Abstract
Confocal laser scanning microscopy (CLSM), transmission electron microscopy (TEM), and soft X-ray scanning transmission X-ray microscopy (STXM) were used to map the distribution of macromolecular subcomponents (e.g., polysaccharides, proteins, lipids, and nucleic acids) of biofilm cells and matrix. The biofilms were developed from river water supplemented with methanol, and although they comprised a complex microbial community, the biofilms were dominated by heterotrophic bacteria. TEM provided the highest-resolution structural imaging, CLSM provided detailed compositional information when used in conjunction with molecular probes, and STXM provided compositional mapping of macromolecule distributions without the addition of probes. By examining exactly the same region of a sample with combinations of these techniques (STXM with CLSM and STXM with TEM), we demonstrate that this combination of multimicroscopy analysis can be used to create a detailed correlative map of biofilm structure and composition. We are using these correlative techniques to improve our understanding of the biochemical basis for biofilm organization and to assist studies intended to investigate and optimize biofilms for environmental remediation applications.
Scanning transmission X-ray microscopy (STXM) (1, 2), which uses near-edge X-ray absorption spectroscopy (NEXAFS) as its contrast mechanism, is a powerful new tool that can be applied to fully hydrated biological materials. This is possible due to the ability of soft X rays to penetrate water, the presence of suitable analytical core edges in the soft X-ray region, and reduced radiation damage (compared to that caused by electron beam techniques). Soft X rays also provide spatial resolution of better than 50 nm, which is suitable for imaging bacteria and bacterial biofilms. The spectral resolution is on the order of 100 meV, which in combination with their intrinsic spectral properties is sufficient to provide good differentiation of classes of biomolecules (26, 43; X. Zhang, T. Araki, A. P. Hitchcock, J. R. Lawrence, and G. G. Leppard, unpublished data). Through the application of tunable soft X rays and appropriate analysis of X-ray absorption spectra in the form of NEXAFS image sequences (15), quantitative chemical mapping at a spatial scale below 50 nm may be achieved. With use of the appropriate spectral range, NEXAFS microscopy provides detailed, quantitative speciation and elemental analysis with parts-per-thousand local and parts-per-million global sensitivities with transmission detection. Soft X-ray microscopy provides a combination of suitable spatial resolution and chemical information at a microscale. In addition, soft X rays interact with nearly all elements and also allow mapping of chemical species based on bonding structure (2). Further, the method uses the intrinsic X-ray absorption properties of the sample, thus eliminating the need for addition of reflective, absorptive, or fluorescent probes and markers that may introduce artifacts or complicate interpretation. It is, however, useful to correlate the results of STXM with those of microscopy methods that allow greater penetration and 3D imaging, as in confocal laser scanning microscopy (CLSM) (18) as well as methods providing higher-resolution imaging, such as transmission electron microscopy (TEM) (22).
The matrix of biofilms is a complex mixture of exopolysaccharides, protein, and nucleic acids (36). The characteristics of transmission X-ray microscopy (scanning of full field) make it potentially ideal for localization and mapping of the distribution of macromolecules in microbial biofilms. Gilbert et al. (7) demonstrated the potential of soft X rays for imaging early-stage Pseudomonas putida biofilms using a full-field transmission X-ray microscope with synchrotron radiation. However, they measured at only a single photon energy and thus did not explore the analytical capability of X-ray microscopy. STXM is superior to full-field TXM because STXM instruments are mounted on high-resolution beam lines that provide spectra of much higher quality (26). The analytical capability of STXM may be useful to address a variety of questions regarding the nature, distribution, and role of protein, carbohydrate, lipid, and nucleic acids in biofilms, particularly in the extracellular matrix. Currently, there is only limited understanding of the roles of fibrils of polysaccharide, protein-coated polysaccharide, and other polymers (e.g., extracellular nucleic acids, extracellular proteins, and associated fibril-lipid structures). Recent work using DNase enzymes (39) indicated that extracellular DNA might have a structural role in biofilm formation and stability. Thus, mapping of macromolecular distributions is highly relevant to understanding biofilm formation, manipulation, and control.
In the present work, we applied a combination of the CLSM, TEM, and STXM methods to examine complex microbial biofilm communities and to map the extent and nature of macromolecules in a biofilm. The biofilms were grown in rotating annular reactors directly on a silicon nitride window. These samples were then removed from the reactor, closed with a second silicon nitride window to form a sealed wet cell, and sequentially subjected to CLSM fluorescence imaging as well as to soft X-ray imaging and analysis. Additional samples created in parallel were treated and analyzed using STXM-TEM techniques. When possible, data sets from different techniques applied to the same material were aligned to allow correlative mapping and improved interpretation of spatial distributions of biological macromolecules in the biofilms. We are developing these correlative techniques in order to improve our understanding of the biochemical basis for biofilm organization and to assist studies intended to investigate and optimize biofilms for environmental remediation applications. This work describes the methods and gives examples to illustrate the complementary nature of the techniques.
MATERIALS AND METHODS
Microbial culture.
Lotic biofilms were grown in rotating annular biofilm reactors as described in detail by Lawrence et al. (21). Natural river water (South Saskatchewan River, Saskatoon, Saskatchewan, Canada [52o10′N latitude and 106o41′W longitude]) was used as inoculum and sole source of nutrients. Methanol (100 mg liter−1) (protein-sequencing grade; Sigma, St. Louis, Mo.) was added to the water as a supplemental carbon source. The bioreactors contained polycarbonate slides that could be removed for microscopic examination of the biofilms. Reactors and slides were custom made in-house as described previously (21). Polycarbonate slides were also modified to allow installation of silicon nitride (Si3N4) windows (Silson, Ltd., Northampton, United Kingdom) required for STXM imaging. Glass coverslips #1 (Fisher Scientific, Toronto, Ontario, Canada) were attached to the polycarbonate surface by one edge using flowable silicon adhesive (RTV; WPI, Inc., Sarasota, Fla.) to provide a hinged clamp to hold the windows in place during biofilm development. The experimental setup included a temperature-controlled reservoir (17 ± 2°C) for the raw water, a peristaltic pump (Watson-Marlowe; Fisher Scientific) for recirculating the water, and the rotating annular reactor with motor and control for adjusting the speed of the rotating inner cylinder. All reactor components were surface sterilized with 1.2% sodium hypochlorite for 1 h. The system was operated at 160 rpm (a surface velocity of 1.9 km h−1). The water in the reservoirs was replaced with fresh river water every 7 days throughout the experimental period.
Confocal laser microscopy and probes.
CLSM was done with an MRC 1024 confocal laser scanning microscope (Bio-Rad, Hemel Hempstead, United Kingdom) attached to a Microphot SA microscope (Nikon, Tokyo, Japan) equipped with water-immersible 63× 0.9 numerical aperture (NA) (Zeiss, Jena, Germany), 40× 0.55 NA (Nikon), and oil immersion 60× 1.4 NA planapochromat lenses (Nikon). Table 1 lists the fluorescent probes that were used to label biofilm samples and to investigate the distribution of specific macromolecules. The following stains were used for staining specific structures in the biofilm community: Syto9 (nucleic acids), SyproOrange (protein) (Molecular Probes, Inc., Eugene Oreg.), selected lectins (Sigma) (glycoconjugate, sugar, and carbohydrate), and Nile Red (Molecular Probes) for lipid-hydrophobic sites. The lectin panel included Arachis hypogaea (CY5), Canavalia ensiformis (tetramethyl rhodamine isothiocyanate), Wisteria floribunda (fluorescein isothiocyanate), Tetragonolobus purpureas lectin (tetramethyl rhodamine isothiocyanate and CY5), Ulex europaeus lectin (fluorescein isothiocyanate and CY5), Datura stramonium-CY5, and Sambucus nigra-CY5 (Table 1). These fluor conjugates were used to assess the distribution of glycoconjugates at the microscale (29, 30). In addition, colloidal gold-labeled lectins (Sigma) were incubated with biofilm material. Lectin staining was performed as described in detail by Neu and Lawrence (29). In brief, for staining, the lectins were dissolved at 100 μg ml−1 in filter-sterilized (pore size, 0.20 μm) river water. The windows and slide pieces of 1 cm2 carrying the biofilm were directly stained with 100 μl of the lectin solution. The samples were then incubated for 20 min at room temperature (22 ± 2°C) in a 100%-humidity chamber. After staining, the samples were carefully rinsed with filter-sterilized (pore size, 0.20 μm; FisherBrand; Fisher Scientific) river water four times to remove unbound lectins. Slide pieces were also prepared for TEM analyses as described below.
TABLE 1.
Fluorescent and fluor-conjugated probes used in this study
| Probe-fluor conjugatea | Supplier | Excitation/emission (nm) | Target |
|---|---|---|---|
| Syto9 | Molecular Probes | 488/522 | Nucleic acids |
| SyproOrange | Molecular Probes | 470/570 | Protein |
| Nile Red | Molecular Probes | 552/636 | Lipids, hydrophobic sites |
| Arachis hypogaea-CY5 | Sigma | 650/670 | β-Gal(1-3)galNAc |
| Canavalia ensiformis-TRITC | Sigma | 554/576 | α-d-Mannose |
| α-d-Glucose | |||
| Wisteria floribunda-FITC | Sigma | 494/520 | N-Acetyl-d-galactosamine |
| Tetragonolobus purpureas-TRITC | Sigma | 554/576 | α-l-Fucose |
| Tetragonolobus purpureas-CY5 | Sigma | 650/670 | α-l-Fucose |
| Ulex europaeus-FITC | Sigma | 494/520 | α-l-Fucose |
| Ulex europaeus-CY5 | Sigma | 650/670 | α-l-Fucose |
| Ulex europaeus-Colloidal Gold | Sigma | Opaque | α-l-Fucose |
| Datura stramonium-CY5 | Sigma | 650/670 | N-Acetylglucosamine |
| N-Acetyllactosamine | |||
| Sambucus nigra-CY5 | Sigma | 650/670 | α-NeuNAc(2-6)gal/galNAc |
Conjugated dye abbreviations: FITC, fluorescein isothiocyanate; TRITC, tetramethylrhodamine isothiocyanate; CY5, pentamethincyanine.
TEM and energy-dispersive spectroscopy (EDS).
Using the correlative TEM preparatory scheme of Liss et al. (25), individual portions of biofilm were embedded in one of two different kinds of resins: the standard low-viscosity epoxy resin of Spurr (34) and the hydrophilic melamine resin Nanoplast FB 101 (6), which were subsequently polymerized by the recommended protocol for each (6, 34). The Spurr's epoxy resin was polymerized by heating (at 70°C for 8 h); the hydrophilic melamine resin was polymerized by heating in two stages taking 4 days total (at 40°C for 48 h in the presence of a desiccant, then at 60°C for another 48 h). Embedding with Spurr's resin was preceded by double fixation (with a glutaraldehyde-based fixative, followed by an osmium-based fixative, both fixatives having been obtained from Marivac Canada, Inc., Montreal, Québec, Canada) as detailed by Liss et al. (25). The resulting blocks of embedded biofilm were sectioned with a diamond knife mounted in an ultramicrotome (Reichert Ultracut E). Ultrathin sections for morphological studies were cut at 50 to 70 nm and mounted on formvar-covered TEM grids (Marivac Canada) with and without counterstaining, for high-resolution examination of internal biofilm structure. Images from the combination of preparatory treatments and embeddings were compared to reveal specific artifacts inherent to each one when used independently (25).
Sections of biofilm were observed and their images were documented in transmission mode using a JEOL JEM 1200 EX TEMSCAN at an accelerating voltage of 80 kV. A Tracor Northern (Noran, Madison, Wis.) X-ray detector and an EDS 2000 microanalysis system (IXRF Systems, Inc., Houston, Tex.) provided EDS spectra of all elements with atomic numbers greater than 10. The element abundances (14) present in measurable amounts were determined using a standardless analysis (33) with measurement times of 60 s each. Sections for EDS were cut at 80- to 100-nm thickness for an optimal compromise of section thickness with image quality; this thickness range improved element detection over the standard range for morphology and minimized beam spread artifacts (4).
Soft X-ray imaging.
In some cases, the STXM measurements were performed on a wet cell constructed from films grown directly on a silicon nitride window (Silson, Inc., Northampton, United Kingdom.) mounted on the reactor strips (see above). In some cases, the biofilm material was removed from the strip by aseptically scraping the slide with a silicon spatula and placing it in a sterile 1-ml centrifuge tube (Fisher Scientific); the biofilm material was subsequently transferred onto a silicon nitride window. The samples were sent via overnight courier from NWRI, Saskatoon, to Advanced Light Source (ALS) at Berkeley, Calif., and recorded as soon as possible. To stabilize the biofilm, the samples were refrigerated (4°C) except during analysis. Soft X-ray spectroscopy and NEXAFS imaging were carried out using several different scanning transmission X-ray microscopes on the ALS beam lines 5.3.2 and 7.0.1. Early work and reference compounds were recorded on the original STXM on ALS beam line 7.0.1 (38). The correlative CLSM-STXM study of the same region was performed on the new STXM on bending magnet 5.3.2 (17, 37) with investigations of the C 1s, Ca 2p, and N 1s inner-shell excitation spectral ranges. Here, the C 1s results are used to investigate the distribution of nucleic acids, protein, carbohydrate, and lipid in the biofilm community. Although the N 1s region provides useful corroboration, only C 1s results are presented in this paper. For C 1s studies of organic material with a density of ∼1 cm3, a sample thickness of a dry sample between 80 and 200 nm gives a peak optical density of 1 to 2, which is optimum with regard to statistical precision and avoiding absorption saturation (which in the STXM532 microscope starts at an optical density of around 3). This is typical of the preparation of solid polymer samples. The same amount of material dispersed in a water column of 2- to 3-μm thickness also gives very good results since that amount of water provides an unstructured optical density of 1 to 1.5 in the C 1s region. In the sample regions examined in this work, the optical densities—including the water and the silicon nitride windows—were between 1.5 and 2.5 in the region of single cells or areas of denser extracellular polymeric substances. However, in all regions there are also quite transparent areas with little or no biofilm components and some areas in which there is cell overlap or inorganic material where the absorption saturates.
STXM data analysis.
STXM was used analytically by acquiring NEXAFS spectra at a single location by using a line scan or through the collection of a sequence of images. The image sequence data were converted to component maps (distributions of distinct chemical species, such as proteins, polysaccharides, nucleic acids, etc.) by spectral fitting using linear-regression procedures (32, 35). When necessary (specifically, for data recorded using the 7.0.1 beam line STXM), spatial alignment of the images was performed using two-dimensional (2-D) Fourier autocorrelation techniques (14) prior to generation of the component maps. The image and spectral processing was carried out using the aXis2000 package (http://unicorn.mcmaster.ca), which is coded in Interactive Data Language (Research Systems, Inc.).
RESULTS AND DISCUSSION
CLM.
Although destructive techniques provide considerable insight regarding biofilms, direct quantitative evaluation of the intact films at the appropriate scale is required. Many techniques have been applied to the analysis of microbial biofilms (19). Recent applications of microscopic approaches have resulted in considerable change and advancement of understanding of these complex biological systems. Of particular interest are the extracellular polymeric substances (EPS), which represent in terms of weight and volume a major structural component of interfacial microbial communities (5, 36). In general, EPS are considered to be largely composed of microbial polysaccharides. However, other EPS may also be associated with biofilm systems, e.g., proteins, nucleic acids, and polymeric lipophilic compounds (29, 41). Typically, destructive analyses have been used to study the exopolymeric matrix of biofilms (31). However, this procedure can eliminate critical information on the spatial distribution, interrelationships, and structure of these constituents. This type of information is highly relevant to understanding fate and effects of contaminants such as metals and organics and for the bioengineering of biofilm systems for environmental recovery and remediation schemes. CLSM has become an indispensable technique for the study of interfacial microbial communities (18, 20). In addition, there are increasingly wide ranges of fluorescent reporter systems and stains that may be applied to gain insight into these structures in fully hydrated, living microbial systems. The techniques include those for detection and quantification of cellular and polymeric compounds in biofilms (18, 29, 30). Here, we have applied these approaches to characterize biofilm materials prior to their analysis by using TEM and STXM.
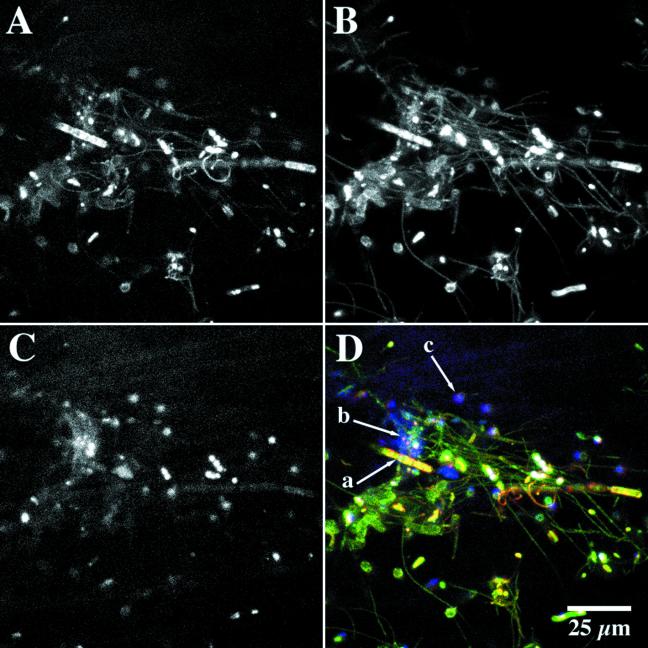
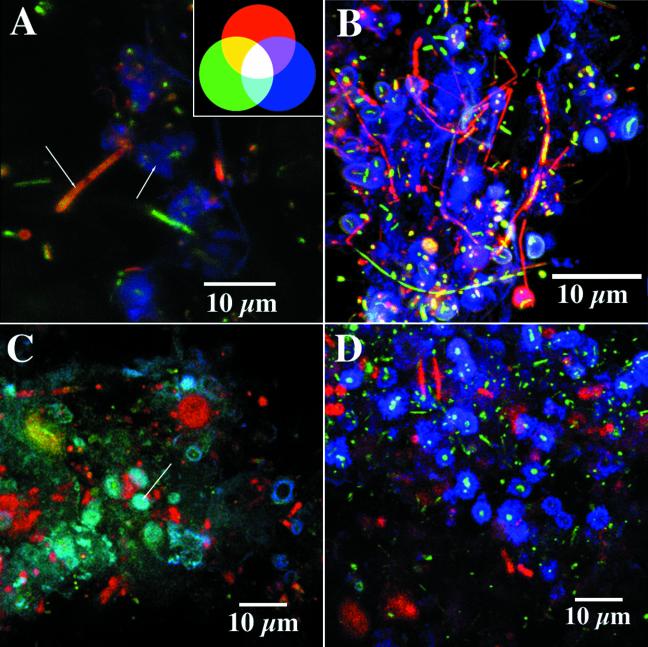
In the present study, combinations of nucleic acid, protein, lipid, and glycoconjugate staining revealed a complex microbial community consisting of numerous cell morphologies and microcolony types (Fig. 1). The biofilm community was heterogeneous in nature, and based on CLSM and light microscopic examination, algal, cyanobacterial, protozoan, and various bacterial morphotypes were present within a chemically variable EPS matrix. Additional staining with nucleic acid-, protein-, lipophilic-, and glycoconjugate-specific fluorescent probes revealed localization and colocalization of these biomolecules within the cells and matrix of the biofilm. Figure 1 shows a three-channel series illustrating the colocalization of nucleic acid-specific, protein-specific, and lipid-specific probes that bind to regions enriched in their target molecules. The biofilm location contains all target biomolecules that were associated with filamentous growth, individual cells, microcolonies, and extensive extracellular polymeric substances in the biofilm. A triple-labeled series of cells can also be observed in Fig. 1D. When these probes were used in combination with glycoconjugate-specific lectins such as those from Datura stramonium or Sambucus nigra with an affinity for N-acetyl-d-glucosamine- and N-acetyl-d-galactosamine-rich regions, respectively, the presence of nucleic acid-, protein-, or lipid-enriched regions associated with polysaccharides could be detected (Fig. 1D and 2). This image series shows both localization and colocalization of the probes associated with cells and their exopolymers. For example, in Fig. 2A, polysaccharide matrix material surrounding nucleic acid-stained cells is shown, as are filaments with both protein and nucleic acid staining and with single bacterial cells and filaments binding only the nucleic acid stain. Figure 2B illustrates the localization of lipids, nucleic acids, and polysaccharides, while Fig. 2C shows proteins, lipids, and polysaccharides present in combination. Figure 2D shows the presence of two types of glycoconjugates (i.e., N-acetyl-d-glucosamine and N-acetyl-d-galactosamine) in extracellular polysaccharides surrounding nucleic acid-rich bacterial cells.
FIG. 1.
Three-channel imaging of biofilm material stained with SyproOrange (protein) (A), Syto9 (nucleic acids) (B), and Nile Red (hydrophobic-lipid rich) (C), and the three-color combination of the channels showing the localization and colocalization of protein (red), nucleic acids (green) and lipid-hydrophobic regions (blue) (D). Arrows indicate protein plus DNA (a), protein plus lipid (b), and regions rich in lipid alone in the biofilm (c).
FIG. 2.
Triple-labeled confocal images of different biofilm samples illustrating the distribution of protein, nucleic acids, lipid, and polysaccharide regions labeled with N-acetylglucosamine- and N-acetyllactosamine-sensitive lectins. Colors denote the following: (A) protein (red), nucleic acids (green), polysaccharide (blue) (arrow indicates filamentous structure with protein plus nucleic acids); (B) lipid (red), nucleic acids (green), polysaccharide (blue); (C) lipid (red), protein (green), and polysaccharide (blue) (arrow indicates cyan region where colocalization of protein and polysaccharide was detected); (D) polysaccharide (red), nucleic acids (green), and polysaccharide (blue)—in this case, the two lectins are localized in distinct polysaccharide regions.
Using CLSM, it has been possible to delineate regions of space, cells, and exopolymeric materials, while optically sectioning and creating 3-D images of biofilms (3, 28, 30, 42). However, as discussed by Neu et al. (30), there are limitations—particularly to the chemical interpretation of fluorescent probe binding patterns in complex environments such as biofilms. Probes such as lectins may recognize specific glycoconjugates, secondary targets, or protein, or they may bind nonspecifically within the biofilm community (30). Other parallel techniques are necessary to confirm the specificity of probes used to define regions of protein, lipid, nucleic acid, and polysaccharide. Thus, there is a requirement for additional approaches to map the macromolecular composition of biofilms and provide confirmation of target specificity.
TEM and EDS.
All kinds of structural entities, which are able to be seen by conventional optical microscopy, CLSM, and STXM, can be examined by TEM at a practical resolution of ca. 1 nm (Nanoplast preparations) and ca. 3 nm (epoxy preparations). The Nanoplast images yield true dimensions for nanoscale particles and native 3-D associations between nanoscale particles (6), but they severely limit the use of selective stains for particle and colloid identification. Such images are used correlatively in conjunction with images from a proxy preparation that permits the classical differentiation of recognizable biological entities in terms of a combination of morphology and grey scale (22). The proxy of choice is a fixative-plus-ruthenium red approach followed by epoxy embedding (25); it provides an optimal compromise of dimensional fidelity and staining characteristics while minimizing extraction and dehydration artifacts. The TEM allows one to search for, at near-nanometer resolution, specific structures of interest (13, 22), as determined previously by the lower spatial resolution CLSM or STXM. TEM and associated EDS provide additional useful detail for a given kind of structure, but TEM does not permit the viewing of exactly the same structure that was examined by both CLSM and STXM.
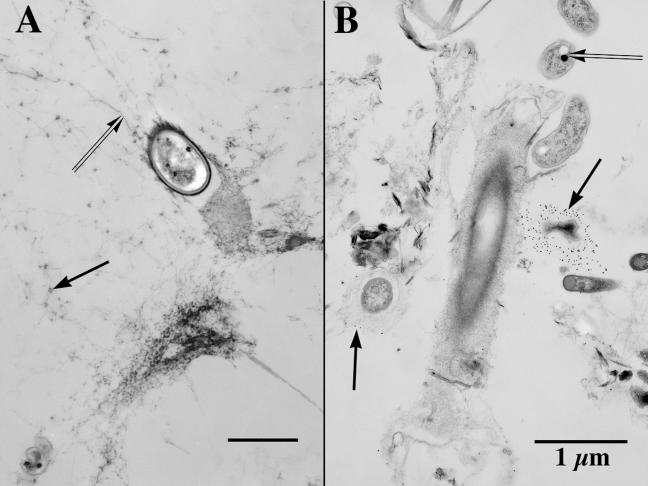
Figure 3A shows a transmission electron photomicrograph of a bacterium in a matrix of fibrillar EPS or “fibrils” (24). Fibrils with a diameter of 2 to 20 nm (but typically 3 to 10 nm) are usually rich in polysaccharide or polysaccharide and protein (23) but can also be composed of DNA (9). Polysaccharide fibrils can be visualized, with or without ruthenium red, by both TEM and atomic force microscopy (40). With the ruthenium red stain applied simultaneously as a probe and as a component of TEM fixatives, polyanionic polysaccharides and DNA are recognizable in TEM images as fibrils having a diameter at the bottom end of the nanoscale (9). The early literature on the connection (chemical-ultrastructural) between polysaccharides and nanoscale fibrils (24) and on the use of ruthenium red with TEM (27) strongly suggested that polysaccharide contributed substantially to ruthenium red binding and to the fibril image. However, in the past two decades, this use of ruthenium red as a probe for polysaccharide has been more inferred than demonstrated, with the result that (i) extracellular DNA may have been misidentified and (ii) polysaccharide fibrils coated with proteins, metals, or small organic molecules have not received sufficient attention.
FIG. 3.
(A) TEM image showing a bacterium held within a biofilm by a very porous (water-rich) matrix of fibrillar EPS, fibrils (double arrow) that cross-connect bacteria, and other colloids. Note the electron-opaque nanoscale agglomerations (arrow) on parts of some fibrils. The scale bar on this image represents 500 nm. (B) TEM image of a portion of biofilm that shows several distinctive bacterial morphotypes (including a sheathed filamentous bacterium, several naked gram-negative bacteria with their enclosed storage granules [double arrow], and an irregularly shaped bacterium surrounded by probes that consist of Ulex europaeus lectins attached to colloidal gold spheres [arrow]). Both images were derived by a double-fixation (glutaraldehyde-ruthenium red followed by osmium tetroxide-ruthenium red) protocol, which preceded an embedding in a low-viscosity epoxy resin.
The bacterium in Fig. 3A is suspended within the biofilm by a very porous 3-D cage of fibrils that surrounds it. Serial sections show that the fibrils are sometimes arranged so as to form the boundary of submicron channels that connect one part of the biofilm to another. Some electron-dense nanoscale particles associated with the EPS matrix contain nickel in quantity sufficient for detection by EDS. One value of TEM is that it can reveal that a particular structure in a biopolymeric matrix consists mainly of voids, whereas CLSM and STXM might suggest that the same structure was solid. Figure 3B shows several rod-shaped bacteria associated with a sheathed filamentous bacterium. Many nanoscale particles are associated with this bacterial community via fibrils. Note that one small bacterium is surrounded by small electron-dense spheres (above the central sheathed bacterium). These spheres are colloidal gold, as confirmed by EDS, and are part of a gold-lectin probe (Ulex europaeus) specific to certain polysaccharide components of the EPS. The TEM image provides additional correlative information on probe localization. With regard to the heterogeneous distribution of biofilm EPS components, TEM can confirm and extend the information from CLSM. In this instance, TEM has confirmed (i) the capacity of such “tagged” molecules to penetrate the biofilm matrix and (ii) the small-scale localization of lectin binding to specific ultrastructural entities within the biofilm. For biofilms having densely packed matrices, there is the possibility of poor penetration by probes, with a concomitant limitation on the utility of TEM. Also, for some chemical probes (especially protein-selective probes), localization sites may be compromised by the cross-linking action of the primary fixative or by extraction (the result of activities by the fixative's buffer, by washes between double fixations, and by dehydrating fluids). Such artifacts have been known for decades (8, 10) and are readily identified, assessed, and minimized (22, 25) through fastidious attention to appropriately modified protocols, as are artifacts of shrinkage (6, 25).
STXM.
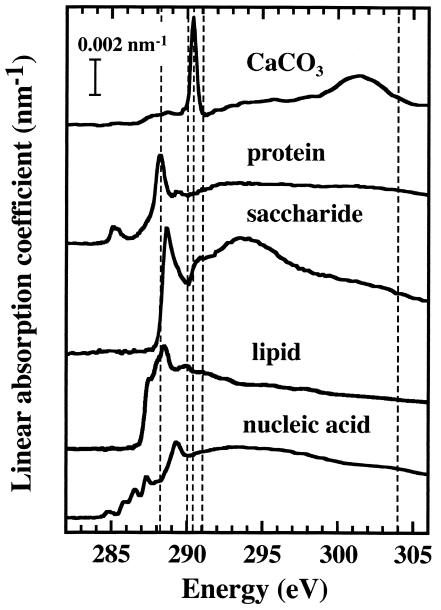
Here, we quantitatively map the biological components of the biofilm (protein, lipid, saccharide, and nucleic acids) by using the C 1s region of their absorption spectra. As a first step, a series of standards were selected to create reference spectra to allow the discrimination of these major biomolecules in the microbial biofilm communities. Figure 4 presents the C 1s spectra of albumin (protein), alginate (polysaccharide), 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine (saturated lipid), and calf thymus DNA. The spectrum of albumin (12, 26) is similar to that of fibrinogen (12) and other proteins (43), as expected from the averaging over the C 1s spectra of the individual amino acids (16). The spectrum of DNA is qualitatively in agreement with that reported by Zhang et al. (43), but the energy scale differs by 0.3 eV. Since the ALS 532 STXM on which this DNA spectrum was recorded has a very stable energy scale that is regularly checked using gas spectroscopy, we are confident the present energy scale is the correct one. The model saccharide and lipid spectra are the first reported NEXAFS of these classes of biological macromolecules to our knowledge. However, just as the protein and DNA spectra are reasonably representative of a class of biomolecules, we believe the C 1s spectra of different saccharides and lipids will be broadly similar. One exception is that the C 1s spectra of lipids will very likely vary in the 285-eV region depending on the degree of unsaturation. The C 1s model spectra plotted in Fig. 4 have been converted to an absolute linear absorption scale by setting the continuum absorption outside the structured spectral regions to match that predicted for the elemental composition using tabulated atomic coefficients (11).
FIG. 4.
C 1s NEXAFS spectra of protein (albumin), polysaccharides (sodium alginate), lipid (1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine), and nucleic acid (calf thymus DNA). All spectra were recorded with the ALS 7.0.1 STXM, with the exception of that of DNA, which was recorded with the ALS 5.3.2 STXM.
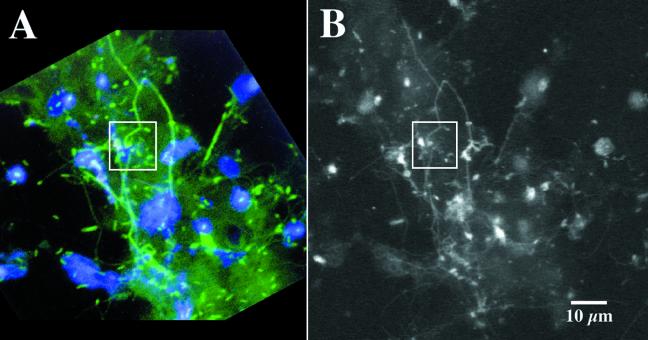
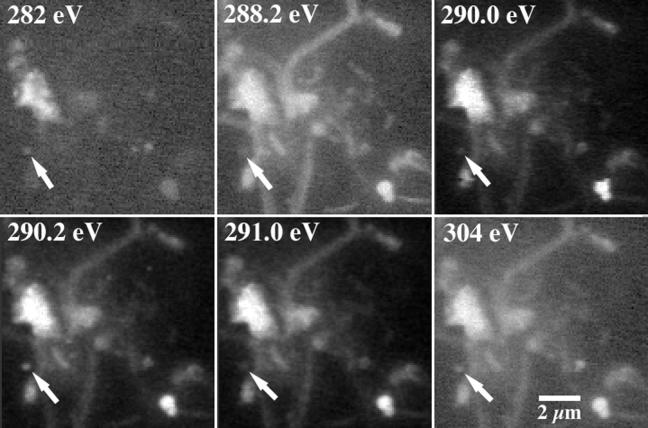
Figure 5 compares CLSM and STXM (288.2 eV) images of the same region of a biofilm. The boxed area is the biofilm region analyzed in detail subsequently by both STXM and CLSM imaging. Figure 6 presents STXM images in optical density format at several characteristic energies (those indicated by dotted lines in Fig. 4) of a region of a biofilm, all recorded in the same area. The image at 282 eV, below the onset of C 1s excitation, shows considerable contrast, which is indicative of thick regions and/or regions containing higher levels of inorganic or oxygen-rich species. The image at 304 eV mainly reflects total density, since all organic species absorb a similar amount (per C atom) at this energy. The difference in the 304- and 282-eV images is a simple means to estimate the carbon density distribution. At intermediate energies, specific organic functional groups absorb to greatly differing extents, thus providing the differential contrast that is the basis for mapping the classes of biomolecules present in the biofilm. Thus, the image at 288.2 eV at the peak of the amide carbonyl π* band highlights protein-rich regions, while the image at 290.2 eV, the peak of the π*CO absorption by the carbonate group, highlights carbonate-rich regions. Also shown in Fig. 6 is a position with a localized, strong absorption at 290.2 eV. When the spectrum of this point is extracted from the image sequence, it is a good match to the spectrum of calcium carbonate, recorded separately.
FIG. 5.
Comparative imaging of the same biofilm location by CLSM with lectin and nucleic acid staining (A) and by STXM-derived optical density image recorded at 288.4 eV (B). The boxed area indicates the region of examination in Fig. 6 and 7.
FIG. 6.
STXM images of a biofilm at selected photon energies: at 282 eV, below C 1s onset; at 288.2 eV, protein π*; at 290.2 eV, CaCO3 π*; at 290.0 and 291.0 eV, adjacent images are shown to demonstrate the sensitivity of STXM to small energy changes; 304 eV, C 1s continuum. The as-recorded transmission images have been converted to optical density (absorbance). The contrast variation is associated with differences in the X-ray absorption of various chemical components at these energies and provides the basis for quantitative chemical mapping.
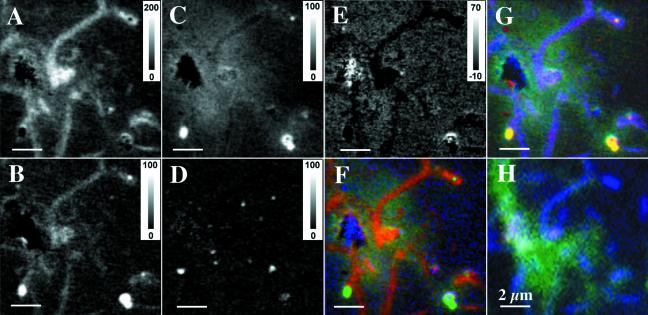
A more powerful technique for mapping the biomolecules than examination of single-energy images is to fit images recorded over a wide range of energies (A. P. Hitchcock, L. M. Croll, H. D. H. Stöver, and E. Kneedler, unpublished data). This allows the overall spectral shape of each component to contribute to the mapping. In addition, each species has some absorption at every photon energy, and thus individual photon energy images cannot be completely specific to single substances. Figure 7 presents quantitative component maps for proteins (A), lipids (B), polysaccharides (C), carbonate (D), and nucleic acid (E) derived from an 81-energy image sequence in the boxed area indicated in Fig. 5, for which selected images are shown in Fig. 6. The grey scale on the five grey-scale component maps is an estimate of the thickness in nanometers of that component. Accuracy is limited by the quality of the reference spectra and the ability to place those reference spectra on a quantitative linear absorption scale (Hitchcock et al., unpublished). The precision of the mapping is largely limited by the statistical quality of the images, although there is also a possibility of systematic error due to overlap of spectra features and instrumental systematic errors. In this case, we estimate the errors to be as large as 20% for the majority components (protein and saccharides) and perhaps 50% or more for the minority components. This fit also included reference spectra for water and “rock,” the spectrum of the region indicated by a shadow in the center left of the images in Fig. 6 and 7, where the absorption is saturated since this is a biofilm-covered inorganic component with an optical density of more than 4. Those latter two maps are not shown, because they are indicative of morphology (“rock” and water) rather than chemistry. In addition to the maps for the individual components, Fig. 7 also shows two color-coded correlative composite maps—for panel F: red (R), protein; green (G), saccharides; blue (B), nucleic acids; for panel G, R, lipids; G, saccharides; B, protein; Fig. 7H also shows the CLSM signal using probes for fucose EPS (green) and nucleic acids (blue). The individual colors in the composite (e.g., R, lipids; G, saccharides; B, protein in Fig. 7G) are byte-scaled maps of each component (i.e., pixels with the maximum concentration of each component represented by full-color intensity [255] and pixels with the minimum concentration of each component represented by black [0]), and thus they give information on location but not relative amounts. If a single intensity scale is used, the maps are dominated by the protein component.
FIG. 7.
Grey-scale STXM maps of protein distribution (A), lipid distribution (B), saccharide distribution (C), carbonate distribution (D), and nucleic acid distribution (E) in the biofilm derived by fitting individual pixels in an 81-image sequence in the C 1s region to the spectra presented in Fig. 4. Color-coded composite maps of proteins (red), saccharides (green), and nucleic acids (blue) (F) and of lipids (red), saccharides (green), and proteins (blue) (G) were all derived from the STXM image sequence of the biofilm. (H) The same region imaged with confocal fluorescence microscopy using probes for fucose EPS (green) and nucleic acids (blue). Note that the strong DNA signal in panel F is most likely an artifact associated with absorption saturation at those points. Bars, 2 μm.
It is interesting to consider in detail the comparison of the color-coded composite maps derived from STXM with those derived by CLSM for exactly the same region (Fig. 7E through H). The color-coded map based on CLSM images shows the presence of fucose-rich polysaccharide regions detected with the lectin from Tetragonolobus purpureas and nucleic acid-rich regions in cells and polymer detected by Syto9 staining (Fig. 7H). The region of polysaccharide detected by the lectin overlaps with that detected by STXM; however, the specificity of the lectin limits it to a subset of the total polysaccharide seen in STXM.
Sample preparation using the silicon nitride windows required for STXM compromises the results of confocal microscopy by adding an additional optical element into the laser light path and restricting observation to the use of longer working distance objective lenses with low numerical apertures (NA < 0.9). In Fig. 7H, the upper half of the window is present and the biofilm was imaged through silicon nitride; thus, the resolution and quality of the image are compromised. However, the images do provide a basis for comparison of results of the two approaches in identical material at similar magnification and resolution.
Comparison of biofilm structures as detected by CLSM, STXM, and TEM.
Fluorescent nucleic acid staining indicated that this component of the biofilm was primarily but not solely cell associated, as was seen using the probe Syto9 with confocal imaging. Although noncellular binding has often been classified as nonspecific staining, it has become apparent (39) that DNA may be a structural component of biofilms. Indeed, extractive studies have often indicated that a considerable fraction of the biofilm EPS is DNA (31). Correlation of fluorescent nucleic acid staining with the results of soft X-ray analyses indicates that both detect the presence of regions of noncellular nucleic acids within the biofilm (Fig. 7). Thus, STXM and CLSM imaging both provide support for the contention that nucleic acids exist extracellularly and may have a structural role in biofilms.
The exopolymeric matrix also bound a wide variety of probes, including Tetragonolobus purpureas, Ulex europaeus, Canavalia ensiformis, Lens culinaris, Datura stramonium, or Sambucus nigra lectins. Binding of these lectins is indicative of the presence of l-fucose (high affinity)-, l-fucose-, d(+)-glucose-, N-acetyl-d-glucosamine- or d(+)-glucose-, sucrose- or d(+)-glucose-, N-acetyl-d-glucosamine-, and N-acetyl-d-galactosamine-rich regions, respectively, in the polysaccharide fraction of the exopolymer matrix of the biofilm. In this instance, there is good correspondence between the mapping of saccharide by probe-based CLSM and the STXM analyses (Fig. 7).
Application of the lipophilic probe Nile red indicated the presence of lipid or highly hydrophobic regions with the biofilms, which were occasionally isolated (Fig. 1 and 2) but were most frequently coincident with regions also binding the protein-sensitive probe SyproOrange. This may be suggestive of the presence of lipoproteins. Consistent with the localization of nucleic acid and lectin probes, the interpretation of parallel CLSM and STXM images indicates colocalization of these biopolymers in cells and extracellular materials of the biofilm. Colocalization may also reflect the presence of lipoproteins and glycoproteins, molecules that have chemistries representative of more than one class of biomolecule. The applicability of STXM to complex biological systems using only natural contrast may be limited by such factors. However, it should be possible to develop labeling approaches analogous to those used in other areas such as CLSM.
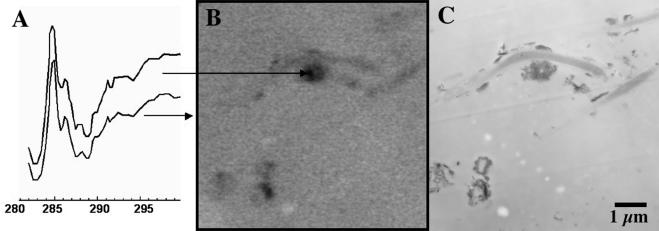
Figure 8 presents a direct comparison of STXM and TEM. In this case, a sample of the same material as examined by CLSM and STXM in Fig. 3 to 7 was fixed by using the methods described above. After preparation and analysis in the TEM, the same grid was measured by STXM. This indicates that we can examine the same areas by all three techniques. This observation clearly demonstrates the large improvement in spatial resolution of TEM relative to STXM (and CLSM). One might have expected the STXM spectroscopy to provide insight into the nature of the cellular and extracellular components. However, the C 1s spectrum of the bacterial region is identical to that of the whole region imaged (Fig. 8A), indicating that there is little of the original organic material present. Loss of organic molecules from bacteria resulting from extractive TEM preparatory techniques is a cause for concern, even though TEM has made an excellent contribution to the fundamental understanding of bacterial ultrastructure. Efforts to improve the situation are under way (8). The ideal sample preparation should preserve the ultrastructure of cells without exchanging the biomolecules with components of fixative solutions; the aldehyde component of primary fixatives is intended to cross-link macromolecules (mainly protein) to keep them in their “living” position, while the overall composition of fixatives should minimize extraction by the buffer. Presently, this is not the case. New sample preparation protocols are needed to complete our capabilities of correlative CLSM, TEM, and STXM microscopic analysis.
FIG. 8.
The spectra on the left (A) are (as indicated by arrows) that of the bacterium in the center and the average of the full region. (B and C) Comparative imaging of the same region of a fixed sample of the biofilm material using STXM (B) and TEM (C).
Advantages and disadvantages of specific microscopy methods.
Comparisons of the results of CLSM and STXM mapping indicate a substantial degree of agreement in the detection of biomolecules within the biofilms and associated with cells and microcolonies. STXM offers quantitative chemical information without the use of fluorescent probes, but the technique is limited by effects such as absorption saturation and inaccuracies in reference spectra. CLSM provides detail on the three-dimensional hydrated structure of the biofilm plus information on biomolecular distributions. The use of lectins may take us past saccharide localization to the identity of some glycoconjugate residues in the EPS. However, component identification in CLSM is reliant not on the properties of the molecule alone as is true in STXM but rather upon the specificity of the probe and the nature of the target molecule. This specificity may be influenced by a wide variety of factors resulting in nonspecific binding and misidentification of structural components. Most fluorescent probes modify the biofilm system to a degree. TEM allows very-high-resolution imaging of dehydrated materials and can give chemical identification when used in conjunction with electron-dense probes such as colloidal gold-labeled lectins. The reliance on secondary binding of probes results in similar limitations to fluorescence but confirms probe penetration and the apparent specificity of the lectins. Limitations to STXM include suitability of the model compounds relative to biofilm material, data acquisition without undue radiation damage, requirement for very thin samples (<200 nm equivalent thickness of dry organic components, <5 μm of water when wet), use of fragile silicon nitride windows, and absorption saturation distortion of analysis in thick regions of a specimen. Both STXM and TEM are limited relative to CLSM in that sample preparation, such as encapsulation in a wet cell (STXM) or fixation and mechanical sectioning (TEM), is needed.
Summary.
We have demonstrated the power of correlative microscopy methods as a tool for studies of biofilms. Specifically we have determined the spatial distribution of macromolecules in a fully hydrated biofilm system. The STXM variant of X-ray spectromicroscopy has been used to map spatial distributions of major biomolecules in complex microbial biofilms. CLSM in combination with fluorescent probes was also able to map major biomolecules and subcomponents in hydrated biofilms, thus showing substantial agreement with STXM results. TEM provided valuable extension and confirmation of data. In addition, we have shown that STXM, CLSM, and TEM may be used in a correlative fashion to define and map biofilm composition. We believe that these correlative techniques will be useful to help understand the biochemical basis for biofilm organization and for investigating and ultimately optimizing biofilms for environmental remediation applications.
Acknowledgments
This work was supported by the National Water Research Institute, Environment Canada, NSERC, and the Canada Research Chair program (A.P.H.). The Advanced Light Source is supported by the Director, Office of Energy Research, Office of Basic Energy Sciences, Materials Sciences Division of the U.S. Department of Energy, under contract no. DE-AC03-76SF00098.
REFERENCES
- 1.Ade, H. 1998. X-ray spectromicroscopy, p. 225-261. In J. A. R. Samson and D. L. Ederer (ed.), Experimental methods in the physical sciences, vol. 32. Academic Press, San Diego, Calif.
- 2.Ade, H., and S. G. Urquhart. 2002. NEXAFS spectroscopy and microscopy of natural and synthetic polymers, p. 285-355. In T. K. Sham (ed.), Chemical applications of synchrotron radiation. World Scientific Publishing, River Edge, N.J.
- 3.Böckelmann, U., W. Manz, T. R. Neu, and U. Szewzyk. 2002. Investigation of lotic microbial aggregates by a combined technique of fluorescent in situ hybridization and lectin-binding-analysis. J. Microbiol. Methods 49:75-87. [DOI] [PubMed]
- 4.Chandler, J. A. 1977. X-ray microanalysis in the electron microscope. North-Holland Publishing Co., Amsterdam, The Netherlands.
- 5.Cooksey, K. E. 1992. Extracellular polymers in biofilms, p. 137-147. In L. F. Melo, T. R. Bott, M. Fletcher, and B. Capdeville (ed), Biofilms—science and technology, vol. 223. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 6.Frosch, D., and C. Westphal. 1989. Melamine resins and their application in electron microscopy. Electron Microsc. Rev. 2:231-255. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert, E. S., A. Khlebnikov, W. Meyer-Ilse, and J. D. Keasling. 1999. Use of soft X-ray microscopy for the analysis of early-stage biofilm formation. Water Sci. Technol. 39:269-272. [Google Scholar]
- 8.Graham, L. L., and T. J. Beveridge. 1990. Evaluation of freeze-substitution and conventional embedding protocols for routine electron microscopic processing of eubacteria. J. Bacteriol. 172:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanke, D. E., and D. H. Northcote. 1975. Molecular visualization of pectin and DNA by ruthenium red. Biopolymers 14:1-17. [DOI] [PubMed] [Google Scholar]
- 10.Hayat, M. A. (ed.). 1970. Principles and techniques of electron microscopy—biological applications. Van Nostrand Reinhold, New York, N.Y.
- 11.Henke, B. L., P. Lee, T. J. Tanaka, R. L. Shimabukuro, and B. K. Fujikawa. 1982. Low-energy X-ray interaction coefficients: photoabsorption, scattering, and reflection. Atomic Nuclear Data Tables 27:1-144.
- 12.Hitchcock, A. P., C. Morin, Y. M. Heng, R. M. Cornelius, and J. L. Brash. 2002. Soft X-ray spectromicroscopy of biomaterials. J. Biomater. Sci. Polym. Ed. 13:919-938. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, T. A., M. M. West, and G. G. Leppard. 1999. Accumulation of heavy metals by individually analyzed bacterial cells and associated nonliving material in polluted lake sediments. Environ. Sci. Technol. 33:3795-3801. [Google Scholar]
- 14.Jackson, T. A., and G. G. Leppard. 2002. Energy dispersive X-ray microanalysis and its applications in biogeochemical research, p. 219-260. In A. Violante, P. M. Huang, J.-M. Bollag, and L. Gianfreda (ed.), Soil mineral-organic matter—microorganism interactions and ecosystem health, vol. 28A. Elsevier Science, Amsterdam, The Netherlands.
- 15.Jacobsen, C., S. Wirick, G. Flynn, and C. Zimba. 2000. Soft X-ray spectroscopy from image sequences with sub-100-nm spatial resolution. J. Microscopy 197:173-184. [DOI] [PubMed] [Google Scholar]
- 16.Kaznacheyev, K., A. Osanna, C. Jacobsen, O. Plashkevych, O. Vahtras, H. Ågren, V. Carravetta, and A. P. Hitchcock. 2002. Inner-shell absorption spectroscopy of amino acids, J. Phys. Chem. A 106:3153-3168.
- 17.Kilcoyne, A. L. D., T. Tylíszczak, W. F. Steele, S. Fakra, P. Hitchcock, K. Franck, E. Anderson, B. Harteneck, E. G. Rightor, G. E. Mitchell, A. P. Hitchcock, L. Yang, T. Warwick, and H. Ade. 2002. Interferometrically controlled scanning transmission microscopes at the advanced light source. J. Synchrotron Radiat. 10:125-136. [DOI] [PubMed]
- 18.Lawrence, J. R., D. R. Korber, G. M. Wolfaardt, D. E. Caldwell, and T. R. Neu. 2002. Analytical imaging and microscopy techniques, p. 39-61. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 19.Lawrence, J. R., T. R. Neu, and K. C. Marshall. 2002. Colonization—adhesion, bioaggregates and biofilms, p. 466-477. In C. J. Hurst, G. R. Knudsen, R. L. Crawford, M. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 20.Lawrence, J. R., and T. R. Neu. 1999. Confocal laser scanning microscopy for analysis of microbial biofilms, p. 131-144. In R. J. Doyle (ed.), Biofilms—methods in enzymology, vol. 310. Academic Press, Orlando, Fla. [DOI] [PubMed]
- 21.Lawrence, J. R., G. D. W. Swerhone, and T. R. Neu. 2000. A simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods 42:215-224. [DOI] [PubMed] [Google Scholar]
- 22.Leppard, G. G. 1992. Evaluation of electron microscope techniques for the description of aquatic colloids, p. 231-289. In J. Buffle and H. P. van Leeuwen (ed.), Environmental particles, vol. 1, IUPAC Environmental Chemistry Series. Lewis Publishers, Chelsea, Mich.
- 23.Leppard, G. G. 1997. Colloidal organic fibrils of acid polysaccharides in surface waters: electron-optical characteristics, activities, and chemical estimates of abundance. Colloids Surf. A 120:1-15. [Google Scholar]
- 24.Leppard, G. G., A. Massalski, and D. R. S. Lean. 1977. Electron-opaque microscopic fibrils in lakes: their demonstration, their biological derivation, and their potential significance in the redistribution of cations. Protoplasma 92:289-309. [DOI] [PubMed] [Google Scholar]
- 25.Liss, S. N., I. G. Droppo, D. T. Flannigan, and G. G. Leppard. 1996. Floc architecture in wastewater and natural riverine systems. Environ. Sci. Technol. 30:680-686. [Google Scholar]
- 26.Loo, B. W., Jr., I. M. Sauerwald, A. P. Hitchcock, and S. S. Rothman. 2001. A new sample preparation method for soft X-ray microscopy: nitrogen-based contrast and radiation tolerance properties of glycol methacrylate-embedded and sectioned tissue. J. Microscopy 204:69-86. [DOI] [PubMed] [Google Scholar]
- 27.Luft, J. H. 1971. Ruthenium red and violet. I. Chemistry, purification, methods of use for electron microscopy and mechanism of action. Anat. Rec. 171:347-368. [DOI] [PubMed] [Google Scholar]
- 28.Neu, T. R. 2000. In situ cell and glycoconjugate distribution in river snow studied by confocal laser scanning microscopy. Aquat. Microb. Ecol. 21:85-95. [Google Scholar]
- 29.Neu, T. R, and J. R. Lawrence. 1999. In situ characterization of extracellular polymeric substances (EPS) in biofilm systems, p. 21-48. In J. Wingender, T. R. Neu, and H.-C. Flemming (ed.), Microbial extracellular substances. Springer-Verlag, Berlin, Germany.
- 30.Neu, T. R., G. D. W. Swerhone, and J. R. Lawrence. 2001. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 147:299-313. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, P. H., and A. Jahn. 1999. Extraction of EPS, p. 49-72. In J. Wingender, T. R. Neu, and H.-C. Flemming (ed.), Microbial extracellular polymeric substances: characterization, structure and function. Springer-Verlag, Berlin, Germany.
- 32.Press, W. H., B. P. Flannery, S. A. Teukolsky, and W. T. Vetterling. 1992. Numerical recipes in C: the art of scientific computing. Cambridge University Press, Cambridge, United Kingdom.
- 33.Reid, D. A., H. C. Ducharme, M. M. West, and J. N. A. Lott. 1998. Iron-rich particles in embryos of seeds from family Pinaceae. Protoplasma 202:122-133. [Google Scholar]
- 34.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 35.Strang, G. 1988. Linear algebra and its applications. Harcourt Brace Jovanovich, San Diego, Calif.
- 36.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 37.Warwick, T., H. Ade, A. L. D. Kilcoyne, M. Kritscher, T. Tylisczcak, S. Fakra, A. P. Hitchcock, P. Hitchcock, and H. A. Padmore. 2002. A new bend magnet beam line for scanning transmission X-ray microscopy at the advanced light source. J. Synchrotron Radiat. 9:254-257. [DOI] [PubMed] [Google Scholar]
- 38.Warwick, T., K. Franck, J. B. Kortwight, G. Meigs, M. Moronne, S. Myneni, E. Rotenberg, S. Seal, W. F. Steele, H. Ade, A. Garcia, S. Cerasari, J. Denlinger, S. Hayakawa, A. P. Hitchcock, T. Tyliszczak, E. G. Rightor, H.-J. Shin, and B. Tonner. 1998. Scanning transmission X-ray microscope for materials science spectromicroscopy at the advanced light source. Rev. Sci. Instrum. 69:2964-2973. [Google Scholar]
- 39.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed]
- 40.Wilkinson, K. J., E. Balnois, G. G. Leppard, and J. Buffle. 1999. Characteristic features of the major components of freshwater colloidal organic matter revealed by transmission electron and atomic force microscopy. Colloids Surf. A 155:287-310. [Google Scholar]
- 41.Wingender, J., T. R. Neu, and H.-C. Flemming. 1999. Microbial extracellular polymeric substances. Springer-Verlag, Berlin, Germany.
- 42.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, and D. E Caldwell. 1998. In situ characterization of biofilm exopolymers involved in the accumulation of chlorinated organics. Microb. Ecol. 35:213-223. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, X., R. Balhorn, J. Mazrimas, and J. Kirz. 1996. Mapping and measuring DNA to protein ratios in mammalian sperm head by XANES imaging. J. Struct. Biol. 116:335-344. [DOI] [PubMed] [Google Scholar]