Abstract
Nonmitochondrial ADP/ATP translocase is an energy parasite enzyme. Its encoding gene, tlc, is found only in Rickettsiales, Chlamydiales, and plant and alga plastids. We demonstrate the presence of tlc in Parachlamydia acanthamoebae. This gene shares more similarity with the tlc1 gene of Chlamydiaceae and the tlc of plant and alga plastids than with the tlc2 gene of Chlamydiaceae. Phylogenetic analysis, including all other tlc homologs found in GenBank, showed that tlc was duplicated in a Chlamydiales ancestor before the appearance of multicellular eukaryotes. A time scale, calibrated with seven independent time points obtained from fossil estimates and from the 16S rRNA molecular clock, was congruent with the molecular clock provided by tlc. Plant and alga plastids acquired tlc approximately when Parachlamydiaceae and Chlamydiaceae diverged, at the eucaryotic radiation time, ca. 1 billion years ago.
Nonmitochondrial ADP/ATP translocase is a very unique enzyme that exchanges bacterial ADP for ATP from the host cell and allows energy parasitism (17, 29, 30). Of its encoding gene, five and two copies were found in the Rickettsiales and Chlamydiaceae genomes, respectively, two clades of obligate intracellular bacteria (4, 23, 24, 27). The presence of the tlc gene in these distantly related bacterial clades was explained by either horizontal transfer, convergent evolution, or as a result of the common origin of Chlamydiales and Rickettsiales (7, 8, 21, 27, 30, 31). Only plant plastids are also known to possess this gene and encode for a protein that exchanges plastid ADP for ATP present in the eukaryotic cytoplasm (19). Since several other proteins of Chlamydiales were phylogenetically related to plant proteins (27), it has been proposed that the ADP/ATP translocase gene (tlc) was acquired from a plant genome by Chlamydiaceae and subsequently transferred to Rickettsiales (30). However, it makes more sense that a protein allowing energy parasitism originated in a clade of obligate intracellular bacteria parasitizing eucaryotes than in the eucaryotic host itself, since the new protein provides ATP to the intracellular bacteria and represents a selective advantage that allows its encoding gene to be fixed in the genome. Moreover, recent studies suggest that the ancestral Chlamydiales may have participated in the ancient chimeric events that led to the formation of the plant lineages and might be related to the cyanobacterium-chloroplast lineage (6, 10).
Parachlamydia is a new genus within Chlamydiales (2) that presents Chlamydia-like developmental stages (15) and shares 80 to 90% similarity of 16S rRNA genes with Chlamydiaceae (12, 14). In contrast to the Chlamydiaceae, which naturally infect multicellular organisms such as mammals and birds, Parachlamydia acanthamoebae naturally infects free-living amoebae and has probably never been a parasite of multicellular organisms. Moreover, since Parachlamydiaceae-Chlamydiaceae divergence was contemporary with the eukaryotic radiation about 1 billion years ago (26), the presence of the tlc gene within the genome of P. acanthamoebae would preclude the hypothesis of a transfer of the tlc gene from plants and might, on the contrary, suggest its transfer from Chlamydiales to plants. Therefore, in the present study, we investigated whether the tlc gene was present within the genome of P. acanthamoebae and evaluated the genetic and phylogenetic relationships of the nonmitochondrial ADP/ATP translocase coding sequences from an evolutionary perspective.
MATERIALS AND METHODS
Parachlamydia culture, purification, and DNA extraction.
P. acanthamoebae strain Hall coccus and Acanthamoeba polyphaga Linc-AP1 were kindly provided by T. J. Rowbotham (Public Health Laboratory, Leeds, United Kingdom). Parachlamydia sp. was grown for 6 days at 32°C within Acanthamoeba polyphaga in peptone-yeast extract-glucose broth (15). Harvested bacteria were purified by centrifugation and ultracentrifugation onto a 10% sucrose barrier. To improve purification, the pellet was suspended in phosphate-buffered saline, loaded onto a discontinuous Gastrografine (Schering, Lys-Lez-Lannoy, France) gradient, and ultracentrifuged at 140,000 × g. Parachlamydiae, which clustered in a large lower band, were collected, centrifuged at 5,800 × g, and resuspended in phosphate-buffered saline twice. DNA was extracted by using the QIAamp DNA Mini-Kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions.
PCR amplification and sequencing of P. acanthamoebae tlc gene.
The strategy for determining the sequence of the gene encoding the ADP/ATP translocase (i.e., tlc) of P. acanthamoebae consisted of (i) amplifying a segment of the targeted gene with degenerate primers and (ii) completing the full-length sequence by the genome-walking approach. adpF1 5′-GAAGC(TA)AAACGTTT(CT)TACGCTCT, adpF11 5′-TGTTCTGGGGGTTAGCCAA, and adpR4 5′-C(AG)TCAATAGC(AG)GCTTTICCTTT-, all designed by alignment of the tlc genes of Chlamydia muridarum, Chlamydia trachomatis, Chlamydophila pneumoniae J138, and Rickettsia conorii, succeeded in amplifying 910-bp (adpF11-R4) and 980-bp (adpF1-R4) nucleotides. These PCR products were purified by using the QIAquick PCR purification kit (Qiagen, Courtaboeuf, France) and sequenced by using the dRhodamine terminator cycle sequencing ready reaction with AmpliTaq DNA (Perkin-Elmer Biosystems, Warrington, United Kingdom). Sequences were determined on 3100 ABI Prism automated sequencer (Applied Biosystems, Courtaboeuf, France). Sequences derived from each primer were aligned, compared, and combined in a single sequence by using Autoassembler software version 2.1 (Applied Biosystems). The unknown 5′ and 3′ ends of this partial sequence were amplified by using the Universal Genome Walker kit (Clontech Laboratories, Palo Alto, Calif.). The validity of the sequence obtained was assessed by comparison with two additional sequences obtained by PCR amplification with primers designed from the sequence immediately flanking the open reading frame: Adp65F (5′-GATCCACGAAAGCACTCTTATT) Adp62R (5′-GGCAATCTATCACGTAATTGAAAAT).
Presence of the tlc gene in additional clades.
The nucleotide sequence of Parachlamydia tlc (GenBank accession number AF490592) and the corresponding amino acid sequence were compared to sequences available in the GenBank database by using the BLASTN and BLASTP 2.2.6 programs available on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov) (1). An iterated profile search was also performed by using position-specific iterated BLAST (PSI-BLAST) (1).
Genetic and phylogenetic analysis.
After alignment with CLUSTAL W (28), genetic distances of nucleotides and amino acids sequences were calculated by using MEGA 2.1 software (18). With the same software, we inferred neighbor-joining (p-distance), minimum evolution (p-distance), and parsimony trees (standard parsimony) by using the amino acid and nucleotide sequences.
Time scale.
The molecular clock of the tlc gene was calibrated with seven points obtained from fossil estimates and from the estimated time of divergence of bacterial species on the basis of the 16S rRNA sequence divergence, assuming a rate of evolution of 1 to 2% per 50 million years (20). The 16S rRNA sequences were edited by removal of the longer 5′ and 3′ ends so that their lengths matched that of the shortest sequence. The percentage of 16S rRNA sequence divergence was calculated by using CLUSTAL W program (28), supported by the PBIL website (http://npsa-pbil.ibcp.fr/cgi-bin/align_clustalw.pl). The time of divergence of green plants and red algae, estimated by Sogin and Silberman (26), the time of divergence of monocotyledons (Oryza sativa) and dicotyledons (Citrus spp., Solanum tuberosum, and Arabidopsis thaliana) estimated by Gale et al. (13), and the estimated times of divergence of bacterial species (assuming a rate of evolution of 1.5% of 16S rRNA sequence per 50 million years) were plotted as a function of the proportion (p) of amino acid sites at which the sequence is different from that of the ancestral ADP/ATP translocase sequence, i.e., the p-distance between each node and the node that separated the more divergent sequences. The equation of the regression line, its standard error, and its r2 coefficient were calculated with Stata 7.0 (Stata Corp., College Station, Tex.) and Microsoft Excel 97 (Microsoft Corp., Redmond, Wash.). To estimate the time of divergence of red algae and green plants and of monocotyledons and dicotyledons, regression was performed similarly, using only the time scale inferred from 16S rRNA genes and without using the times estimated by Sogin and Silberman (26) and by Gale et al. (13). Similar analyses were performed by using a minimum evolution tree inferred from amino acid sequences. To further test the reliability of the time scale, we performed an omit test, which assessed how the time estimates were modified by the omission of each of the calibration point.
RESULTS
Presence of the tlc gene in P. acanthamoebae.
We succeeded in amplifying a segment of the tlc gene of P. acanthamoebae with degenerate primers and completed the full-length sequence by the genome-walking approach (25). The tlc gene sequence of P. acanthamoebae strain Hall coccus has been deposited in the GenBank database under accession number AF490592.
Presence of tlc gene in Galdieria sulfuraria (a red algae), Citrus hybrid cultivar (citrus), Oryza sativa (rice), Holospora obtusa and Caedibacter caryophilus (endosymbionts of Paramecium), Encephalitozoon cuniculi (microsporidia), and Medicago sativa (alfalfa).
By using the basic local alignment search tool (BLAST) with the amino acid sequence of the Parachlamydia ADP/ATP translocase as input, we found tlc gene sequences in additional clades, including G. sulfuraria (a red algae; GenBank accession number AJ251356 [236 of 468 identities]), Citrus hybrid cultivar (GenBank accession number AY098893 [231 of 480 identities]), O. sativa (rice; GenBank accession number AP003234 [236 of 496 identities]), H. obtusa (GenBank accession number AY120885 [183 of 465 identities]), Caedibacter caryophilus (GenBank accession number AJ441310 [45 of 53 identities]), and M. sativa (GenBank accession number AF416339 [52 of 81 identities]). However, when gap and extension penalties of 11 and 1, respectively, were used, BLAST analysis did not detect the tlc gene sequences in Wolbachia sp. (an endosymbiont of Drosophila melanogaster), cyanobacteria, protozoa, or animals. A BLAST search also identified four E. cuniculi proteins of unknown functions (GenBank accession numbers NP_586157 to NP_597260 [93 of 443 to 111 of 473 identities]). With the exception of the short tlc sequence of M. sativa (317 bp), we used all of these tlc sequences in the genetic and phylogenetic analyses.
Proteins of unknown functions related to the ADP/ATP translocase in Chlamydiales, cyanobacteria, and plant pathogen (Xylella fastidiosa).
By PSI-BLAST, we identified proteins of unknown function with significant alignments within the proteomes of two cyanobacteria (Nostoc sp. and Trichodesmium erythraeum), within that of Chlamydiales, and within different γ-proteobacteria (including X. fastidiosa, a plant pathogen). We performed phylogenetic analysis with the amino acid sequences of all of these proteins of unknown function and with the four microsporidium proteins (which exhibit sequence similarity with ADP/ATP translocase but whose functions are also unknown) and with the Chlamydiales, Rickettsiales, and plant plastid ADP/ATP translocase. The tree topology was similar to that shown in Fig. 1, except that the E. cuniculi proteins and the other proteins of unknown function rooted deeply, being phylogenetically far from the ADP/ATP translocase (data not shown).
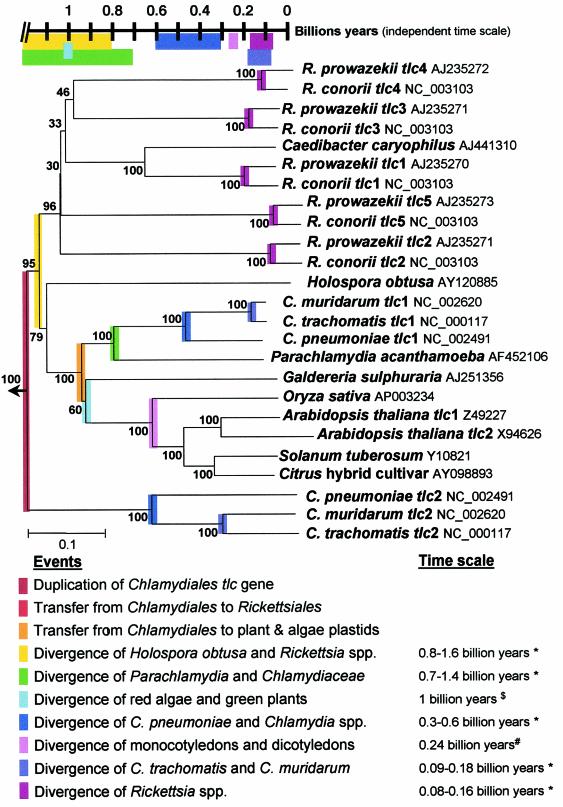
FIG. 1.
p-Distance neighbor-joining tree inferred from amino acid sequences of the ADP/ATP translocase of P. acanthamoebae, Chlamydiaceae, Rickettsiales, and plant and alga plastids. Bootstrap values resulting of 100 replications are present at branch points. The tree was rooted with four proteins present in E. cuniculi, whose functions are unknown and were identified by BLAST of nonmitochondrial ADP/ATP translocase. The time scale was derived from estimates obtained from the literature (13, 26) and on the basis of 16S rRNA gene sequence divergence (20). Note the congruence between the node of a given divergence (see neighbor-joining tree) and its estimated time (see time scale). ✽, Estimated from 16S rRNA divergence (20); $, estimated by Sogin et al. (26); #, estimated by Gale et al. (13).
Genetic analysis of amino acid and nucleotide sequences.
P. acanthamoebae tlc shared greater amino acid sequence similarity with tlc1 of Chlamydiaceae (62 to 63%) and with tlc of plant plastids (53 to 54%) than with Rickettsiales (34 to 43%) and tlc2 of Chlamydiaceae (36 to 38%) (Table 1). Similarly, P. acanthamoebae tlc shared greater nucleotide sequence similarity with tlc1 of Chlamydiaceae and with that of plant plastids than with Rickettsiales and tlc2 of Chlamydiaceae (data not shown). Conversely, plant plastid tlc shared greater nucleotide and amino acid sequences similarity with tlc of P. acanthamoebae and tlc1 of Chlamydiaceae than with those of Rickettsiales and tlc2 of Chlamydiaceae. Alignment of the amino acid sequences also showed that the ADP/ATP translocase of green plants and red alga plastids present a N-terminal peptide transit of about 90 and 150 amino acids, respectively, whereas that of bacteria do not present this peptide. Such transit peptides are present in most plastids proteins and are necessary to target the protein encoded by nuclear genes to plastids (5).
TABLE 1.
Comparison of similarities of complete amino acid sequences (excluding alignment gaps) of ADP/ATP-translocase-encoding genes (tlc) of Chlamydiales, Rickettsiales, and plant and alga plastids
| Sequence no. | Species | % Similarity of sequencea:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | ||
| 1 | Parachlamydia sp. | — | ||||||||||||||||||
| 2 | C. muridarum 1b | 63 | — | |||||||||||||||||
| 3 | Chlamydophila pneumoniae 1 | 62 | 80 | — | ||||||||||||||||
| 4 | C. trachomatis 1 | 63 | 96 | 80 | — | |||||||||||||||
| 5 | C. muridarum 2 | 36 | 38 | 39 | 37 | — | ||||||||||||||
| 6 | Chlamydophila pneumoniae 2 | 38 | 40 | 41 | 40 | 66 | — | |||||||||||||
| 7 | C. trachomatis 2 | 37 | 39 | 39 | 38 | 84 | 68 | — | ||||||||||||
| 8 | R. conorii 1c | 36 | 34 | 36 | 34 | 28 | 29 | 27 | — | |||||||||||
| 9 | R. conorii 2 | 38 | 37 | 38 | 37 | 31 | 31 | 31 | 41 | — | ||||||||||
| 10 | R. conorii 3 | 40 | 40 | 40 | 40 | 31 | 30 | 31 | 39 | 45 | — | |||||||||
| 11 | R. conorii 4 | 34 | 33 | 34 | 33 | 30 | 29 | 29 | 36 | 40 | 40 | — | ||||||||
| 12 | R. conorii 5 | 43 | 43 | 43 | 43 | 31 | 31 | 30 | 39 | 42 | 47 | 41 | — | |||||||
| 13 | H. obtusa | 40 | 40 | 40 | 40 | 31 | 28 | 30 | 34 | 35 | 36 | 33 | 36 | — | ||||||
| 14 | A. thaliana 1 | 53 | 52 | 52 | 52 | 34 | 33 | 33 | 33 | 36 | 36 | 32 | 40 | 38 | — | |||||
| 15 | A. thaliana 2 | 46 | 46 | 45 | 46 | 29 | 30 | 29 | 28 | 30 | 30 | 27 | 34 | 32 | 84 | — | ||||
| 16 | S. tuberosum | 54 | 52 | 53 | 52 | 34 | 33 | 34 | 34 | 37 | 38 | 33 | 39 | 39 | 86 | 76 | — | |||
| 17 | Citrus hybrid cultivar | 53 | 51 | 52 | 51 | 35 | 34 | 34 | 33 | 36 | 37 | 33 | 39 | 38 | 86 | 77 | 91 | — | ||
| 18 | O. sativa | 53 | 52 | 53 | 51 | 35 | 34 | 35 | 33 | 37 | 38 | 34 | 39 | 38 | 80 | 72 | 82 | 83 | — | |
| 19 | G. sulphuraria | 53 | 52 | 52 | 52 | 33 | 33 | 33 | 36 | 36 | 39 | 33 | 41 | 38 | 59 | 51 | 59 | 59 | 61 | — |
The number at the top of each column corresponds to the sequence as defined in column 1. —, 100% similarity.
Isoenzyme 1.
Similar rates obtained for R. prowazekii.
Phylogenetic analysis.
Phylogenetic analysis generated robust trees with significant bootstrap values. Figure 1 shows a neighbor-joining tree inferred from amino acid sequences and rooted with the four E. cuniculi proteins identified by BLAST. Its topology was similar to that of the neighbor-joining tree inferred from the nucleotides sequences and from both minimum evolution and parsimony trees, with the only exception being that in both parsimony trees the tlc of Holospora sp. clustered with that of Rickettsiales. In neighbor-joining, minimum-evolution, and parsimony trees inferred from amino acid sequences, the node separating the tlc1 of Chlamydiales and the tlc of plant and alga plastids from the tlc of the Rickettsia spp. was supported by bootstrap values of 95, 92, and 44%, respectively, whereas the node separating the tlc1 of Chlamydiales from the tlc of plant and alga plastids was strongly supported by bootstrap values of 100, 100, and 65%, respectively. In all trees inferred from amino acid sequences, the node separating tlc1 of Chlamydiales from tlc2 of Chlamydiales was strongly supported by bootstrap values of 100%.
Time scale.
The divergences of the 16S rRNA sequence between Chlamydiales and Rickettsiales, P. acanthamoebae and Chlamydiaceae, H. obtusa and Rickettsia spp., Chlamydophila pneumoniae and Chlamydia spp., and R. conorii and Rickettsia prowazekii were of 55, 14, 16, 6, and 1.59%, respectively. Thus, if we assume a rate of evolution of 1 to 2% per 50 million years (20), the respective divergences of these organisms may have occurred more than 2.75 billions years ago, 0.7 to 1.4 billion years ago, 0.8 to 1.6 billion years ago, 300 to 600 million years ago, and 80 to 160 million years ago (see Fig. 1).
Congruence of p-distance and time scale.
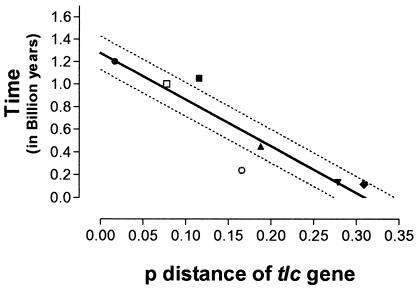
The dependence of the p-distance of the neighbor-joining tree inferred from the amino acid sequences of the tlc genes on the time estimated for bacterial species on the basis of 16S rRNA sequence divergence (20) and for plastids from the time of divergence of monocotyledons and dicotyledons (13) and of G. sulfuraria (a red algae) and Viridiplantae (green plants) was nearly linear, as shown by the low extent of deviation from the regression line (r2 = 0.84) (Fig. 2). The equation of the regression line is: t = −4.11p + 1.28, where t is the time from now in billion years and “p” is the proportion of amino acid sites at which the sequence is different from that of the ancestral tlc sequence at the time of its duplication in Chlamydiales, i.e., the p-distance from a given node to the middle of the neighbor-joining tree (see Fig. 1 and 2). If p = 0, the estimated time of duplication of the tlc gene in Chlamydiales will be ca. 1.28 billion years. The reliability of this estimated time was supported by the omit test. By omitting any of each of the calibration point, the time of duplication of the tlc gene ranged from 1.20 to 1.33 billion years, with a mean of 1.28 ± 0.04 billion years. The estimated times of transfer of the tlc gene from Chlamydiales to Rickettsiales and to plant and alga plastids would be ca. 1.16 and 0.98 billion years, respectively. Similarly, the estimated times of duplication of the tlc gene in Rickettsia spp. and in A. thaliana would be about 1,017 to 1,095 and 223 million years, respectively. The extent of deviation of the regression line was lower (r2 = 0.94) when only the time of divergence of bacterial species estimated on the basis of 16S rRNA sequence divergence was used as a time scale (20). The equation of the regression line was then: t = −4.13p + 1.34. The p-distance was 0.089 between the node separating the G. sulfuraria from Viridiplantae and the midpoint of the neighbor-joining tree (Fig. 1); thus, their time of divergence is about 0.97 billion years. Similarly, the time of divergence for monocotyledons and dicotyledons was estimated to be about 507 million years. Using the minimum evolution tree inferred from the amino acid sequence, the date of duplication of the tlc gene, the date of transfer of the gene from Chlamydiales to plants, the date of divergence of algae from green plants, and the date of divergence of monocotyledons and dicotyledons are close to those estimated with the neighbor-joining tree (Table 2). By omitting any of the calibration points and using the minimum-evolution tree inferred from the amino acid sequence, the time of duplication of the tlc gene ranged from 1.19 to 1.33 billion years, with a mean of 1.27 ± 0.05 billion years, further supporting the reliability of the estimated times.
FIG. 2.
Congruence (r2 = 0.88) of the time scale and the p-distance of the tlc gene. The time scale was derived from independent estimates obtained from the literature (13, 26) and on the basis of 16S rRNA gene sequence divergence (20). The p-distance between each node and the midpoint of the neighbor-joining tree inferred from the amino acid sequences of the ADP/ATP translocase represent the proportion (p) of amino acid sites at which the sequence is different from that of the ancestral sequence, at the time of duplication of the Chlamydiales ADP/ATP-translocase-encoding gene (tlc). Symbols: •, divergence of H. obtusa and Rickettsia spp.; ▪, divergence of Parachlamydia spp. and Chlamydiaceae; □, divergence of red algae and green plants; ▴, divergence of C. pneumoniae and Chlamydia spp.; ○, divergence of monocotyledons and dicotyledons; ▾, divergence of C. trachomatis and C. muridarum; ⧫, Rickettsia spp. Lines: solid line, regression line (t = −4.11p + 1.28); dotted lines, confidence intervals (t = −4.11p + 1.13 and t = −4.11p + 1.43).
TABLE 2.
Date of duplication of Chlamydiales tlc gene and transfers of the tlc gene from Chlamydiales to Rickettsiales and from Chlamydiales to plant and alga plastidsa
| Event | Date (in billion yr)b estimated by:
|
|
|---|---|---|
| Neighbor joining | Minimum evolution | |
| Duplication of Chlamydiales tlc gene | 1.28 | 1.27 |
| Transfer from Chlamydiales to Rickettsiales | 1.16 | 1.15 |
| Transfer from Chlamydiales to plant and alga plastids | 0.98 | 0.97 |
| Divergence of red algae and green plants | 0.97 | 0.95 |
| Divergence of monocotyledons and dicotyledons | 0.51 | 0.50 |
Estimated with ADP-ATP translocase amino acid-sequences by using neighbor-joining and minimum-evolution trees with a time scale derived from the literature (13, 26) and on the basis of 16S rRNA sequence divergence.
The dates of divergence of red algae from green plants and of monocotyledons from dicotyledons were estimated by using a time scale calibrated only on the basis of 16S rRNA sequence divergence.
DISCUSSION
The sequence of the tlc gene in P. acanthamoeba was detected in the present study and demonstrated to have been present about 1 billion years ago when Parachlamydiaceae and Chlamydiaceae are estimated to have diverged. By BLAST analysis, we also detected the sequences of the tlc genes in G. sulfuraria (red algae), Citrus hybrid cultivar (dicotyledons), O. sativa (monocotyledons), H. obtusa (Rickettsiales exclusively growing within the nucleus of some Paramecium spp.), and Caedibacter caryophilus (another endosymbiont of Paramecium spp. belonging to Rickettsiales). We also detected four proteins of unknown function in E. cuniculi (microsporidia), which shared some degree of similarity with the tlc gene. With these sequences, those of the five tlc paralogs of R. conorii (23) and those used by Wolf et al. (30), we performed genetic analysis and generated robust trees with significant bootstrap values. The topology of these trees is similar to that recently published by Amiri et al. (3). The deep branching of the four E. cuniculi proteins suggests that the ADP/ATP translocase of Chlamydiales, Rickettsiales, and plant plastids share a common ancestor with these proteins. The presence of additional proteins of unknown function in the genome of Chlamydiales, cyanobacteria, and plant pathogens (such as X. fastidiosa), which exhibited similar sequences profiles with ADP/ATP translocase and, like E. cuniculi proteins, branched deep in the phylogenetic tree, suggests that the ADP/ATP translocase shares a common ancestor with another transmembrane transport protein that is present in both cyanobacteria and Chlamydiales. The function of these proteins, like the one found by BLAST in E. cuniculi genome, remains to be determined. We focused here on analysis of the evolutionary relationship of the ADP/ATP translocase, whose function has been confirmed only for Rickettsiales, Chlamydiales, and plant plastids. The closer genetic similarity of the tlc of red algae and plant plastids with that of Chlamydiales compared to those of Rickettsiales suggested that the tlc gene has been exchanged between eucarya and the ancestral Chlamydiales. The tree shown in Fig. 1 supports our hypothesis that tlc originated in the Chlamydiales ancestor and not in plants. Indeed, if the two tlc genes found in Chlamydiaceae resulted from gene duplication, this event predates the divergence of the Parachlamydiaceae and Chlamydiaceae and the transfer of tlc1 to eucaryotes (Fig. 1). This tree inferred from amino acid sequences was representative of the topology obtained with parsimony and with other datasets.
Our study provides a scale of time derived from the divergence of 16S rRNA gene sequences (20), with or without additional time estimates obtained from the literature (13, 26). The congruence of the p-distance derived from amino acid sequences of the ADP/ATP translocase with these time scales (r2 coefficients of 0.84 and 0.94, respectively) confirms the value of the tree presented in Fig. 1. This congruence confirms the value of the calibration of the molecular clock, performed based on both a fossil estimate and another molecular clock. The congruence of the p-distance derived only from tlc nucleotide sequences with the scale of time derived from the divergence of 16S rRNA sequences is further confirmed by the estimated time of divergence of red algae from green plants of 0.97 billion years that is similar to the 1 billion years estimate of Sogin and Silberman (26). The reliability of these results is further confirmed by the fact that dates estimated by using the neighbor-joining tree inferred from the ADP/ATP translocase amino acid sequences were really close to those estimated by using the minimum-evolution tree (Table 2) and by the fact that omission of any of the calibration point of the time scale only slightly modified the estimated time of duplication of the tlc genes in the Chlamydiales (standard deviations of 0.04 and 0.05 billions years, respectively).
The time scale analysis suggests that the ancestral sequence duplicated in Chlamydiales 1.27 to 1.28 billion years ago (see Table 2). Since the estimated time of transfer of the tlc gene to Rickettsiales was about 1.15 to 1.16 billion years, the presence of tlc genes is probably due to horizontal transfer and not to the speciation of Chlamydiales from Rickettsiales, which is estimated to have occurred more than 2.75 billion years ago. Although horizontal transfer of genes between Rickettsiales and Chlamydiaceae is unlikely to occur today, since these clades do not share common host cells, such a transfer may have occurred more than one billion years ago, at a time when these bacteria may have been facultatively intracellular and may have shared a common ancestral cell host, such as a free-living amoeba. The latter hypothesis is supported by the fact that a branch of evolution of both clades still parasitizes Acanthamoeba spp. (2, 11). The absence of the tlc gene in Wolbachia sp. (a clade that diverged from Rickettsia spp. 100 to 200 million years ago) is probably due to subsequent gene loss (22). The tlc1 gene was apparently transferred to eucarya 0.97 to 0.98 billion years ago, i.e., around the time when Chlamydiaceae and Parachlamydiaceae diverged (0.7 to 1.4 billion years ago). The presence of tlc1 in red algae and in higher plant plastids and its absence in the sequenced genomes of protozoan and animals show that transfer occurred after the divergence of red algae and plants from other eucaryotes but before that of Rhodophyta (red algae) from Viridiplantae (green plants). The latter divergence is contemporary with the eukaryotic radiation, which has also been estimated to have occurred about 1 billion years ago (9, 26). Since these time evaluations are congruent with our tree representations, we believe that they reflect the true time of transfer of tlc1 to eukaryotes.
Everett et al. (10) suggested that the common Chlamydiales ancestor might be related to the cyanobacterium-chloroplast lineage; the absence of the tlc gene in the genome of Synechocystis sp., Nostoc sp., and T. erythraeum (three cyanobacteria) (16), being then explained by a subsequent gene loss (22) or by the appearance of that gene in the Chlamydiales genome after their speciation from cyanobacteria. Brinkman et al. (6) showed that the vast majority of plant-like genes in Chlamydiales correspond to plant genes that are derived from and function in the chloroplast, suggesting that the ancestral Chlamydiales might have been involved in the endosymbiotic origin of chloroplasts or have at least played a role in the chimeric events that led to the formation of plant lineages.
The existence of the tlc gene and its duplication, long before the radiation of eukaryotes, including plants and animals, demonstrates the long history of parasitism of Chlamydiales with unicellular eukaryotes and is the oldest evidence of bacterial parasitism. One branch of Chlamydiales still parasitizes protozoa, whereas the other is associated with multicellular animals.
Acknowledgments
We thank the Swiss National Science Foundation for funding the postodoctoral fellowship of Gilbert Greub, and we thank J. S. Dumler (Baltimore, Md.) and P. Pontarotti (Marseille, France) for reviewing the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., N. Springer, W. Schönhuber, W. Ludwig, E. N. Schmid, K. D. Müller, and R. Michel. 1997. Obligate intracellular bacterial parasites of Acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amiri, H., O. Karlberg, and S. E. Andersson. 2003. Deep origin of plastid/parasite ATP/ADP translocases. J. Mol. Evol. 56:137-150. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 5.Archibald, J. M., and P. J. Keeling. 2002. Recycled plastids: a “green movement” in eukaryotic evolution. Trends Genet. 18:577-584. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman, F. S., J. L. Blanchard, A. Cherkasov, Y. Av-Gay, R. C. Brunham, R. C. Fernandez, B. B. Finlay, S. P. Otto, B. F. Ouellette, P. J. Keeling, A. M. Rose, R. E. Hancock, and S. J. Jones. 2002. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 12:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavalier-Smith, T. 2000. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5:174-182. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle, R. F. 2002. Biodiversity: microbial genomes multiply. Nature 416:697-700. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle, R. F., D. F. Feng, S. Tsang, G. Cho, and E. Little. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271:470-477. [DOI] [PubMed] [Google Scholar]
- 10.Everett, K. D. E., S. Kahane, R. M. Bush, and M. G. Friedman. 1999. An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria. J. Bacteriol. 181:4734-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K. H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K. H. Schleifer, and R. K. Gautom. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale, M. D., and K. M. Devos. 1998. Plant comparative genetics after 10 years. Science 282:656-659. [DOI] [PubMed] [Google Scholar]
- 14.Greub, G., and D. Raoult. 2002. Parachlamydiaceae, potential emerging pathogens. Emerg. Infect. Dis. 8:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greub, G., and D. Raoult. 2002. Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl. Environ. Microbiol. 68:3076-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko, T., and S. Tabata. 1997. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 38:1171-1176. [DOI] [PubMed] [Google Scholar]
- 17.Krause, D. C., H. H. Winkler, and D. O. Wood. 1985. Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Mohlmann, T., J. Tjaden, C. Schwoppe, H. H. Winkler, K. Kampfenkel, and H. E. Neuhaus. 1998. Occurence of two plastidic ATP/ADP transporters in Arabidopsis thaliana L: molecular characterisation and comparative structural analysis of similar ATP/ADP translocators from plastids and Rickettsia prowazekii. Eur. J. Biochem. 252:353-359. [DOI] [PubMed] [Google Scholar]
- 20.Ochman, H., S. Elwyn, and N. A. Moran. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 96:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 22.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1098. [DOI] [PubMed] [Google Scholar]
- 23.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 24.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renesto, P., D. Gautheret, M. Drancourt, and D. Raoult. 2001. Determination of the rpoB gene sequences of Bartonella henselae and Bartonella quintana for phylogenic analysis. Res. Microbiol. 151:831-836. [DOI] [PubMed] [Google Scholar]
- 26.Sogin, M. L., and J. D. Silberman. 1998. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int. J. Parasitol. 28:11-20. [DOI] [PubMed] [Google Scholar]
- 27.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler, H. H. 1976. Rickettsial permeability: an ADP-ATP transport system. J. Biol. Chem. 251:389-396. [PubMed] [Google Scholar]
- 30.Wolf, Y. I., L. Aravind, and E. V. Koonin. 1999. Rickettsiae and chlamydiae evidence of horizontal gene tranfer and gene exchange. Trends Genet. 15:173-175. [DOI] [PubMed] [Google Scholar]
- 31.Zomorodipour, A., and S. G. E. Andersson. 1999. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett. 452:11-15. [DOI] [PubMed] [Google Scholar]