | 53.1 ± 7.3 | 41.5 ± 12.4 | 4.7 × 10−9 ± 1.1 × 10−9 | 3.9 × 10−9 ± 1.5 × 10−9 |
| D. africanus (7-hr incubation) | 373.3 ± 140.4 | 495.2 ± 54.5 | 64.4 ± 28.1 | 88.8 ± 10.9 | 5.6 × 10−10 ± 2.3 × 10−10 | 6.2 × 10−10 ± 1.8 × 10−10 |
| D. africanus (3 day incubation) | 220.6 ± 100.2 | 190.9 ± 66.3 | 42.4 ± 22.0 | 36.4 ± 15.7 | 1.5 × 10−10 ± 7.9 × 10−10 | 2.3 × 10−10 ± 9.3 × 10−10 |
Values are average ± standard deviation of three replicate 5-h Hg assays.
Specific rates were corrected for average blank MeHg concentration.
Normalized to total cell density at end of assay.
Average assay detection limit (DL) = 6.2 pM MeHg h−1 (n = 10). There was a high average blank (117.4 pM MeHg h−1) because of estuarine medium.
NG, no growth.
Acetyl-CoA pathway enzyme assays.
To determine the presence of the acetyl-CoA pathway in all eight SRB strains, permeabilized cell enzyme assays were performed for two key acetyl-CoA enzymes, FDH and CODH. CO oxidation in SRB has been found to be catalyzed only by CODH (14, 45, 48, 49, 52), so that assaying CO oxidation gives a measure of CODH activity. FDH's role in SRB appears to be more varied, participating in other pathways besides the acetyl-CoA pathway (23). Therefore, CODH activity is the most diagnostic test for the existence of the acetyl-CoA pathway. Assays for PDH, a fairly ubiquitous enzyme in SRB, and α-KETO, a citric acid cycle enzyme, were conducted as positive controls, to ensure the biochemical integrity of enzymes in the permeabilized cells (45).
All strains studied displayed PDH activity (Table 2). Among the two Desulfobacter strains, only D. curvatus exhibited α-KETO activity. The three Desulfovibrio strains and the complete-oxidizing D. multivorans displayed FDH activity; neither Desulfobulbus nor Desulfobacter strains had measured FDH activity above the average method detection limit. Only one species tested, D. multivorans, had CODH activity above the average method detection limit, and it thus appears to be the only one to have the acetyl-CoA pathway.
TABLE 2.
Acetyl-CoA and positive control enzyme activities
| Species | Enzyme activity (U [μmol min−1 mg of protein−1])a
|
|||
|---|---|---|---|---|
| PDH | FDH | CODH | α-KETO | |
| Incomplete oxidizers | ||||
| D. africanus | 0.244 ± 0.019 | 0.083 ± 0.002 | <DL | NAb |
| D. desulfuricans subsp. desulfuricans | 0.329 ± 0.127 | 1.655 ± 0.086 | <DL | NA |
| D. vulgaris subsp. vulgaris Marberg | 0.263 ± 0.013 | 1.202 ± 0.159 | <DL | NA |
| D. propionicus 1pr3 | 0.302 ± 0.025 | <DL | <DL | NA |
| D. propionicus MUD | 0.045 ± 0.004 | <DL | <DL | NA |
| Desulfobacter strain BG-8 | 0.253 ± 0.005 | <DL | <DL | <DL |
| Complete oxidizers | ||||
| D. multivorans 1be1 | 0.127 ± 0.015 | 4.667 ± 0.336 | 1.910 ± 0.049 | NA |
| D. curvatus | 0.061 ± 0.009 | <DL | <DL | 0.494 ± 0.054 |
Values are averages ± standard deviation of three replicate enzyme assays. Average method detection limit (DL) for PDH assay = 0.036 U (n = 11), DL for FDH assay = 0.024 U (n = 8), DL for CODH assay = 0.027 U (n = 12), and DL for α-KETO = 0.025 U (n = 2).
NA, not available.
The enzyme activities measured for the eight sulfate-reducing species are comparable to rates previously measured in other sulfate-reducing species. The PDH and CODH enzymatic activities we measured were on the high end compared to those in studies of complete-oxidizing SRB (45), perhaps because we did not freeze the samples before performing the assays. Our FDH rates varied widely among species, similar to the variability in rates observed by Shauder et al. (45). The variability in rates observed between studies is likely due to differences in assay optimization among researchers, but is also possibly due to intrinsic differences in enzyme expression and activity.
Inhibition assays.
On the basis of the Hg methylation and enzyme assay experiments, it appears that of the four species found to methylate mercury, only one—the complete oxidizer D. multivorans—possesses the acetyl-CoA pathway. To verify that mercury methylation in other species is due to other biochemical pathways and not to acetyl-CoA activity below the limit of detection of our enzyme assay, we performed additional inhibition experiments with chloroform (CHCl3), a known inhibitor of the acetyl-CoA pathway (11, 44).
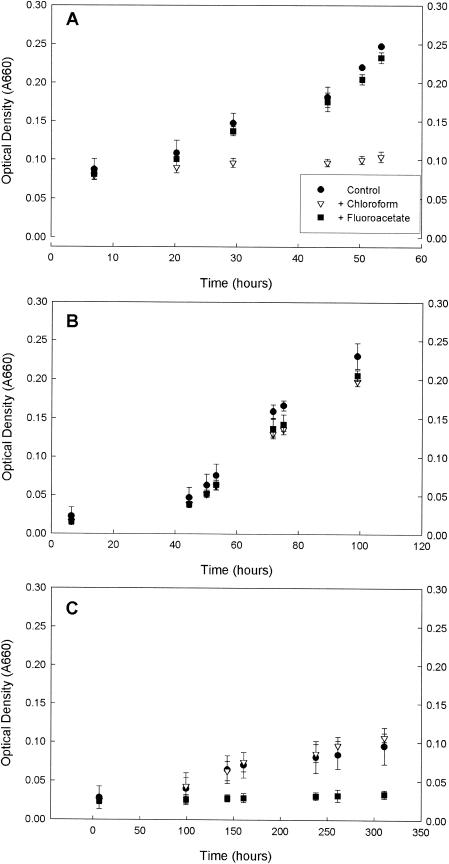
We first evaluated the effectiveness and specificity of chloroform as an inhibitor of the acetyl-CoA pathway in three SRB strains (Fig. 2) by comparing bacterial growth in control cultures, cultures amended in CHCl3, and cultures amended with fluoroacetate, a known inhibitor of the citric acid cycle (11). D. multivorans, which depends on the acetyl-CoA pathway for major carbon metabolism, was inhibited by chloroform, while fluoroacetate had minor effects compared to control assays (Fig. 2A). Neither chloroform nor fluoroacetate (Fig. 2B) affected growth of D. africanus, an incomplete-oxidizing SRB that depends on neither the acetyl-CoA pathway nor the citric acid cycle for major carbon metabolism. D. curvatus, a complete-oxidizing, citric acid cycle-utilizing SRB, was inhibited by fluoroacetate but not by chloroform (Fig. 2C). Therefore, it appears that chloroform is effective and at least partly specific in its inhibition of the acetyl-CoA pathway.
FIG. 2.
Growth of D. multivorans (A), D. africanus (B), and D. curvatus (C) when exposed to 50 μM chloroform (▿), 500 μM fluoroacetate (▪), and no addition (•). Error bars represent the standard deviation of three replicate assays.
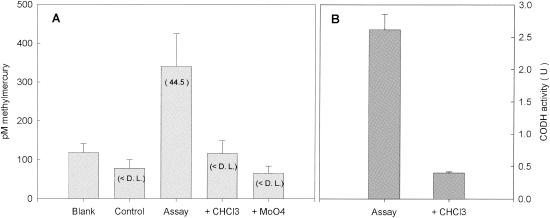
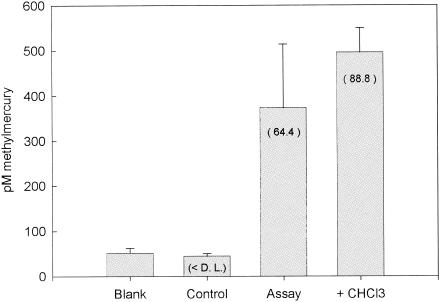
As expected, chloroform was as effective in inhibiting Hg methylation in D. multivorans cultures as in inhibiting growth. The concentrations of methylmercury in chloroform-amended assays was less than the average method detection limit (Fig. 3A and Table 3). A similar inhibition of growth and Hg methylation was observed in cultures amended with molybdate, a known inhibitor of sulfate reduction. Simultaneous CODH activity experiments indicated that CODH activity was seven times lower in cultures incubated with chloroform than in uninhibited cultures (Fig. 3B).
FIG. 3.
(A) Methylmercury concentrations produced after a 5-h HgCl2 incubation in D. multivorans inhibition experiment. Blank corrected methylation rates (picomolar concentration per hour) are in parentheses. D.L., detection limit. (B) CODH activity measured for noninhibited and inhibited cultures during methylation assay. Error bars represent the standard deviation of three replicate assays.
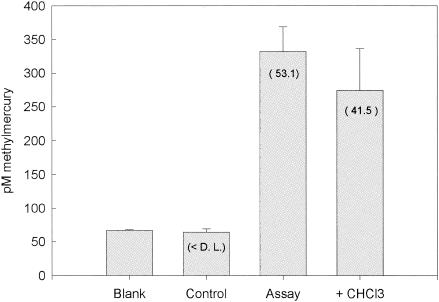
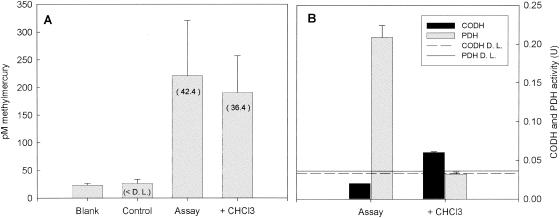
Similar chloroform inhibition experiments with D. propionicus 1pr3 and D. africanus, which are able to methylate Hg but do not appear to use the acetyl-CoA pathway, showed no significant difference in mercury methylation rates or growth rates between the control assay and chloroform additions for either strain (P > 0.05) (Fig. 4 and 5). Interestingly, measurements of PDH activity in D. africanus showed strong inhibition by CHCl3, demonstrating perhaps that at the concentration we used, chloroform may not be completely specific for the acetyl-CoA pathway. To ensure that enough time was given for chloroform to inhibit the acetyl-CoA pathway, another experiment was run in which D. africanus cultures were incubated with chloroform for 3 days prior to the experiment (Fig. 6). Due to the long length of inhibitor incubation, the cultures in the assays with CHCl3 grew at approximately half the rate as those in the assays with no inhibitor. However, no significant difference (P > 0.05) was observed between the inhibited and noninhibited normalized Hg methylation rates (Table 3).
FIG. 4.
Methylmercury concentrations produced after a 5-h HgCl2 incubation from D. propionicus 1pr3 inhibition assay. D.L., detection limit. Blank corrected methylation rates (picomolar concentration per hour) are in parentheses. Error bars represent the standard deviation of three replicate assays.
FIG. 5.
Methylmercury concentrations produced after 5-h HgCl2 incubation from D. africanus inhibition assay (7-h inhibitor incubation). D.L., detection limit. Blank corrected methylation rates (picomolar concentration per hour) are in parentheses. Error bars represent the standard deviation of three replicate assays.
FIG. 6.
(A) Methylmercury concentrations produced after a 5-h HgCl2 incubation from D. africanus inhibition experiment (3-day inhibitor incubation). D.L., detection limit. Blank corrected methylation rates (picomolar concentration per hour) are in parentheses. (B) CODH activity (in black) and PDH activity (in gray) for noninhibited and inhibited cultures during methylation assay. Error bars represent the standard deviation of three replicate assays.
DISCUSSION
To evaluate a possible relationship between the presence of the acetyl-CoA pathway and the ability to methylate mercury in a wide variety of SRB, we compiled our results with published work (Table 4). There is no consistent relationship between the existence of the acetyl-CoA pathway and the ability to methylate Hg among all SRB tested. Only complete-oxidizing genera (Desulfococcus, Desulfosarcina, and Desulfobacterium) that require the acetyl-CoA pathway for basic metabolism and previously studied D. desulfuricans strain LS have both the acetyl-CoA pathway and mercury methylation capacity. The other complete-oxidizing strains, which use the citric acid cycle, do not methylate mercury. None of the incomplete oxidizer strains we tested has an active acetyl-CoA pathway (contrary to D. desulfuricans LS), but four of these strains methylate mercury nonetheless. 16S rRNA sequence homology (97%) (42) suggests that BG-8 is closely related to the complete oxidizer D. curvatus; however, our enzyme assays indicate that it is an incomplete oxidizer. Although it has been classified in the genus Desulfobacter, it possesses neither the acetyl-CoA nor the citric acid pathway and methylates mercury. Chloroform inhibition experiments confirmed that the acetyl-CoA pathway is not present in D. africanus and D. propionicus 1pr3 nor is it active in Hg methylation.
TABLE 4.
Relationship between Hg methylation capacity and presence of the acetyl-CoA pathway
| Species | Result for:
|
|
|---|---|---|
| Hg methylationa | Acetyl-CoA pathwaya,b | |
| Incomplete oxidizer | ||
| D. africanus | Yes | <DL |
| D. desulfuricans LS | Yes (14) | Yes (14) |
| D. desulfuricans subsp. desulfuricans | <DLc | <DL |
| D. vulgaris subsp. vulgaris Marberg | <DL | <DL |
| Desulfobulbus propionicus 1pr3 | Yes | <DL |
| D. propionicus MUD | Yes | <DL |
| Desulfobacter BG-8 | Yes (29) | <DL |
| Complete oxidizer | ||
| D. multivorans 1be1 | Yes | Yes |
| D. variabilis 3be13 | Yesc | Yes (45) |
| D. autotrophicum | Yesc | Yes (45) |
| D. hydrogenophilus | <DLc | <DL (45) |
| D. curvatus | <DL | <DL |
Associated references follow in parentheses. <DL, less than detection limit.
The presence of the acetyl-CoA pathway was determined based on positive CODH activity.
Gilmour et al., unpublished.
Our finding that the acetyl-CoA pathway is not the major Hg-methylating mechanism in the majority of incomplete-oxidizing SRB strains demands a reexamination of the role of the carbon metabolism pathway and of vitamin B12 in mercury methylation in SRB. In view of the results of Choi, Bartha, and coworkers with D. desulfuricans LS and in view of the fact that, so far, all species that have been found with the acetyl-CoA pathway do methylate mercury, it seems plausible that the acetyl-CoA pathway is sufficient to enable mercury methylation in SRB. This is not absolutely certain, however, since the previous studies of D. desulfuricans LS provided no direct evidence linking the acetyl-CoA pathway with Hg methylation. 14C-labeling studies demonstrated that radiolabeled serine (when added as a spike) was the most effective methyl donor to mercury, with 95% of the carbon group on MeHg originating from serine (14). Serine is not directly involved in the acetyl-CoA pathway, but instead provides a methylene group to tetrahydrofolate, a methyl-transferring coenzyme found in numerous pathways. When the SRB strain was grown on formate, the 14C group from formate was found on only 50% of the MeHg produced (14). It is thus possible that D. desulfuricans LS does not use the acetyl-CoA pathway for Hg methylation. In complete-oxidizing SRB that use the acetyl-CoA pathway for carbon metabolism, it is difficult to unravel the link between acetyl-CoA pathway and Hg methylation. As exemplified by our experiments with D. multivorans, inhibition of the acetyl-CoA pathway inhibits mercury methylation, but this of course could be the indirect result of the overall metabolic and growth inhibition of the organism.
Methionine synthase, which transfers a methyl group to homocysteine, has been hypothesized as a possible alternative site for mercury methylation (14, 46), and this enzyme has been implicated in N. crassa (33). End product inhibition of methionine synthase by the addition of up to 5 mM methionine in D. desulfuricans LS culture (14) and 5 and 10 mM methionine in D. africanus culture (unpublished data) caused no inhibition of Hg methylation. However, the effectiveness of end product inhibition on methionine synthase activity has not been determined in SRB.
Recently, Siciliano and Lean (46) have reported a correlation between mercury methylation and methionine synthase activity in Escherichia coli cell extracts and in sediments. However, the results are difficult to interpret since a methylating agent (s-adensylmethionine) and vitamin B12 were added to the samples and may have lead to abiotic methylation of mercury under the high-temperature conditions of the assay. The correlation obtained in the field may simply reflect overall patterns of biological activity.
Like the corrinoid-containing protein in the acetyl-CoA pathway, most methionine synthase enzymes contain vitamin B12. However, in N. crassa, methionine synthase is thought to be B12 independent (33, 35), perhaps containing a zinc active site as identified in E. coli (for review, see reference 1). The presence and form of methionine synthase in SRB are not known. Our inhibition experiments with chloroform may shed light on whether B12 is involved in mercury methylation, if, as implied by previous studies, chloroform functions as an inhibitor via its reaction with B12. Studies of various alkyl halide inhibitors in chemical assays (32) and cell extract assays (20) indicate that CHCl3 inhibition occurs by directly reacting with the corrinoid. Mechanistic studies by Wood et al. (50) and Krone et al. (32) found that chloroform and other alkyl halides bind to the cobalt center of B12, with one of the chlorine atoms acting as a leaving group. Using partially purified methionine synthase from E. coli, Wood et al. (50) also found that chloroform can inhibit methionine formation by the corrinoid-containing enzyme. Thus, our results showing no CHCl3 inhibition of mercury methylation in D. propionicus 1pr3 and D. africanus imply that Hg methylation is independent of a B12-containing methionine synthase and may not require a cobalamin enzyme at all.
Interestingly, three strains that methylate mercury but lack the acetyl-CoA pathway, D. propionicus 1pr3, D. propionicus MUD, and isolate BG-8, have the unique capacity to utilize propionate as a carbon source. Desulfobulbus species utilize a propionate metabolism pathway with a B12-containing enzyme, methylmalonyl-CoA mutase (31, 47). Although this enzyme does not directly transfer a methyl group in the conversion of propionate to acetate, and thus is not an obvious source of methylmercury formation, further study of this enzyme may be warranted. Environmental isolate BG-8, which has been identified as belonging to the genus Desulfobacter based on 16S phylogeny, grows on propionate and presumably utilizes a similar propionate degradation pathway.
Since not all SRB methylate mercury, the activity of biochemical pathways specific to certain SRB appears to be required for the methylation of Hg. We have shown that some incomplete oxidizers (the majority of those tested) are able to methylate mercury through a pathway that is independent of the acetyl-CoA pathway. From previous studies and our own data, it seems likely, although not certain, that SRB strains that have the acetyl-CoA pathway, including many complete oxidizers, use it to methylate Hg. If this is the case, there are at least two distinct pathways for mercury methylation in SRB, one of which is independent of the acetyl-CoA pathway and possibly does not involve vitamin B12. The biochemistry of mercury methylation in SRB and its implication for methylation rates in nature clearly deserve further study.
Acknowledgments
This work was supported by EPA grant R-827915-01-0.
We thank Richard Devereux for the generous gift of strain BG-8, Elizabeth Malcolm and Chad Hammerschmidt for technical assistance, Bess Ward for review of the manuscript, and Tom Spiro and Allen Milligan for helpful discussions.
REFERENCES
- 1.Banerjee, R. 1997. The yin-yang of cobalamin biochemistry. Chem. Biol. 4:175-186. [DOI] [PubMed] [Google Scholar]
- 2.Barkay, T., M. Gillman, and R. R. Turner. 1997. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Appl. Environ. Microbiol. 63:4267-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit, J. M., C. C. Gilmour, R. P. Mason, and A. Heyes. 1999. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ. Sci. Technol. 33:951-957. [Google Scholar]
- 4.Benoit, J. M., R. P. Mason, and C. C. Gilmour. 1999. Estimation of mercury-sulfide speciation in sediment pore waters using octanol-water partitioning and implications for availability to methylating bacteria. Environ. Toxicol. Chem. 18:2138-2141. [DOI] [PubMed] [Google Scholar]
- 5.Benoit, J. M., C. C. Gilmour, and R. P. Mason. 2001. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3). Appl. Environ. Microbiol. 67:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benoit, J. M., C. C. Gilmour, and R. P. Mason. 2001. The influence of sulfide on solid phase mercury bioavailability for methylation by pure cultures of Desulfobulbus propionicus (1pr3). Environ. Sci. Technol. 35:127-132. [DOI] [PubMed] [Google Scholar]
- 7.Benoit, J. M., C. C. Gilmour, A. Heyes, R. P. Mason, and C. L. Miller. 2003. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems, p. 262-297. In Y. Cai and O. C. Braids (ed.), Biogeochemistry of environmentally important trace elements. American Chemical Society, Washington, D.C.
- 8.Bloom, N. S. 1989. Determination of picogram levels of methylmercury by aqueous phase ethylation followed by cryogenic gas chromatography with cold vapor atomic fluorescence detection. Can. J. Fish Aquat. Sci. 46:1131-1140. [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Branfireun, B. A., K. Bishop, N. T. Roulet, G. Granberg, and M. Nilsson. 2001. Mercury cycling in boreal ecosystems: the long-term effect of acid rain constituents on peatland pore water methylmercury concentrations. Geophys. Res. Lett. 28:1227-1230. [Google Scholar]
- 11.Chidthaisong, A., and R. Conrad. 2000. Specificity of chloroform, 2-bromoethanesulfonate and fluoroacetate to inhibit methanogenesis and other anaerobic processes in anoxic rice field soil. Soil Biol. Biochem. 32:977-988. [Google Scholar]
- 12.Choi, S.-C., and R. Bartha. 1993. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 59:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, S.-C., T. Chase, Jr., and R. Bartha. 1994. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 60:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, S.-C., T. Chase, and R. Bartha. 1994. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 60:4072-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi, S.-C., and R. Bartha. 1994. Environmental-factors affecting mercury methylation in estuarine sediments. Bull. Environ. Contam. Toxicol. 53:805-812. [DOI] [PubMed] [Google Scholar]
- 16.Compeau, G. C., and R. Bartha. 1985. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 50:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Compeau, G. C., and R. Bartha. 1987. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl. Environ. Microbiol. 53:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 19.Devereux, R., M. R. Winfrey, J. Winfrey, and D. A. Stahl. 1996. Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment. FEMS Microbiol. Ecol. 20:23-31. [Google Scholar]
- 20.Ghambeer, R. K., H. G. Wood, M. Schulman, and L. Ljungdahl. 1971. Total synthesis of acetate from CO2, methyltetrahydrofolate, and methyl-B12 by Clostridium thermoaceticum. Arch. Biochem. Biophys. 143:471-484. [DOI] [PubMed] [Google Scholar]
- 21.Gilmour, C. C., and E. A. Henry. 1991. Mercury methylation in aquatic systems affected by acid deposition. Environ. Pollut. 71:131-169. [DOI] [PubMed] [Google Scholar]
- 22.Gilmour, C. C., E. A. Henry, and R. Mitchell. 1992. Sulfate stimulation of mercury methylation in fresh-water sediments. Environ. Sci. Technol. 26:2281-2287. [Google Scholar]
- 23.Hansen, T. A. 1993. Carbon metabolism of sulfate-reducing bacteria, p. 21-40. In J. M. Odom and R. Singleton (ed.), The sulfate-reducing bacteria: contemporary perspectives. Springer-Verlag, New York, N.Y.
- 24.Henry, E. A. 1992. The role of sulfate-reducing bacteria in environmental mercury methylation. Ph.D. dissertation. Harvard University, Cambridge, Mass.
- 25.Horvat, M., N. S. Bloom, and L. Liang. 1993. Comparison of distillation with other current isolation methods for the determination of MeHg compounds in low level environmental samples. 1. Sediments. Anal. Chim. Acta 281:135-152. [Google Scholar]
- 26.Horvat, M., and L. Liang, and N. S. Bloom. 1993. Comparison of distillation with other current isolation methods for the determination of methyl mercury compounds in low level environmental samples. 2. Water. Anal. Chim. Acta 282:153-168. [Google Scholar]
- 27.Jay, J. A., F. M. M. Morel, and H. F. Hemond. 2000. Mercury speciation in the presence of polysulfides. Environ. Sci. Technol. 34:2196-2200. [Google Scholar]
- 28.King, J. K., F. M. Saunders, R. F. Lee, and R. A. Jahnke. 1999. Coupling mercury methylation rates to sulfate reduction rates in marine sediments. Environ. Toxicol. Chem. 18:1362-1369. [Google Scholar]
- 29.King, J. K., J. E. Kostka, M. E. Frischer, and F. M. Saunders. 2000. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl. Environ. Microbiol. 66:2430-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koizumi, Y., S. Takii, M. Nishino, and T. Nakajima. 2003. Vertical distribution of sulfate-reducing bacteria and methane-producing archaea quantified by oligonucleotide probe hybridization in the profundal sediments of a mesotrophic lake. FEMS Microbiol. Ecol. 44:101-108. [DOI] [PubMed] [Google Scholar]
- 31.Kremer, D. R., and T. A. Hansen. 1988. Pathway of propionate degradation in Desulfobulbus propionicus. FEMS Microbiol. Lett. 49:273-277. [Google Scholar]
- 32.Krone, U. E., R. K. Thauer, H. P. C. Hogenkamp, and K. Steinbach. 1991. Reductive formation of carbon monoxide from CCl4 and FREONs 11, 12, and 13 catalyzed by corrinoids. Biochemistry 30:2713-2719. [DOI] [PubMed] [Google Scholar]
- 33.Landner, L. 1971. Biochemical model for biological methylation of mercury suggested from methylation studies in-vivo with Neurospora crassa. Nature 230:452-454. [DOI] [PubMed] [Google Scholar]
- 34.Macalady, J. L., E. E. Mack, D. C. Nelson, and K. M. Scow. 2000. Sediment microbial community structure and mercury methylation in mercury-polluted Clear Lake, California. Appl. Environ. Microbiol. 66:1479-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens, J. H., H. Barg, M. J. Warren, and D. Jahn. 2002. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58:275-285. [DOI] [PubMed] [Google Scholar]
- 36.Morel, F. M. M., A. M. L. Kraepiel, and M. Amyot. 1998. The chemical cycle and bioaccumulation of mercury. Annu. Rev. Ecol. Syst. 29:543-566. [Google Scholar]
- 37.National Research Council. 2000. Toxicological effects of methylmercury. National Academy Press, Washington, D.C.
- 38.Oremland, R. S., and D. G. Capone. 1988. Use of “specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10:285-383. [Google Scholar]
- 39.Postgate, J. R. 1984. The sulfate-reducing bacteria, 2nd ed., p. 12-33. Cambridge University Press, Cambridge, United Kingdom.
- 40.Ragsdale, S. W. 1991. Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit. Rev. Biochem. Mol. Biol. 26:261-300. [DOI] [PubMed] [Google Scholar]
- 41.Ridley, W. P., L. J. Dizikes, and J. M. Wood. 1977. Biomethylation of toxic elements in the environment. Science 197:329-332. [DOI] [PubMed] [Google Scholar]
- 42.Rooney-Varga, J. N., B. R. S. Genthner, R. Devereux, S. G. Willis, S. D. Friedman, and M. E. Hines. 1998. Phylogenetic and physiological diversity of sulfate-reducing bacteria isolated from a salt marsh sediment. Syst. Appl. Microbiol. 21:557-568. [DOI] [PubMed] [Google Scholar]
- 43.Sahm, K., B. J. MacGregor, B. B. Jorgensen, and D. A. Stahl. 1999. Sulfate reduction and vertical distribution of sulfate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ. Microbiol. 1:65-74. [DOI] [PubMed] [Google Scholar]
- 44.Scholten, J. C. M., R. Conrad, and A. J. M. Stams. 2000. Effect of 2-bromo-ethane sulfonate, molybdate and chloroform on acetate consumption by methanogenic and sulfate-reducing populations in freshwater sediments. FEMS Microbiol. Ecol. 32:35-42. [DOI] [PubMed] [Google Scholar]
- 45.Shauder, R., B. Eikmanns, R. K. Thauer, F. Widdel, and G. Fuchs. 1986. Acetate oxidation to CO2 in anaerobic bacteria via a novel pathway not involving reactions of the citric acid cycle. Arch. Microbiol. 145:162-172. [Google Scholar]
- 46.Siciliano, S. D., and D. R. S. Lean. 2002. Methyltransferase: an enzyme assay for microbial methylmercury formation in acidic soils and sediments. Environ. Toxicol. Chem. 21:1184-1190. [PubMed] [Google Scholar]
- 47.Stams, A. J. M., D. R. Kremer, K. Nicolay, G. H. Weenk, and T. A. Hansen. 1984. Pathway of propionate formation in Desulfobulbus propionicus. Arch. Microbiol. 139:167-173. [Google Scholar]
- 48.Tarasova, N. B., and M. I. Belyaeva. 1998. The CO dehydrogenase activity of Desulfovibrio desulfuricans growing under chemoorganotrophic and chemolithoheterotrophic conditions. Microbiology 67:504-508. [Google Scholar]
- 49.Tasaki, M., Y. Kamagata, K. Nakamura, K. Okamura, and K. Minami. 1993. Acetogenesis from pyruvate by Desulfotomaculum thermobenzoicum and differences in pyruvate metabolism among 3 sulfate-reducing bacteria in the absence of sulfate. FEMS Microbiol. Lett. 106:259-264. [Google Scholar]
- 50.Wood, J. M., F. S. Kennedy, and R. S. Wolfe. 1968. The reaction of multihalogenated hydrocarbons with free and bound reduced vitamin B12. Biochemistry 7:1707-1713. [DOI] [PubMed] [Google Scholar]
- 51.Wood, J. M. 1974. Biological cycles for toxic elements in the environment. Science 183:1049-1052. [DOI] [PubMed] [Google Scholar]
- 52.Yagi, T. 1958. Enzymatic oxidation of carbon monoxide. Biochim. Biophys. Acta 30:194-195. [DOI] [PubMed] [Google Scholar]
- 53.Zeikus, J. G., G. Fuchs, W. Kenealy, and R. K. Thauer. 1977. Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J. Bacteriol. 132:604-613. [DOI] [PMC free article] [PubMed] [Google Scholar]