Abstract
The microbial and enzymatic degradation of a new energetic compound, 2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane (CL-20), is not well understood. Fundamental knowledge about the mechanism of microbial degradation of CL-20 is essential to allow the prediction of its fate in the environment. In the present study, a CL-20-degrading denitrifying strain capable of utilizing CL-20 as the sole nitrogen source, Pseudomonas sp. strain FA1, was isolated from a garden soil. Studies with intact cells showed that aerobic conditions were required for bacterial growth and that anaerobic conditions enhanced CL-20 biotransformation. An enzyme(s) involved in the initial biotransformation of CL-20 was shown to be membrane associated and NADH dependent, and its expression was up-regulated about 2.2-fold in CL-20-induced cells. The rates of CL-20 biotransformation by the resting cells and the membrane-enzyme preparation were 3.2 ± 0.1 nmol h−1 mg of cell biomass−1 and 11.5 ± 0.4 nmol h−1 mg of protein−1, respectively, under anaerobic conditions. In the membrane-enzyme-catalyzed reactions, 2.3 nitrite ions (NO2−), 1.5 molecules of nitrous oxide (N2O), and 1.7 molecules of formic acid (HCOOH) were produced per reacted CL-20 molecule. The membrane-enzyme preparation reduced nitrite to nitrous oxide under anaerobic conditions. A comparative study of native enzymes, deflavoenzymes, and a reconstituted enzyme(s) and their subsequent inhibition by diphenyliodonium revealed that biotransformation of CL-20 is catalyzed by a membrane-associated flavoenzyme. The latter catalyzed an oxygen-sensitive one-electron transfer reaction that caused initial N denitration of CL-20.
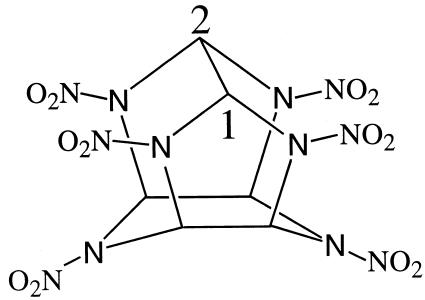
2,4,6,8,10,12-Hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane (CL-20) is a high-energy polycyclic nitramine compound (17) with a rigid caged structure (Fig. 1). Due to its high energy content and superior explosive properties, it may replace conventionally used explosives such as hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX) in the future. The environmental, biological, and health impacts of this energetic chemical and its metabolic products are not known. The severe environmental contamination and biological toxicity of the widely used monocyclic nitramine explosives RDX and HMX are already well documented (11, 13, 16, 22). It is likely that due to its structural similarity with RDX and HMX, CL-20 may also pose a serious threat to the environment by contaminating soils, sediments, and groundwater. Therefore, the microbial degradation of CL-20 should be studied under in vitro and in vivo conditions in order to determine the reaction products and to gain insights into the mechanisms involved in its degradation.
FIG. 1.
Molecular structure of CL-20.
Previous reports on the biodegradation and biotransformation of RDX and HMX by a variety of microorganisms (aerobic, anaerobic, and facultative anaerobes) and enzymes have shown that initial N denitration can lead to ring cleavage and decomposition (3, 5-6, 9, 12-15, 21, 26). In a recent study, Trott et al. (24) reported the aerobic biodegradation of CL-20 by the soil isolate Agrobacterium sp. strain JS71. The isolate utilized CL-20 as the sole nitrogen source and assimilated 3 mol of nitrogen per mol of CL-20. However, no information was provided about the mechanism of CL-20 biodegradation.
In the present study, a denitrifying Pseudomonas sp. strain, FA1, that utilized CL-20 as a sole nitrogen source was isolated from a garden soil sample. The CL-20 biotransformation conditions were optimized in aqueous medium. The nature and function of the enzyme(s) responsible for the biotransformation of CL-20 by strain FA1 were studied. Stoichiometries of the products formed during the biotransformation of CL-20 by the membrane-associated enzyme(s) from Pseudomonas sp. strain FA1 were determined, and an initial enzymatic N denitration reaction mechanism is proposed.
MATERIALS AND METHODS
Chemicals.
CL-20 in ɛ form and at 99.3% purity was provided by ATK Thiokol Propulsion, Brigham City, Utah. NADH, NADPH, diphenyliodonium chloride (DPI), flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), NaNO2, dicumarol, 2,2-dipyridyl, 2-methyl-1,2-di-3-pyridyl-1-propanone (metyrapone), and phenylmethanesulfonyl fluoride were purchased from Sigma Chemicals, Oakville, Ontario, Canada.
Nitrous oxide (N2O) was purchased from Scott specialty gases, Sarnia, Ontario, Canada. Carbon monoxide (CO) was purchased from Aldrich Chemical Company, Milwaukee, Wis. All other chemicals were of the highest purity available.
Isolation and identification of the CL-20-degrading strain.
One gram of garden soil was suspended in 20 ml of minimal medium (ingredients per liter of deionized water: K2HPO4, 1.22 g; KH2PO4, 0.61 g; NaCl, 0.20 g; MgSO4, 0.20 g; and succinate, 8.00 g [pH 7.0]) supplemented with CL-20 at a final concentration of 4.38 mg liter−1 added from a 10,000-mg liter−1 stock solution made in acetone. The inoculated medium was incubated under aerobic conditions at 30°C on an orbital shaker (150 rpm) in the dark. The disappearance of CL-20 was monitored over several days. The enriched culture was plated periodically onto the same medium with 1.8% agar (Difco, Becton Dickinson and Co., Sparks, Md.), and surfaces of solidified agar plates were layered with 10 μM CL-20. The isolated colonies were subcultured three times with the same agar plates and were tested for their ability to biotransform CL-20 in liquid medium. Of the few isolated bacterial strains, a denitrifying strain capable of utilizing CL-20 as a sole nitrogen source, FA1, was selected for further study.
For identification and characterization of strain FA1, we used the standard biochemical techniques reported in Bergey's Manual of Systematic Bacteriology (19). Total cellular fatty acids (fatty acid methyl ester) analysis and 16S rRNA gene analysis were performed and analyzed by MIDI Laboratories (Newark, Del.).
Biotransformation studies with strain FA1.
In biotransformation studies, CL-20 was added to the medium in concentrations above saturation levels (i.e., ≥10 μM or 4.38 mg liter−1) from a 10,000-mg liter−1 stock solution made in acetone. The aqueous solubility of CL-20 has been reported as 3.6 mg liter−1 at 25°C (10). Higher CL-20 concentrations were used in order to detect and quantify the metabolites which are otherwise produced in trace amounts during biotransformation. To determine the residual CL-20 during biotransformation studies, the media were inoculated in multiple identical batches of serum bottles. At each time point, the total CL-20 content in one serum bottle was solubilized in 50% aqueous acetonitrile and analyzed by a high-performance liquid chromatography (HPLC) (mentioned below).
A minimal medium (MM) was used for the CL-20 biotransformation studies and was composed of (per liter of deionized water) 1.22 g of K2HPO4, 0.61 g of KH2PO4, 0.20 g of NaCl, 0.20 g of MgSO4, 8.00 g of succinate, and 10 ml of trace elements (pH 7.0). Modified Wolfe's mineral solution was used as the trace element solution and was composed of (per liter of deionized water) 0.20 g of MnSO4·H2O, 0.10 g of CaCl2·2H2O, 0.10 g of CoCl2·6H2O, 0.15 g of ZnCl2, 0.01 g of CuSO4·5H2O, 0.10 g of FeSO4·7H2O, 0.05 g of Na2MoO4, 0.05 g of NiCl2·6H2O, and 0.05 g of Na2WO4·2H2O.
A comparative-growth experiment was performed with (NH4)2SO4 and CL-20 as sole nitrogen sources to determine the number of nitrogen atoms from CL-20 that were incorporated into the biomass. Cells were grown in MM containing increasing concentrations of either (NH4)2SO4 or CL-20 as a sole nitrogen source at 30°C under aerobic conditions on an orbital shaker (150 rpm) in the dark for 16 h. After the incubation period, the microbial growth yield in the form of total viable-cell counts were determined by a standard plate count method. In this method, the cultures were serially diluted in sterile phosphate-buffered saline (PBS) and spread plated onto Luria-Bertani agar plates (per liter of deionized water, 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, and 15 g of agar). All ingredients, except NaCl, were purchased from Becton Dickinson and Company. The plates were incubated at 30°C overnight. After incubation, the number of bacterial colonies grown in the plates was considered to determine the total viable-cell count per ml of the culture.
In order to determine the effect of alternate cycles of aerobic and anaerobic growth conditions on CL-20 biotransformation by the isolate FA1, cells were grown in MM containing 10 mM (NH4)2SO4 and 25 μM CL-20 in two serum bottles under aerobic conditions up to a late log phase (optical density at 600 nm [OD600], ∼0.60), and then anaerobic conditions were created in one of the two growing cultures by flushing the headspace with argon for 30 min. The cultures were further grown to stationary phase. Growth and CL-20 disappearance in both serum bottles were monitored over the course of the experiment.
To determine whether the enzyme system responsible for CL-20 biotransformation was induced or constitutive, two batches of cells were grown in MM containing 10 mM (NH4)2SO4 in the presence and absence of CL-20 (10 μM). At mid-log phase, the cells were harvested by centrifugation at 4°C and washed three times with PBS, pH 7.0. The washed cells (5 mg of wet biomass/ml) were tested for their ability to biotransform CL-20 under aerobic and anaerobic conditions.
Preparation of cytosolic and membrane-associated enzymes.
Bacterial cells were cultured in 2 liters of MM containing 10 mM (NH4)2SO4 up to a mid-log phase (8 to 9 h; OD600, 0.45) at 30°C and then induced with 10 μM CL-20. After induction, the cells were further incubated up to 12 to 16 h (OD600, 0.95). Cells were harvested by centrifugation, washed three times with PBS (pH 7.0), and then suspended in 50 mM potassium phosphate buffer (pH 7.0) containing 1 mM phenylmethanesulfonyl acid and 100 mM NaCl. The washed cell biomass (0.2 g/ml) was subjected to disruption with a French press at 20,000 lb/in2. The disrupted cell suspension was centrifuged at 9,000 × g for 30 min at 4°C to remove cell debris and undisrupted cells. The supernatant was centrifuged at 165,000 × g for 1 h at 4°C. The pellet (membrane protein fraction) and supernatant (soluble-protein fraction) thus obtained were separated and mixed with 10% glycerol, and aliquots were prepared and stored at −20°C until further use. The protein content was determined with a bicinchoninic acid protein assay kit from Pierce Chemical Company, Rockford, Ill.
Total flavin (FMN and FAD) contents in the crude extract, the membrane fraction, and the soluble-protein fractions were determined by a spectrophotometric method described by Aliverti et al. (1). Deflavoenzyme(s) and reconstituted deflavoenzyme(s) were prepared as described before (3).
Biotransformation assays.
Enzyme-catalyzed biotransformation assays were performed under aerobic as well as anaerobic conditions in 6-ml glass vials. Anaerobic conditions were created by purging all the solutions with argon gas three times (10 min each time at 10-min intervals) and replacing the headspace air with argon in sealed vials. Each assay vial contained, in 1 ml of assay mixture, CL-20 (25 μM), NADH or NADPH (150 μM), a soluble-enzyme or membrane enzyme preparation (1.0 mg), and potassium phosphate buffer (50 mM, pH 7.0). Reactions were performed at 30°C. Different controls were prepared by omitting enzyme, CL-20, or NADH from the assay mixture. Boiled enzyme was also used as a negative control. Residual NADH or NADPH was measured as described before (3). Samples from the liquid and gas phases in the vials were analyzed for residual CL-20 and biotransformed products. The CL-20 biotransformation activity of the enzyme(s) was expressed as nanomoles per hour per milligram of protein unless otherwise stated.
The bioconversion of nitrite to nitrous oxide was determined by incubating 20 μM NaNO2 with a membrane enzyme preparation using NADH as the electron donor. The disappearance of nitrite and the formation of nitrous oxide were measured periodically. Results were compared with those for a control without NaNO2.
Enzyme inhibition studies.
Inhibition with DPI, an inhibitor of flavoenzymes that acts by forming a flavin-phenyl adduct (7), was assessed by incubating the enzyme preparation with DPI at different concentrations (0 to 2.0 mM) at room temperature for 30 min before CL-20 biotransformation activities were determined. Other enzyme inhibitors, such as dicumarol, carbon monoxide (60 s of bubbling through the enzyme solution), metyrapone, and 2,2-dipyridyl, were incubated with the enzyme preparation at different concentrations for 30 min at room temperature. Thereafter, the CL-20 biotransformation activity of the treated enzyme was determined.
Analytical procedures.
CL-20 was analyzed with an HPLC connected to a photodiode array detector (λ, 230 nm). Samples (50 μl) were injected into a Supelcosil LC-CN column (4.6 mm [inside diameter] by 25 cm) (Supelco, Oakville, Ontario, Canada), and the analytes were eluted with an isocratic mobile phase of 70% methanol in water at a flow rate of 1.0 ml/min.
Nitrite (NO ), nitrous oxide (N2O), and formaldehyde (HCHO) were analyzed by previously reported methods (3-5).
), nitrous oxide (N2O), and formaldehyde (HCHO) were analyzed by previously reported methods (3-5).
Formic acid (HCOOH) was measured using an HPLC from Waters (pump model 600 and autosampler model 717 plus) equipped with a conductivity detector (model 430). The separation was made on a DIONEX IonPac AS15 column (2 by 250 mm). The mobile phase was 30 mM KOH, with a flow rate of 0.4 ml/min at 40°C. The detection of formic acid was enhanced by reducing the background with an autosuppressor from ALTECH (model DS-Plus), and the detection limit was 100 ppb.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of Pseudomonas sp. strain FA1 was deposited in GenBank under accession number AY312988.
RESULTS AND DISCUSSION
Isolation and identification of CL-20-degrading strain FA1.
The standard enrichment techniques were used to isolate CL-20-degrading strains from garden soil samples. The enrichment experiments were carried out over a period of 3 weeks, and four CL-20-degrading strains designated FA1 to FA4 were isolated. Strain FA1 biotransformed CL-20 at a higher rate than those of the other isolates (data not shown) and was capable of utilizing CL-20 as a sole nitrogen source; therefore, it was selected for further study.
Strain FA2 was identified as a Bacillus species by 16S rRNA gene analysis, while strains FA3 and FA4 remained unidentified. FA1 was characterized by standard biochemical tests mentioned in Bergey's Manual of Systematic Bacteriology (19). Strain FA1 was a non-spore-forming, gram-negative, motile bacterium with a small rod structure (approximately 1.5 to 2.0 μm). Biochemically, it showed positive results for oxidase, catalase, and nitrite reductase and utilized succinate, fumarate, acetate, glycerol, and ethanol as sole carbon sources. It utilized CL-20, ammonium sulfate, ammonium chloride, and sodium nitrite as sole nitrogen sources. Total cellular fatty acid methyl ester analysis of strain FA1 showed a similarity index of 0.748 with Pseudomonas putida biotype A. On the other hand, 16S rRNA gene analysis showed that strain FA1 was 99% similar to Pseudomonas sp. strain C22B (GenBank accession number AF408939) isolated from a soil sample in a shipping container. No published data are available with regard to strain C22B. On the basis of the above data, we identified and named strain FA1 Pseudomonas sp. strain FA1.
Growth of strain FA1 on CL-20 as a nitrogen source.
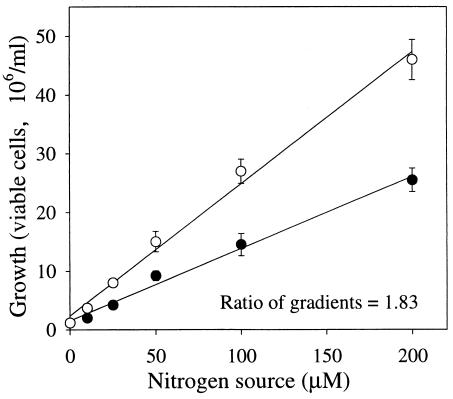
As mentioned above, strain FA1 was capable of utilizing CL-20, ammonium sulfate, ammonium chloride, and sodium nitrite as sole nitrogen sources. In order to determine the number of nitrogen atoms from CL-20 that were incorporated into biomass, cells were grown in MM containing different concentrations of either (NH4)2SO4 or CL-20. After incubation, the growth yield in the form of total viable-cell counts was determined. The growth yield using CL-20 as the nitrogen source was about 1.83-fold higher than that observed with (NH4)2SO4 (Fig. 2). No growth was observed in the control experiment without any nitrogen source. The ratio of growth yields in (NH4)2SO4 to those in CL-20 (Fig. 2) indicated that of the 12 nitrogen atoms per CL-20 molecule, approximately 4 nitrogen atoms were assimilated into the biomass. In a previous report, a soil isolate, Agrobacterium sp. strain JS71, utilized CL-20 as a sole nitrogen source and assimilated 3 mol of nitrogen per mol of CL-20 (24).
FIG. 2.
Growth of Pseudomonas sp. strain FA1 at various concentrations of CL-20 (○) and (NH4)2SO4 (•). The viable-cell count in early-stationary-phase culture (16 h) was determined for each nitrogen concentration. The linear-regression curve for (NH4)2SO4 has a gradient of 0.122 and an r2 of 0.990. The linear-regression curve for CL-20 has a gradient of 0.224 and an r2 of 0.992. Data are means of results from duplicate experiments, and error bars indicate standard errors. Some error bars are not visible due to their small size.
Biotransformation of CL-20 by intact cells.
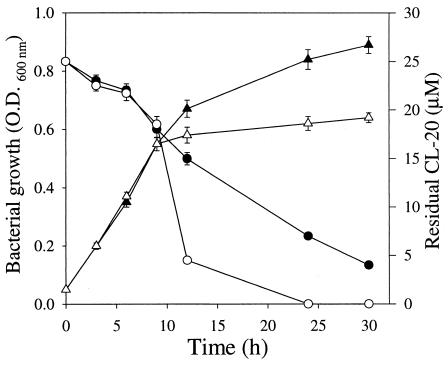
In a study of the effect of an alternate cycle of aerobic and anaerobic growth conditions on CL-20 biotransformation, we observed that after anaerobic conditions were created in one of the two growing cultures at 9 h of growth, most of the CL-20 was biotransformed in the subsequent 2 h of incubation but that under aerobic conditions, it took more than 20 h to biotransform the same amount of CL-20 (Fig. 3). This experimental finding indicated that the growth of Pseudomonas sp. strain FA1 was faster under aerobic conditions and that CL-20 biotransformation by the mid-log-phase (8- to 9-h) culture was more rapid under anaerobic conditions.
FIG. 3.
Effects of an alternating cycle of aerobic and anaerobic growth conditions on the biotransformation of CL-20 by Pseudomonas sp. strain FA1. Shown are levels of growth (▴) and CL-20 degradation (•) under aerobic conditions. Open triangles and circles show the levels of growth and CL-20 biotransformation, respectively, under aerobic conditions (for the first 9 h) and then under anaerobic conditions. Data are means of results from triplicate experiments, and error bars indicate standard errors. Some error bars are not visible due to their small size.
An experiment with uninduced and CL-20 (10 μM)-induced cells showed CL-20 biotransformation activities of 1.4 ± 0.05 and 3.2 ± 0.1 nmol h−1 mg of protein−1, respectively, indicating that CL-20 was biotransformed at a 2.2-fold-higher rate by the induced cells than by the uninduced cells. This experimental finding indicated that there may have been an up-regulation of an enzyme in the induced cells that might have been responsible for CL-20 biotransformation. In addition, the increase in activity may have been due to an improved uptake of CL-20 following induction of the cells with CL-20.
Localization of the enzyme(s) responsible for CL-20 biotransformation.
The CL-20 biotransformation activities of cell crude extract, the cytosolic soluble enzyme(s), and the membrane enzyme(s) were determined under aerobic as well as anaerobic conditions. We found that all three enzyme fractions exhibited higher activities under anaerobic conditions (Table 1) than those observed under aerobic conditions (data not shown). In the case of the membrane enzyme(s), CL-20 biotransformation was about fivefold higher under anaerobic conditions (11.5 ± 0.4 nmol h−1 mg of protein−1) than under aerobic conditions (2.5 ± 0.1 nmol h−1 mg of protein−1), indicating the involvement of an initial oxygen-sensitive step during the biotransformation of CL-20. As a result, the subsequent study was carried out under anaerobic conditions.
TABLE 1.
Effect of flavin contents in native- and deflavoenzyme preparations on the CL-20 biotransformation activities of various enzyme fractions from Pseudomonas sp. strain FA1 under anaerobic conditionsa
| Enzyme(s) | Total flavin content in native enzyme(s) (nmol/mg of protein) | CL-20 biotransformation activity of native enzyme(s) (nmol h−1 mg of protein−1) | Total Flavin content in deflavoenzyme(s) (nmol/mg of protein) | CL-20 biotransformation activity of deflavoenzyme(s) (nmol h−1 mg of protein−1) |
|---|---|---|---|---|
| Cell crude extract | 22.6 ± 1.3 | 15.6 ± 0.7 | ND | ND |
| Cytosolic soluble enzyme(s) | 5.5 ± 0.2 | 2.3 ± 0.05 | 1.2 ± 0.2 | 0.5 ± 0.05 |
| Membrane-associated enzyme(s) | 12.6 ± 0.6 | 11.5 ± 0.4 | 3.8 ± 0.3 | 2.7 ± 0.1 |
Data are means ± standard errors from triplicate experiments. ND, not determined.
The CL-20 biotransformation activity of the membrane enzyme(s) using NADH or NADPH as an electron donor was 11.5 ± 0.4 or 2.1 ± 0.1 nmol h−1 mg of protein−1, respectively, indicating that the responsible enzyme was mainly NADH dependent.
The CL-20 biotransformation activities of membrane and soluble-enzyme fractions were 11.5 ± 0.4 and 2.3 ± 0.05 nmol h−1 mg of protein−1, respectively (Table 1), which clearly indicated that the enzyme(s) responsible for CL-20 biotransformation was membrane associated. The CL-20 biotransformation activities observed in the soluble-enzyme fraction presumably leached out from the membrane enzyme fraction during the cell disruption process.
Enzymatic biotransformation of CL-20 and product stoichiometry.
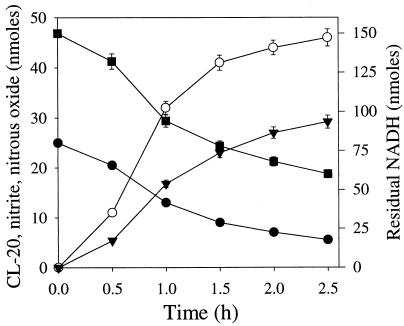
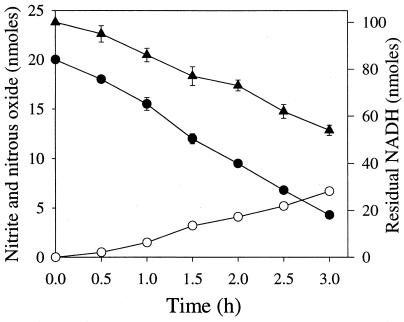
The membrane enzyme(s) catalyzed the biotransformation of CL-20 optimally at pH 7.0. Activity remained unchanged between pHs 6.0 and 7.5, but higher or lower pHs caused reduction in activity (data not shown). A time course study carried out with the membrane enzyme(s) showed that CL-20 disappearance was accompanied by the formation of nitrite and nitrous oxide at the expense of the electron donor NADH (Fig. 4). After 2.5 h of reaction, each reacted CL-20 molecule produced about 2.3 nitrite ions, 1.5 molecules of nitrous oxide, and 1.7 molecules of formic acid (Table 2). Of the total 12 nitrogen atoms (N) and 6 carbon atoms (C) per reacted CL-20 molecule, we recovered approximately 5 N (as nitrite and nitrous oxide) and 2 C (as HCOOH) atoms, respectively. The remaining seven N and four C atoms may be present in an unidentified intermediate(s).
FIG. 4.
Time course study of NADH-dependent biotransformation of CL-20 by a membrane-associated enzyme(s) from Pseudomonas sp. strain FA1 under anaerobic conditions. Symbols indicate the levels of CL-20 (•), NADH (▪), nitrite (○), and nitrous oxide (▾). Data are means of results of triplicate experiments, and error bars indicate standard errors. Some error bars are not visible due to their small size.
TABLE 2.
Stoichiometries of reactants and products during biotransformation of CL-20a
| Reactant or product | Amt (nmol) | Molar ratio of reactant to product per reacted CL-20 molecule |
|---|---|---|
| Reactants | ||
| CL-20 | 20 | 1.0 |
| NADH | 90 | 4.5 |
| Products | ||
| Nitrite (NO2−) | 46 | 2.3 |
| Formate (HCOOH) | 34 | 1.7 |
| Nitrous oxide (N2O) | 29 | 1.5 |
CL-20 was biotransformed by a membrane-associated enzyme(s) (1 mg/ml) from Pseudomonas sp. strain FA1 at pH 7.0 and 30°C for 2.5 h under anaerobic conditions. The data are means of results of triplicate experiments.
Pseudomonas sp. strain FA1 was a denitrifying bacterium; hence, nitrite was observed as a transient intermediate during CL-20 biotransformation and was partially converted to nitrous oxide. This observation was proved by incubating the membrane enzyme(s) with inorganic NaNO2 under the same reaction conditions as those used for CL-20. The results showed an NADH-dependent reduction of nitrite (used as NaNO2) to nitrous oxide (Fig. 5).
FIG. 5.
Time course study of the NADH-dependent reduction of nitrite to nitrous oxide by a membrane-associated enzyme(s) from Pseudomonas sp. strain FA1 under anaerobic conditions. Symbols indicate the levels of nitrite (•), nitrous oxide (○), and NADH (▴). Data are means of results from triplicate experiments, and error bars indicate standard errors. Some error bars are not visible due to their small size.
In biological systems, the enzymatic conversion of nitrite to nitrous oxide occurs via a transient formation of nitric oxide (NO), and this process involves two enzymes, i.e., nitrite reductase (converts nitrite to nitric oxide) and nitric oxide reductase (converts nitric oxide to nitrous oxide). Since Pseudomonas species are known to produce these two reductase enzymes (2, 8), we assume that the membrane preparation from strain FA1 may contain these two enzymes.
Involvement of a flavoenzyme(s) in the biotransformation of CL-20.
The total flavin contents were measured in crude extract, cytosolic soluble enzymes, and membrane enzymes. The membrane enzyme(s) contained about 56% of the total flavin content and retained about 74% of the total CL-20 biotransformation activity present in the crude extract (Table 1). In the deflavoenzyme preparation there was a corresponding decrease in flavin content as well as CL-20 biotransformation activity (Table 1), which indicated the involvement of a flavin moiety in CL-20 biotransformation. Furthermore, the CL-20 biotransformation activity of the deflavoenzyme was restored up to 75% after reconstitution with equimolar concentrations of FAD and FMN (100 μM each). The comparison of CL-20 biotransformation activities of the native enzyme (11.5 ± 0.4 nmol h−1 mg of protein−1), deflavoenzyme (2.7 ± 0.1 nmol h−1 mg of protein−1), and reconstituted enzyme(s) (8.90 ± 0.5 nmol h−1 mg of protein−1) clearly showed the involvement of a flavoenzyme(s) in the biotransformation of CL-20 by Pseudomonas sp. strain FA1. The free FAD and FMN also biotransformed CL-20 in the presence of NADH; however, the biotransformation rate was about fivefold lower than that of the native membrane enzyme(s). This finding additionally supported the involvement of a flavin-containing enzyme in CL-20 biotransformation and also indicated that the flavin moieties have to be in an enzyme-bound form in order to function efficiently.
Study with DPI showed a 62% inhibition of CL-20 biotransformation (Table 3). Analogously with previous reports which proved that DPI targets flavin-containing enzymes that catalyze one-electron transfer reactions (7, 18), the present study suggested the involvement of such an enzyme during the biotransformation of CL-20 by strain FA1. The involvement of a flavoenzyme in the biotransformation of RDX (3) and HMX (5) via one-electron transfer has already been established. In a previous study with diaphorase (a FMN-containing flavoenzyme from Clostridium kluyveri), an oxygen-sensitive one-electron transfer reaction that caused the N denitration of RDX, leading to its decomposition, was reported (3). However, a xanthine oxidase catalyzed an oxygen-sensitive, initial single N denitration of HMX at the FAD site, leading to the spontaneous decomposition of the molecule (5).
TABLE 3.
Effects of enzyme inhibitors on the CL-20 biotransformation activity of a membrane-associated enzyme(s)c
| Inhibitor (2 mM) | % CL-20 biotransformation activitya |
|---|---|
| None (control) | 100 ± 1.7 |
| Diphenyliodonium | 38 ± 2.3 |
| Dicumarol | 86 ± 3.5 |
| Metyrapone | 91 ± 2.8 |
| Carbon monoxideb | 87 ± 2.2 |
| 2,2-Dipyridyl | 92 ± 3.4 |
One hundred percent CL-20 biotransformation activity was equivalent to 11.5 nmol h−1 mg of protein−1. Data are mean percentages of CL-20 biotransformation activity ± standard errors (n = 3).
Carbon monoxide was bubbled through the aqueous phase and headspace for 60 s in sealed vials.
The CL-20 biotransformation activity of the membrane-associated enzyme(s) (1 mg/ml) was determined at pH 7.0 and 30°C after 1 h under anaerobic conditions.
On the other hand, enzyme inhibitors such as dicumarol (a diphosphopyridine nucleotide-triphosphopyridine nucleotide-diaphorase inhibitor) (23), metyrapone, and CO (cytochrome P450 inhibitors) (6) and the metal chelator 2,2-dipyridyl did not show effective inhibition of the CL-20 biotransformation activity of the membrane enzyme(s) from strain FA1 (Table 3). The inhibition study ruled out the possibility of involvement of the above-mentioned enzymes or a similar type of enzyme during the biotransformation of CL-20 by Pseudomonas sp. strain FA1.
Proposed initial reaction of CL-20 biotransformation.
According to the time course study described above, the disappearance of CL-20 was accompanied by the formation of nitrite (Fig. 4) and this reaction was oxygen sensitive. Additionally, the DPI-mediated inhibition of CL-20 degradation activity (Table 3) showed the involvement of a flavoenzyme catalyzing one-electron transfer. The evidence suggests that the CL-20 molecule undergoes enzyme-catalyzed one-electron reduction to form an anion radical of CL-20. This anion radical undergoes denitration to form a free radical, which eventually undergoes spontaneous ring cleavage and decomposition to produce nitrous oxide, nitrite, and formic acid. Previously, the one-electron transfer reaction catalyzed by a diaphorase, a flavoenzyme from C. kluyveri, which caused the N denitration of RDX, leading to its decomposition, was reported (3). The present study analogously with the results of initial biotransformations of other cyclic nitramine compounds, such as RDX (3) and HMX (5), supports an initial enzymatic N denitration of CL-20 prior to ring cleavage. On the other hand, thermolysis (20) and photolysis (J. Hawari, unpublished results) of CL-20 also suggested that an initial homolysis of a N—NO2 bond in CL-20 leads to the formation of a N-centered free-radical that undergoes rapid ring cleavage and decomposition.
The source of nitrite ions in the present study is probably the four nitro groups bonded to the two cyclopentane rings in the CL-20 structure (Fig. 1). Nitrous oxide can be produced in two different ways: first, by enzymatic reduction of nitrite, and second, during secondary decomposition of the CL-20 free radical as it was previously suggested by Patil and Brill (20). Analogously, nitrous oxide was also produced during the secondary decomposition of RDX (3) and HMX (5, 14). Formic acid was presumably formed following denitration and the cleavage of the C—C bond (C1—C2 in Fig. 1). The C1—C2 bond bridging the two cyclopentanes in the CL-20 structure is relatively longer (25) and thus weaker than the other C—C bonds and would cleave more rapidly. Formaldehyde (HCHO), a major carbon compound produced during the biotransformation of RDX and HMX (3-5, 12-15), was not observed in the present study.
In conclusion, a Pseudomonas sp. strain capable of utilizing CL-20 as the sole nitrogen source, FA1, was isolated and identified from a soil sample. Strain FA1 grew well under aerobic conditions but biotransformed CL-20 under anaerobic conditions to produce nitrite, nitrous oxide, and formic acid. Studies with the deflavo form of the enzyme and its subsequent reconstitution and the inhibition of holoenzyme by DPI evidently support the involvement of a flavoenzyme in CL-20 biotransformation. Several lines of evidence in the present study have proved that the enzyme responsible for the biotransformation of CL-20 by strain FA1 is an NADH-dependent, membrane-associated flavoenzyme. The present study has provided the insights into the initial microbial and enzymatic biotransformation of CL-20 and some of its products that were not known before. However, further work is necessary to identify the intermediates and end products from CL-20 to supplement the mass balance study, which would help in determining the complete biodegradation pathway of CL-20. A vast literature available online (http://www.ncbi.nlm.nih.gov) revealed that Pseudomonas and the bacteria belonging to the family Pseudomonadaceae are prevalent in almost all types of environments, e.g., soils and marine and fresh water sediments. The present study therefore helps in understanding the environmental fate (biotransformation, biodegradation, and/or natural attenuation) of cyclicnitramine explosive compounds such as CL-20.
Acknowledgments
We thank the Natural Sciences and Engineering Research Council and the National Research Council of Canada for a visiting fellowship to B. Bhushan and the Strategic Environmental Research and Development Program for funding the project (grant CU 1256). We also thank the Department of National Defense, Val Belair, Quebec, Canada, for its support.
Special thanks are due to Tara Hooper and Dominic Manno for their assistance. We sincerely acknowledge the analytical and technical support of A. Corriveau, C. Beaulieu, A. Halasz, S. Deschamps, and C. Groom. We also acknowledge the helpful discussions of V. Balakrishnan, D. Fournier, and J. S. Zhao and the critical suggestions of the editor and anonymous reviewers for improving the manuscript.
REFERENCES
- 1.Aliverti, A., B. Curti, and M. A. Vanoni. 1999. Identifying and quantifying FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol. Biol. 131:9-23. [DOI] [PubMed] [Google Scholar]
- 2.Arese, M., W. G. Zumft, and F. Cutruzzola. 2003. Expression of a fully functional cd1 nitrite reductase from Pseudomonas aeruginosa in Pseudomonas stutzeri. Protein Expr. Purif. 27:42-48. [DOI] [PubMed] [Google Scholar]
- 3.Bhushan, B., A. Halasz, J. C. Spain, and J. Hawari. 2002. Diaphorase catalyzed biotransformation of RDX via N-denitration mechanism. Biochem. Biophys. Res. Commun. 296:779-784. [DOI] [PubMed] [Google Scholar]
- 4.Bhushan, B., A. Halasz, J. Spain, S. Thiboutot, G. Ampleman, and J. Hawari. 2002. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) catalyzed by a NAD(P)H:nitrate oxidoreductase from Aspergillus niger. Environ. Sci. Technol. 36:3104-3108. [DOI] [PubMed] [Google Scholar]
- 5.Bhushan, B., L. Paquet, A. Halasz, J. C. Spain, and J. Hawari. 2003. Mechanism of xanthine oxidase catalyzed biotransformation of HMX under anaerobic conditions. Biochem. Biophys. Res. Commun. 306:509-515. [DOI] [PubMed] [Google Scholar]
- 6.Bhushan, B., S. Trott, J. C. Spain, A. Halasz, L. Paquet, and J. Hawari. 2003. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a rabbit liver cytochrome P450: insight into the mechanism of RDX biodegradation by Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 69:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty, S., and V. Massey. 2002. Reaction of reduced flavins and flavoproteins with diphenyliodonium chloride. J. Biol. Chem. 277:41507-41516. [DOI] [PubMed] [Google Scholar]
- 8.Forte, E., A. Urbani, M. Saraste, P. Sarti, M. Brunori, and A. Giuffre. 2001. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur. J. Biochem. 268:6486-6490. [DOI] [PubMed] [Google Scholar]
- 9.Fournier, D., A. Halasz, J. C. Spain, P. Fiurasek, and J. Hawari. 2002. Determination of key metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 68:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groom, C. A., A. Halasz, L. Paquet, P. D'Cruz, and J. Hawari. 2003. Cyclodextrin-assisted capillary electrophoresis for determination of the cyclic nitramine explosives RDX, HMX and CL-20: comparison with high-performance liquid chromatography. J. Chromatogr. A 999:17-22. [DOI] [PubMed] [Google Scholar]
- 11.Haas, R., E. von Löw Schreiber, and G. Stork. 1990. Conception for the investigation of contaminated munitions plants. 2. Investigation of former RDX-plants and filling stations. Fresenius' J. Anal. Chem. 338:41-45. [Google Scholar]
- 12.Halasz, A., J. Spain, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Insights into the formation and degradation of methylenedinitramine during the incubation of RDX with anaerobic sludge. Environ. Sci. Technol. 36:633-638. [DOI] [PubMed] [Google Scholar]
- 13.Hawari, J. 2000. Biodegradation of RDX and HMX: from basic research to field application, p. 277-310. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.
- 14.Hawari, J., A. Halasz, S. Beaudet, L. Paquet, G. Ampleman, and S. Thiboutot. 2001. Biotransformation routes of octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine by municipal anaerobic sludge. Environ. Sci. Technol. 35:70-75. [DOI] [PubMed] [Google Scholar]
- 15.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal sludge. Appl. Environ. Microbiol. 66:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myler, C. A., and W. Sisk. 1991. Bioremediation of explosives contaminated soils (scientific questions/engineering realities), p. 137-146. In G. S. Sayler, R. Fox, and J. W. Blackburn (ed.), Environmental bio/technology for waste treatment. Plenum Press, New York, N.Y.
- 17.Nielsen, A. T., A. P. Chafin, S. L. Christian, D. W. Moore, M. P. Nadler, R. A. Nissan, and D. J. Vanderah. 1998. Synthesis of polyazapolycyclic caged polynitramines. Tetrahedron 54:11793-11812. [Google Scholar]
- 18.O'Donnell, V. B., G. C. Smith, and O. T. Jones. 1994. Involvement of phenyl radicals in iodonium inhibition of flavoenzymes. Mol. Pharmacol. 46:778-785. [PubMed] [Google Scholar]
- 19.Palleroni, N. J. 1984. Gram-negative aerobic rods and cocci: family I Pseudomonadaceae, p. 140-198. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 20.Patil, D. G., and T. B. Brill. 1991. Thermal decomposition of energetic materials. 53. Kinetics and mechanisms of thermolysis of hexanitrohexaazaisowurtzitane. Combust. Flame 87:145-151. [Google Scholar]
- 21.Seth-Smith, H. M. B., S. J. Rosser, A. Basran, E. R. Travis, E. R. Dabbs, S. Nicklin, and N. C. Bruce. 2002. Cloning, sequencing, and characterization of the hexahydro-1,3,5-trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl. Environ. Microbiol. 68:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talmage, S. S., D. M. Opresko, C. J. Maxwel, C. J. E. Welsh, F. M. Cretella, P. H. Reno, and F. B. Daniel. 1999. Nitroaromatic munition compounds: environment effects and screening values. Rev. Environ. Contam. Toxicol. 161:1-156. [DOI] [PubMed] [Google Scholar]
- 23.Tedeschi, G., S. Chen, and V. Massey. 1995. DT-diaphorase: redox potential, steady-state, and rapid reaction study. J. Biol. Chem. 270:1198-1204. [DOI] [PubMed] [Google Scholar]
- 24.Trott, S., S. F. Nishino, J. Hawari, and J. C. Spain. 2003. Biodegradation of the nitramine explosive CL-20. Appl. Environ. Microbiol. 69:1871-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xinqi, Z., and S. Nicheng. 1996. Crystal and molecular structures of ɛ-HNIW. Chin. Sci. Bull. 41:574-576. [Google Scholar]
- 26.Zhao, J. S., A. Halasz, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Biodegradation of RDX and its mononitroso derivative MNX by Klebsiella sp. strain SCZ-1 isolated from an anaerobic sludge. Appl. Environ. Microbiol. 68:5336-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]