Abstract
Recent concern that the increased use of triclosan (TCS) in consumer products may contribute to the emergence of antibiotic resistance has led us to examine the effects of TCS dosing on domestic-drain biofilm microcosms. TCS-containing domestic detergent (TCSD) markedly lowered biofouling at 50% (wt/vol) but was poorly effective at use levels. Long-term microcosms were established and stabilized for 6 months before one was subjected to successive 3-month exposures to TCSD at sublethal concentrations (0.2 and 0.4% [wt/vol]). Culturable bacteria were identified by 16S rDNA sequence analysis, and their susceptibilities to four biocides and six antibiotics were determined. Microcosms harbored ca. 10 log10 CFU/g of biofilm, representing at least 27 species, mainly gamma proteobacteria, and maintained dynamic stability. Viable cell counts were largely unaffected by TCSD exposure, but species diversity was decreased, as corroborated by denaturing gradient gel electrophoresis analysis. TCS susceptibilities ranged widely within bacterial groups, and TCS-tolerant strains (including aeromonads, pseudomonads, stenotrophomonads, and Alcaligenes spp.) were isolated before and after TCSD exposure. Several TCS-tolerant bacteria related to Achromobacter xylosoxidans became clonally expanded during dosing. TCSD addition did not significantly affect the community profiles of susceptibility to the test biocides or antibiotics. Several microcosm isolates, as well as reference bacteria, caused clearing of particulate TCS in solid media. Incubations of consortia and isolates with particulate TCS in liquid led to putative TCS degradation by the consortia and TCS solubilization by the reference strains. Our results support the view that low-level exposure of environmental microcosms to TCS does not affect antimicrobial susceptibility and that TCS is degradable by common domestic biofilms.
Triclosan (TCS; 2,4,4′-trichloro-2′-hydroxydiphenylether) is a commonly deployed antibacterial compound (15). Since its introduction in the 1960s, it has been used variously as an antiseptic (44) in medicated soaps, hand washes (4), and therapeutic baths for methicillin-resistant Staphylococcus aureus-infected patients (12, 56). Over the past decade, the broad spectrum of antimicrobial activity displayed by TCS has led to its application in a range of formulations intended for domestic use, including a variety of shampoos (26), toothpastes (40), deodorants (7), detergents, molded plastic containers, and chopping boards. A survey of liquid soaps in the United States revealed that 45% contained antibacterial agents, many of which included TCS (51). Recently, the particular use of hardware products that contain TCS has increased considerably (7).
Studies have demonstrated that TCS (Irgasan) interacts with an enoyl-acyl carrier protein (ACPR) reductase (FabI) in Escherichia coli (19, 35). This is an essential enzyme in the fatty acid biosynthetic pathway of many different bacterial species (22), including gram-positive organisms (20, 21). TCS is a potent inhibitor of the ACPR (22, 28, 35). Much of the research into this aspect of TCS activity has focused upon enzyme systems derived from E. coli (35) and Pseudomonas aeruginosa (46). There is, however, a considerable degree of intergeneric conservation of this enzyme, with homologous target enzymes (InhA) being functionally important in various mycobacteria (34, 48), Bacillus subtilis (21), and S. aureus (20). Naturally occurring TCS-resistant ACPRs (InhK) have been documented (19), and P. aeruginosa possesses both TCS-sensitive and TCS-resistant FabI homologues. Of concern is that at sublethal concentrations, TCS can select for mutations in the FabI gene of E. coli (35, 46). These mutations confer a heightened lack of susceptibility in this already relatively insensitive bacterium. The ACPR of Mycobacterium tuberculosis is the target for isoniazid, which is currently the most widely used antituberculosis drug (37). McMurry et al. (34) have demonstrated that partial isoniazid resistance in Mycobacterium smegmatis may be conferred by mutations in the InhA gene, the gene homologous to that encoding FabI. Interestingly, isoniazid-resistant M. tuberculosis mutants retain their susceptibility to TCS, suggesting separate interactive sites on the ACPR enzyme (48). TCS is also a substrate for the MexCD-OprJ efflux system in P. aeruginosa, and exposure of a susceptible Delta (mexAB oprM) strain to TCS selected multidrug-resistant bacteria at high frequencies (10).
The massive increase in TCS use, together with recent discoveries concerning its mechanism of action, has called into question the possible long-term consequences of resistance selection and bioaccumulation. Indeed, trace levels of TCS have been detected in aquatic environments (29, 38), human milk (1), fish bile, and human plasma (23).
The use of TCS for domestic and clinical applications may expose a wide range of environmental and potentially pathogenic nosocomial bacteria to TCS, often at low concentrations, which may theoretically select for organisms with reduced TCS susceptibility. The concern has been that, hypothetically, these organisms may also be less sensitive to unrelated therapeutic compounds (30).
The aim of this study was to investigate the effects of short- and long-term TCS use on the bacterial composition and antibacterial resistance properties of domestic-drain biofilm ecosystems. These were identified as high-risk ecosystems (33, 39, 47) in which chronic TCS exposure is likely to occur in relation to consumer product use. The occurrence of highly heterogeneous bacterial communities within such sink drain biofilms has previously been demonstrated, with viable cell counts of enteric species and pseudomonads ranging from 9.8 to 11.3 log10 cells/g of biofilm (31). Stable long-term laboratory models for the culture of species in these biofilms have been developed.
MATERIALS AND METHODS
Biofilm samples.
Biofilm material was excised from the horizontal pipe section of a polyvinyl chloride kitchen drain outlet of a house situated in Greater Manchester, United Kingdom (31). The samples were taken from a conventional sink drain outlet with attached waste disposal unit that had been in situ for 15 years. This household had not used biocidal detergent products other than bleach since the sink drain was installed. The pipe joints were separated, and biofilm was excised with a sterile scalpel. Samples were transported to the laboratory for processing within 2 h under a normal air atmosphere in plastic universal bottles.
Domestic-drain microcosms.
Domestic-drain microcosms were established according to methods described previously (31). Briefly, drain biofilm (2.5 g) was macerated with a sterile mortar and pestle and diluted 1:10 in sodium phosphate buffer (22.5 ml, 0.1 M, pH 6.5) containing 0.45% (wt/vol) NaCl which had been prereduced (boiled for 5 min and cooled under a constant stream of anaerobic gas, a 5:95 mixture of CO2 and N2). The samples were homogenized for 1 min in a flask shaker (Griffin, London, United Kingdom) in the presence of approximately five glass beads (3.5 to 5.5 mm; BDH, Poole, United Kingdom). The diluted material from samples was used to inoculate two constant-depth film fermenters (CDFFs). Initially, short-term microcosms were established to study the lethality of TCS. For these experiments, growth medium was continuously added to the fermenters by a peristaltic pump (Gilson) at a rate of 5 ml/h. Composition of medium intended to simulate dishwashing water was as follows (in grams per liter): starch, 1.0; peptone, 0.5; tryptone, 0.5; yeast extract, 0.5; NaCl, 1.0; margarine (Flora; Unilever, Crawley, United Kingdom), 0.05; hemin, 0.001; and tomato ketchup, (Heinz, Uxbridge, United Kingdom), 0.05. For long-term microcosm studies, the fermenters were inoculated twice, with a 7-day interval between inoculations, by using additional, resampled, homogenized but undiluted drain biofilm. Anaerobiosis was maintained for the first 48 h by continuous gassing with oxygen-free gas (CO2 and N2 at a 5:95 ratio) at 1 liter/h, and temperature was uncontrolled (ambient lab temperatures ranged from 18 to 24°C). Biofilms were shielded from light by covering the CDFFs with aluminum foil shrouds. Throughout, the microcosms were maintained on a feast-famine regimen (20-min perfusion four times daily of 0.5 ml of synthetic dishwater/min) as described above but supplemented with domestic detergent (Fairy Original; Procter and Gamble, Newcastle Upon Tyne, United Kingdom) at 0.05 g/liter.
Discontinuous feeding regimens were controlled by using programmable electronic timers (Micromark, London, United Kingdom). In order to model more accurately the open nature of a domestic sink drain, the fermenter pans were continuously wetted with untreated tap water (1 ml/h). CDFFs enable the continuous culture of biofilms at an accurately set depth (31, 32, 43). In all cases, the biofilm (Teflon plug) depth was set at 5.0 mm. Developed communities were characterized periodically over the course of the investigation, both by immediate use of samples for culture by archiving of samples for subsequent denaturing gradient gel electrophoresis (DGGE) and residual TCS analysis (31).
Addition of TCS to the microcosms.
For short-term lethality studies, dilutions of a TCS-containing detergent (TCSD; Palmolive Ultra concentrated dishwashing liquid and hand soap; Colgate Palmolive Company, New York, N.Y.) were added to established drain microcosms by a peristaltic pump for 10 min at 6-h intervals at a flow rate of 55.2 ml/h. For long-term studies, duplicate microcosms were stabilized for 6 months, after which Palmolive Ultra dishwashing liquid was added as the domestic detergent in the artificial dishwater at 0.2% (wt/vol). After 3 months, the concentration of detergent was doubled and the incubations were extended for a further 3 months.
Bacterial characterization by culture.
Drain biofilm material (1.0 g) or CDFF plugs (two) were macerated with a sterile mortar and pestle, homogenized, and diluted 1:10 (as described above). For enumeration, dilutions of macerated drain or model biofilm (1:10) were serially diluted by using prereduced half-strength peptone water (7.5 g/liter). In order to minimize variation due to the sampling of immature biofilms, only those CDFF pans that had been in situ for at least 1 month were removed for analysis. Aliquots (0.1 ml) of appropriate dilutions were plated in triplicate onto a variety of selective and nonselective media (Oxoid, Basingstoke, United Kingdom) as follows: Wilkins-Chalgren agar (anaerobic and facultative heterotrophs and gram-positive cocci), R2A (aerobic and facultative heterotrophs), Pseudomonas isolation agar with cetrimide-sodium nitrate selective supplements (P. aeruginosa), mannitol salts agar (gram-positive cocci), and MacConkey agar no. 3 (enteric organisms).
Since bacterial characterization of complex communities by culture is often complicated by dominant strains' obscuring the presence of numerically important but less-dominant bacteria, these bacteria were also isolated by culturing over gradients of antimicrobial compounds on R2A agar. Gradients were generated by using a model CU spiral plater (Spiral Systems, Cincinnati, Ohio). This approach increases the diversity of species that can be recovered. The following compounds were used as selective agents: TCS, chlorhexidine, penicillin V, cetrimide, chloro-3,5-dimethyl-phenol, vancomycin, chlortetracycline, ciprofloxacin, erythromycin, and fusidic acid. Plates were incubated for up to 5 days, both aerobically and in an anaerobic cabinet (in which the atmosphere was a 10:10:80 mix of H2, CO2, and N2). Criteria used for selecting bacterial populations for use as markers of population change in the biofilm communities included numerical dominance, ease of selective cultivation, and visual recognition. Morphologically distinct colonies were subcultured and archived (−60°C) for subsequent identification on the basis of morphology, oxidase test results, Gram reaction results, and rDNA sequencing.
Direct bacterial cell counts.
The proportion of the viable bacterial communities that could be cultured by using the methods employed was estimated by comparison by using vital staining and direct microscopy. A subsample (100 μl) of the 10−2 or 10−3 dilution (prepared for viable cell counting) was stained by using a live-dead bacterial viability kit (BacLight; Molecular Probes, Leiden, The Netherlands), and cells were counted by using an improved Neubauer counting chamber in conjunction with fluorescence microscopy with a 100-W mercury vapor lamp. Live (green fluorescent) and dead (red fluorescent) cells were visualized separately by using fluorescein and Texas red band-pass filters, respectively, according to the manufacturer's instructions.
Detemination of CDFF bioburden.
Bioburdens were measured by recording the residual mass of biofilm within CDFF sample pans.
Antimicrobial susceptibilities of isolated drain bacteria.
Stock solutions (4 mg/ml) of the following compounds were prepared as indicated by the manufacturers in deionized, distilled water: tetracycline, penicillin V, erythromycin, chlortetracycline, chlorhexidine, and cetrimide. TCS was dissolved in 25% ethanol. All solutions were sterilized by filtration through cellulose acetate filters (0.2-μm pore size; Millipore, Watford, United Kingdom) and stored at −70°C. A model CU spiral plater (Spiral Systems) was used for spiral gradient endpoint determinations of MICs. This apparatus can deposit accurate volumes of fluids onto the surfaces of agar plates such that either a uniform deposition density or a continuously varying dilution is established across the radius of the plate, with a very high degree of reproducibility (55). Petri dishes (10-cm diameter) were filled with 27.5 ± 1.0 ml of R2A agar to produce a mean agar depth of approximately 3.5 mm. The plates were kept for 2 days at room temperature prior to use to ensure dryness of the agar surface. Stock solutions (50 μl) of the antimicrobial compounds were then deposited onto the agar surface by using the variable cam of the spiral plater. This establishes a concentration gradient covering a ca. 1,000-fold dilution series. Control plates comprised identical agar plates without antimicrobial. Plates were dried for up to 1 h at room temperature prior to deposition of inoculation material along the spiral track by using the uniform cam of the spiral plater. After further drying (1 h), plates were inverted and incubated aerobically overnight at 37°C. The radial distance of the growth endpoint from the center of the spiral was measured by using calibrated callipers. MICs were determined based on inhibition zones by using the following equation: P = (RP × DF)/(h × SR), where P (potency) is the effective concentration (μg/ml) of antimicrobial at the interface between growth and inhibition, RP is the original concentration (μg/ml) of antimicrobial deposited, DF is the deposition factor, h is the agar height (mm), and SR is the surface ratio.
Degradation of particulate TCS.
Initially, as a preliminary screen for TCS degradation, samples of biofilm removed from the drain microcosms (0.1 g) or pure cultures were tested. Isolated drain bacteria (0.1 ml) were spread directly onto solid media supplemented with TCS at 2 g/liter. Since this concentration is considerably greater than the aqueous solubility of TCS (ca. 10 mg/liter), the TCS forms a dense precipitate within the agar. Clearing of this precipitate around a bacterial colony could result from either solubilization or degradation. The following agar types were used: mineral salts agar containing (per liter in distilled water) 2.30 g of K2HPO4, 0.40 g of NaHPO4, 1.30 g of (NH4)SO4, and 0.25 g of MgSO4; mineral salts glucose agar comprising mineral salts agar supplemented with glucose (2 g/liter); mineral salts TCS agar prepared as mineral salts glucose agar but with TCS substituting for glucose; and R2A TCS agar comprising R2A agar supplemented with TCS (2 g/liter).
In order to differentiate between TCS solubilization and degradation and to allow an analysis of the fate of TCS in consortium samples, a liquid culture screening method was also used. In this method, slurries of microcosm biofilm material (1.0% [wt/vol]) and reference strains or drain isolates in pure culture were prepared in nutrient broth containing a colloidal suspension of TCS at 0.5 g/liter. These preparations were incubated at 30°C for 56 h. Anaerobic incubations of microcosm biofilm were run as described above, with prereduced Wilkins-Chalgren broth substituting for nutrient broth, and broths were incubated in an anaerobic cabinet. Samples were then harvested and centrifuged for 10 min (10,000 × g), and supernatants were stored at −60°C for subsequent analysis.
Analysis of residual TCS.
Drain samples were archived throughout the period of TCS addition and stored at −60°C for subsequent analysis of TCS. TCS analysis was done by using liquid chromatography with negative ion electrospray mass spectrometry for detection. Samples were shaken to mix, and a 200-μl aliquot was diluted with an equal volume of acetonitrile containing 30 μg of 2-phenylphenol/ml as an internal standard. Resulting solutions were filtered (0.2-μm-pore-size Acrodisc CR Teflon filters) and analyzed by using a model 2695 Alliance liquid chromatograph equipped with a UV photodiode array detector and a Micromass ZMD mass spectrometer operated in scan mode (Waters Corporation, Milford, Mass.). Both UV and mass spectral signals were collected. Liquid chromatographic analysis used a Luna phenyl-hexyl column (150 mm [length] by 2.0 mm [internal diameter]; particle size, 3 μm; Phenomenex, Torrance, Calif.) with an acetonitrile-water-0.01% ammonium acetate mobile phase. TCS was eluted at approximately 11 min with an acetonitrile gradient that increased linearly from 50 to 90% over 12 min. Quantitation was based on the area responses at m/z's of 287 and 289 relative to the 2-phenylphenol internal standard area (m/z = 169) by using a linear calibration which bracketed the observed sample concentrations. Negative ion electrospray mass spectrometry was approximately five times more sensitive (limit of detection, 0.16 mg/liter) than UV detection and provides positive identification of TCS in the complex matrix.
Partial 16S rDNA gene sequencing of bacterial isolates and excised DGGE gel bands.
Strains were subcultured on R2A agar until pure cultures were obtained, and then bacterial colonies (two to three) were aseptically removed from the surface of the plate and homogenized in a reaction tube containing nanopure water (100 μl). The bacterial suspension was heated to 100°C in a boiling water bath for 10 min and then centrifuged for 10 min (10,000 × g). The supernatant was used as a template for PCR. Partial 16S rRNA gene sequences were amplified by using the primers 8FPL1 (5′-GAG TTT GAT CCT GGC TCA G-3′) and 806R (5′-GGA CTA CCA GGG TAT CTA AT-3′) at 5 μM each. Each PCR consisted of Red Taq DNA polymerase ready mix (25 μl; Sigma, Dorset, United Kingdom), forward and reverse primers (2 μl), nanopure water (16 μl), and template DNA (5 μl). A Perkin-Elmer thermal DNA cycler model 480 was used to run 35 thermal cycles as follows: 94°C (1 min), 53°C (1 min), and 72°C (1 min). The final cycle incorporated a 15-min chain elongation step (72°C). For analysis of the major resolved DGGE amplicons, selected resolved bands were cut out of the polyacrylamide gels under UV illumination and incubated at 4°C for 20 h together with 20 μl of nanopure water in nuclease-free universal bottles. Portions (5 μl) were then removed and used as a template for a PCR identical to that outlined in “DGGE analysis.” The reverse (non-GC clamp) primer (HDA2) was used in subsequent sequencing reactions for excised DGGE gel bands (31, 32). Amplified products were purified by using Qiaquick PCR purification kits (Qiagen Ltd., West Sussex, United Kingdom) and sequenced by using the primers described above. DNA sequences were compiled by using GENETOOL LITE 1.0 (http://www.biotools.com) to obtain consensus sequences or to check and edit unidirectional sequences. For excised DGGE band PCRs, the presence of a GC clamp upon sequence analysis confirmed that the correct target, rather than an extraneous contaminant, had been reamplified.
Construction of neighbor-joining trees.
BLAST (http://www.ncbi.nlm.nih.gov/BLAST) searches were performed with each compiled sequence against the sequences in the EMBL prokaryote database. Closest relative species were assigned based on comparisons of compiled partial 16S rRNA gene sequences against sequences in EMBL by using FASTA3 and BLAST. Unambiguous positions of representative sequences of closely related strains were then aligned by using CLUSTALX version 1.64b. Neighbor-joining analysis was conducted with the correction of Jukes and Cantor (25) by using TREECON 1.3b (52) with Methanobacterium thermautotrophicum as the outgroup and showing bootstrap values as percentages of 100 replications.
Community DNA extraction.
Archived biofilm material (0.2 to 0.5 g) was mixed with 1 ml of sodium phosphate buffer (0.12 M, pH 8.0), vortex mixed, and subjected to two cycles of freeze heating (−60°C for 10 min, 60°C for 2 min). Samples were then transferred to a bead-beater vial containing 0.3 g of sterile zirconia beads (0.1 mm diameter). Tris-equilibrated phenol (pH 8.0, 150 μl) was added, and the suspension was shaken three times for 80 s at maximum speed (Mini-Bead-Beater; Biospec Products, Bartlesville, Okla.). After 10 min of centrifugation at 13,000 × g, the supernatant was extracted three times with an equal volume of phenol-chloroform and once with chloroform-isoamyl alcohol (24:1 [vol/vol]). The DNA was precipitated from the aqueous phase with 3 volumes of ethanol, air dried, and resuspended in 100 μl of deionized water. The amount and quality of DNA extracted were estimated by electrophoresis of 5-μl aliquots on a 0.8% agarose gel and by comparison to a molecular weight standard (stained with ethidium bromide). DNA extracts were stored at −60°C prior to analysis.
PCR amplification for DGGE analysis.
The V2-V3 region of the 16S rDNA (rRNA gene), corresponding to positions 339 to 539 of E. coli, was amplified with the eubacterium-specific primers HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) as used by Walter et al. (54). The reactions were performed in 0.2-ml tubes by using a Perkin-Elmer DNA thermal cycler (model 480; Cambridge, United Kingdom). In all cases, reactions were carried out using Red Taq DNA polymerase ready mix (25 μl; Sigma), HDA primers (2 μl each and 5 μM), nanopure water (16 μl), and extracted community DNA (5 μl). Optimization studies, as described by Muyzer et al. (41), showed that extracted community DNA required a minimum 1:10 dilution to ensure reliable PCRs. Quantification and standardization of extracted DNA was achieved by using a fluorescence assay (DNA quantitation kit; Sigma) according to the manufacturer's instructions. The thermal program was as follows: 94°C (4 min) followed by 30 thermal cycles of 94°C (30 s), 56°C (30 s), and 68°C (60 s). The final cycle incorporated a 7-min chain elongation step (68°C).
DGGE analysis.
Biofilm samples were analyzed by DGGE by using a D-Code universal mutation detection system (Bio-Rad, Hemel Hempstead, United Kingdom). Polyacrylamide (8%) gels of 16 by 16 cm (1-mm deep) were run with 1× TAE buffer diluted from 50× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, and 1 mM EDTA). Initially, separation parameters were optimized by running PCR products from selected pure cultures of drain bacteria and PCR amplicons from extracted drain DNA on gels with a 0-to-100% denaturation gradient, perpendicular to the direction of electrophoresis. (A 100% denaturing solution contained 40% [vol/vol] formamide and 7.0 M urea.) Denaturing gradients were formed with two 10% acrylamide (acrylamide-bis, 37.5:1) stock solutions (Sigma). On this basis, a denaturation gradient for parallel DGGE analyses, ranging from 20 to 60%, was selected, as well as PCR amplicons from the isolates Pseudomonas nitroreducens MBRG 4.6, Aeromonas sp. strain MBRG 4.2, Stenotrophomonas maltophilia MBRG 4.17, Pseudoxanthomonas sp., Alcaligenes xylosoxidans MBRG 4.31, Citrobacter sp., Eubacterium sp. strain MBRG 7.1, and Microbacterium phyllosphaerae MBRG 4.30. For community analyses, the gels also contained a 30-to-60% denaturing gradient. DNA for loading onto gels was quantified, and where necessary, amounts were standardized between samples by using a fluorescence assay (see above). Electrophoresis was carried out at 150 V at 60°C for approximately 4.5 h. All gels were stained with SYBR Gold stain (diluted to 10−4 in 1× TAE; Molecular Probes, Leiden, The Netherlands) for 30 min. Gels were viewed and images were documented by using a BioDoc-It system (UVP, Upland, Calif.).
Bacteria.
P. aeruginosa PAO1 (ATCC 15692) was obtained from the American Type Culture Collection (http://www.atcc.org). E. coli 2500 was a TCS-insusceptible strain and was produced by repeated passage of E. coli ATCC 8729 in the presence of increasing TCS concentrations.
Chemicals.
Unless otherwise stated, chemicals and antimicrobial agents were obtained from Sigma. Formulated bacteriological media were purchased from Oxoid, Basingstoke, United Kingdom. TCS (Irgasan DP300) was obtained from Oils and Soaps Ltd., Bradford, United Kingdom.
Nucleotide sequence accession numbers.
Sequences for isolated cell clones were deposited into the EMBL sequence database. The accession numbers are given in parentheses: Klebsiella planticola MBRG 1.1 (AJ508364), Hafnia alvei MBRG 1.2 (AJ508360), Klebsiella oxytoca MBRG 1.3 (AJ508361), Lactobacillus paracasei MBRG 1.4 (AJ508362), Bacillus subtilis MBRG 1.5 (AJ508358), Enterobacter asburiae MBRG 1.6 (AJ508359), Pseudomonas sp. strain MBRG 1.7 (AJ508363), E. coli MBRG 2.1 (AJ508367), Moraxella osloensis MBRG 2.3 (AJ508366), Bacillus subtilis MBRG 2.4 (AJ508365), Staphylococcus epidermidis MBRG 2.5 (AJ508368), E. coli MBRG 3.1 (AJ508369), Aeromonas hydrophila MBRG 4.1 (AJ508693), Aeromonas sp. strain MBRG 4.2 (AJ508692), Aeromonas hydrophila MBRG 4.3 (AJ508690), Aeromonas caviae MBRG 4.4 (AJ508691), Pseudomonas putida MBRG 4.5 (AJ508696), Pseudomonas nitroreducens MBRG 4.6 (AJ508698), Pseudomonas sp. strain MBRG 4.7 (AJ508697), Serratia proteamaculans MBRG 4.8 (AJ508694), Stenotrophomonas maltophilia MBRG 4.10 (AJ508703), Stenotrophomonas acidaminiphila MBRG 4.11 (AJ508701), Stenotrophomonas acidaminiphila MBRG 4.12 (AJ508700), Ralstonia sp. strain MBRG 4.13 (AJ508607), unidentified eubacterium MBRG 4.14 (AJ508699), Klebsiella pneumoniae MBRG 4.15 (AJ508695), Delftia acidovorans MBRG 4.16 (AJ508611), Stenotrophomonas maltophilia MBRG 4.17 (AJ508702), Bacillus cereus MBRG 4.19 (AJ508707), Bacillus cereus MBRG 4.21 (AJ508706), unidentified alpha proteobacterium MBRG 4.22 (AJ508610), unidentified alpha proteobacterium MBRG 4.24 (AJ508612), Bacillus cereus MBRG 4.25 (AJ508705), Flavobacterium sp. strain MBRG 4.26 (AJ508710), Haloanella gallinarum MBRG 4.27 (AJ508708), Flavobacterium sp. strain MBRG 4.28 (AJ508709), Haloanella gallinarum MBRG 4.29 (AJ508711), Microbacterium phyllosphaerae MBRG 4.30 (AJ508704), and Achromobacter xylosoxidans MBRG 4.31 (AJ508608).
RESULTS
Effects of TCS on drain microcosms.
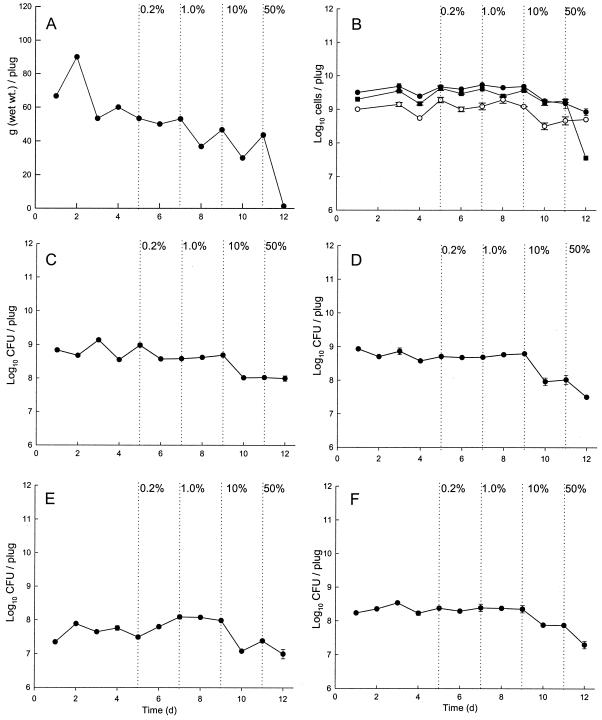
Fig. 1 shows data for short-term (14-day) exposure of microcosms to TCSD. TCSD lowered biofouling of CDFF plugs at concentrations above 10% (vol/vol) and markedly so at 50% (vol/vol). Direct and differential viable cell counts showed that the microcosms achieved dynamic stability prior to TCSD addition. Viable and live-dead cell counts showed that TCSD was moderately bactericidal against the biofilm communities at concentrations of 10% and above. At 50%, total viable direct cell counts were reduced by two orders of magnitude. Table 1 shows that in long-term experiments (336 days), baseline drain microcosms harbored ca. 10 log10 CFU of culturable bacteria/g of biofilm. Enteric species formed a large proportion of the bacterial types selectively cultured, with lesser proportions of gram-positive cocci and P. aeruginosa (Table 1). These unstressed control communities remained largely unchanged over 160 days. Successive 3-month exposures of the microcosms to TCS, four times daily, at concentrations of 0.2 and 0.4% (wt/vol) did not affect the total culturable cell counts. The total CFU count represented only ca. 4% of total cell counts on selective agars, while live-dead direct counting in the short-term experiments showed that numbers of total live cells were up to one order of magnitude higher than those of total culturable cells.
FIG. 1.
Bioburdens, direct counts, and viable cell counts of selected groups of drain bacteria in the simulator before and during product addition. Data are means ± standard deviations of results from two separate sample pans analyzed in triplicate. The schedule of TCSD addition is indicated. Graphs show numbers 5 days prior to dosing and then after successive 2-day exposures to 0.2, 1.0, 10, and 50% TCSD. (A) Bioburden. (B) Live-dead direct counts. Closed circles, total count; closed squares, live cell count; open circles, dead cell count. (C) Total culturable aerobes and facultative species. (D) Total culturable anaerobes. (E) Total culturable pseudomonads. (F) Culturable enteric species.
TABLE 1.
Effects of TCSD exposure on culturable bacteria in drain biofilm ecosystems grown in drain simulator
| Bacterial group | Mean no. of log10 CFU per gram (wet wt) of biofilm ± SD at TCSD concna:
|
||
|---|---|---|---|
| Baseline | 0.2% (vol/vol) | 0.4% (vol/vol) | |
| Total culturable heterotrophsb | 10.2 ± 0.1 | 10.1 ± 0.4 | 10.1 ± 0.1 |
| Enteric species | 8.8 ± 0.1 | 8.2 ± 0.5 | 9.1 ± 0.1 |
| P. aeruginosa | 7.0 ± 0.4 | 6.8 ± 0.5 | 7.2 ± 0.2 |
| Gram-positive cocci | 7.6 ± 0.4 | 7.6 ± 0.3 | 8.2 ± 0.4 |
n = 3. Baseline, numbers of log10 CFU per gram of biofilm in microcosm samples removed at steady state prior to addition of TCSD. Samples exposed to TCSD at 0.2% were removed 3 months after the addition of TCSD. Samples exposed to TCSD at 0.4% were removed after 3 additional months.
Total putative culturable heterotrophs based on sum of aerobic, facultative, and strictly anaerobic organisms cultured on R2A and Wilkins-Chalgren agar.
Phylogenetic characterization of biofilms before and after TCS exposure.
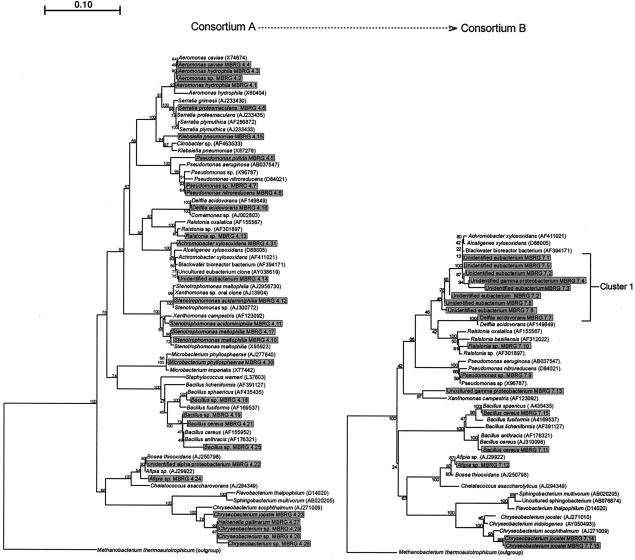
Isolation and identification of distinct morphotypes from the control, unstressed consortium (A) revealed considerable bacterial diversity. Since it was not possible to unambiguously identify all of the isolates from the control or stressed microcosms, phylogenetic trees were constructed for the major, phylogenetically distinct species isolated from both communities (Fig. 2).
FIG. 2.
Phylogenetic trees representing drain microcosm consortia (culturable species) before (consortium A) and after (consortium B) 6 months of exposure to TCSD. Cluster 1 comprised bacteria able to degrade particulate TCS. Isolated strains are displayed in shaded boxes, and reference strains are unshaded. The tree is based on 540-bp sequences of 16S RNA genes and was constructed by using the neighbor-joining method of Jukes and Cantor (25). The scale bar indicates 0.10 estimated substitutions per nucleotide. TREECON for Windows (52) was used to construct the tree. Bootstrap values denote confidence limits on the phylogenies, based on percentages of 100 replications. Methanobacterium thermautotrophicum was used to root the trees.
The trees unambiguously show a reduction of microbial diversity after exposure to TCS. In particular, the numbers of aeromonads, bacilli, chryseobacteria, klebsiellae, stenotrophomonads, and Microbacterium phyllosphaerae cells were all reduced or fell below detection thresholds following TCS exposure. The bacterial species represented after stress were in most cases isolatable without stress, albeit at a lower level. Diversity and cell numbers within a group of unidentified eubacteria closely related to Alcaligenes xylosoxidans (cluster 1), however, increased (Fig. 2). In unstressed microcosms, the phylogenetic trees demonstrated that the unidentified eubacterium MBRG 4.14 was most closely related to the Black water bioreactor bacterium (AY394171) and to the uncultured eubacterium clone (AY038619). The unidentified alpha proteobacterium MBRG 4.22 was closely related to both Achromobacter xylosoxidans (AF411021) and Bosea thiooxidans (AJ250798).
Effects of TCS on antimicrobial susceptibilities of drain microcosms.
Data in Table 2 show MIC ranges of a number of antimicrobials (determined by spiral gradient endpoint), before and after 6 months of exposure to TCS, for bacterial clones isolated from the microcosms and grouped on the basis of phylogenetic relatedness. These data show that while there was considerable intrageneric and some intraspecies variation in susceptibilities to most of the agents, this variation was less marked after TCS exposure. With all antimicrobials, insusceptible bacteria were isolated in high numbers both from control microcosms and from TCS-exposed samples. Overall, ciprofloxacin was the most effective agent, while penicillin V was relatively ineffective. Tolerance to the biocides cetrimide, TCS, chloro-3,5-dimethyl-phenol, and chlorhexidine was common. Generally, the least susceptible bacterial group was related to the Alcaligenes group (cluster 1), of which several members were innately insusceptible to all of the test agents (Table 2). With respect to TCS-mediated changes in susceptibilities, there were few if any alterations in susceptibilities that were either significant in extent or that could be attributed to TCS exposure.
TABLE 2.
Dominant cultivable microcosm isolates and their susceptibilities to selected antimicrobial agents, before and after 6 months of TCS exposure
| Type of isolate | No. of strains
|
MIC (μg/ml) ofa:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CE
|
CP
|
CT
|
DP
|
FA
|
V
|
PV
|
ER
|
TCS
|
CH
|
|||||||||||||
| Before | After | Before | After | Before | After | Before | After | Before | After | Before | After | Before | After | Before | After | Before | After | Before | After | Before | After | |
| Aeromonads | 3 | 0 | 0.3-15.3 | NA | 0.3-60 | NA | 0.3 | NA | 0.3-60 | NA | 0.3-3.0 | NA | 0.3-13.0 | NA | 0.3-60 | NA | 0.3-6.5 | NA | 0.3-T | NA | 0.3-5.2 | NA |
| Pseudomonads | 3 | 3 | 4.1-R | R | 0.3-1.6 | 0.3 | 0.3-0.9 | 1.1-1.8 | R | R | 9.9-R | R | 3.3-R | R | R | R | 0.3-R | R | 0.3-T | T | 1.0-T | NA |
| Serratia spp. | 1 | 0 | 0.3-R | NA | 0.3 | NA | 0.3-13.0 | NA | 6.8-R | NA | R | NA | 3.3-3.5 | NA | R | NA | 5.6-R | NA | T | NA | 2.3-7.9 | NA |
| Stenotrophomonads | 4 | 1 | 4.2-R | NA | 0.3-R | NA | 0.3-R | NA | 0.3-R | NA | 0.3-R | NA | 3.5-R | NA | 2.3-R | NA | 2.9-R | NA | T | NA | 2.2-T | NA |
| Ralstonia spp. | 0 | 2 | NA | 0.3-R | NA | 0.3-R | NA | 0.3-R | NA | R | NA | R | NA | R | NA | 4.3-R | NA | 0.3-R | NA | 1.3-3.1 | NA | 1.3-3.6 |
| Alcaligenes clade | 3 | 7 | 1.7-R | 0.3-R | 0.3-R | 0.3-R | 0.3-R | 0.3-4.3 | 1.1-R | 0.3-R | 8.5-R | R | 0.3-R | 1.4-R | 12.0-R | 0.3-R | 3.9-R | 0.3-R | 0.3-T | 1.9-T | 1.2-T | 0.9-T |
| Bacilli | 4 | 2 | 1.0-R | 0.3-1.9 | 1.4-R | 0.3 | 0.3-R | 0.3 | 0.9-R | R | 0.8-R | 0.8-1.6 | 0.3-R | 0.3 | 0.3-R | 0.3-1.2 | 0.3-R | 0.3 | 0.8-T | 0.8-3.0 | 0.6-T | 0.6-6.0 |
| Afipia spp. | 1 | 1 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 2.2-2.5 | 2.2-2.5 | 1.9-2.2 | 1.9-2.2 | 0.3 | 0.3 |
| Chryseobacterium spp. | 3 | 1 | 3.8-R | 11.1 | 0.3-R | 0.3 | 2.1-5.8 | R | 0.8-R | R | 0.3-R | R | R | 3.3 | 2.6-R | R | 2.6-R | R | 0.3-T | 2.8 | T | T |
| Microbacterium spp. | 1 | 0 | 6.9 | NA | R | NA | 2.7 | NA | 5.1 | NA | 1.4 | NA | R | NA | 6.2 | NA | 2.7 | NA | 4.3 | NA | T | NA |
| α Proteobacteria | 1 | 0 | 6.3 | NA | R | NA | R | NA | 2.7 | NA | 5.7 | NA | 3.1 | NA | 5.71 | NA | 0.3 | NA | 0.3 | NA | 2.5 | NA |
Data show susceptibility ranges. T, tolerant (>2 mg/ml); R, resistant (>200 μg/ml); NA, not applicable. Bacterial groupings are based on the phylogenetic tree (Fig. 1). CE, cetrimide; CP, ciprofloxacin; CT, chlortetracycline; DP, chloro-3,5-dimethyl-phenol; A, fusidic acid; V, vancomycin; PV, penicillin V; ER, erythromycin; CH, chlorhexidine.
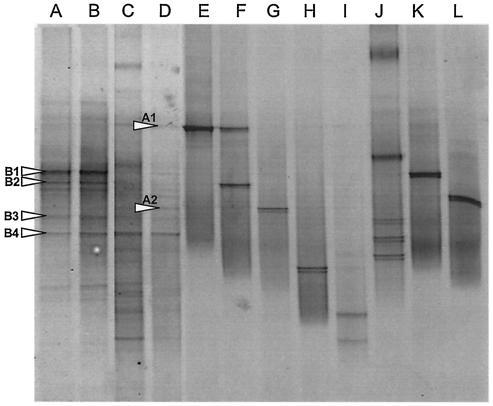
DGGE analysis of long-term microcosms.
Fig. 3 shows DGGE fingerprints from extracted community DNA. PCR-DGGE, using eubacterium-specific primers (16S rRNA gene), indicated that microcosms were stable before addition of an antimicrobial and that eubacterial diversity was reduced following TCS exposure. Figure 3 also showed that most reference strains produced single major DGGE bands, although secondary bands were observed, particularly with Aeromonas sp. strain MBRG 4.2.
FIG. 3.
Negative image of parallel DGGE gels showing community fingerprints for biofilm samples and pure cultures of reference strains. (A and B) Long-term drain microcosm samples taken 4 weeks apart from steady-state fermenters (before TCSD addition). (C) Drain microcosms after 3 months of exposure to TCSD (0.2%). (D) Drain microcosms following a further 3 months of exposure to TCSD (0.4%). (E) Pseudomonas nitroreducens MBRG 4.6. (F) Aeromonas sp. strain MBRG 4.2. (G) Stenotrophomonas maltophilia MBRG 4.17. (H) Pseudoxanthomonas sp. (I) Achromobacter xylosoxidans MBRG 4.31. (J) Citrobacter sp. (K) Eubacterium MBRG 7.1. (L) Microbacterium phyllosphaerae.
Sequence analysis of selected DGGE amplicons.
Data in Table 3 show closest relatives based on results of BLAST searches with DNA sequences obtained from major DGGE gel bands. The dominant phylotypes before TCSD addition were related to beta proteobacteria (bands B2 and B3). The sequences obtained from bands B1 and B5 did not give significant database matches. Following TCSD dosing, amplicons with homology to an alpha proteobacterium (AF236002) and an uncultured Sphingomonas sp. increased in apparent abundance (Table 3).
TABLE 3.
Characterization of dynamic changes in microcosms: sequences of dominant PCR amplicons derived from DGGE gels before and after TCSD exposure
| Time point | DNA amplicon | Total no. of bp | Ambiguity (%) | Closest relative (% sequence similarity)a |
|---|---|---|---|---|
| Before TCSD exposure | B1 | 173 | 25 | No significant match |
| B2 | 176 | 24 | Uncultured beta proteo- bacterium AF388886 (96) | |
| B3 | 174 | 20 | Beta proteobacterium AY146666 (94) | |
| B4 | 186 | 26 | No significant match | |
| After TCSD exposure | A1 | 188 | 11 | Uncultured Sphingomonas sp. AF388890 (92) |
| A2 | 153 | 15 | Alpha proteobacterium AF236002 (94) |
Identities based on information from BLAST database.
Fate of TCS in drain biofilm microcosms.
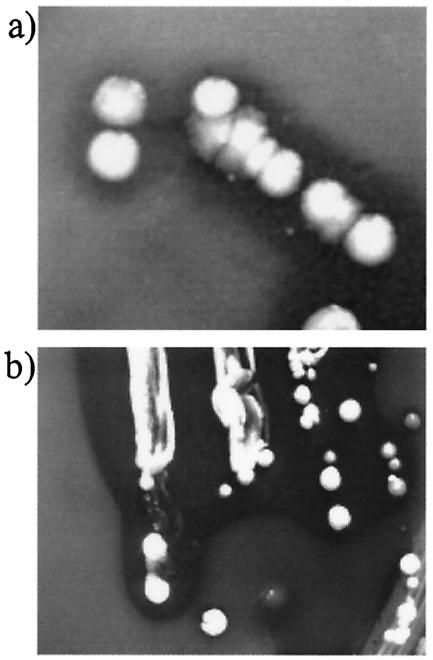
Fig. 4 shows clearing zones of particulate TCS within R2A agar plates. No colonies formed on mineral salts TCS agar, where TCS was the sole C source, although all of the selected bacteria formed colonies on saturated TCS agar and produced clearing zones. Data in Table 4 show that the unidentified eubacteria MBRG 7.2 and MBRG 7.6 were the most effective TCS-degrading isolates in pure culture. Microcosm consortia, however, degraded TCS in liquid culture more effectively than did any of the individual isolates, and degradation by the consortia was total in in situ microcosms and greatest under anaerobic conditions with ex situ consortia. Incubations with P. aeruginosa PAO1 showed an apparent increase in levels of TCS.
FIG. 4.
Clear zones produced on agar containing particulate TCS (2 g/liter). (a) Unidentified eubacterium MBRG 7.6 on R2A agar. (b) E. coli P2500 on MacConkey agar. Opacity is due to the presence of TCS at concentrations considerably higher than its aqueous solubility.
TABLE 4.
Degradation of TCS in incubations of drain consortia and isolated bacteria
| Samplea | Oxygenation status | % TCS degradationb (SD) |
|---|---|---|
| In situ consortium | Aerobic | 100 |
| Baseline consortium (A) | Aerobic | 66.7 (13.0) |
| Anaerobic | 83.7 (2.9) | |
| TCSD-exposed consortium (B) | Aerobic | 69.8 (32.3) |
| Anaerobic | 78.7 (14.1) | |
| Eubacterium MBRG 7.1 | Aerobic | 41.4 (12.1) |
| Eubacterium MBRG 7.5 | Aerobic | 28.5 (32.0) |
| Eubacterium MBRG 7.2 | Aerobic | 48.7 (11.0) |
| Eubacterium MBRG 7.6 | Aerobic | 67.3 (14.3) |
| Stenotrophomonas maltophilia MBRG 4.17 | Aerobic | 34.4 (10.9) |
| Serratia proteamaculans MBRG 4.8 | Aerobic | 13.6 (42.9) |
| E. coli P2500 | Aerobic | 15.3 (75.8) |
| P. aeruginosa PAO1 | Aerobic | −158 (149) |
Consortium samples A and B consisted of material excised from these consortia and incubated in vitro. All samples produced clear zones in saturated TCS agar. Samples of TCSD-exposed consortia were removed from microcosms after 6 months of exposure to antibacterial domestic detergent.
Values are percentages of TCS disappearance as measured by high-performance liquid chromatography and are means of results from duplicate bioreactors analyzed in duplicate for each sample.
DISCUSSION
The microcosms detailed here were previously developed and validated as a stable laboratory model of domestic-drain biofilms. They provide for the long-term maintenance and laboratory analysis of this ecosystem, which is a major site of bacterial colonization in the home (8, 11, 14, 31, 47). The possible adverse effects of TCS use are currently receiving considerable research attention (9, 10, 35). Most notably, this attention regards the possibility that the effectiveness of TCS might be compromised and that susceptibility to other antimicrobials might decrease via nonspecific mechanisms (30, 46). Importantly, the observation that susceptibility of E. coli to TCS is rapidly decreased following passage in the presence of TCS has not been shown to be true for unrelated bacteria (6, 27, 49).
Most research into development of resistance to TCS has utilized pure cultures. Few researchers have studied the effect of this biocide upon the dynamics of bacterial communities and mixed cultures where competition may dictate against susceptibility changes associated with fitness costs. Kitchen sink drains are arguably one of the most highly TCS-exposed environments, due to the widespread use of TCS in consumer products (51). Accordingly, we have studied the effects of chronic, sublethal exposure of biofilm communities to TCS in a domestic-drain biofilm simulator upon bacterial vitality, population dynamics, and bacterial susceptibilities to a range of biocides and antibiotics.
We used selective culture to characterize community effects of TCS in this study. Only ca. 4% of total counts on selective agars could be accounted for by the total CFU count, while live-dead direct counting in the short-term experiments showed that numbers of total live cells were up to one order of magnitude higher than those of total culturable cells. These observations suggest either a failure to culture bacteria from the drains or variable plating efficiencies of the culturable bacteria. This has been shown previously by DGGE of community 16S rDNA to relate to variable CFU recoveries of culturable species rather than the inability to culture certain species (31).
Exposure of drain microcosms to use levels of TCSD (0.2 to 0.4%) did not significantly lower total counts of drain biofilm bacteria for a number of possible reasons. Biofilms are considerably less susceptible to antimicrobials than are planktonic cells (2, 16, 17, 50), suggesting that lack of significant lethality could be simply due to the insusceptible biofilm phenotype. In addition, many of the bacteria isolated from the baseline drain microcosm were innately tolerant to TCS and could grow in saturated solutions of the biocide or on saturated TCS agar plates. Innate tolerance to TCS has been described previously (27) and relates to physiology of the bacteria, particularly the expression of efflux pumps (9, 10). Indeed, agars selective for pseudomonads (10) and aeromonads (3) contain TCS as a selective agent, while similar media formulated to be selective for Yersinia enterocolitica (45) also select for aeromonads (3). The innate insusceptibility of the drain bacteria might also account for the lack of changes attributable to TCS exposure in community antimicrobial susceptibility (Table 4).
Since changes in bacterial composition were not apparent from selective counting, the observed dynamic changes must relate to changes at the species or clonal level. The trees (Fig. 2) demonstrate that the diversity of the drain consortium became markedly reduced or that the numbers of certain species fell below detection thresholds following TCS exposure. The species represented after stress were in most cases present before the stress, albeit at a lower level. This was notable for the TCS-tolerant eubacteria (cluster 1) (Fig. 2). Importantly, these bacteria are notable as efficient TCS degraders (Table 4). These data show that losses or failure to detect organisms poststress in certain cases is related to the inhibition or killing of particularly sensitive clones, with a coincidental expansion of preexisting less-susceptible strains. Not all of the population changes, however, could be explained on this basis. For example, aeromonads were not detected in the stressed fermenters (Fig. 1), and these bacteria exhibit variable but low susceptibilities to TCS (Table 2) (3). Similarly, stenotrophomonads have low TCS susceptibilities, and these species also became undetectable following TCS exposure. DGGE analysis (Fig. 3) gave further evidence for decreases in bacterial diversity and showed that the fermenters were in steady state during the preantimicrobial phase. For the analysis of reference strains, there was generally good separation of different bacterial species, although Pseudomonas nitroreducens MBRG 4.6 and Aeromonas sp. strain MBRG 4.2 were not separated. Secondary and tertiary bands may have resulted from rRNA heterogeneity within individual species. The qualities of sequences obtained from DGGE amplicons were variable, and two gel bands (B1 and B4) failed to give significant database matches. Sequence analysis, however, did show that two major DGGE bands, before TCSD addition, corresponded to beta proteobacteria, and bands that appeared during TCSD dosing had homology to Sphingomonas sp. (AF388890) and an unidentified alpha proteobacterium, which are unlikely to be overtly pathogenic species.
Perturbation of ecosystems frequently results in effects that cannot be simply extrapolated from pure culture. For example, loss of the aeromonads might have effected a cross-feeding relationship or resulted in the clonal expansion of an inhibitory bacterium. By such mechanisms, degradation of an ecosystem may rearrange the competitive hierarchy, making a normally less competitive species more competitive under the altered selective environment.
TCS degradation was suggested by the failure to detect this compound within TCS-exposed microcosms. Running TCS incubations of an ex situ microcosm gave further evidence for degradation (Table 4). Importantly, in agreement with the findings of Hay et al. (18), we were unable to isolate species that could degrade TCS when it was the sole carbon source. Consortium samples and, to a lesser degree, several isolates (MBRG 7.5, MBRG 7.2, and and MBRG 7.6) apparently degraded TCS when alternative carbon sources were provided, however (Table 4). The greater degradation in consortium samples occurred presumably due to synergy between multiple species. The production of clear zones on particulate TCS agar was not associated with degradation. All test strains, including the type cultures P. aeruginosa PAO1 and E. coli P2500, produced this clearing. P. aeruginosa PAO1, for example, produced considerable clearing of particulate TCS, but this, together with the apparent increases in TCS levels in liquid culture incubations, was due to TCS solubilization, probably resulting from the elaboration of a biosurfactant (Table 4). This bacterium is known to produce a number of such compounds, including rhamnolipid (42).
The detoxification of TCS by bacterial (18) and fungal (24) consortia has been recently reported to occur in activated sludge (13, 18) and compost (36) samples. Earlier studies demonstrated environmental degradation of up to 50% of TCS added to sewage sludge under both aerobic and anaerobic conditions (53). Interestingly, Alcaligenes xylosoxidans subsp. denitrificans, previously implicated in TCS degradation (36), is closely related to the bacteria in cluster 1, as evidenced by phylogenetic placements of these species (Fig. 2). The demonstration of TCS-degradative activities in domestic-drain-type biofilms may be of significance when considering the effects and environmental fate of TCS used in the kitchen. In view of the apparent widespread nature of TCS-degradative activities, it is somewhat surprising that trace levels of TCS have been detected in aquatic environments (29, 38), human milk (1), fish bile, and human plasma (23). This is especially notable since there is no apparent residue from its daily use in dentifrices (5). It is possible, however, that TCS degradation is an energy-dependent process and as such may be more active in environments such as domestic-drain biofilms and activated sludge than in highly oligotrophic freshwater environments. Furthermore, TCS degradation appears to be relatively slow and incomplete, even in nutrient-rich environments, as evidenced by data in Table 4 and in previous investigations (18, 53).
In conclusion, long-term exposure of domestic-drain biofilms to sublethal levels of TCS did not affect bacterial vitality or significantly alter antimicrobial susceptibility. This lack of effect may reflect (i) the general insusceptibility of drain biofilm bacteria to TCS, (ii) lack of TCS penetration into the biofilm, and (iii) the biodegradation of TCS by the drain biofilm consortium. Insusceptibility of drain isolates to a range of antimicrobials appears to relate to innate resistance or insusceptibility as opposed to developed, transferable resistance. Since the sample used to inoculate the microcosm system had been obtained from a household that had not used biocide-containing products since the installation of the sink, intrinsic biocide or drug insusceptibility of the associated microcosm cannot be attributed to past exposure to TCS. We conclude therefore that the emergence of antibiotic resistance through TCS use in the kitchen is highly improbable.
Acknowledgments
A.J.M., R.G.L., and P.G. are grateful to Procter and Gamble, Cincinnati, Ohio, for their support.
REFERENCES
- 1.Adolfsson-Erici, M., M. Pettersson, J. Parkkonen, and J. Sturve. 2002. Triclosan, a commonly used bactericide found in human milk and in the aquatic environment in Sweden. Chemosphere 46:1485-1489. [DOI] [PubMed] [Google Scholar]
- 2.Allison, D. G., A. J. McBain, and P. Gilbert. 2000. Microbial biofilms: problems of control, p. 310-327. In H. Lappin-Scott, P. Gilbert, M. Wilson, and D. Roberts (ed.), Community structure and cooperation in biofilms. Cambridge University Press, Cambridge, England.
- 3.Altorfer, R., M. Altwegg, J. Zollinger-Iten, and A. von Graevenitz. 1985. Growth of Aeromonas spp. on cefsulodin-Irgasan-novobiocin agar selective for Yersinia enterocolitica. J. Clin. Microbiol. 22:478-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayliffe, G. A., J. R. Babb, J. G. Davies, and H. A Lilly. 1988. Hand disinfection: a comparison of various agents in laboratory and ward studies. J. Hosp. Infect. 11:226-243. [DOI] [PubMed] [Google Scholar]
- 5.Bagley, D. M., and Y. J. Lin. 2000. Clinical evidence for the lack of triclosan accumulation from daily use in dentifrices. Am. J. Dent. 13:148-152. [PubMed] [Google Scholar]
- 6.Bamber, A. I., and T. J. Neal. 1999. An assessment of triclosan susceptibility in methicillin-resistant and methicillin-sensitive Staphylococcus aureus. J. Hosp. Infect. 41:107-109. [DOI] [PubMed] [Google Scholar]
- 7.Bhargava, H. N., and P. A. Leonard. 1996. Triclosan applications and safety. Am. J. Infect. Control 24:209-218. [DOI] [PubMed] [Google Scholar]
- 8.Charaf, U. K. 1997. Biofilms in our drains, p. 175-181. In J. Wimpenny, P. Handley, P. Gilbert, H. Lappin-Scott, and M. Jones (ed.), Biofilms: community interactions and control. Bioline Press, Cardiff, United Kingdom.
- 9.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuanchuen, R., K. Beinlich, T. T. Hoang, A. Becher, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cogan, T. A., J. Slader, S. F. Bloomfield, and T. J. Humphrey. 2002. Achieving hygiene in the domestic kitchen: the effectiveness of commonly used cleaning products. J. Appl. Microbiol. 92:885-892. [DOI] [PubMed] [Google Scholar]
- 12.Cookson, B. D., H. Farrelly, P. Stapleton, R. P. J. Garvey, and M. R. Price. 1991. Transferable resistance to triclosan in MRSA. Br. Med. J. 337:1548-1549. [DOI] [PubMed] [Google Scholar]
- 13.Federle, T. W., S. K. Kaiser, and B. A. Nuck. 2002. Fate and effects of triclosan in activated sludge. Environ. Toxicol. Chem. 21:1330-1337. [PubMed] [Google Scholar]
- 14.Finch, J. E., J. Prince, and M. Hawkworth. 1978. A bacteriological survey of the domestic environment. J. Appl. Microbiol. 45:357-364. [DOI] [PubMed] [Google Scholar]
- 15.Furia, T. E., and A. G. Schenkel. 1968. A new, broad spectrum bacteriostat. Soap Chem. Spec. 44:47-50 and 116-122. [Google Scholar]
- 16.Gilbert, P., J. R. Das, M. V. Jones, and D. G. Allison. 2001. Assessment of resistance towards biocides following the attachment of micro-organisms to, and growth on, surfaces. J. Appl. Microbiol. 91:248-254. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert, P., T. Maira-Litran, A. J. McBain, A. H. Rickard, and F. W. Whyte. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46:202-256. [PubMed] [Google Scholar]
- 18.Hay, A. G., P. M. Dees, and G. S. Sayler. 2001. Growth of a bacterial consortium on triclosan. FEMS Microbiol. Ecol. 36:105-112. [DOI] [PubMed] [Google Scholar]
- 19.Heath, R. J., and C. O. Rock. 1995. Enoyl-acyl carrier protein reductase (FabI) plays a determinant role in completing cycles of fatty-acid elongation in Escherichia coli. J. Biol. Chem. 270:26538-26542. [DOI] [PubMed] [Google Scholar]
- 20.Heath, R. J., G. E. Roland, and C. O. Rock. 2000. Inhibition of the Staphylococcus aureus NADPH-dependant enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 275:4654-4659. [DOI] [PubMed] [Google Scholar]
- 21.Heath, R. J., N. Su, C. K. Murphy, and C. O Rock. 2000. The enoyl-acyl-carrier-protein reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 275:40128-40133. [DOI] [PubMed] [Google Scholar]
- 22.Heath, R. J., Y. T. Yu, M. A. Shapiro, E. Olson, and C. O. Rock. 1998. Broad-spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316-30320. [DOI] [PubMed] [Google Scholar]
- 23.Hovander, L., T. Malmberg, M. Athanasiadou, I. Athanassiadis, S. Rahm, A. Bergman, and E. Wehler. 2002. Identification of hydroxylated PCB metabolites and other phenolic halogenated pollutants in human blood plasma. Arch. Environ. Contam. Toxicol. 42:105-117. [DOI] [PubMed] [Google Scholar]
- 24.Hundt, K., D. Martin, E. Hammer, U. Jonas, M. K. Kindermann, and F. Schauer. 2000. Transformation of triclosan by Trametes versicolor and Pycnoporus cinnabarinus. J. Antimicrob. Chemother. 49:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jukes, T. H., and C. P. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 26.Kwochka, K. W., and J. J. Kowalski. 1991. Prophylactic efficacy of four antibacterial shampoos against Staphylococcus intermedius in dogs. Am. J. Vet. Res. 52:115-118. [PubMed] [Google Scholar]
- 27.Lear, J. C., J. Y. Maillard, P. W. Dettmar, P. A. Goddard, and A. D. Russell. 2002. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources. J. Ind. Microbiol. Biotechnol. 29:238-242. [DOI] [PubMed] [Google Scholar]
- 28.Levy, C. W., A. Roujeinikova, S. Sedelnikova, P. J. Baker, A. R. Stuitje, A. R. Slabas, D. W. Rice, and J. B. Rafferty. 1999. Molecular basis of triclosan activity. Nature 398:383-384. [DOI] [PubMed] [Google Scholar]
- 29.Lindstrom, A., I. J. Buerge, T. Poiger, P. A. Bergqvist, M. D. Muller, and H. R. Buser. 2002. Occurrence and environmental behavior of the bactericide triclosan and its methyl derivative in surface waters and in wastewater. Environ. Sci. Technol. 36:2322-2329. [DOI] [PubMed] [Google Scholar]
- 30.McBain, A. J., and P. Gilbert. 2001. Biocide tolerance and the harbingers of doom. Int. Biodeterior. Biodegrad. 47:55-61. [Google Scholar]
- 31.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, A. H. Rickard, S. A. Symmons, and P. Gilbert. 2003. Microbial characterization of biofilm in domestic drains and the establishment of stable biofilm microcosms. Appl. Environ. Microbiol. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, and P. Gilbert. 2003. Growth and molecular characterization of dental plaque microcosms. J. Appl. Microbiol. 94:655-664. [DOI] [PubMed] [Google Scholar]
- 33.McGeer, A., D. E. Low, J. Penner, J. Ng, C. Goldman, and A. E. Simor. 1990. Use of molecular typing methods to study the epidemiology of Serratia marcescens. J. Clin. Microbiol. 28:55-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMurry, L. M., P. F. McDermott, and S. B. Levy. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob. Agents Chemother. 43:711-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Triclosan targets lipid synthesis. Nature 394:531-532. [DOI] [PubMed] [Google Scholar]
- 36.Meade, M. J., R. L. Waddell, and T. M. Callahan. 2001. Soil bacteria Pseudomonas putida and Alcaligenes xylosoxidans subsp. denitrificans inactivate triclosan in liquid and solid substrates. FEMS Microbiol. Lett. 204:45-48. [DOI] [PubMed] [Google Scholar]
- 37.Miesel, L., D. A. Rozwarski, J. C. Sacchettini, and W. R. Jacobs, Jr. 1998. Mechanisms for isoniazid action and resistance. Novartis Found. Symp. 217:209-220. [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki, T., T. Yamagishi, and M. Matsumoto. 1984. Residues of 4-chloro-1-(2,4-dichlorophenoxy)-2-methoxybenzene(triclosan methyl) in aquatic biota. Bull. Environ. Contam. Toxicol. 32:227-232. [DOI] [PubMed] [Google Scholar]
- 39.Moore, J. E., I. Thompson, M. Crowe, J. Xu, A. Shaw, B. C. Millar, A. O. B. Redmond, A. J. M. Reid, C. Clarke, and J. S. Elborn. 2002. Burkholderia cepacia from a sink drain. J. Hosp. Infect. 50:235-237. [DOI] [PubMed] [Google Scholar]
- 40.Moran, J., M. Addy, R. G. Newcombe, and I. Marlow. 2001. A study to assess the plaque inhibitory action of a newly formulated triclosan toothpaste. J. Clin. Periodontol. 28:86-89. [DOI] [PubMed] [Google Scholar]
- 41.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 42.Noordman, W. H., and D. B. Janssen. 2002. Rhamnolipid stimulates uptake of hydrophobic compounds by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 68:4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters, A., and J. Wimpenny. 1988. A constant-depth laboratory model film fermenter. Biotechnol. Bioeng. 32:263-270. [DOI] [PubMed] [Google Scholar]
- 44.Regos, J., O. Zak, R. Solf, W. A. Vischer, and E. G. Weirich. 1979. Antimicrobial spectrum of triclosan, a broad-spectrum antimicrobial agent for topical application. II. Comparison with some other antimicrobial agents. Dermatologica 158:72-79. [DOI] [PubMed] [Google Scholar]
- 45.Schiemann, D. A. 1979. Synthesis of a selective agar medium for Yersinia enterocolitica. Can. J. Microbiol. 25:1298-1304. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer, H. P. 2001. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 202:1-7. [DOI] [PubMed] [Google Scholar]
- 47.Scott, E., S. F. Bloomfield, and C. G. Barlow. 1982. An investigation of microbial contamination in the home. J. Hyg. (Cambridge) 89:279-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slayden, R. A., R. E. Lee, and C. E. Barry. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38:514-525. [DOI] [PubMed] [Google Scholar]
- 49.Sreenivasan, P., and A. Gaffar. 2002. Antiplaque biocides and bacterial resistance: a review. J. Clin. Periodontol. 29:965-974. [DOI] [PubMed] [Google Scholar]
- 50.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 51.Susman, E. 2001. Too clean for comfort. Environ. Health Perspect. 109:A18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van de Peer, Y., and R. De Wachter. 1997. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput. Appl. Biosci. 13:227-230. [DOI] [PubMed] [Google Scholar]
- 53.Voets, J. P., P. Pipyn, P. van Lancker, and W. Verstraete. 1976. Degradation of microbiocides under different environmental conditions. J. Appl. Bacteriol. 40:67-72. [DOI] [PubMed] [Google Scholar]
- 54.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wexler, H. M., E. Molitoris, P. R. Murray, J. Washington, R. J. Zabransky, P. H. Edelstein, and S. M. Finegold. 1996. Comparison of spiral gradient endpoint and agar dilution methods for susceptibility testing of anaerobic bacteria: a multilaboratory collaborative evaluation. J. Clin. Microbiol. 34:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zafar, A. B., R. C. Butler, D. J. Reese, L. A. Gaydos, and P. A. Mennonna. 1995. Use of 0.3% triclosan (Bacti-Stat) to eradicate an outbreak of methicillin-resistant Staphylococcus aureus in a neonatal nursery. Am. J. Infect. Control 23:200-208. [DOI] [PubMed] [Google Scholar]