Abstract
The plasma membrane proton pump ATPase (H+-ATPase) plays a major role in the activation of ion and nutrient transport and has been suggested to be involved in several physiological processes, such as cell expansion and salt tolerance. Its activity is regulated by a C-terminal autoinhibitory domain that can be displaced by phosphorylation and the binding of regulatory 14-3-3 proteins, resulting in an activated enzyme. To better understand the physiological consequence of this activation, we have analyzed transgenic tobacco (Nicotiana tabacum) plants expressing either wild-type plasma membrane H+-ATPase4 (wtPMA4) or a PMA4 mutant lacking the autoinhibitory domain (ΔPMA4), generating a constitutively activated enzyme. Plants showing 4-fold higher expression of wtPMA4 than untransformed plants did not display any unusual phenotype and their leaf and root external acidification rates were not modified, while their in vitro H+-ATPase activity was markedly increased. This indicates that, in vivo, H+-ATPase overexpression is compensated by down-regulation of H+-ATPase activity. In contrast, plants that expressed ΔPMA4 were characterized by a lower apoplastic and external root pH, abnormal leaf inclination, and twisted stems, suggesting alterations in cell expansion. This was confirmed by in vitro leaf extension and curling assays. These data therefore strongly support a direct role of H+-ATPase in plant development. The ΔPMA4 plants also displayed increased salt tolerance during germination and seedling growth, supporting the hypothesis that H+-ATPase is involved in salt tolerance.
The H+-ATPase transports protons out of the cell across the plasma membrane, thus establishing the proton electrochemical gradient that contributes to the maintenance of the intracellular and extracellular pHs and drives secondary transport of ions and metabolites. As solute transport is directly related to osmotic water movement, the H+-ATPase is also a key player in turgor regulation and thus regulates the cell size, e.g. during stomatal aperture (for review, see Sussman, 1994; Morsomme and Boutry, 2000; Palmgren, 2001; Lefebvre et al., 2003). The H+-ATPase has also been proposed to play a direct role in the regulation of growth and development. It is regulated at both the transcriptional and posttranslational levels by auxin, a major growth hormone, and so has been proposed to be a key player in cell elongation. According to the acid growth theory, upon activation by auxin, the H+-ATPase acidifies the apoplasm and thus activates enzymes involved in cell wall loosening (for review, see Rayle and Cleland, 1992; Hager, 2003). The transducing pathway leading from auxin to H+-ATPase activation is unknown. The H+-ATPase might be involved in another way, as acidification of the extracellular space is expected to facilitate transport of the protonated form of auxin into the cell and so further stimulate the cell response (Leyser, 2005). However, whether the effect of auxin on cell elongation actually requires cell wall acidification by H+-ATPase is still a matter of controversy (Grebe, 2005, 2006; Kutschera, 2006).
Plant H+-ATPases are encoded by a multigene family organized into five subfamilies (Arango et al., 2003; Baxter et al., 2003). The expression of several members in various species has been examined and shown to have different cell or tissue specificities, sometimes with important overlap. For example, the plasma membrane H+-ATPase genes NpPMA2 and NpPMA4 from Nicotiana plumbaginifolia are both highly expressed in several cell types from various plant organs and both are presumably involved in various transport functions, such as in root hairs and epidermal cells, phloem companion cells, and guard cells (Moriau et al., 1999). The simultaneous expression of these two isoforms, which belong to two different subfamilies, raised the possibility of different kinetics or regulatory properties. This was addressed by heterologous expression of these isoforms and Arabidopsis (Arabidopsis thaliana) H+-ATPase isoforms in yeast (Saccharomyces cerevisiae). Expression of three Arabidopsis H+-ATPase isoforms (AHA1, 2, and 3) showed that they had different kinetic parameters, such as Km, Vmax, and optimal pH (Palmgren and Christensen, 1993). Expression of N. plumbaginifolia PMA2 and PMA4 allowed replacement of the yeast H+-ATPases, but conferred different growth properties, PMA2 allowing yeast growth at external pHs above 5.5, but PMA4 allowing growth down to pH 4.0. These functional differences were linked to a different proton pumping capability (Luo et al., 1999).
Although the H+-ATPase has been suggested to be involved in various and sometimes complex physiological processes, genetic evidence is still limited. In Arabidopsis, Young et al. (1998) analyzed transgenic plants expressing AHA3 with an altered C terminus. When grown in vitro, the Arabidopsis transformants were more resistant to acid medium, suggesting a role for this plasma membrane H+-ATPase in cytoplasmic pH homeostasis. However, no effect was reported for plants grown under normal conditions. Plants homozygous for AHA4 disruption showed a dramatic reduction in rosette growth when grown under salt stress conditions and this correlated with a 4- to 5-fold increase in the Na to K ratio in leaf tissues (Vitart et al., 2001). Gene disruption of AHA10 resulted in seed coats with a transparent testa, a marked reduction in proanthocyanidin levels, and fragmentation of the vacuole in seed coat endothelial cells, suggesting a role of this plasma membrane H+-ATPase in endomembrane biogenesis (Baxter et al., 2005). We previously showed that preventing N. plumbaginifolia PMA4 expression (by cosuppression) had pleiotropic effects on plant growth and development: Sugar transport from the mature leaves was reduced and development retarded, the flowers were male sterile, and guard cells were affected, with a reduced stomatal aperture (Zhao et al., 2000). These results demonstrated that PMA4 is actively involved in several physiological traits that rely on transport activity.
Enzymatic regulation of H+-ATPase has been well documented following the identification of an autoinhibitory domain in its C-terminal region that keeps enzyme activity at a low level (Palmgren et al., 1991). The H+-ATPase can be stimulated by displacement of the autoinhibitory domain upon phosphorylation of the penultimate residue, a Thr, and the subsequent binding of 14-3-3 regulatory proteins (Fuglsang et al., 1999; Svennelid et al., 1999; Camoni et al., 2000; Maudoux et al., 2000). It was recently shown that this activation results in the formation of a complex of six H+-ATPases and six 14-3-3 proteins (Kanczewska et al., 2005; Ottmann et al., 2007). However, there is little information on how this regulatory system operates during plant growth and development. A notable exception is the formation of the H+-ATPase/14-3-3 complex in guard cells upon blue light activation (Kinoshita and Shimazaki 1999, 2002); this complex can also be seen in the plant upon addition of fusicoccin, a fungal toxin, that irreversibly prevents dissociation of the H+-ATPase/14-3-3 complex (Baunsgaard et al., 1998; Fullone et al., 1998; Olsson et al., 1998; Piotrowsky et al., 1998; Oecking and Hagemann, 1999; Kanczewska et al., 2005). The crystal structure of the complex of a plant 14-3-3 protein and the last 52 amino acid residues of PMA2 revealed a previously unidentified mode of interaction in which a 14-3-3 dimer simultaneously binds two H+-ATPase C termini (Ottmann et al., 2007).
In spite of these various genetic and biochemical data, a role of H+-ATPase in plant cell expansion and organ development has not yet been directly demonstrated. Here, we report the analysis of tobacco (Nicotiana tabacum) plants that ectopically expressed either wild-type NpPMA4 (wtPMA4) or NpPMA4 lacking the last 103 residues (ΔPMA4), corresponding to the C-terminal autoinhibitory region. We provide evidence that, in contrast to wtPMA4 overexpression, which did not induce any phenotypic modification, expression of the constitutively activated ΔPMA4 resulted in greater in vivo proton pumping activity, increased salt tolerance, and altered plant development, possibly related to cell expansion.
RESULTS
ΔPMA4 Has Higher Activity and Sustains Yeast Growth Better Than Wild-Type wtPMA4
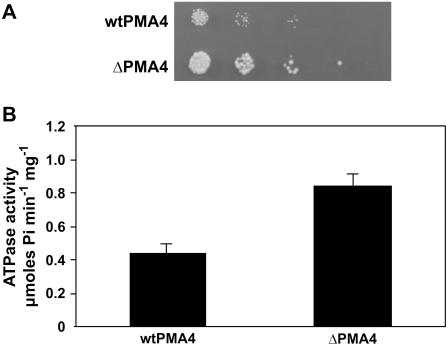
We previously produced transgenic tobacco plants expressing two different PMA4 constructs, one consisting of the full-length N. plumbaginifolia PMA4 (wtPMA4) and the other PMA4 lacking the last 103 C-terminal codons (ΔPMA4; Zhao et al., 2000). The latter construct was expected to code for a constitutively activated enzyme and was used because we thought that overexpression of an activated isoform might result in a stronger effect on the plant. Both were placed under the control of the PMA4 transcription promoter reinforced by the enhancer from the cauliflower mosaic virus 35S promoter (Zhao et al., 1999). Before characterizing these plants, we used heterologous expression in yeast to determine whether the truncated PMA4 had a higher ATPase activity as expected. The wild-type and truncated PMA4 cDNAs were placed under the control of the strong constitutive transcriptional promoter of PMA1, the major yeast H+-ATPase gene, and were introduced into the yeast strain YAK2, deleted of its own two H+-ATPase genes (de Kerchove d'Exaerde et al., 1995). N. plumbaginifolia wtPMA4 and ΔPMA4 were able to sustain the growth of these yeast cells lacking their own H+-ATPases. However, ΔPMA4 allowed faster growth than PMA4 (Fig. 1A). The specific ATPase activity, determined on a purified plasma membrane fraction, was 1.9-fold higher in ΔPMA4-expressing cells than in wtPMA4-expressing cells (Fig. 1B). Because the amounts of the two enzymes in the plasma membrane fraction determined by western blotting were not significantly different (data not shown), we conclude that the molecular activity of ΔPMA4 was clearly increased.
Figure 1.
Growth and ATPase activity of yeast cells expressing wild-type or truncated N. plumbaginifolia H+-ATPase PMA4. A, Growth of yeast strain YAK2 transformed with the wild-type PMA4 gene (wtPMA4) or the truncated PMA4 gene (ΔPMA4). Serial 2-fold dilutions were spotted on a plate at pH 4.0. B, Specific activity of plasma membrane H+-ATPase from YAK2 transformed with the complete PMA4 gene (wtPMA4) or truncated PMA4 gene (ΔPMA4).
Identification of wtPMA4-Overexpressing or ΔPMA4-Expressing Transgenic Tobacco Plants
Several transgenic tobacco plants expressing wtPMA4 or ΔPMA4 had been obtained, but not characterized at the physiological level (Zhao et al., 2000). They were brought to the next generation and the expression of wtPMA4 or ΔPMA4 was confirmed for most of them (data not shown). While wtPMA4 plants did not show any abnormal phenotype, several ΔPMA4 plants showed abnormal inclination of leaves and twisted stems (examples will be shown below). Five lines (wtPMA4 28 and 29, ΔPMA4 41, 51, and 72) were chosen randomly and brought to the third and fourth generation for detailed characterization. All data reported here were obtained with third or fourth generation plants and were observed for all lines retained for each construct.
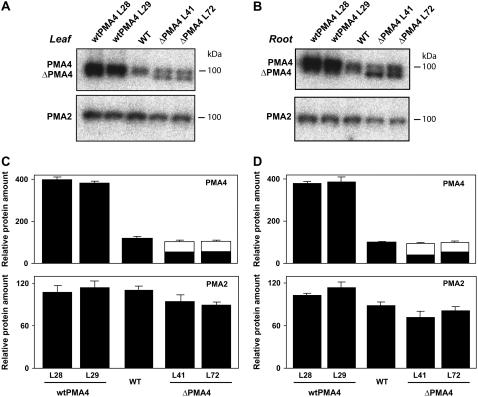
PMA4 and ΔPMA4 levels in leaf and root microsomal fractions were compared to those in untransformed plants by western blotting using anti-PMA4 antibodies. The wtPMA4 plants showed a 3- to 4-fold increase in PMA4 levels in leaf (Fig. 2, A and C) and root tissues (Fig. 2, B and D). In the ΔPMA4 plants, the truncated form (white columns) was present at a level close to that of the endogenous PMA4 (black columns), which, however, was present at only about half the levels seen in an untransformed plant. PMA2, the other major H+-ATPase isoform in N. plumbaginifolia (Arango et al., 2003), was present at a similar level in transgenic and control plants, showing that ectopic expression of wtPMA4 or ΔPMA4 had no effect on PMA2 expression.
Figure 2.
PMA4 protein levels in transgenic and untransformed tobacco plants. Microsomal proteins (15 μg) from the leaves (A and C) and roots (B and D) of wtPMA4 (lines 28 and 29), ΔPMA4 (lines 41 and 72), or untransformed (wild type) plants were separated on 10% SDS-PAGE, blotted, and subjected to immunodetection with anti-PMA4 or anti-PMA2 antibodies and I125 conjugated protein A for quantification. A and B, Phosphorimage of a representative immunoblot performed as described above. C and D, Graphical representation of the phosphorimager counts corresponding, respectively, to A and B, expressed as a percentage of that for the untransformed plant. The white and black bars for ΔPMA4 indicate, respectively, ΔPMA4 and endogenous PMA4. Error bars indicate the sem.
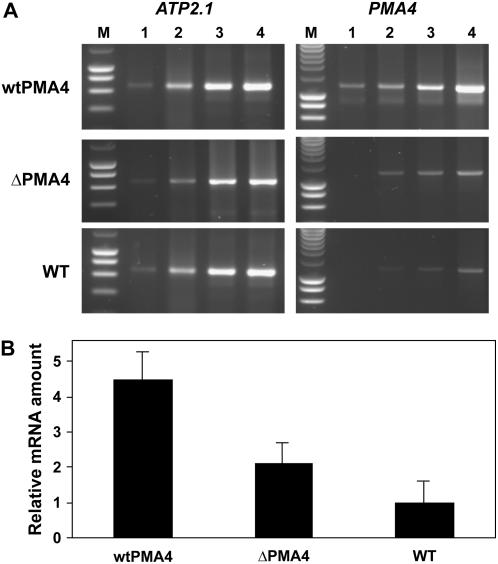
We then examined whether the lower expression of ΔPMA4 compared to wtPMA4 was also observed at the transcription level. We performed a semiquantitative reverse transcriptase (RT)-PCR experiment on RNA extracted from matures leaves of a wtPMA4 and a ΔPMA4 line to determine the PMA4 expression level. The fragment amplified was common for the endogenous PMA4, wtPMA4, and ΔPMA4, to follow the total transcript level and encompassed an exon-intron junction to rule out nuclear DNA amplification. The data were normalized to the RNA level of an internal control (the ATP2.1 gene encoding the β-subunit of mitochondrial ATP synthase). In wtPMA4 plants, PMA4 transcript levels were 4.5-fold higher that in untransformed plants, while, in ΔPMA4 plants, PMA4 transcript levels were 2.1-fold higher than in untransformed plants (Fig. 3B). This lower increase for ΔPMA4 compared to wtPMA4 is in agreement with the observed lower protein expression although the difference was larger in the latter case, suggesting a different posttranscriptional fate.
Figure 3.
RT-PCR analysis of PMA4 mRNA from transgenic and untransformed plants. A, Typical ethidium bromide-stained gel of RT-PCR assays, performed with single-strand cDNA synthesized from RNA extracted from mature leaves of wtPMA4 (line 28), ΔPMA4 (line 41), and untransformed (wild type) plants. Two pairs of PCR primers were used, one for the PMA4 gene and the other for the internal control gene ATP2.1. PCR products are expected at 1,185 (PMA4) and 652 bp (ATP2.1). The PCR products were electrophoresed on an agarose gel after 24 (lanes 1), 26 (lane 2), 28 (lane 3), or 30 (lane 4) reaction cycles. M indicates the lane containing a SmartLadder marker (Eurogentec). B, Graphical representation of RT-PCR analysis showing PMA4 mRNA levels compared to those in control plants. Bands were quantified with a PhosphorImager. The PMA4/ATP2.1 (internal control) ratio is expressed relative to that for the untransformed plant (wild type). Data represent the mean ± sem for three independent experiments.
Measurement of the External pH
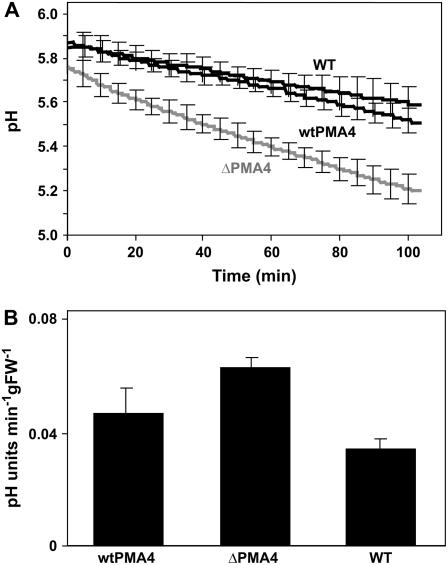
To estimate H+-ATPase activity in vivo, we monitored the time course of changes in the external pH of leaf discs incubated in medium (Amborabé et al., 2001). The initial pH was similar for untransformed (wild type) and wtPMA4 leaf discs, but lower for ΔPMA4 discs (Fig. 4), suggesting that the apoplastic pH of the latter was lower. The pH decreased slightly faster for wtPMA4 than for wild-type leaf discs, but fell more rapidly with leaf discs from ΔPMA4 plants (acidification rate increased by 80%).
Figure 4.
Time course of pH variation in the bathing medium of leaf discs. A, The lower epidermis of leaf discs of wtPMA4 (line 28), ΔPMA4 (line 41), and untransformed (wild type) plants was stripped off and the discs floated on medium containing 250 mm mannitol, 0.5 mm CaCl2, and 0.25 mm MgCl2, and the pH was recorded over time. B, Rate of pH change. Values represent the means ± sem for two independent experiments each performed as in A with lines 28 and 29 (wtPMA4) and lines 41 and 72 (ΔPMA4).
Most of this external acidification depended on H+-ATPase activity, since it was abolished by erythrosin B, an H+-ATPase inhibitor (data not shown).
We then determined whether the lower starting pH and higher acidification activity seen with ΔPMA4 plant leaf discs correlated with a lower apoplastic pH. We measured the leaf apoplastic pH using an infiltration technique (Husted and Schjoerring, 1995; Table I). The apoplastic pH of wild-type and wtPMA4 plants was not significantly different (5.79 and 5.74, respectively). These values were very similar to those found by López-Millán et al. (2000). However, the apoplastic pH of ΔPMA4 leaves was significantly lower at pH 5.35, whereas the apoplastic air and water volumes were unchanged (Table I). We therefore conclude that the higher acidification rate observed in ΔPMA4 leaf discs results in a lower apoplastic pH.
Table I.
Apoplastic pH, air, and water volume of control and transgenic tobacco leaves
Data are the means ± sd of two separate experiments each with two wtPMA4 lines (28 and 29) and two ΔPMA4 lines (41 and 72).
| Line | Apoplastic pH | Apoplastic Air Volume | Apoplastic Water Volume |
|---|---|---|---|
| μL g−1 fresh weight−1 | μL g−1 fresh weight−1 | ||
| wtPMA4 | 5.74 ± 0.08 | 214 ± 20 | 51 ± 8 |
| ΔPMA4 | 5.35 ± 0.05a | 203 ± 28 | 49 ± 12 |
| Control | 5.79 ± 0.03 | 195 ± 22 | 47 ± 6 |
Significant difference (P < 0.05).
Root External Acidification
The pH changes induced by plant roots are mainly caused by H+-ATPase-catalyzed proton release combined with, among other things, nutrient uptake in the form of cations (e.g. NH4+) or the release of HCO3−, organic acid anions, and other ions (Hinsinger et al., 2003). To monitor the root external pH, the root systems from plants grown in hydroponic conditions were trapped within a thin (1.5 mm) agar film containing the pH-sensitive dye, bromocresol purple. After 1 h, the dye changed color from purple (alkaline environment) to yellow (acidic environment) around the roots of ΔPMA4 plants, but a much smaller change was seen for the roots of the wild-type and wtPMA4 plants (Fig. 5). We therefore conclude that ΔPMA4 expression results in a lowering of the root external pH.
Figure 5.
Visualization of the external pH along the roots of wtPMA4 (line 29), ΔPMA4 (lines 72 and 41), and untransformed (wild type) plants. Plants were grown in hydroponic conditions for 10 d, then incubated for 1 h in an agarose gel film containing bromocresol purple. The yellow color corresponds to zones where the pH has been acidified.
In Vitro Plasma Membrane H+-ATPase Activity
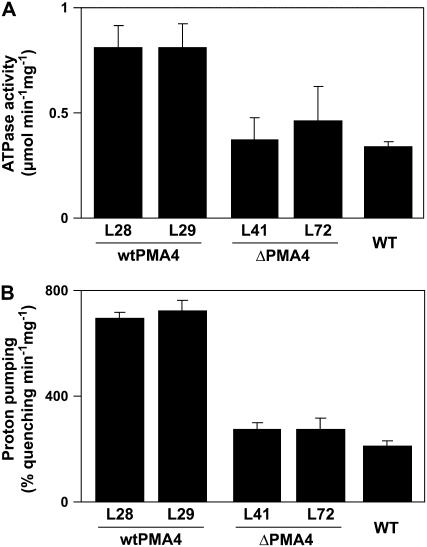
To compare the in vitro ATPase activity of the wtPMA4 and ΔPMA4 plants, ATPase-specific activity was determined on a purified leaf plasma membrane fraction. In contrast to the in vivo data, the wtPMA4 plants showed a 2.3-fold higher activity than the untransformed wild-type and ΔPMA4 plants (Fig. 6A).
Figure 6.
ATPase and proton transport activity of plasma membranes from wtPMA4 (lines 28 and 29), ΔPMA4 (lines 41 and 72), and untransformed (wild type) leaves. A, Vanadate-sensitive ATPase activity determined by measuring Pi release colorimetrically. B, Initial rate of H+ pumping of the H+-ATPase determined by the ACMA fluorescence quenching technique.
H+-ATPase proton pumping activity was estimated by measuring the rate of ATP-dependent quenching of the fluorescence of 9-amino-6-chloro-2-methoxyacridine (ACMA), a well-known pH probe. As shown in Figure 6B, vanadate-sensitive proton pumping activity was more than 3-fold higher in wtPMA4 plants than in wild-type plants. These ATPase and proton pumping data therefore contrast with those observed in vivo and possibly result from release of inhibition during subcellular fractionation (see “Discussion”).
Alteration of Plant Development in ΔPMA4 Transgenic Plants
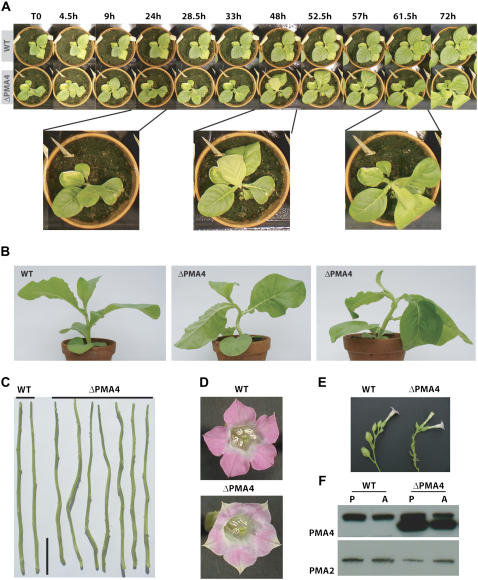
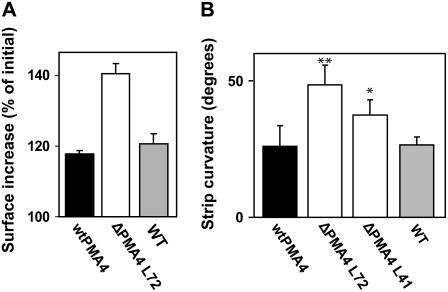
Analysis of transgenic plants over four generations showed that, in contrast to the untransformed and wtPMA4 plants, ΔPMA4 plants displayed several developmental abnormalities. Soon after transfer to soil, young ΔPMA4, but not wtPMA4, plants usually showed abnormal inclination of developing leaves, some of which had temporary their lower surface facing upwards (Fig. 7, A and B). After a few weeks, the emerging stem did not grow straight but bent and some leaves were bumpy and twisted (Fig. 7B). At a later stage, the plants had a more normal leaf architecture, but the stems remained twisted due to an increased internode curvature (Fig. 7C). These data indicate that the activated PMA4 interferes with developmental processes. This phenotype might reflect a deregulated cell expansion and is in agreement with the role of H+-ATPase in cell expansion proposed by the acid growth theory (Hager, 2003). We therefore determined whether the ΔPMA4 leaf had a higher capacity for cell expansion. When expansion of leaf discs was measured after 24 h incubation in medium, it was twice as high for ΔPMA4 plants than for wtPMA4 or untransformed plants (Fig. 8A). Another phenotype sometimes associated with cell expansion is leaf strip curvature or epinasty (Keller and Van Volkenburgh, 1997). When leaf strips were incubated for 24 h in medium and their curvature measured, it was more than 2-fold higher in ΔPMA4 leaf strips than in untransformed plants (Fig. 8B), indicating a propensity of this material to expansion in an asymmetrical way, the adaxial face growing faster than the abaxial face.
Figure 7.
Morphological phenotypes of ΔPMA4 and untransformed (wild type) plants. A, Abnormal leaf bending. An untransformed (wild type) plant and a ΔPMA4 plant (line 41) were transferred to soil. After 3 weeks, the same plants were photographed at the indicated time. The wild-type plant shows normal development, while the ΔPMA4 plant displays intermittently bended leaves. Below is an enlargement of the ΔPMA4 plant showing an upsidedown leaf. Images are representative of several plants of lines 41 and 72. B, Twisted stem and leaves. The untransformed (wild type) plant grows straight and its leaves display normal bending. The two ΔPMA4 plants (lines 41 and 72) have a twisted stem and abnormally bending leaves. C, Twisted stem. The leaves were removed from the stems of mature plants to display their curvature. Stems from untransformed (wild type) plants display a low and regular node bending. Stems from ΔPMA4 plants (lines 41 and 72) display large and irregular bending. Bar = 10 cm. D, The ΔPMA4 flower (line 72) is pale and the ends of the petals show no indentation. The pollen yield is lower than for the wild-type plant. E, Pods are formed on the wild-type flower stem while ΔPMA4 flowers drop off. F, Western-blotting analysis of a microsomal fraction (15 μg proteins) from the upper (colored) part of the petal (P) and from anthers (A) of an untransformed plant (wild type) and a ΔPMA4 plant (line 72). Immunodetection was performed with anti-PMA4 or anti-PMA2 antibodies. Similar data (D–F) were obtained for line 41.
Figure 8.
In vitro expansion of leaf discs and curvature of leaf strips. A, Area increase of leaf discs. Leaf discs (1 × 1 cm) from mature leaves were incubated for 24 h as indicated in “Materials and Methods” and the leaf disc area expressed as a percentage of the initial leaf disc area was calculated (mean + sem, n = 5, three independent experiments). B, Curvature of leaf strips. Leaf strips were incubated for 24 h as indicated in “Materials and Methods.” Images were taken after 24 h and the strip curvature measured (mean + sem, n = 10, five independent experiments).
At the flowering stage, ΔPMA4 flowers were characterized by pale and joined petals (Fig. 7D). Anthocyanin content was reduced by up to 60% (data not shown). The pollen yield was generally severely reduced (Fig. 7D) and this pollen had a reduced viability according to the Alexander's staining: 46.6% ± 12.5% (line 72) or 54.8% ± 4.9% (line 41) stained pollen for ΔPMA4 compared to 86.7% ± 4.8% for the untransformed (n = 11 flowers × 200 pollen grains).
Western-blotting analysis indicated that the ΔPMA4 expression was very high in the upper part of the petals (corresponding to the anthocyanin-stained region) as well as in the anther tissues (Fig. 7F).
The yield of seed pods was generally reduced and some plants did not give any mature seed pod (Fig. 7E). Flower abortion varied between 20% to 100%. As a consequence, while the number of flowers was typically between 50 and 70 for wild-type plants, the flowering period was extended for the ΔPMA4 plants and the number of flowers increased generally above 100 and sometimes up to 200 per plant (data not shown). Reciprocal crosses between ΔPMA4 and untransformed plants showed that the seed yield was restored when ΔPMA4 plants were pollinated with wild-type pollen. In contrast, pollination of wild-type plants with ΔPMA4 pollen generally produced a very low number of seeds. These results strongly indicate that the function of the male, but not the female, organ is compromised in ΔPMA4 plants.
In contrast to wtPMA4 plants, in the progeny of self-pollinated ΔPMA4 plants when seed pods were obtained, homozygous plants for the ΔPMA4 phenotype, and the coexpressed kanamycin marker gene were never obtained, indicating that homozygous ΔPMA4 plants are not viable, possibly due to the higher ΔPMA4 expression. Furthermore, even when pollinated with wild-type pollen, the number of seeds that germinated varied from pod to pod and could be less than 20%, indicating that lethality could also affect heterozygous seeds. All experiments involving ΔPMA4 plants, including those above, were therefore performed on heterozygous plants selected on kanamycin.
Increased Salt Stress Resistance of ΔPMA4 Plants
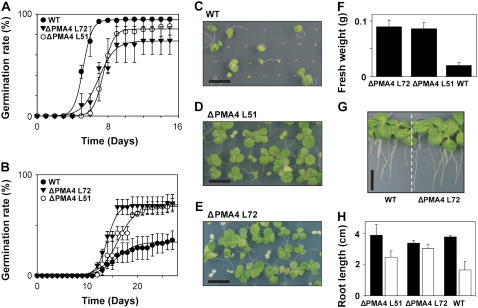
Involvement of the proton pump in plant resistance to salt stress has been suggested by the up-regulation of H+-ATPase genes seen in the presence of salts (for review, see Morsomme and Boutry, 2000; Palmgren, 2001). However, direct evidence is still lacking. To investigate this, we sowed seeds from ΔPMA4, wtPMA4, and untransformed plants in standard Murashige and Skoog medium with or without NaCl supplementation. We mentioned before that no homozygous ΔPMA4 seeds could be obtained. The tests were therefore performed on plates supplemented with kanamycin; the untransformed plants that germinated, but showed sensitivity to kanamycin once the cotyledons developed, were eliminated from the calculations. On standard Murashige and Skoog medium, all the lines germinated synchronously to a level close to 100% for the wild type (Fig. 9A) and wtPMA4 plants (data not shown). Under these conditions, the germination yield of ΔPMA4 seeds was reduced to various extents according to the seed lot, as mentioned before. In the presence of 200 mm NaCl, germination was delayed. However, ΔPMA4 seeds germinated faster and with a better yield than wild-type seeds (Fig. 9B). The better salt resistance of ΔPMA4 plants was also shown by the fact that, after germination, only plantlets from these lines were able to develop green leaves in addition to cotyledons in medium containing 150 mm NaCl (Fig. 9, C–F).
Figure 9.
Phenotype analysis of transgenic and control plants grown on salt medium. A and B, Cumulative germination rates of ΔPMA4 (lines 51 an 72) and untransformed (wild type) seeds in a medium with (B) or without (A) 200 mm NaCl. Means ± sem (n = 50, four independent experiments) and sigmoidal regression curves (R2 > 0,98) are presented. For the heterozygous ΔPMA4 line, kanamycin-sensitive seedlings were not taken into account. C to E, Phenotype of wild type (C) and ΔPMA4 line 51 (D) and line 72 (E) seedlings on 200 mm NaCl. Plants were photographed 25 d after the first plants germinated (bar, 1 cm). ΔPMA4 plants were grown on medium supplemented with kanamycin. Kanamycin-sensitive plants are white. F, Fresh weight of wild-type and ΔPMA4 plantlets grown in the same conditions as in C to E. G and H, Root growth under saline conditions. Ten days after germination on standard Murashige and Skoog medium, the plantlets were transferred for 14 d to Murashige and Skoog medium with or without 200 mm NaCl. G, Representative picture of the phenotype (bar, 1 cm). H, Means ± sem (n = 5, three independent experiments); the black bars are the results in Murashige and Skoog and the white bars those in Murashige and Skoog + 200 mm NaCl. [See online article for color version of this figure.]
Root growth in saline conditions was also examined. To avoid any artifact due to the germination delay, seeds were germinated synchronously in the absence of salt, then transferred to Murashige and Skoog plates with or without 200 mm NaCl. The untransformed (Fig. 9G) and wtPMA4 (data not shown) plants were able to grow in the presence of salt, indicating that the accumulation of salt in germinating plantlets is crucial and that avoiding salt stress during this early stage might help the plants to cope with highly saline media. Nevertheless, ΔPMA4 plant roots showed better growth in saline conditions than those of untransformed plants (Fig. 9H). Thus, expression of ΔPMA4 increased resistance to salt stress compared to untransformed plants.
DISCUSSION
Regulation of H+-ATPase Activity
We designed a constitutively activated PMA4 H+-ATPase isoform by deleting the entire C-terminal region. Expression in yeast demonstrated that this truncated PMA4 (ΔPMA4) had higher ATPase and proton pumping activity and conferred better growth to yeast cells than the full-length form. Ectopic ΔPMA4 expression in tobacco led to transgenic plants that expressed ΔPMA4 at a lower level than that seen for full-length wtPMA4. Comparison of the transcript and protein levels showed a lower enzyme to RNA ratio for ΔPMA4 plants than for wtPMA4 plants. This might be explained by a lower stability of ΔPMA4 protein. However, no ΔPMA4 transgenic plant had ΔPMA4 mRNA levels as high as the wtPMA4 mRNA levels seen in wtPMA4 plants, suggesting that high H+-ATPase activity could be detrimental for plant cells and that counter selection of plants showing high ΔPMA4 expression possibly occurred during transformation or regeneration.
Although ΔPMA4 expression was not particularly high in leaves and roots of transgenic plants and was compensated to some extent by a reduction in endogenous wtPMA4, the in vivo H+-ATPase activity was clearly enhanced. This was evidenced by the lower leaf apoplastic pH and the higher external acidification rate in leaves and roots. In contrast, several-folds overexpression of wtPMA4 did not modify the plant phenotype or increase the external acidification. We can rule out the possibility that the overexpressed PMA4 was misfolded or kept in internal membranes, since purified plasma membranes from wtPMA4 plants had higher ATPase and proton pumping activity than those from untransformed and ΔPMA4 plants, thus contrasting with the in vivo results. We can therefore propose that, in vivo, the increased levels of wtPMA4 were compensated by down-regulation of H+-ATPase activity. This raises the question of how this could occur. A first hypothesis might be that this compensation is due to less H+-ATPase activation by phosphorylation of the penultimate residue, a Thr, and the subsequent binding of 14-3-3 proteins. This would illustrate the importance of this regulatory mechanism. However, this hypothesis might be questioned by several observations. First, taking into account the increase of PMA4 enzyme in wtPMA4, this compensation effect implies that in wild-type plants, H+-ATPase is activated to a large extent in most cell types by this mechanism under normal growth conditions. Yet, H+-ATPase/14-3-3 complex formation has only been observed in guard cells upon blue light activation (Kinoshita and Shimazaki, 1999, 2002), in different tissues upon addition of the toxin, fusicoccin (Baunsgaard et al., 1998; Fullone et al., 1998; Piotrowsky et al., 1998; Oecking and Hagemann, 1999), and in metabolically activated culture cells (Kanczewska et al., 2005). Moreover, ΔPMA4 expression in the ΔPMA4 plants was low and at the same level as the endogenous PMA4 which itself was reduced by half (Fig. 2). Yet, this moderate expression of an activated H+-ATPase was sufficient to induce various phenotypes. Therefore, we cannot expect that in the wild type, the H+-ATPase is activated to a large extent in most tissues because this would give an activity similar to that of ΔPMA4 and thus lead to an abnormal phenotype. Finally, if the higher ATPase amount in wtPMA4 plants was compensated by a lower activation by phosphorylation of the penultimate Thr and lower binding of 14-3-3 proteins, we should have observed the same H+-ATPase activity ratio in vitro with isolated membranes since the membrane-bound H+-ATPase/14-3-3 complex is stable (Kanczewska et al., 2005) and thus the phosphorylated Thr is protected from phosphatases in vitro. Yet, in vitro, we observed higher ATPase activity for the wtPMA4 lines.
An alternative hypothesis to explain the compensation effect is that down-regulation in wtPMA4 plants is caused by a still unidentified negative regulatory mechanism. Fine tuning of H+-ATPase activity is expected. Indeed, this enzyme is a major component of the plasma membrane and therefore a high ATP consumer, and has many physiological roles that might require more than a single regulatory mechanism. In addition, it has been shown that PMA2 that is not activated by Thr phosphorylation and binding of 14-3-3 proteins still keeps approximately half the activity of the activated form (Maudoux et al., 2000). Thus, there is still room for down-regulation. Actually, there is evidence that, in addition to the now well characterized activating phosphorylation of the penultimate Thr residue, inhibitory phosphorylation events also occur. For instance, in vivo H+-ATPase dephosphorylation in tomato (Solanum lycopersicum) cells activates the enzyme (Vera-Estrella et al., 1994; Xing et al., 1996). Systemin, a primary wounding signal, is suggested to inhibit H+-ATPase activity via Ca2+-dependent phosphorylation of H+-ATPase in cultured tomato cells (Schaller and Oecking, 1999). The Ca2+-dependent phosphorylation of H+-ATPase and the resulting inhibition of H+ pumping have been reported in root cells of oat (Avena sativa) and beet (Beta vulgaris) (Schaller and Sussman, 1988; Suzuki et al., 1992; Lino et al., 1998). Desbrosses et al. (1998) have suggested that the plasma membrane H+-ATPase in tobacco cells is activated through dephosphorylation. Recently, a new phosphorylation site (Ser-931) has been identified in the C-terminal region of AHA2, an Arabidopsis H+-ATPase (Fuglsang et al., 2007). However, phosphorylation of this residue results in preventing binding of 14-3-3 proteins and therefore ATPase activation, but does not seem to inhibit the enzyme. This residue is therefore unlikely to correspond to the phosphorylated residue hypothesized here in wtPMA4 plants.
The fact that this putative inhibitory regulation in wtPMA4 plants was not observed in vitro might result from phosphatase action during cell homogenization and fractionation. Another alternative is that the binding of a still unknown regulatory protein causes H+-ATPase down-regulation in wtPMA4 plants.
If the hypothesis of phosphorylation- or regulatory protein-dependent H+-ATPase inactivation is correct, how can we explain that this inhibition was not observed in the ΔPMA4 plants, which were characterized by a constitutively activated enzyme? The simplest explanation is that the putative negative regulation involves residues in the C-terminal region and thus absent in ΔPMA4. Alternatively, it could involve residues elsewhere, but resulting in enhancement of the inhibitory action of the C-terminal regulatory domain, e.g. by increasing its interaction with the rest of the enzyme. In this case, the removal of the inhibitory domain would prevent any effect of the hypothesized inhibitory regulation.
Evaluating this hypothesis of a PMA2 negative regulation will require more in-depth analysis, for instance by analyzing phosphorylated residues of overexpressed PMA4 provided with a tag allowing its purification and characterization.
A previous report showed that Arabidopsis transgenic plants expressing the AHA3 isoform activated by deletion within its inhibitory C-terminal region displayed reduced growth inhibition when seedlings were grown in vitro at a pH below 5.0 (Young et al., 1998). These authors suggested that, in these young seedlings lacking a cuticle, the alkaline phloem might be more sensitive than other cells within the stem, leaves, and roots to the steeper pH gradient across the plasma membrane. However, no unusual phenotype other than this lower sensitivity to acid stress was reported for these plants expressing activated AHA3. This difference with the data reported here for ΔPMA4 plants might be explained in at least two ways. First, in the Arabidopsis plants, the AHA3 transcription promoter used directs expression exclusively to the phloem companion cells (De Witt et al., 1991), whereas the promoter used in our study for ΔPMA4 has a broad range of expression (Zhao et al., 1999). Second, the deletion in the activated AHA3 consisted only of the last 46 C-terminal residues, while that in ΔPMA4 was 103 residues, i.e. the whole C-terminal region following the last transmembrane span. The first half of the C-terminal region was therefore still present in the activated AHA3. As this region still contains residues belonging to the autoinhibitory domain (Dambly and Boutry, 2001), it is possible that this deleted AHA3 form is less activated than ΔPMA4.
Alteration of Plant Development
In addition to cell division, plant growth is accomplished by an irreversible increase in cell size. For cell wall expansion to occur, three conditions must be met: (1) adequate turgor must exist inside the cell; (2) extensibility must be achieved through rearrangement or loosening of the existing cell wall; and (3) synthesis and deposition of newly formed wall components must occur (Cosgrove, 1997). The so-called acid growth theory suggests hydrogen ions as the cell wall-loosening factor. Secreted protons decrease the apoplastic pH and wall-loosening processes are thereby activated (Rayle and Cleland, 1992; Hager, 2003). This theory is supported by several observations: (1) external acidification of some plant tissues, notably coleoptiles, induces their elongation; (2) addition of auxin induces tissue expansion correlated with H+-ATPase activation and decreased external pH; and (3) fusicoccin, a toxin that strongly activates H+-ATPase, induces cell elongation. However, several aspects of the acid growth theory are still controversial (e.g. Grebe, 2005, 2006; Kutschera, 2006). For instance, decreasing the external pH does not always induce cell expansion. One reason might be the difficulty in crossing the cuticular barrier. This usually requires removing or damaging the epidermis so that the lower pH external medium can access the internal tissues. The reverse genetics approach adopted in this article is original in several ways. First, the lower external pH did not require damaging the epidermis and was not induced by external application of any chemicals, but by acting directly on the enzyme that generates the transmembrane pH difference. Second, the effect of a lower external pH can be studied in the whole plant at any developmental stage without harming any tissue. Finally, activating the H+-ATPase not only increases the transmembrane pH difference, but is also expected to increase the transmembrane potential difference. This might be important when considering cell expansion, since, besides wall loosening, it also requires sustained osmotic pressure, which depends, in part, on active ion and nutrient transport. Some transport, such as K+ through channels, depends more on the transmembrane potential difference than on the transmembrane pH difference.
The various ΔPMA4 phenotypic properties were consistently observed for three lines for which third and fourth generation seeds were available. We can therefore conclude that the phenotypic properties of the ΔPMA4 plants are directly linked to ΔPMA4 expression and are not an artifact due to genomic insertional events. What did we learn from the ΔPMA4 phenotype? The size of ΔPMA4 plant organs was not modified compared to untransformed plants. This does not exclude the possibility that H+-ATPase has an important role to play in this aspect, since, as mentioned above, high ΔPMA4 expression might have been counter selected if linked to a lethal phenotype. However, ΔPMA4 expression induced several developmental modifications of the plant, namely bent leaves, twisted stems, pale petals, and male sterility. Upon transfer of ΔPMA4 plants to soil, intermittent abnormal leaf inclination and chaotic stem bending were observed. The former phenotype disappeared in more mature plants, but stem bending remained during the whole growth. Sometimes leaves were bumpy, due to an interveinal surface larger than the surface provided by the veinal network. Microscopic observation of stems or leaves did not show any consistent cell morphological change in ΔPMA4 plants. Determining statistically significant differences in cell size or number was not possible, since the phenotype easily identified at the macroscopic level might result from the accumulated effect of a small increase in cell expansion or division in each of many cells. However, the observation that mature ΔPMA4 leaf discs had an increased expansion rate in vitro is a strong indication that the phenotype is related to deregulated cell expansion.
How leaf expansion is coordinated is still debated, but it is generally thought to result from expansion promotion by some cell types and expansion restriction by others. Because of their loose organization, the mesophyll cells probably do not play a direct role. Based on various observations, the epidermis has been considered to restrict the expansion of underlying tissues, while the veins drive expansion (Van Volkenburgh, 1999). Although there was an interveinal surface increase in some ΔPMA4 leaves, interveinal leaf discs incubated in vitro were still capable of expanding more than wild-type leaf discs, suggesting that, in vivo, the expansion capacity was not fully used. Together with the observation that the surface of the whole leaf was not significantly modified, this suggests that, in vivo, the network of the main veins, rather than the epidermis, limits leaf expansion. Another and nonexclusive hypothesis is that a low external pH is not the only factor responsible for activating cell expansion. For instance, proteins, such as expansins and endotransglycosylases, are required to weaken the wall (Van Volkenburgh, 1999). Finally, it is also possible that the apoplastic pH of ΔPMA4 plants is not low enough to have a full effect on cell elongation. As mentioned above, the constitutive expression of ΔPMA4 might well have made it impossible to obtain transgenic calli or plants with high ΔPMA4 expression because of lethal growth or developmental problems. In this respect, the use of inducible transcription promoters would be an interesting approach.
The in vitro leaf strip expansion observed for ΔPMA4 was not random, but epinastic, due to faster growth of the upper (adaxial) than the lower (abaxial) leaf surface. The epinasty observed in ΔPMA4 leaf strips is reminiscent of the phenotype observed with plants that overexpress auxin binding protein 1 (Jones et al., 1998) and supports the hypothesis that auxin-induced cell expansion and leaf epinasty are mediated by H+-ATPase activation. This does not exclude the possibility that auxin, in addition to activating H+-ATPase, might influence cell expansion by an additional effect. Indeed, auxin-induced growth of tobacco leaf tissues has been observed without cell wall acidification (Keller and Van Volkenburgh, 1998).
While auxin is known to stimulate H+-ATPase, H+-ATPase also has a direct effect on auxin transport. Auxin is a weak acid and therefore H+-ATPase activation, which results in a lowering of the external pH, should facilitate uptake of the protonated auxin into the cell. Li et al. (2005) showed that a change in the expression of a vacuolar pyrophosphatase alters the amounts of both H+-ATPase and PIN1, an auxin transporter, in the plasma membrane, and results in altered morphogenesis. In this case, it was difficult to distinguish the respective effects of modification of PIN1 or H+-ATPase. Likewise, the phenotype of the ΔPMA4 plants might indirectly result from improved auxin uptake. However, this hypothesis is not supported by the in vitro observation of ΔPMA4 leaf disc expansion and strip curvature, which were readily observed in the absence of auxin added to the medium. At least in this case, a more direct effect of the transmembrane pH difference is probably involved.
Another developmental problem observed in ΔPMA4 plants was male sterility. Pollen formation is a critical developmental stage and male sterility has been observed when PMA4 expression is prevented by cosuppression (Zhao et al., 2000). Failure in pollen formation might be correlated with the high ΔPMA4 expression observed in the anther tissues. However, contrary to other phenotypic properties displayed by ΔPMA4 plants, seed pod development and seed viability varied among ΔPMA4 plants and also among flowers from the same plant. This variation might be due to a threshold hypothesis often suggested to explain leaf variegation (Wang et al., 2000). In the case where ΔPMA4 expression would pass a threshold, interference with cell processes might contribute, in a snowball effect, to irreversible developmental alteration.
The reduced petal pigmentation seen in ΔPMA4 flowers was unexpected, as anthocyanins accumulate in the vacuole. However, an indirect role of H+-ATPase in the formation of flavonoids has been recently supported by the observation that disruption of AHA10 in Arabidopsis results in much reduced levels of proanthocyanidin in the seed coat endothelium (Baxter et al., 2005). These authors proposed that AHA10 might be recruited to acidify endosomes or a provacuolar compartment and either influence the uptake or transformation of protoanthocyanidin precursors or control membrane trafficking. Although analysis of a PMA4-GFP fusion protein in tobacco did not give any indication that PMA4 or ΔPMA4 can be recruited into internal membranes (Lefebvre et al., 2004), we cannot rule this out in particular cell types or under particular circumstances.
H+-ATPase and Salt Tolerance
Sodium is highly toxic when it accumulates within the cell. Several exclusion mechanisms exist to maintain the sodium concentration low within the cytosol. One of these involves exclusion within the vacuole, which relies on Na+/H+ antiport energized by the tonoplast H+-ATPase and pyrophosphatase. Another involves Na+ efflux out of the cell through a Na+/H+ antiporter (SOS1 in Arabidopsis) thought to be activated by the pH gradient generated across the plasma membrane by H+-ATPase (for review, see Blumwald et al., 2000; Zhu, 2003; Yamaguchi and Blumwald, 2005). The role of the latter has been inferred from the observation of increased H+-ATPase activity under salt stress conditions (for review, see Morsomme and Boutry, 2000; Palmgren, 2001). More direct evidence was provided by the observation that an Arabidopsis mutant disrupted in the H+-ATPase AHA4 gene has increased sensitivity to salt stress (Vitart et al., 2001). Here, we showed that expressing a constitutively activated H+-ATPase increased salt tolerance during the germination and growth of seedlings. However, considering the altered development of ΔPMA4 plants, the ubiquitous expression of an activated H+-ATPase is not an interesting solution for improving salt tolerance of crop plants. This would require the use of a more specific transcription promoter, e.g. active in the root epidermis or induced by salt stress.
MATERIALS AND METHODS
Plant Materials and Transgene Constructs
Tobacco (Nicotiana tabaccum) ‘SR1’ was used as the wild-type plant. Gene constructions and the production of wtPMA4 and ΔPMA4 transgenic plants have been described previously (Zhao et al., 2000). Third or fourth generation plants were used in the experiments reported here. Wild-type and transgenic seeds were germinated on agar medium containing Murashige and Skoog salts (Murashige and Skoog, 1962), 3% Suc, and, for transgenic seeds, 100 mg L−1 of kanamycin. Plants at the 4 to 5 leaf stage were transferred to peat pellet (Jiffy Products International AS) for 2 weeks, then to soil and grown in the greenhouse under 16 h light (185 μmol m−2 s−1). The plants were fed with 6-6-6 NPK once a week. All the data reported in this article were obtained with, in addition to the nontransformed parental line, two wtPMA4 lines (lines 27 and 28) and two or three ΔPMA4 lines (41, 51, and 72).
Yeast Transformation and Cell Fractionation
The PMA4 or ΔPMA4 cDNA used for plant transformation was introduced into the plasmid 2μp(PMA1) under the control of the yeast (Saccharomyces cerevisiae) PMA1 transcription promoter (Luo et al., 1999). Transformation of the YAK2 yeast strain was followed by growth on 5-fluoro-orotic acid to counter select plasmids bearing the yeast H+-ATPase gene and to determine whether the plant H+-ATPase allowed growth of yeast (Luo et al., 1999). Plasma membrane purification and ATPase assays were performed as described previously (Dambly and Boutry, 2001).
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from tobacco leaves by acid guanidinium thiocyanate/phenol/chloroform extraction (Chomczynski and Sacchi, 1987). The RNA pellet was suspended in 1% diethylpyrocarbonate and the concentration estimated spectrophotometrically at 260 nm.
A two-step semiquantitative RT-PCR method was used. The first cDNA strand was synthesized using 2.5 μg of total RNA as template in 25 μL of reaction mixture, which included 0.5 μg of oligo-(dT)18n primer and 200 units of Moloney murine leukemia virus RT (Promega). Three microliters of the first-strand cDNA was used to amplify fragments specific for PMA4 or ATP2-1 (encoding the β-subunit of mitochondrial ATP synthase and used as an internal control; Boutry and Chua, 1985). The primers (0.5 μm each) were: 5′-ATGGCAAAAGCTATCAGC-3′ (PMA4 5′), 5′-GATACCGGCTCGTGCCTC-3′ (PMA4 3′), 5′-TGAGCTCATCCATACCCAAA-3′ (ATP2 5′), and 5′-TCTTTGCTGGTGTTGGTGAA-3′ (ATP2 3′). The cDNA yield was measured according to the PCR signal generated from the internal standard from 24 to 30 cycles starting with 2 μL of cDNA solution. The volume of each cDNA pool was adjusted to give the same exponential phase PCR signal for ATP2 after 24 cycles. An aliquot (10 μL) of each PCR reaction mixture was subjected to electrophoresis in an agarose gel and the ethidium bromide-stained band quantified using a Molecular Imager system (Bio-Rad Laboratories).
Preparation of Plant Microsomal and Plasma Membrane Fractions
Fresh leaf tissues (100 g) of fully expanded leaves were homogenized at 2°C in a Waring blender (Waring Laboratory) in 400 mL of homogenization buffer (50 mm Tris-HCl, pH 8.0, 250 mm sorbitol, 2 mm EDTA, 0.7% [w/v] polyvinylpyrrolidone, 5 mm dithiothreitol) and the protein inhibitor cocktail (1 mm phenylmethylsulfonyl fluoride, 0.1 μg mL−1 of leupeptin, and 1 μg mL−1 each of pepstatin, chymostatin, antipain, and aprotinin), and the homogenate filtered through two layers of Miracloth (Calbiochem) and centrifuged twice at 10,000g for 15 min at 4°C, then the supernatant was centrifuged at 20,800g for 60 min at 4°C. The final pellet was suspended in 500 μL of 5 mm KH2PO4, 3 mm KCl, 330 mm Suc, and the protease inhibitor cocktail, pH 7.8. Plasma membranes were prepared by phase partition (Larsson et al., 1987), the membrane pellet being finally suspended in 400 μL of 10 mm Tris-HCl, pH 6.5, and 330 mm Suc.
Protein concentrations were measured as described by Bradford (1976), using bovine serum albumin as a standard.
Western-Blotting Analysis
Microsomal proteins (20 μg) were separated by SDS-PAGE (10% acrylamide) and electrophoretically transferred to a nitrocellulose membrane (Immobilon; Millipore), which were then probed with rabbit antibodies against PMA4 or PMA2 (Moriau et al., 1999) and I125-conjugated protein A (Amersham Pharmacia Biotech) for quantitative immunodetection using a GS-525 Molecular Phosphorimager system (Bio-Rad Laboratories).
Plant ATPase Assays
ATPase assays was performed by measuring the release of phosphate in 15 min at 30°C in 100 μL of 1 mm MgATP, 1 mm free Mg2+ (MgCl2), 50 mm MES-NaOH (pH 6.5), 1.25 mm phosphoenol pyruvate, 25 mm K2SO4, 5.2% (w/v) glycerol, 0.15% (w/v) Brij 58, 7.5 mm NaN3, 75 mm KNO3, 0.3 mm sodium molybdate, and 2 μg of protein.
ATP-Dependent Proton Transport Assay
Proton transport into plasma membrane vesicles was measured by the quenching of ACMA using an AMINCO-Bowman Series II spectrofluorometer (Thermo Spectronic). The reaction mixture (1.5 mL) contained 50 mm MES-NaOH (pH 6.5), 25 mm K2SO4, 5.2% (w/v) glycerol, 0.15% Brij 58, 0.2 μm ACMA, and 75 μg of protein. After equilibration of the vesicles with the dye for 3 min at 30°C, 1 mm of MgATP was added, and the fluorescence monitored. After 5 min, the ionophore, nigericin, was added to a final concentration of 130 μm, producing an immediate reversal of fluorescence quenching. The addition of vanadate before MgATP completely prevented the fluorescence quenching by MgATP. Fluorescence was measured with an excitation wavelength of 410 nm and an emission wavelength of 480 nm.
Determination of Apoplastic Volumes and pH
Leaf apoplastic air and water volumes and the apoplastic pH were measured as described previously (Husted and Schjoerring, 1995).
Measurement of External Acidification
Tobacco leaf discs (around 100 mg fresh weight) were excised from the leaves after peeling off the lower epidermis and preincubated (peeled face down) for 30 min in 300 mm mannitol, 0.5 mm CaCl2, 0.25 mm MgCl2 (Amborabé et al., 2001). The discs were then transferred to 6 mL of fresh medium in a vial (3 cm) on a magnetic stirrer for aeration. The pH was measured with a pH microelectrode linked to a C832 multiparameter analyzer (Consort).
Pollen Viability Staining
Pollen viability was estimated using Alexander's stain (Alexander, 1980).
Root External Acidification
Plants were transferred for 2 weeks to half-concentrated Hoagland nutrient solution (Sigma Chemical). Spatial localization of acidification was determined by positioning the root system between two glass sheets separated by 1.5 mm thick insets filled with the same nutrient solution containing 0.009% bromocresol purple and 1% agarose (pH 6.2; Vansuyt et al., 2003).
Determination of Leaf Surface Expansion and Strip Curvature
The methods for leaf disc surface expansion and leaf strip curvature measurements were adapted from those of Keller and Van Volkenburgh (1998). Fully developed leaves from plants at the early stage of flowering were used. Leaf strips (2 × 10 mm) and discs (1 × 1 cm) were cut from the interveinal region in the middle of the leaf and incubated for 24 h at room temperature in 10 mm Suc, 10 mm KCl, and 0.5 mm MES, pH 6.0. Leaf discs were photographed with a digital camera and their surfaces estimated as their pixel content divided by the pixel content of a 1 cm2 control surface imaged on the same picture. Leaf strip curvature was estimated as the angle made by the two tangents to the two terminal parts of each strip.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number X66737.
Acknowledgments
We thank Anne-Marie Faber and Pierre Gosselin for their excellent technical assistance.
This work was supported by grants from the Interuniversity Poles of Attraction Program (Belgian State, Scientific, Technical, and Cultural Services), the European Community, the Belgian Fund for Scientific Research, and the Human Frontier Science Program. F.G. held an individual Marie Curie European fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Marc Boutry (boutry@fysa.ucl.ac.be).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alexander MP (1980) A versatile stain for pollen, fungi, yeast and bacteria. Stain Technol 55 13–18 [DOI] [PubMed] [Google Scholar]
- Amborabé BE, Fleurat-Lessard P, Bonmort J, Roustan JP, Roblin G (2001) Effects of eutypine, a toxin from Eutypa lata, on plant cell plasma membrane: possible subsequent implication in disease development. Plant Physiol Biochem 39 51–58 [Google Scholar]
- Arango M, Gévaudant F, Oufattole M, Boutry M (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216 355–365 [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Fuglsang AT, Jahn T, Korthout HAAJ, de Boer AH, Palmgren MG (1998) The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J 13 661–671 [DOI] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren M, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102 2649–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP (2000) Sodium transport in plant cells. Biochim Biophys Acta 1465 140–151 [DOI] [PubMed] [Google Scholar]
- Boutry M, Chua NH (1985) Nuclear gene encoding the beta subunit of the mitochondrial ATP synthase in Nicotiana plumbaginifolia. EMBO J 4 2159–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Camoni L, Iori V, Marra M, Aducci P (2000) Phosphorylation-dependent interaction between plant plasma membrane H+-ATPase and 14-3-3 protein. J Biol Chem 275 9919–9923 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162 156–159 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell 9 1031–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambly S, Boutry M (2001) The two major plant plasma membrane H+-ATPases display different regulatory properties. J Biol Chem 276 7017–7022 [DOI] [PubMed] [Google Scholar]
- de Kerchove d'Exaerde A, Supply P, Dufour JP, Bogaerts P, Thinès D, Goffeau A, Boutry M (1995) Functional complementation of a null mutation of the yeast Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene. J Biol Chem 270 23828–23837 [DOI] [PubMed] [Google Scholar]
- De Witt ND, Harper J, Sussman MR (1991) Evidence for a plasma membrane proton pump in phloem cells of higher plants. Plant J 1 121–128 [DOI] [PubMed] [Google Scholar]
- Desbrosses G, Steling J, Renaudin JP (1998) Dephosphorylation activates the purified plant plasma membrane H+-ATPase: possible function of phosphothreonine residues in a mechanism not involving the regulatory C-terminal domain of the enzyme. Eur J Biochem 251 496–503 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 proteins. Plant Cell 19 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drummn K, Jahn T, Stensballe A, Mattei B, Jensen ON, Palmgren MG (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J Biol Chem 274 36774–6780 [DOI] [PubMed] [Google Scholar]
- Fullone MR, Visconti S, Marra M, Fogliano V, Aducci P (1998) Fusicoccin effect on the in vitro interaction between plant 14-3-3 proteins and plasma membrane H+-ATPase. J Biol Chem 273 7698–7702 [DOI] [PubMed] [Google Scholar]
- Grebe M (2005) Growth by auxin: when a weed needs acid. Science 310 60–61 [DOI] [PubMed] [Google Scholar]
- Grebe M (2006) Response. Science 311 953–954 [Google Scholar]
- Hager A (2003) Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res 116 483–505 [DOI] [PubMed] [Google Scholar]
- Hinsinger P, Plassard C, Tang C, Jaillard B (2003) Origins of root-mediated pH changes in the rhizosphere and their responses to environmental constraints: a review. Plant Soil 248 43–59 [Google Scholar]
- Husted S, Schjoerring JK (1995) Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol 109 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito MA, Binns AN (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282 1114–1117 [DOI] [PubMed] [Google Scholar]
- Kanczewska J, Marco S, Vandermeeren C, Maudoux O, Rigaud JL, Boutry M (2005) Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimmer into a hexamer. Proc Natl Acad Sci USA 102 11675–11680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CP, Van Volkenburgh E (1997) Auxin-induced epinasty of tobacco leaf tissues: a non ethylene-mediated response. Plant Physiol 113 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CP, Van Volkenburgh E (1998) Evidence that auxin-induced growth of tobacco leaf tissues does not involve cell wall acidification. Plant Physiol 118 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard. EMBO J 18 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol 43 1359–1365 [DOI] [PubMed] [Google Scholar]
- Kutschera U (2006) Acid growth and plant development. Science 311 952–953 [DOI] [PubMed] [Google Scholar]
- Larsson C, Widell S, Kjellbom P (1987) Preparation of high-purity plasma membranes. Methods Enzymol 148 558–568 [Google Scholar]
- Lefebvre B, Batoko H, Duby G, Boutry M (2004) Targeting of a Nicotiana plumbaginifolia H+-ATPase to the plasma membrane is not by default and requires cytosolic structural determinants. Plant Cell 16 1772–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Boutry M, Morsomme P (2003) The yeast and plant plasma membrane H+ pump ATPase: divergent regulation for the same function. Prog Nucleic Acid Res 74 203–237 [DOI] [PubMed] [Google Scholar]
- Leyser O (2005) Auxin distribution and plant pattern formation: how many angels can dance on the point of PIN? Cell 121 819–822 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310 121–125 [DOI] [PubMed] [Google Scholar]
- Lino B, Baizabal-Aguirre VM, Gonzalez de la Vara LE (1998) The plasma-membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta 204 352–359 [DOI] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Abadía A, Abadía J (2000) Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet: implications for iron and carbon transport. Plant Physiol 124 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Morsomme P, Boutry M (1999) The two major types of plant plasma membrane H+-ATPases show different enzymatic properties and confer differential pH sensitivity of yeast growth. Plant Physiol 119 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J Biol Chem 275 17762–17770 [DOI] [PubMed] [Google Scholar]
- Moriau L, Michelet B, Bogaerts P, Lambert L, Oufattole M, Boutry M (1999) Expression analysis of two gene subfamilies encoding the plasma membrane H+-ATPase in Nicotiana plumbaginifolia reveals the major transport functions of this enzyme. Plant J 19 31–41 [DOI] [PubMed] [Google Scholar]
- Morsomme P, Boutry M (2000) The plant plasma membrane H+-ATPase: structure, function and regulation. Biochim Biophys Acta 1465 1–16 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Oecking C, Hagemann K (1999) Association of 14-3-3 proteins with the C-terminal autoinhibitory domain of the plant plasma membrane H+-ATPase generates a fusicoccin-binding complex. Planta 207 480–482 [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol 118 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, Marco S, Jaspert N, Marcon C, Schauer N, Weyand M, Vandermeeren C, Duby G, Boutry M, Wittinghofer A, et al (2007) Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell 25 427–440 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52 817–845 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Christensen G (1993) Complementation in situ of the yeast plasma membrane H+-ATPase gene pma1 by an H+-ATPase gene from a heterologous species. FEBS Lett 317 216–222 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266 20470–20475 [PubMed] [Google Scholar]
- Piotrowsky M, Morsomme P, Boutry M, Oecking C (1998) Complementation of the Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene generates a highly abundant fusicoccin binding site. J Biol Chem 273 30018–30023 [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE (1992) The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol 99 1271–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Oecking C (1999) Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Sussman MR (1988) Phosphorylation of the plasma-membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta 173 509–518 [DOI] [PubMed] [Google Scholar]
- Sussman MR (1994) Molecular analysis of proteins in the plant plasma membrane. Annu Rev Plant Physiol Plant Mol Biol 45 211–234 [Google Scholar]
- Suzuki YS, Wang Y, Takemoto JY (1992) Syringomycin-stimulated phosphorylation of the plasma membrane H+-ATPase from red beet storage tissue. Plant Physiol 99 1314–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Volkenburgh E (1999) Leaf expansion—an integrated plant behaviour. Plant Cell Environ 22 1463–1473 [Google Scholar]
- Vansuyt G, Souche G, Straczek A, Briat JF, Jaillard B (2003) Flux of protons released by wild type and ferritin over-expressor tobacco plants: effect of phosphorus and iron nutrition. Plant Physiol Biochem 41 27–33 [Google Scholar]
- Vera-Estrella R, Barkla BJ, Higgins VJ, Blumwald E (1994) Plant defense response to fungal pathogens-activation of host-plasma membrane H+-ATPase by elicitor-induced enzyme dephosphorylation. Plant Physiol 104 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart V, Baxter I, Doerner P, Harper JF (2001) Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J 27 191–201 [DOI] [PubMed] [Google Scholar]
- Wang Y, Duby G, Purnelle B, Boutry M (2000) Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. Plant Cell 12 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing T, Higgins VJ, Blumwald E (1996) Regulation of plant defense response to fungal pathogens: two types of protein kinases in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell 8 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Blumwald E (2005) Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10 615–620 [DOI] [PubMed] [Google Scholar]
- Young JC, DeWitt ND, Sussman MR (1998) A transgene encoding a plasma membrane H+-ATPase that confers acid resistance in Arabidopsis thaliana. Genetics 149 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Dielen V, Kinet JM, Boutry M (2000) Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth and male fertility. Plant Cell 12 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Moriau L, Boutry M (1999) Expression analysis of the plasma membrane H+-ATPase pma4 transcription promoter from Nicotiana plumbaginifolia activated by the CaMV 35S promoter enhancer. Plant Sci 149 157–165 [Google Scholar]
- Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6 441–445 [DOI] [PubMed] [Google Scholar]