Abstract
More than 40 bacterial strains belonging to the cosmopolitan Polynucleobacter necessarius cluster (Betaproteobacteria) were isolated from a broad spectrum of freshwater habitats located in three climatic zones. Sequences affiliated with the freshwater P. necessarius cluster are among the most frequently detected in studies on bacterial diversity in freshwater ecosystems. Despite this frequent detection with culture-independent techniques and the cosmopolitan occurrence of members affiliated with this cluster, no isolates have been reported thus far. The isolated strains have been obtained from lakes, ponds, and rivers in central Europe, the People's Republic of China, and East Africa by use of the filtration-acclimatization method. The 16S rRNA gene sequences of the isolates are 98.8 to 100% identical to reference sequences obtained by various authors by use of culture-independent methods. The isolates, aerobic heterotrophs, grew on a wide range of standard complex media and formed visible colonies on agar plates. Thus, the previous lack of isolates cannot be explained by a lack of appropriate media. Most of the isolates possess, under a wide range of culture conditions, very small cells (<0.1 μm3), even when grown in medium containing high concentrations of organic substances. Thus, these strains are obligate ultramicrobacteria. The obtained strains have a C-shaped cell morphology which is very similar to that of recently isolated ultramicrobacterial Luna cluster strains (Actinobacteria) and the SAR11 cluster strains (Alphaproteobacteria).
In recent years, diversity studies employing culture-independent methods have strongly expanded the knowledge about the phylogenetic composition of freshwater bacterioplankton (7, 15, 32). In a comparative study, Zwart and colleagues (32) identified 34 putative phylogenetic clusters of bacteria which seem to contain typical freshwater inhabitants. Seventy percent of these clusters were reported to contain no cultivated representatives. Each of the other clusters contained usually only a few cultivated strains, which are far from covering the whole range of diversity represented by the available environmental sequences. Zwart and coauthors also analyzed a set of 14 diversity studies for the detection of each of the 34 putative freshwater clusters. All of these analyzed studies have used random cloning of 16S rRNA genes. One of the most frequently detected putative clusters was the betaproteobacterial Polynucleobacter necessarius cluster (the ACK-1 cluster [14], or the beta II cluster [7]). This cluster was discovered by Hirons and colleagues in a study on diversity of bacterioplankton in the Adirondack mountain lakes (14). Meanwhile, more than 100 environmental 16S rRNA gene sequences affiliated with the P. necessarius cluster have been deposited in the public databases during the past eight years. Most of the sequences are partial sequences of cloned genes. The sequences have been obtained from a broad spectrum of lakes and river ecosystems widely distributed over the northern hemisphere, as well as from one lake in the southern hemisphere. This set of ecosystems included, for instance, the arctic Toolik Lake (2, 5), the antarctic Sombre Lake (19), the predominately acidic Adirondack mountain lakes (14, 18), the deep oligotrophic Lake Baikal (7, 24), and rivers ranging from oligotrophic to eutrophic (4, 22, 23, 32). Only one published P. necessarius cluster sequence has been obtained from a nonsurface freshwater system (20). This short sequence was obtained in an investigation on the bacterial diversity in boreholes along a tunnel 626 m below ground surface. No P. necessarius cluster sequences have been reported from freshwater sediments or marine or soil systems.
Pernthaler and colleagues applied a cluster-specific probe for detection of P. necessarius cluster members in the acidic Lake Fuchskuhle by way of fluorescence in situ hybridization. Up to 50% of the bacterioplankton consisted of bacteria affiliated with the P. necessarius cluster (3a).
No pure-culture isolates affiliated with the P. necessarius cluster have been reported thus far. Bruns and colleagues systematically studied the influence of signal compounds (e.g., cyclic AMP) and different incubation conditions on the culturability of freshwater bacterioplankton (3). They grew diluted samples of bacterioplankton in synthetic medium and analyzed the obtained mixed cultures by 16S rRNA gene fingerprinting techniques. Analysis of sequences obtained from the fingerprints demonstrated that they successfully enriched members of the P. necessarius cluster in their mixed cultures.
The species after which the P. necessarius cluster was named is an obligate endosymbiont of the hypotrichous ciliate Euplotes aediculatus (13, 28). This bacterium was never grown in pure culture but was described as a species in 1987 (13). The type strain of this species is still growing as an endosymbiont in a ciliate culture.
In this paper, the first isolation and cultivation of strains affiliated with the P. necessarius cluster is reported, and insights into the ecology of this group of representative freshwater bacteria are presented.
MATERIALS AND METHODS
Sampling sites.
Samples from nine ecologically contrasting freshwater habitats located in three climatic zones were processed in order to isolate representative freshwater bacteria (Table 1). Samples taken from Lake Mondsee were immediately processed after sampling. All of the other samples were transported to the laboratory in sealed tubes and processed within a few days after sampling. All treatments after sampling were carried out under aseptic conditions.
TABLE 1.
Characteristics of aquatic habitats from which P. necessarius cluster strains were isolated
| Sampling site | No. of samples | Habitat | Location | Geographic coordinates | Countrya | Climatic zone | Trophic statusb | pH |
|---|---|---|---|---|---|---|---|---|
| Lake Mondsee | 4 | Deep submontane lake in prealpine region | Near Salzburg | 47°50′N, 13°20′E | Austria | Temperate | Oligomeso- trophic | 8.7-9.1 |
| Le Canal de Roanne à Digoin | 1 | Small channel | Avrilly | 46°15′N, 4°10′E | France | Temperate | NK | NK |
| Pond in Beijing | 1 | Small pond | Beijing | 39°50′N, 116°25′E | P.R. China | Temperate | NK | NK |
| Yangtze (Chang) River | 1 | Large polluted river | Near Nanjing | 32°05′N, 118°30′E | P.R. China | Subtropical | Eutrophicc | 7.7 |
| Lake Tai hud | 1 | Large shallow lake | Near Wuxi | 31°30′N, 120°20′E | P.R. China | Subtropical | Hypertrophicd | 8.3 |
| Humble Park pond 1 | 1 | Small pond with macro- phytes (Lotus) | Suzhou | 31°20′N, 120°40′E | P.R. China | Subtropical | Eutrophicc | 9.0 |
| Humble Park pond 2 | 1 | Small pond with macro- phytes (Lotus) | Suzhou | 31°20′N, 120°40′E | P.R. China | Subtropical | Eutrophicc | 9.0 |
| Tiger Hill (Huqiu) pond | 1 | Small pond without macrophytes | Suzhou | 31°20′N, 120°40′E | P.R. China | Subtropical | Eutrophicc | 7.4 |
| Lake Victoria | 1 | Large tropical lake | Near Kampala | 0°20′N, 32°40′E | Uganda | Tropical | Mesotrophic | ∼8 |
P.R. China, People's Republic of China.
NK, not known.
Estimation based on a single measurement of total phosphorus.
Meiling Bay.
Isolation and maintenance of strains.
For isolation, the filtration-acclimatization method was used (12). Briefly, 5 to 10 ml of sample was filtered through 0.2-μm-pore-size filters (Minisart syringe filters; Sartorius, Göttingen, Germany). Subsamples of 100 μl of filtrate were transferred to wells of sterile 24-well cell culture plates and diluted with 900 μl of sterile inorganic basal medium (12). One to four culture plates were set up per sample. The bacteria contained in the filtrated subsamples were stepwise acclimatized to higher substrate concentrations. Therefore, each well received stepwise-increasing doses of NSY (nutrient broth, soytone, yeast extract) medium (12). In most of the isolation experiments performed, screening of wells for growth of bacteria was done by dropping 5-μl samples of the cultures onto agar plates with concentrations of NSY of 3 g liter−1 (hereafter referred to as 3-g liter−1 NSY agar plates) and subsequently incubating the plates at room temperature. In a few isolation experiments, the wells were screened microscopically before plating of samples onto agar plates (12). In all of the experiments, samples of colonies grown on plates were suspended in inorganic basal medium, stained with DAPI (4′,6′-diamidino-2-phenylindole [Sigma]; 0.1 mg ml−1 final concentration), and screened by epifluorescence microscopy for the presence of small C-shaped cells. Positive colonies were subcultured in liquid 3-g liter−1 NSY medium and plated again onto NSY agar plates. This procedure was repeated until pure cultures were established. Pure cultures were preserved by deep freezing (3 g-liter−1 NSY medium plus 10% glycerol and storage at −70°C).
Sequencing of 16S rRNA genes and phylogenetic analysis.
Amplification and sequencing of the 16S rRNA genes of the isolates were performed as described previously (12), but PCR products were sequenced by MWG-Biotech (Ebersberg, Germany). The program BLAST (http://www.ncbi.nlm.nih.gov/BLAST/; 1) was used to find all publicly available sequences closely related to the sequences obtained from the isolates, as well as all sequences affiliated with the P. necessarius cluster. Therefore, complete and partial sequences of the isolates and other members of the cluster were submitted for comparison. Self-owned as well as publicly available reference sequences were aligned by using the ARB software package (http://www.arb-home.de; 30). For construction of phylogenetic trees, the aligned sequences were exported to other programs. Neighbor-joining trees were constructed with the software Mega version 2.1 (http://www.megasoftware.net; 17). Evolutionary distances were corrected for multiple substitutions according to the algorithm of Jukes and Cantor (16). Maximum likelihood trees were constructed with the software Treefinder (written by G. Jobb; http://www.treefinder.de).
Growth of isolates on various complex media.
Ten different complex media were used in order to reveal the range of media which support growth of the isolates. The media used for this purpose were Bacto Yeast Extract, Bacto Peptone, soytone peptone, Bacto Tryptic Soy Broth (without dextrose), Bacto Luria-Bertani Broth, Bacto Nutrient Broth, R2A agar (Remel), standard method agar (Remel), eugonic broth (Remel), and Bacto Brain Heart Infusion (Becton Dickinson). Unless stated otherwise, media were purchased from Difco. For better comparability, all media were adjusted to substrate concentrations of 3 g liter−1 and agar concentrations of 1.5%. The test was restricted to a subset of 10 isolates representing almost all of the isolated 16S rRNA genotypes. Ten microliters of precultures (liquid 3-g liter−1 NSY medium) were spread onto the agar plates by use of an inoculating loop, and the plates were incubated at room temperature (ca. 20°C) for 10 days. Incubation of the isolates on 3-g liter−1 NSY agar plates served as a positive control for bacterial growth. Formation of visible colonies within 10 days was assessed as support of growth by a particular medium.
Determination of bacterial growth rates.
For a subset of 10 isolates, the growth rates in liquid 3-g liter−1 NSY medium were determined by measurement of the optical density at 575 nm during growth at a constant temperature of 20°C.
Determination of bacterial cell sizes.
The cell sizes of several isolates grown under various culture conditions were determined as described previously (12).
Nucleotide sequence accession numbers.
The nearly complete 16S rRNA gene sequences of the 25 isolates were deposited in the EMBL nucleotide sequence database under the accession numbers AJ550649 to AJ550673.
RESULTS
Isolation of strains.
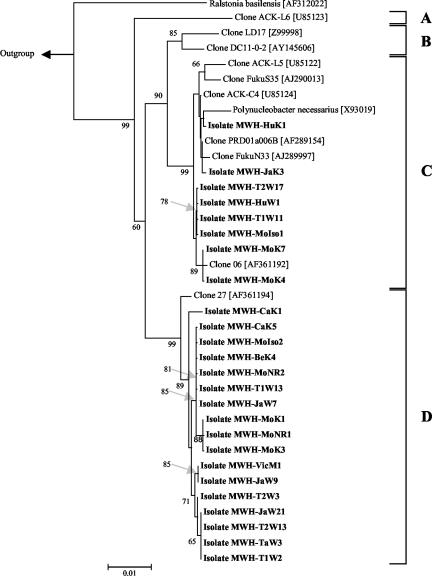
Forty-five strains with partial 16S rRNA gene sequences affiliated with the P. necessarius cluster were isolated from 12 samples obtained from nine habitats (Table 1). In the cases in which coisolates obtained from the same water samples shared identical partial (ca. 500-bp) sequences, only one representative was included in further sequencing. The remaining 25 isolates (Table 2) represent 11 different 16S rRNA genotypes (Fig. 1). All of the 25 isolates sharing identical sequences were isolated either from different habitats or from samples from the same habitat (Lake Mondsee) taken on different dates.
TABLE 2.
Isolates affiliated with the P. necessarius cluster and partial or nearly complete environmental 16S rRNA gene sequences with highest similarities
| Isolate | Accession no. | Source | Closest relativea | Accession no. | Sourceb | Similarity (%) | Reference |
|---|---|---|---|---|---|---|---|
| MWH-MoNR1 | AJ550649 | Lake Mondsee | Clone SC17 (p) | AY187480 | Mula-Mutha River, India | 99.8c | Satoor et al., unpublished |
| MWH-MoNR2 | AJ550650 | Lake Mondsee | Clone CRE-FL11 (p) | AF141441 | Columbia River, United States | 100.0 | 4 |
| MWH-MoIso1 | AJ550671 | Lake Mondsee | Clone FukuN66 (p) | AJ290078 | Fuchskuhle, Germany | 99.3 | 7 |
| MWH-MoIso2 | AJ550672 | Lake Mondsee | Clone CRE-FL11 (p) | AF141441 | Columbia River, United States | 100.0 | 4 |
| MWH-MoK1 | AJ550652 | Lake Mondsee | Clone SC17 (p) | AY187480 | Mula-Mutha River, India | 99.8c | Satoor et al., unpublished |
| MWH-MoK7 | AJ550673 | Lake Mondsee | Clone 06 | AF361192 | Rimov Reservoir, Czech Republic | 99.6 | 25 |
| MWH-MoK3 | AJ550653 | Lake Mondsee | Clone SC17 (p) | AY187480 | Mula-Mutha River, India | 99.8c | Satoor et al., unpublished |
| MWH-MoK4 | AJ550654 | Lake Mondsee | Clone 06 | AF361192 | Rimov Reservoir, Czech Republic | 99.6 | 25 |
| MWH-CaK1 | AJ550667 | Canal Roanne à Digoin | DGGE band wj 16 (p) | AF530937 | Walchensee, Germany | 98.8 | A. Bruns et al., unpublished |
| MWH-CaK5 | AJ550655 | Canal Roanne à Digoin | Clone CR-FL13 (p) | AF141398 | Columbia River, United States | 99.8 | 4 |
| MWH-VicM1 | AJ550651 | Lake Victoria | Clone CR99-2-58 (p) | AF429244 | Yangtze River, P. R. China | 100.0 | 22 |
| MWH-TaW3 | AJ550661 | Lake Tai hu | Clone CR99-2-49 (p) | AF429235 | Yangtze River, P.R. China | 99.8 | 22 |
| MWH-BeK4 | AJ550656 | Pond in Beijing | Clone CR-FL13 (p) | AF141398 | Columbia River, United States | 99.8 | 4 |
| MWH-JaW7 | AJ550658 | Yangtze River | Clone CR-FL13 (p) | AF141398 | Columbia River, United States | 99.8 | 4 |
| MWH-JaW21 | AJ550660 | Yangtze River | Clone CR99-2-49 (p) | AF429235 | Yangtze River, P.R. China | 99.8 | 22 |
| MWH-JaW9 | AJ550659 | Yangtze River | Clone CR99-2-58 (p) | AF429244 | Yangtze River, P.R. China | 100.0 | 22 |
| MWH-JaK3 | AJ550657 | Yangtze River | Clone FukuN33 | AJ289997 | Fuchskuhle, Germany | 99.8 | 7 |
| MWH-T1W13 | AJ550662 | Humble Park pond 1 | Clone CR-FL13 (p) | AF141398 | Columbia River, United States | 99.8 | 4 |
| MWH-T1W11 | AJ550670 | Humble Park pond 1 | Clone FukuN66 (p) | AJ290078 | Fuchskuhle, Germany | 99.3 | 7 |
| MWH-T1W2 | AJ550669 | Humble Park pond 1 | Clone CR99-7-39 (p) | AF429073 | Yangtze River, P.R. China | 99.8 | 22 |
| MWH-T2W13 | AJ550663 | Humble Park pond 2 | Clone CR99-7-39 (p) | AF429073 | Yangtze River, P.R. China | 99.8 | 22 |
| MWH-T2W3 | AJ550668 | Humble Park pond 2 | DGGE band WL8-0 (p) | AF497887 | Weser River, Germany | 100.0 | 23 |
| MWH-T2W17 | AJ550664 | Humble Park pond 2 | Clone FukuN66 (p) | AJ290078 | Fuchskuhle, Germany | 99.3 | 7 |
| MWH-HuK1 | AJ550665 | Tiger Hill pond | Clone ACK-C4 (p) | U85124 | Carry Pond, United States | 98.9 | 14 |
| MWH-HuW1 | AJ550666 | Tiger Hill pond | Clone FukuN66 (p) | AJ290078 | Fuchskuhle, Germany | 99.3 | 7 |
P, partial sequence; DGGE, denaturing gradient gel electrophoresis.
P.R. China, People's Republic of China.
Sequence seems to contain a 36-nucleotide-long sequence stretch from the vector at the 5′ end. This part of the sequence was omitted from the analysis.
FIG. 1.
Neighbor-joining tree showing the phylogenetic relationships of isolated and uncultured strains affiliated with the P. necessarius cluster. The tree was calculated with 1,240 nucleotide sequence stretches of the 16S rRNA genes. The tree is based on all publicly available sequences affiliated with the P. necessarius cluster which have sequence lengths of ≥1,292 nucleotides. The length limit has been defined by the maximum sequence length available for representatives of the various subclusters. Four proposed subclusters (A to D) are indicated by brackets. Alcaligenes faecalis (M22508) served as the outgroup (data not shown). R. basilensis is the closest known relative of the P. necessarius cluster bacteria. Bootstrap (1,000 replicates) values of ≥60% are shown. The scale bar indicates 1% estimated sequence divergence.
In total, six samples taken from Lake Tai hu at three different locations have been processed, but despite the large number of samples it was possible to isolate only a single strain belonging to the P. necessarius cluster. In the case of all six samples, large filamentous bacteria occurred in the majority of the 144 established enrichment cultures. These filamentous bacteria are able to overgrow P. necessarius cluster strains, as well as other ultramicrobacteria (12). The same filamentous morphotypes also occurred in cultures inoculated with samples taken from other habitats. These bacteria occurred with lower frequencies in the other enrichment cultures than in the enrichment cultures obtained from Lake Tai hu samples. Therefore, isolation experiments with the other investigated samples were more successful in terms of the number of P. necessarius cluster isolates obtained than the trials with the Lake Tai hu samples.
All isolated strains possess a C-shaped morphology and small cells (Fig. 2). The strains formed tiny, nonpigmented, convex colonies with entire margins on NSY agar plates. In comparison to most of the isolates usually growing on NSY plates inoculated with lake water, the tested strains grew slowly and formed only small colonies (≤1 mm after 10 days of incubation). A few isolates never grew to colony size (>0.1 mm), which strongly complicated the handling of single colonies.
FIG. 2.
Photomicrograph of DAPI-stained cells of isolate MWH-VicM1 grown in 3-g liter−1 NSY medium.
Cell sizes of the isolated strains.
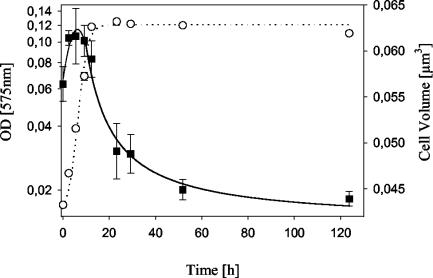
During the isolation procedure, the established enrichment cultures were screened microscopically for the presence of small C-shaped cells. During this process, no C-shaped cells larger than 0.1 μm3 were observed. All finally isolated strains possessed small cells when grown in low-substrate-concentration media. Most isolates even kept this small cell size when growing exponentially in a medium with high substrate concentrations (Table 3; Fig. 3).
TABLE 3.
Growth of isolates on different solidified complex media in comparison to growth on NSY agara
| Strain | Growth of isolates on:
|
Doubling time (h) | Cell size (μm3) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yeast extract | Peptone | Soytone peptone | Tryptic soy | Luria- Bertani broth | R2A | Nutrient broth | Standard method agar | Eugonic broth | Bacto Brain Heart Infusion | |||
| MWH-JaK3 | +++ | + | + | + | + | + | +++ | ++ | + | W | 4.4 | 0.057 |
| MWH-T2W17 | W | + | W | − | + | + | W | − | W | + | 3.7 | 0.097 |
| MWH-MoIso1 | + | + | W | − | + | + | + | − | + | + | 6.7 | 0.135 |
| MWH-MoK4 | ++ | +++ | W | W | + | + | − | − | − | + | 6.3 | 0.126 |
| MWH-CaK1 | + | + | + | + | ++ | ++ | + | ++ | − | +++ | 5.9 | 0.051 |
| MWH-MoIso2 | − | + | − | + | + | + | + | + | − | + | 5.7 | 0.051 |
| MWH-BeK4 | − | + | + | + | + | ++ | W | W | W | + | 8.6 | ND |
| MWH-MoK1 | W | ++ | W | ++ | +++ | +++ | + | − | − | − | 6.7 | 0.056 |
| MWH-VicM1 | + | + | + | + | ++ | +++ | + | +++ | W | + | 4.5 | 0.062 |
| MWH-T2W13 | + | + | + | + | + | + | + | W | − | W | 7.6 | 0.062 |
NSY agar was the medium used for isolation of the strains. Data on doubling times and cell sizes of the isolates growing exponentially in liquid 3-g liter−1 NSY medium at 20°C are also presented. Symbols: +++, colonies larger than those on NSY medium; ++, colonies of the same size as those on NSY medium; +, colonies smaller than those on NSY medium; W, weak growth; −, no growth; ND, not determined.
FIG. 3.
Growth of the isolate MWH-VicM1 in liquid 3-g liter−1 NSY medium at 20°C and cell volumes observed during exponential and stationary stages. Data represent means and standard deviations of results from three replicates. Open circles, optical density (OD); closed squares, cell volume.
Growth on various complex media.
The 10 tested strains grew on a wide range of complex media (Table 3). Each of the 10 tested complex media supported the growth of at least a few strains, and the majority of the media supported the growth of most tested strains. Best results were obtained with NSY, R2A, Luria-Bertani, and peptone agar (Table 3).
16S rRNA gene phylogeny.
The phylogenetic analysis of the 16S rRNA gene sequences obtained from the isolated strains revealed that all isolates are more closely related to uncultured bacteria than to any bacterial strain previously obtained from pure cultures (Table 2). Some of the isolates share identical sequences with uncultured bacteria, and sequences of most of the isolates differ in only a few nucleotide positions from sequences obtained by culture-independent techniques. The closest related previously described species is the obligate endosymbiont P. necessarius (96 to 99% similarity), and the closest related bacteria available as pure cultures are Ralstonia basilensis (93 to 94% similarity), R. campinensis (93 to 94% similarity), and R. paucula (93 to 94% similarity) (8, 29).
The minimum sequence similarity within the whole P. necessarius cluster is 95.7%. The whole P. necessarius cluster can be subdivided into four subclusters (labeled A, B, C, and D) which contain sequences separated from the other sequences by sequence similarities of ≤98% (Fig. 1). Sequence similarities between members of subclusters C and D are in the range of 96.4 to 97.8%. Subcluster A contains only one sequence long enough to be considered in the phylogenetic tree shown in Fig. 1. In analysis with data sets including shorter sequences, other sequences clustered within subcluster A, and results from bootstrap analysis supported this subcluster (data not shown). Furthermore, other shorter sequences clearly clustered within the subclusters B, C, and D. The subclusters were consistent in the neighbor-joining tree (Fig. 1), the maximum likelihood tree (data not shown), and in a minimum evolution tree published by Zwart and colleagues (32). On the other hand, all three trees show some differences in the branching orders. Analysis with larger data sets including shorter sequences indicated that more than the four described major subclusters may exist. All described subclusters contain several environmental sequences, but all isolates analyzed thus far fall into two of the four subclusters. Both of these subclusters contain identical sequences obtained from isolates originating from distantly located ecosystems (Fig. 1). On the other hand, sequences obtained from organisms inhabiting the same ecosystem are affiliated with different subclusters or occur in part at different positions within the same subclusters. For instance, eight strains were isolated from Lake Mondsee, which represent four different genotypes distributed over subclusters C and D.
DISCUSSION
Isolation.
Given the observed cosmopolitan distribution of P. necessarius cluster bacteria, the occurrence in a broad spectrum of freshwater habitats, the expected significant contribution to freshwater bacterioplankton, and the ability of isolates to form visible colonies on agar plates, it is highly surprising that isolation of strains belonging to this cluster has not been reported before. Clearly, the previous lack of isolates cannot be explained by a lack of suitable media supporting the growth of these bacteria. With certainty, there have been many research projects which included plating of freshwater samples onto diverse solid media. When isolates obtained in such projects have been screened microscopically, the P. necessarius cluster members should have attracted attention because of their unusually small cells and their unusual morphology. Thus, the previous lack of isolates clearly indicates that members of this cluster are not readily cultivable.
I assume that one crucial step in the isolation of these bacteria is the acclimatization procedure. The applied slow transition from the low environmental substrate concentrations to the high concentrations of standard microbiological media may enable the bacteria to acclimatize their physiology to the artificial conditions existent in these media. Another crucial step, at least in the case of culturing in liquid media, seems to be the exclusion of strains able to overgrow the slowly growing P. necessarius cluster members in the cultures.
The filtration-acclimatization method employed in this study has been recently developed and employed for the isolation of ultramicrobial Actinobacteria from freshwater (12). Interestingly, only one isolate (MWH-VicM1) affiliated with the P. necessarius cluster has been obtained during this previous isolation effort. On the other hand, only a few pure cultures of members of the actinobacterial Luna-1 and Luna-2 clusters were obtained in the study presented here. The two series of isolation experiments differ in only two seemingly minor points. In the first series, screening of cell culture plate wells was performed by spreading diluted samples onto agar plates, while in the second set of isolation efforts, undiluted samples were dropped onto the plates and incubated without spreading. Secondly, the first series of isolation experiments included samples taken at in situ temperatures of <10°C, while all samples used in the second series of isolation efforts were taken at in situ temperatures of >10°C. Both series of isolation efforts, however, included several samples taken from Lake Mondsee, which is inhabited by both groups of freshwater ultramicrobacteria. Currently it is not clear which factors caused the markedly different results of the two series of isolation efforts.
Phylogenetic structure of the P. necessarius cluster.
The phylogenetic analyses presented by others (32) as well as that in this study clearly show that the P. necessarius cluster is organized into at least four different subclusters. The sequence dissimilarities separating these subclusters may indicate that these subclusters represent different species. Isolates belonging to subclusters C and D, however, have similar morphologies and cell sizes. Likewise, the investigated strains belonging to the two subclusters do not show any consistent differences in growth on the 10 complex media (Table 3). Further detailed studies may reveal that strains belonging to different subclusters differ in physiological and ecological traits.
Ecology of the P. necessarius cluster bacteria.
All investigated isolates affiliated with the P. necessarius cluster can be characterized as aerobic heterotrophic bacteria with small cells. The only other well-investigated representative of the P. necessarius cluster is the obligate endosymbiont P. necessarius which was found in cytoplasmic vesicles of the hypotrichous ciliate Euplotes aediculatus but has never been cultured in pure culture (13, 28). This endosymbiont is closely related to the isolates (Fig. 1) but is distinctively different in morphology (slightly curved rods versus C-shaped cells) and cell size. Cell volumes of all the isolates are <0.2 μm3 (Table 3) while the endosymbiotic strain possesses larger cell volumes of 0.2 to 0.5 μm3 (calculated from data from reference 6). Furthermore, P. necessarius cells stained with DNA-specific dyes show usually 3 to 9, but in some cases up to 12, intensely stained and regularly spaced dots. Some of the isolates affiliated with subcluster C also show such nucleoid-like structures when stained with DAPI. In contrast to those of the endosymbiont P. necessarius, almost all cells of these strains show only a single nucleoid, but in cases of dividing stages two nucleoid-like structures have been observed. All of these differences between the endosymbiont and the isolates, as well as the small sample volumes (≤10 ml) used for isolation experiments, the filtration step (with 0.2-μm-pore-size filters) applied during the isolation process, and the observed occurrence of high numbers of free-living cells affiliated with the P. necessarius cluster by in situ hybridization (3a), indicate that it is highly unlikely that the isolates are endosymbionts of planktonic ciliates. Thus, it is much more likely that the P. necessarius cluster represents an important fraction of the free-living freshwater bacterioplankton. On the other hand, it seems that a free-living ancestor of the endosymbiont P. necessarius successfully invaded a ciliate cell and subsequently adapted to the life strategy of an obligate endosymbiont.
A highly important ecological trait which is shared by almost all isolated P. necessarius cluster members is their small (ultramicrosize) cell size (<0.1 μm3), which is even retained under favorable growth conditions (Fig. 2). In this trait, all of the isolates differ markedly from the obligate endosymbiont P. necessarius, as well as from the large-celled R. basilesis, R. campinensis, and R. paucula (8), which are the closest known relatives of the P. necessarius cluster. In the case of free-living, non-particle-associated pelagic bacteria, this trait seems to be one of the prerequisites necessary for establishing populations contributing large fractions to total bacterioplankton. Larger cells are often disadvantageous in the interaction with bacterivorous predators (9, 10, 27). In some cases, large bacterial morphotypes, e.g., bacterial filaments, may receive protection against grazing by some groups of predators (11, 26), but the only way to minimize vulnerability to grazing by all predators seems to be the possession of small cells. This appears to be one of the reasons for the dominance of small bacterial cells in marine and freshwater bacterioplankton.
Interestingly, the P. necessarius cluster isolates share their C-shaped morphology and ultramicrosize with isolates affiliated with the ubiquitous marine SAR11 cluster (21), as well as with actinobacterial Luna cluster members (12). This C-shaped morphology may be a specific adaptation to planktonic life or may provide advantages in the interaction with predators. In a previous experiment with one strain affiliated with the Luna-2 cluster which possesses such a C-shaped cell morphology, a complete protection against grazing by a nanoflagellate was observed (12).
The occurrence of bacteria of the P. necessarius cluster is not restricted to specific types of freshwater ecosystems or to certain climatic zones. They occur in acidic (e.g., Adirondack mountain lakes [14]) and alkaline (e.g., Lake Mondsee and Lake Tai Hu [Table 2]) lakes; they were detected in large lakes (e.g., Lake Baikal [7, 24]) and in small artificial ponds (e.g., Tiger Hill pond in the People's Republic of China [Table 2]); they were observed in arctic (e.g., Toolik Lake [2, 5]), temperate (e.g., Fuchskuhle [7]), and tropical (e.g., Lake Victoria [Table 2]) habitats. Furthermore, they inhabit freshwater ecosystems covering almost the whole range of trophic statuses. The range spans from oligotrophic (e.g., Toolik Lake) over oligomesotrophic (e.g., Lake Mondsee) and eutrophic (e.g., Lake Loosdrecht [32]) to hypertrophic (Lake Tai Hu, Meiling Bay [Table 2]) freshwater ecosystems. Furthermore, the cluster has been detected in several running water systems (e.g., Columbia River [4], Weser River [23], and Yangtze River [22] [Table 2]). On the other hand, there are a few lakes in which the P. necessarius cluster has not been detected (e.g., Crater Lake [31], Lake Gossenköllesee [7], and Lake IJssel [32]). Currently, it is not known whether this lack of detection was caused by low numbers of P. necessarius members combined with methodological limitations or whether this bacterial group is really absent in these lakes. Some of these lakes have very large surface-to-catchment-area ratios and sparse coverage of the catchment areas with vegetation. Thus, import of allochthonous organic substances can be assumed to play only a minor role in these ecosystems. Pernthaler and colleagues observed a higher relative abundance of P. necessarius cluster bacteria in that half of the artificially divided Lake Fuchskuhle which has higher concentrations of humic substances (3a). Both the observation from Lake Fuchskuhle and the lack of detection in lakes with low impact of the catchment areas on the availability of organic carbon in the water columns may indicate that the P. necessarius cluster bacteria utilize allochthonous humic substances. Besides this hypothesis, the currently available data do not indicate that the P. necessarius cluster has a preference for specific types of freshwater ecosystems. When detailed in situ data on abundance and potential seasonal fluctuations of the cluster members become available, it may turn out that the whole cluster or single subgroups prevail in some types of ecosystems more successfully than in others.
Acknowledgments
I am grateful to Peter Stadler and Matthias Pöckl for their skillful lab assistance, to Qinglong L. Wu for guiding and assisting me during sampling of habitats in the People's Republic of China, to Grace Ssanyu Asiyo, Kampala, Uganda, for water samples from Lake Victoria, and to Pan Hongxi (NIGLAS) and Liselotte Eisl for their chemical analysis of samples. Doris and Axel Pitt, Avrilly, France, enabled the sampling of Canal de Roanne à Digoin with the research vessel Bateaux Bleu. Michael Schauer is acknowledged for providing valuable comments on an earlier version of the manuscript. Sampling of sites in the People's Republic of China was made possible by the Chinese Academy of Sciences, the Nanjing Institute of Geography & Limnology (NIGLAS), and the Austrian Academy of Sciences.
This study was supported by the Austrian Science Fund (project P15655).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahr, M., J. E. Hobbie, and M. L. Sogin. 1996. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat. Microb. Ecol. 11:271-277. [Google Scholar]
- 3.Bruns, A., U. Nubel, H. Cypionka, and J. Overmann. 2003. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Burkert, U., J. Pernthaler, F. Warnecke, D. Babenzien, E. Zwirnmann, and R. Amann. Members of a readily enriched β-proteobacterial clade are common in the surface waters of a humic lake. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 4.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteve, I., and N. Gaju. 1999. Bacterial symbioses. Predation and mutually beneficial associations. Int. Microbiol. 2:81-86. [PubMed] [Google Scholar]
- 7.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goris, J., P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters, and P. Vandamme. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. E vol. Microbiol. 51:1773-1782. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, M. W., and M. G. Höfle. 1999. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl. Environ. Microbiol. 65:4863-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heckmann, K., and H. J. Schmidt. 1987. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int. J. Syst. Bacteriol. 37:456-457. [Google Scholar]
- 14.Hirons, W. D., E. A. Methé, S. A. Nierzwickibauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y.
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Methé, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition—analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 19.Pearce, D. A., C. J. van der Gast, B. Lawley, and J. C. Ellis-Evans. 2003. Bacterioplankton community diversity in a maritime antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol. Ecol. 45:59-70. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen, K., J. Arlinger, S. Ekendahl., and L. Hallbeck. 1996. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Aespoe hard rock laboratory, Sweden. FEMS Microbiol. Ecol. 19:249-262. [Google Scholar]
- 21.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 22.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selje, N., and M. Simon. 2003. Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat. Microb. Ecol. 30:221-237. [Google Scholar]
- 24.Semenova, E. A., and K. D. Kuznedelov. 1998. A study of the biodiversity of Baikal picoplankton by comparative analysis of 16S rRNA gene 5′-terminal regions. Mol. Biol. 32:754-760. [PubMed] [Google Scholar]
- 25.Simek, K., J. Pernthaler, M. G. Weinbauer, K. Hornak, J. R. Dolan, J. Nedoma, M. Masin, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simek, K., J. Vrba, J. Pernthaler, T. Posch, P. Hartman, J. Nedoma, and R. Psenner. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simek, K., and T. H. Chrzanowski. 1992. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl. Environ. Microbiol. 58:3715-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Springer, N., R. Amann, W. Ludwig, K. H. Schleifer, and H. Schmidt. 1996. Polynucleobacter necessarius, an obligate bacterial endosymbiont of the hypotrichous ciliate Euplotes aediculatus, is a member of the beta-subclass of Proteobacteria. FEMS Microbiol. Lett. 135:333-336. [DOI] [PubMed] [Google Scholar]
- 29.Steinle, P., G. Stucki, R. Stettler, and K. W. Hanselmann. 1998. Aerobic mineralization of 2,6-dichlorophenol by Ralstonia sp. strain RK1. Appl. Environ. Microbiol. 64:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. 1998. ARB: a software environment for sequence data. Department of Microbiology, Technische Universität München, Munich, Germany.
- 31.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 32.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]