Abstract
The bryophyte Physcomitrella patens is unlike any other plant identified to date in that it possesses a gene that encodes an ENA-type Na+-ATPase. To complement previous work in yeast (Saccharomyces cerevisiae), we determined the importance of having a Na+-ATPase in planta by conducting physiological analyses of PpENA1 in Physcomitrella. Expression studies showed that PpENA1 is up-regulated by NaCl and, to a lesser degree, by osmotic stress. Maximal induction is obtained after 8 h at 60 mm NaCl or above. No other abiotic stress tested led to significant increases in PpENA1 expression. In the gametophyte, strong expression was confined to the rhizoids, stem, and the basal part of the leaf. In the protonemata, expression was ubiquitous with a few filaments showing stronger expression. At 100 mm NaCl, wild-type plants were able to maintain a higher K+-to-Na+ ratio than the PpENA1 (ena1) knockout gene, but at higher NaCl concentrations no difference was observed. Although no difference in chlorophyll content was observed between ena1 and wild type at 100 mm NaCl, the impaired Na+ exclusion in ena1 plants led to an approximately 40% decrease in growth.
Soil salinity represents a growing problem in agriculture because high sodium concentrations are toxic to most plants, causing reduced growth and productivity (Munns et al., 1999). With more than 20% of irrigated land worldwide affected by increasing salinity, it is important to understand the basis for the different levels of salinity tolerance that are commonly observed in plants and to discover how certain plants are able to maintain growth in saline conditions. Plants are sessile and have to adapt and thrive in many different types of soils with varying ionic compositions. This is in contrast to animal cells, which are more or less exposed to a constant ionic environment. The membrane potential across the plasma membrane is approximately −150 mV for most plant cells (Benito et al., 2002). When this negative potential is combined with a difference in pH, the resulting electrochemical potential difference for H+ energizes H+-coupled transport catalyzed by the many symporters and antiporters in the plasma membrane and hence allows the uptake of essential nutrients, such as K+, and efflux of toxic solutes, such as Na+.
Na+ is the main toxic ion in saline soils for most plants, including the major cereals (Tester and Davenport, 2003). Many cytosolic enzymes are inactivated by Na+ and high Na+ concentrations are detrimental to cellular metabolism. In addition, excess Na+ creates hyperosmotic stress, causing a range of effects, such as secondary oxidative damage (Mittler, 2002). To survive in a saline environment, plants have evolved a range of protective mechanisms. Recent studies, which have identified and characterized key enzymes in these protective pathways, especially in salt-tolerant species, have allowed the engineering of crop plants with increased salt tolerance. Several strategies for improvement of salt tolerance have been employed (Chinnusamy et al., 2005). The initial strategies were to increase the production of small osmolytes like mannitol, Glybetaine, Pro, and trehalose (Tarczynski et al., 1993; Kishor et al., 1995; Xu et al., 1996; Sakamoto and Murata, 2002). A more recent strategy has involved the overexpression of vacuolar and plasma membrane ion transporters, increasing Na+ exclusion from the cytosol. Overexpression of two Na+/H+ antiporters, NHX1 and SOS1 (for salt overly sensitive), has conferred salt tolerance to transgenic plants by sequestering Na+ in the vacuole or transporting Na+ across the plasma membrane out of the cell, respectively (Apse et al., 1999; Shi et al., 2002, 2003). AtNHX1 was identified in Arabidopsis (Arabidopsis thaliana) based on homology to the yeast (Saccharomyces cerevisiae) NHX1 transporter and was able to complement the salt-sensitive nhx1 yeast strain (Gaxiola et al., 1999). AtNHX1 is ubiquitously expressed in the plant and expression is induced by NaCl and abscisic acid (ABA; Shi and Zhu, 2002). AtSOS1 was initially identified as a locus required for salt tolerance in Arabidopsis and loss-of-function mutations in AtSOS1 caused plants to be extremely salt sensitive and to accumulate more Na+ than wild type in the shoots under salt stress (Wu et al., 1996; Shi et al., 2000). AtSOS1 transcription is specifically up-regulated upon NaCl stress and expression is mainly found in parenchyma cells bordering the xylem and in the epidermal cells surrounding the root tip (Shi et al., 2002).
Soil fungi are like plants in that they are exposed to similar challenges arising from close contact to a changing ionic environment. Phylogenetic analyses have shown that plant and fungal cells share many ion transporters; however, one major difference is the apparent absence of the ENA-type Na+-ATPases in vascular plants. In plants, maintaining nontoxic levels of Na+ appears to rely solely upon Na+/H+ antiporters. A surprising exception was the recent identification of two Na+-ATPases (PpENA1 and PpENA2) in the bryophyte, Physcomitrella patens (Benito and Rodríguez-Navarro, 2003). PpENA1 was shown to act as a Na+ pump when expressed heterologously in yeast and complemented a salt-sensitive yeast strain deficient in Na+ and K+ efflux (Benito and Rodríguez-Navarro, 2003). This implies that Physcomitrella has either gained the gene or the gene has been lost during the evolution of higher plants. In contrast, PpENA2 was unable to complement the salt-sensitive yeast strain.
Although PpENA1 activity has been characterized in a heterologous system by Benito and Rodríguez-Navarro (2003), little is known about the functional importance of Na+-ATPases in bryophytes. Furthermore, heterologous expression of membrane transporters can alter the properties of the transporter (e.g. the ion selectivity of the transporter; Pardo et al., 2006). For this reason, study of this gene's properties in situ provides a valuable test of function. Physcomitrella is an excellent plant model system because it performs homologous recombination with high efficiency. This allowed us to study the physiological role of PpENA1 in planta under sodium stress. From expression analyses and by generating knockouts via homologous recombination, we determined that PpENA1 plays an essential role in Physcomitrella under moderate salt stress by improving K+/Na+ homeostasis and allowing normal growth.
RESULTS
Induction of PpENA1 under NaCl Stress
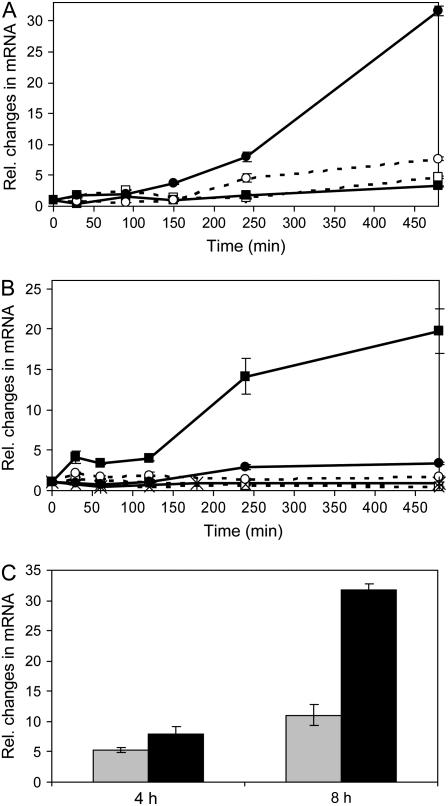
To determine over what time frame and under which salt concentrations PpENA1 plays a physiologically important role, quantitative reverse transcription (qRT)-PCR was performed on protonemata exposed to various NaCl concentrations (5–400 mm) for different times (30 min to 3 d). Exposing protonemata to low NaCl concentrations (530 mm) caused a 3- to 8-fold increase in expression. However, increasing the NaCl concentration to 60 mm caused a much higher, 30-fold, induction in PpENA1 mRNA levels after 8 h (Fig. 1A). Longer exposure to higher NaCl concentrations caused no further changes in PpENA1 expression (data not shown). The response to increasing Na+ was nonlinear, suggesting the induction response is saturated by 60 mm NaCl (Fig. 1, A and B). In contrast, exposure to KCl had no effect on PpENA1 expression (data not shown).
Figure 1.
Transcription pattern of PpENA1 under abiotic stress. A, Ten-day-old protonemata were exposed to different concentrations of NaCl. Tissue was harvested at different time points and mRNA extracted for qRT-PCR; 5 mm (white square), 10 mm (black square), 30 mm (white circle), and 60 mm (black circle). B, PpENA1 expression in protonemata exposed to different abiotic stresses. Protonemata were either not treated (hatched, white square), or exposed to 100 mm NaCl (black square), UV-B (black, cross), 50 μm ABA (hatched, cross), cold (hatched, white circle), or 50 μm methyl viologen (solid, black circle). C, The level of PpENA1 up-regulation after protonemata were exposed for 4 or 8 h to either 30 mm PEG (gray) or 60 mm NaCl (black), which impose a similar change in the osmotic potential.
qRT-PCR was also performed for PpENA2, but the level of expression was too low in the protonemata to allow proper analysis (Table I).
Table I.
mRNA copy number of PpENA1, PpENA2, and selected control genes in 10−9 L of cDNA
Total mRNA was extracted from either 10-d-old protonemata or 5-week-old gametophytes, quantified, adjusted to the same RNA concentration for cDNA synthesis, and used for RT-PCR.
| Tissue | PpTub1 | PpEF1α | PpHis-3 | PpMdhar2 | PpENA1a | PpENA2 |
|---|---|---|---|---|---|---|
| Protonema | 215 ± 59 | 312 ± 129 | 74 ± 11 | 30 ± 2 | 10 ± 2 | 1.5 ± 0.9 |
| Gametophyte | 309 ± 53 | 205 ± 49 | 127 ± 41 | 31 ± 2 | 49 ± 11 | 2.6 ± 1.4 |
Genes are expressed significantly different in the protonema compared to the gametophyte (n = 4; P < 0.01).
To determine whether the induction of PpENA1 was specific to Na+ stress, protonemata were exposed to NaCl, osmotic stress, cold, oxidative stress, or ABA. Only NaCl treatment caused significant induction of PpENA1 mRNA (Fig. 1B). To test whether the Na+-induced up-regulation of PpENA1 was due to ionic or osmotic effects, protonemata were exposed to a similar decrease in water potential using either 60 mm NaCl or 30 mm polyethylene glycol (PEG) 1,500 for 4 and 8 h (Fig. 1C). After 8 h, osmotic treatment led to a 10-fold induction, compared to a 30-fold induction with NaCl, indicating that the induction of PpENA1 expression under Na+ stress is due to a combination of osmotic and ionic effects.
Tissue-Specific Expression of PpENA1 and PpENA2
The expression of PpENA1 and PpENA2 in protonema and gametophyte were analyzed using RT-PCR and mRNA levels were compared to a selection of control genes (Table I). BLAST searches in the recently released draft of the genomic sequences (http://moss.nibb.ac.jp) confirmed that single copies of PpENA1 and PpENA2 are present in the Physcomitrella genome. The primers used for RT-PCR were designed to the 3′ end of the genes based on publicly available EST sequences. Besides the nonspliced PpENA2 mRNA, three putative splice variants have been identified (Benito and Rodríguez-Navarro, 2003). The primers used to detect PpENA2 expression were therefore designed to anneal upstream of the alternative splice sites to ensure detection of all mRNA variants. Very low expression of PpENA2 was detected in both protonema and gametophytes. In contrast, expression of PpENA1 was 7- or 19-fold higher than PpENA2 in protonema and gametophytes, respectively. A 5-fold higher level of expression of PpENA1 in gametophytes compared to protonema was observed. Neither PpENA2 nor the control genes showed this significant difference in expression levels between protonemal and gametophytic tissues (Table I).
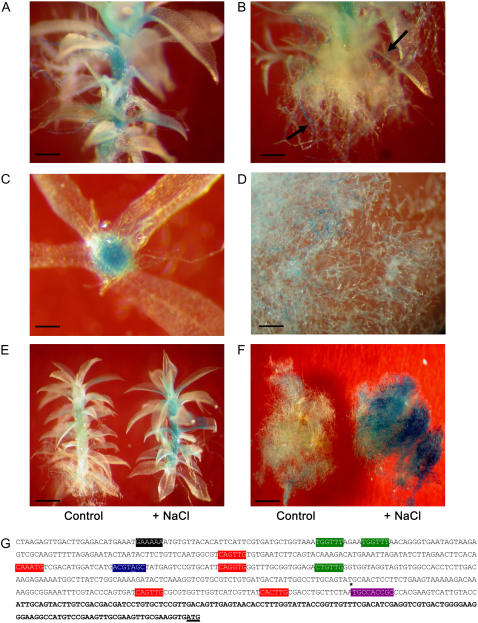
To determine whether PpENA1 expression was confined to certain cell types, wild-type Physcomitrella was transformed with a construct containing a PpENA1 promoter-GUS fusion. The promoter-GUS construct consisted of 1,311 bp upstream of the putative transcription start site and a 137-bp 5′-untranslated region (UTR). The publicly available EST sequences originate from a cDNA library with mainly full-length cDNAs and 5′ UTR is defined as the sequence included in the ESTs (BJ188922 and BJ191872; Nishiyama et al., 2003). Twelve independent stable transformants were obtained, of which eight showed consistent, but variable, levels of GUS staining. Differences in GUS levels are a common phenomenon observed in transgenic plants, including Physcomitrella (Kamisugi and Cuming, 2005). In the nonstressed gametophyte, GUS staining was confined to the stem, the basal part of the leaves, and to a small number of rhizoids originating from the base of the gametophyte (Fig. 2, A–C). No staining was present in the apical part of the leaf and staining was less pronounced in the top leaves compared to the leaves near the base of the gametophyte. GUS staining was ubiquitous in the protonemata, although some apical and subapical protonematal cells showed more intense staining than others (Fig. 2D). The expression pattern in the gametophyte under NaCl stress was determined by dividing colonies of gametophytic tissue and exposing one-half to medium containing 100 mm NaCl for 24 h. Only gametophytes originating from the same transformation event were compared. In all eight transformants, GUS staining became much more pronounced after NaCl stress, but the staining was still confined to the same tissue types as in the untreated control (Fig. 2E). When protonemata were exposed to 100 mm NaCl for 24 h, staining became very intense throughout the whole colony (Fig. 2F).
Figure 2.
Tissue-specific expression of PpENA1. The PpENA1 promoter (1,451 bp) was fused to a GUS-reporter gene and the construct transformed into Physcomitrella. A, GUS staining of a representative 5-week-old gametophyte grown on standard medium without NaCl, showing GUS staining in the stem and the basal part of the leaves. Bar = 0.4 mm. B, GUS staining of rhizoids from a nonstressed gametophyte. Arrows indicate the strongly stained filaments. Bar = 0.4 mm. C, Cross section of a gametophyte showing staining in the hydroids, cortex, and epidermis. Bar = 0.2 mm. D, Ubiquitous GUS staining in nonstressed protonemata. Bar = 0.4 mm. E, Comparison of GUS staining in two gametophytes originating from the same colony either left untreated or exposed to 100 mm NaCl for 24 h before GUS staining. The staining is significantly stronger, but still confined to the same tissues after salt stress. Bar = 0.6 mm. F, The effect of salt treatment on protonemata. A 10-d-old protonematal colony was divided in two and one-half of the colony was exposed to 100 mm NaCl for 24 h and the other half not treated. The strong GUS staining throughout the tissue is seen after NaCl stress. Bar = 1.5 mm. G, In silico PpENA1-promoter analysis. Putative cis-regulatory elements in the PpENA1 promoter were identified searching PLACE or NSITE-PL. Because the importance of regulatory elements generally decreases with increasing distance from the transcription start site, only the first 500 bp upstream of the putative transcription start site were included in the analysis. Putative GT-1 box (GAAAAA, black), Myb (WAACCA, CTGTTG, green), Myc (CANNTG, red), ABRE (ACGTGGC, blue), and CE-1 coupling elements (TGCCACCGC*, purple) are indicated. Mismatches in the consensus sequence are underlined and elements with an expected mean frequency <0.05 in 500 bp of random DNA sequence are marked with an asterisk. The putative 5′ UTR based on available EST sequences is shown in bold.
In silico analysis of the promoter was performed to identify putative cis-regulatory elements, which could be responsible for the observed expression pattern (Fig. 2G). Several elements were identified that are known to be up-regulated under dehydration stress (Myb, Myc, and CE-1 coupling elements; Yamaguchi-Shinozaki and Shinozaki, 2005). The only putative ABA regulatory element contained a mismatch at a conserved position, which is likely to make it nonfunctional (Shen et al., 2004). In addition, the consensus sequence for a GT-1 element shown to be involved in salt-dependent up-regulation was identified in the promoter (Park et al., 2004).
K+/Na+ Homeostasis in Wild-Type and ena1 Knockouts
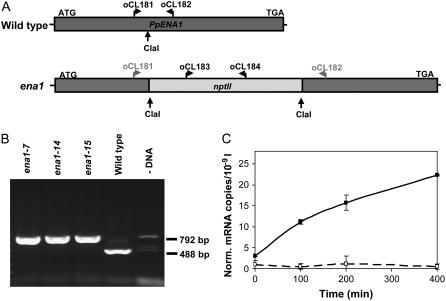
To determine the importance of PpENA1 in Physcomitrella, mutants lacking Na+-ATPase were generated. The nptII selective gene was inserted into a cDNA clone of PpENA1 using a ClaI site situated 1,189 bp downstream of the start ATG. This knockout cassette was used for transformation of wild-type Physcomitrella. After selection, 15 stable transformants were obtained. The stable transformants were PCR screened using genomic DNA as a template and primers specific to either side of the ClaI restriction site (Fig. 3A). A short PCR fragment (488 bp) would be generated if the wild-type PpENA1 gene was still present. A primer pair annealing to the nptII cassette was also included in the PCR reaction (792 bp) as a positive control. For three independent lines (ena1-7, -14, and -15), PCR reactions consistently gave the 792-bp nptII fragment and no 488-bp PpENA1 fragment (Fig. 3B). To confirm that the knockout cassette had replaced the PpENA1 gene, mRNA was purified from wild-type and ena1 protonemata exposed to 100 mm NaCl. A clear increase in PpENA1 mRNA was observed over time in the wild type, whereas the PpENA1 transcript levels remained at the background level in the ena1 mutants (Fig. 3C).
Figure 3.
Generating PpENA1 knockouts by homologous recombination. A, cDNA clone of PpENA1 was restriction enzyme digested at a unique ClaI site and the nptII gene inserted to generate a knockout cassette. B, The transformants were screened by PCR on genomic DNA using a mix of primers specific to PpENA1 (oCL181, oCL182) and nptII (oCL183, oCL184). A 488-bp band is expected in the wild type and a 792-bp band in the transformants where the PpENA1 gene has been replaced by the knockout cassette (ena1). Although all four primers anneal to the PpENA1 knockout cassette, the short fragment will be favored during the PCR. If the selective cassette is inserted in the genome, without replacing the PpENA1 gene, both the 488- and 792-bp PCR fragments will be present. C, PpENA1 mRNA levels. To ensure that PpENA1 was not expressed in the ena1 lines, mixed protonemata and gametophytes from wild type (black line) and ena1 (stippled line) were exposed to 100 mm NaCl and tissue harvested at different time points used for qRT-PCR. PpENA1 expression was normalized against four control genes (see “Materials and Methods”).
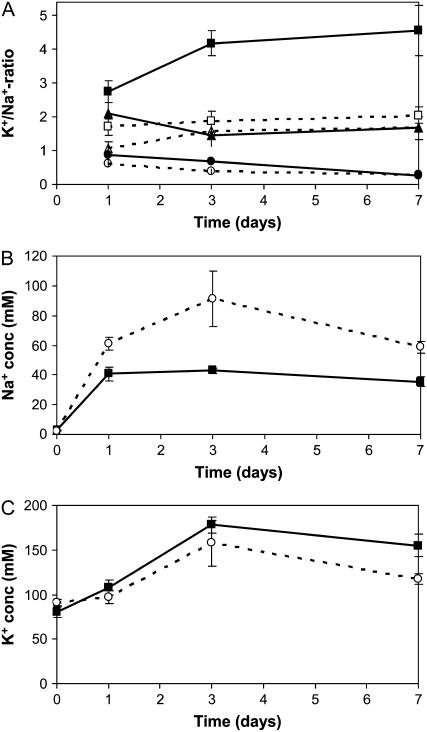
One of the reasons Na+ is toxic to plants is that Na+ can replace K+ in certain enzymes, rendering them nonfunctional. To test whether the lack of PpENA1 affected the K+-to-Na+ ratio in Physcomitrella under NaCl stress, gametophytes were placed on medium containing 100, 200, or 400 mm NaCl for 1, 3, or 7 d. The K+-to-Na+ ratio in the untreated wild type was 30 ± 6 (n = 6), whereas the ena1 lines had a slightly higher K+-to-Na+ ratio of 39 ± 1 (n = 9; P < 0.05). Within 1 d of exposure to high levels of NaCl, the K+-to-Na+ ratio decreased dramatically (Fig. 4A). At 200 and 400 mm NaCl, the K+-to-Na+ ratio remained low and, even after 7 d, no significant recovery was observed. Upon exposure to 100 mm NaCl, however, the wild type was able to improve the K+-to-Na+ ratio by 50% after 3 d. In contrast, no significant recovery was observed in the ena1 lines. To determine whether the ability of the wild-type plants to improve the K+-to-Na+ ratio at 100 mm was due to exclusion of Na+, the intracellular Na+ and K+ concentrations were measured. Both wild type and ena1 mutants experienced a dramatic increase in intracellular Na+ concentration in the first 24 h after transfer to 100 mm NaCl. In the wild-type gametophytes, however, no further increase was observed (Fig. 4B). In contrast, the intracellular Na+ concentration increased further by 60% to 100% in the ena1 mutants (Fig. 4C; P < 0.001) to be almost the same as the external Na+ concentration. The intracellular K+ concentration increased to the same extent in both wild type and ena1 mutants.
Figure 4.
The intracellular K+-to-Na+ ratio of wild-type and ena1 plants exposed to varying NaCl concentrations. A, Five-week-old gametophytes of wild type (black; n = 6) and ena1 (stippled; n = 9) were exposed to 100 (square), 200 (triangle), or 400 mm (circle) NaCl for up to 7 d. The data for ena1 represent the average for three independent ena1 lines. Intracellular Na+ (B) and K+ (C) concentrations in wild-type (black) and ena1 gametophytes (stippled) were calculated after exposure to 100 mm NaCl. Water content of the tissue was determined by weighing the gametophytes before and after drying.
Growth of Wild-Type and ena1 Plants under Salt Stress
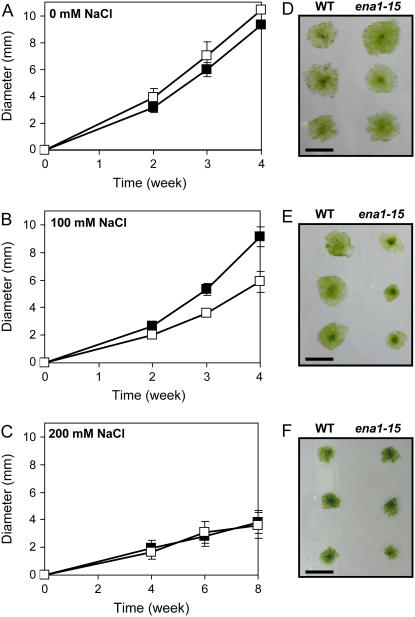
To establish whether the inability to improve the K+-to-Na+ ratio had a negative effect on fitness, wild-type and ena1 plants were grown on standard or salt-containing medium. The growth experiment was initiated using protoplasts to ensure that the starting material was homogeneous. Protoplasts were allowed to regenerate for 2 weeks before being transferred to medium containing 0, 100, or 200 mm NaCl. Growth was followed for 4 to 8 weeks by measuring the diameter of moss colonies.
On standard medium without added NaCl, no statistically significant difference in growth was observed between wild-type and ena1 plants, although ena1 appeared to grow marginally better (Fig. 5, A and D). When tissue was transferred to medium containing 100 mm NaCl, ena1 grew significantly slower and, after 4 weeks, the diameter of the ena1 colonies were 36% smaller than wild type (Fig. 5, B and C; P < 0.005). No growth impairment was observed for the wild type, which grew at a similar rate on standard or 100 mm NaCl medium. When tissue was transferred to medium containing 100 mm KCl and growth was followed for 4 weeks, no significant difference was observed between wild-type and ena1 plants (colony diameter of wild type = 6.8 ± 0.9 mm [n = 47]; ena1 = 6.9 ± 0.8 mm [n = 44]). When wild-type and ena1 tissue was transferred to medium containing 200 mm of NaCl or KCl, both lines grew significantly slower than on standard medium, but no difference in growth was observed. Even after an extended growth period, none of the colonies exposed to 200 mm NaCl or KCl formed thallus, but remained in the protonemal state.
Figure 5.
Effect of salt stress on growth of wild-type and ena1 plants. Protoplasts from wild type and ena1 were generated from 1-week-old protonemata to obtain identical starting material for the growth experiment. Protoplasts were allowed to recover for 2 weeks and then transferred to either standard medium (A) or medium with 100 (B) or 200 (C) mm NaCl. Growth of wild-type (black square) and ena1 plants (white square) was followed by measuring the diameter of 40 to 60 colonies from each treatment. Representative wild-type and ena1-15 colonies after 4 weeks of growth on standard medium (D) or medium with 100 mm NaCl (E) or after 8 weeks of growth on 200 mm NaCl (F). Bar = 1 cm.
Exposure to stressful levels of NaCl often leads to loss of chlorophyll and development of chlorotic areas. Although ena1 growth was impaired at 100 mm NaCl, the colonies did not appear paler or more chlorotic than wild type under these conditions. Total chlorophyll was extracted from wild-type and ena1 plants grown on standard or 100 mm NaCl medium to determine more precisely whether the total level or composition of chlorophyll was changed. The chlorophyll in nonstressed tissue was 21 ± 3 and 19 ± 1 mg chl/g dry weight for ena1 and wild type, respectively. No significant difference was observed between ena1 and wild-type plants grown at 100 mm NaCl having 18 ± 2 and 19 ± 1 mg chl/g dry weight, respectively. However, a small decrease in the chl a/b ratio was observed in the ena1 tissue compared to wild type grown at 100 mm being 2.01 ± 0.03 and 2.14 ± 0.09, respectively (P < 0.05).
DISCUSSION
Physcomitrella is tolerant of high levels of NaCl in the surrounding medium and can maintain growth at Na+ concentrations that would be detrimental to most vascular plants (Benito and Rodríguez-Navarro, 2003; Frank et al., 2005). A gene encoding an AtSOS1 homolog and two ENA-type ATPases has previously been identified in Physcomitrella (Benito and Rodríguez-Navarro, 2003). The physiological importance of having two Na+ efflux systems in plants is not known, but it is possible that PpENA1, like Na+-pumping ATPases in fungi, is responsible for the extrusion of Na+ into the surrounding medium and hence contributes to the high NaCl tolerance of Physcomitrella. In yeast, expression of the plasma membrane Na+/H+ antiporter sod2 from Schizosaccharomyces pombe can complement the salt-sensitive S. cerevisiae ena knockout line and the expression of ScENA can complement the sod2∷ura4 mutation in S. pombe (Bañuelos et al., 1995; Hahnenberger et al., 1996). Hence, under certain stress conditions, the two systems appear to be functionally redundant.
How Is PpENA1 Regulated?
The large (25- to 30-fold) transcriptional up-regulation, specifically in response to NaCl, implies an important functional role of PpENA1 in salinity tolerance (Fig. 1). This up-regulation is much more substantial than the 2- to 5-fold up-regulation of the salt-responsive Na+/H+ antiporters observed in vascular plants (Shi et al., 2002; Shi and Zhu, 2002; Yokoi et al., 2002; Kant et al., 2006). Except for osmotic stress, none of the other abiotic stresses tested led to increased PpENA1 transcript levels, confirming the specificity of the response. It is intriguing that exogenously added ABA had no effect on PpENA1 expression. One explanation could be that the regulation of PpENA1 has to be highly controlled and specific to salt stress. In silico analysis of the PpENA1 promoter revealed the presence of several putative regulatory elements. Although the elements used for the search mainly originated from vascular plants, it has previously been shown that ABA regulatory elements are conserved through plant evolution (Knight et al., 1995).
A 6-nucleotide element (GT-1) was recently identified in the promoter of a calmodulin from soybean (Glycine max; Park et al., 2004). The promoter was not responding to ABA, but the presence of this GT-1 element led to strong up-regulation upon salt treatment. An identical element was identified within the first 500 bp of the PpENA1 promoter, suggesting that this element could be the core-regulating cis-element.
In vascular plants, regulation of the two Na+/H+ antiporters (NHX1 and SOS1) appears to be very similar to PpENA1, although differences are also apparent. In Arabidopsis, AtNHX1 is up-regulated after exposure to high levels of NaCl or KCl (Gaxiola et al., 1999; Shi and Zhu, 2002; Yokoi et al., 2002). A similar induction of OsNHX1 is observed in rice (Oryza sativa) after treating plants with NaCl or with isoosmotic levels of mannitol. In accordance with PpENA1 results, osmotic stress only caused partial induction of OsNHX1 compared to ionic stress (Fukuda et al., 2004). Although this could suggest a similar type of regulation, a significant difference is that NHX1 up-regulation is induced by ABA (Shi and Zhu, 2002; Yokoi et al., 2002). ABA dependence could, however, be species specific because no increase in Na+-dependent H+ efflux was observed in the tonoplasts from Mesembryanthemum crystallinum after ABA treatment (Barkla et al., 1999). Like PpENA1, AtSOS1 expression is up-regulated by NaCl, but no up-regulation is observed after ABA treatment (Shi et al., 2000).
In vascular plants, the salt-specific response is also posttranslationally coordinated via the SOS pathway consisting of a Ser-Thr kinase (SOS2) and a calcium-binding protein (SOS3; Shi et al., 2000; Gong et al., 2004). It was recently shown that the SOS pathway not only regulates the activity of SOS1, but is also involved in regulating the vacuolar ion-exchange activity of AtNHX (Qiu et al., 2004). The finding of Physcomitrella ESTs showing 69% and 67% identity at the amino acid level to AtSOS2 (BJ187011, BJ589574) and AtSOS3 (BJ201990, BJ609228) opens the possibility that a SOS pathway also coordinates the salt response in Physcomitrella. If similar coordination occurs in Physcomitrella, it will be interesting in future studies to see whether PpENA1 activity is regulated by the SOS pathway.
What Is the Physiological Role of PpENA1?
The capacity of plants to maintain a high K+-to-Na+ ratio is likely to be a key determinant in salt tolerance. The steady-state level of both ions will ultimately reflect the sum of the many transporters involved in uptake, efflux, and compartmentation. To determine the physiological advantage of having Na+-ATPase in planta, the K+-to-Na+ ratio was determined at different NaCl concentrations (Fig. 4A). The salt stress experiment clearly showed that wild-type Physcomitrella was able to maintain a higher K+-to-Na+ ratio at 100 mm NaCl than the ena1 mutant. However, at higher NaCl concentrations, the advantage of having PpENA1 disappeared and the K+-to-Na+ ratio in wild type and ena1 was similar. Under severe stress (200–400 mm NaCl), it appears that PpENA1 is overwhelmed and no difference in the growth or K+-to-Na+ ratio of ena1 and wild-type plants could be observed (Figs. 4A and 5C).
Growth impairment correlated well with the intracellular K+-to-Na+ ratio; when the ratio was 3 or above, growth was similar to the nonstressed control, but at 2 or below, growth was impaired. A similar observation has been made in yeast, where normal growth was observed only when the K+-to-Na+ ratio was above 2 (Bañuelos et al., 1998). The K+ concentration increased in both wild-type and ena1 plants upon exposure to NaCl. This effect is not due to shrinking of the tissue because no changes in water content per unit dry weight were observed (data not shown) and the increased uptake or decreased efflux of K+ could be a compensating response to the increased intracellular levels of Na+. The growth difference was observed at pH 6.5, where PpSOS1 would be expected to be active and yet the presence of PpENA1 gave the wild type a growth advantage. A similar complementary advantage of having both the ENA-type ATPases (ENA1-4) and Na+/H+ antiporter (NHA1) under salt stress has been observed in yeast (Bañuelos et al., 1998). At alkaline pH, ENA1-4 became more important for salt tolerance. A growth experiment at pH 8 was also attempted with wild-type and ena1 plants in the presence of 100 or 200 mm NaCl. However, both lines were stressed and grew poorly even in the absence of NaCl (data not shown). The difficulty of cultivating and performing stress experiments with Physcomitrella at alkaline pH has also been noted by Benito and Rodríguez-Navarro (2003).
One of the reasons for Na+ toxicity is that Na+ can replace K+ in essential enzymes and thereby inactivate them. Under severe Na+ toxicity, metabolism and photosynthesis will be affected, leading to decreased levels of chlorophyll. No difference in chlorophyll content per unit dry weight was observed between stressed and nonstressed tissue or between ena1 and wild-type plants. Hence, it appears that, although ena1 plants contain more intracellular Na+, it is able to protect the sensitive components either by sequestering the Na+ in the vacuole, synthesizing compatible osmolytes, or up-regulating levels of protective proteins. Up-regulation of a dehydrin-like protein essential for the recovery of Physcomitrella after salt stress and of an enzyme involved in protection against reactive oxygen species has recently been shown (Lunde et al., 2006; Saavedra et al. 2006). The lack of visual signs of stress implies that, even in ena1 plants, Na+ does not build up to toxic concentrations in the cytosol and that both wild-type and ena1 plants are able to manage the intracellular Na+. However, the impaired growth of ena1 plants on salt suggests that the metabolic price for doing so is higher in ena1 than wild type.
In Arabidopsis, AtSOS1 is mainly expressed in the epidermis cells of the root tip, the pericycle, and the parenchyma cells bordering the xylem, suggesting that AtSOS1 is involved in xylem loading-unloading of Na+ and some exclusion into the soil (Shi et al., 2002). The tissue in Physcomitrella is generally only one cell layer thick and mineral transport does not occur in a vascular system, but either from cell to cell or in specialized cells, called hydroids, found in the stem of the gametophyte (Reski, 1998). GUS staining suggested that Na+ exclusion is particularly important in the rhizoids, which are in direct contact with the medium. Strong expression was also observed in the mature stem, in the central hydroids, the surrounding cortex, and the epidermal cell layer. In addition, the basal part of the one cell-layer-thick leaves was GUS stained. Although vascular plants and mosses are very different in their morphology, PpENA1 is expressed like AtSOS1 in tissue involved in nutrient transport, thereby minimizing the amount of Na+ reaching the photosynthetically active cells. The strong and ubiquitous expression of PpENA1 in protonemata, suggesting a more efficient Na+ extrusion from this tissue, could explain why Physcomitrella does not differentiate into gametophytes at high salt concentrations.
What Is the Functional Role of PpENA2?
The functional role, if any, of the PpENA1 homolog, PpENA2, is still unknown. However, it is clear that PpENA2 expression is significantly lower than PpENA1 and this low expression level is not able to complement the PpENA1 knockout at 100 mm NaCl (Table I; Fig. 5, B and E). This is in line with previous experiments showing that PpENA2, in contrast to PpENA1, could not complement a salt-sensitive yeast strain (ena1-4, nha1) on high-salt medium (Benito and Rodríguez-Navarro, 2003). BLAST searches among more than 100,000 Physcomitrella EST sequences reveal four EST sequences (BJ580267, BJ581369, BJ583232, BJ594559) representing the 3′ end of PpENA2. These sequences align with the genomic sequences and do not suggest that the PpENA2 gene contains any introns. However, three splice variants were identified in the 3′ end by Benito and Rodríguez-Navarro (2003) and, whereas the nonspliced PpENA2 gives rise to a protein of similar length to PpENA1 (972 and 967 amino acids, respectively), the three splice variants cause PpENA2 to be either truncated or extended. Although this needs to be investigated further, it is possible that the low level of PpENA2 expression combined with the alternative splicing of the 3′ end gives rise to very little functional product. Redundancy of the Na+-ATPases has also been observed in yeast (Garciadeblas et al., 1993).
Why Has PpENA1 Been Maintained in Physcomitrella?
When PpENA1 apparently gives Physcomitrella a clear selective advantage under moderate salt stress, it is intriguing that Na+-ATPases are absent in vascular plants. The presence of both a Na+,K+-ATPase and a SOS1 homolog in the red alga, Porphyra, living in the sea at high salinity and alkaline pH also suggests that having both types of pumps is important. The early land on which plants evolved was likely to be oligotrophic and low in Na+ (Rodríguez-Navarro and Rubio, 2006), so it is possible that Na+-ATPases were lost due to the low selective pressure. Although not statistically significant, wild-type plants appear to grow slightly more slowly than ena1 plants under nonstressful conditions. Hence, under these conditions, there might be a small growth penalty of having PpENA1, which could explain the evolutionary loss. Under nonstressful conditions, the wild type had a slightly lower K+-to-Na+ ratio than ena1 plants and it is possible that PpENA1 at low Na+ concentrations excludes essential K+ instead. From complementation studies in yeast, it appears that PpENA1, like ScENA, has a certain affinity for K+ (Benito and Rodríguez-Navarro, 2003). In yeast, the dual affinity for both Na+ and K+ was shown to lead to concomitant efflux of K+ with Na+ under low Na+ conditions (Benito et al., 2002). In contrast, no K+ efflux was observed at high Na+ concentrations. A similar scenario could be envisioned for PpENA1 in Physcomitrella.
One explanation for why PpENA1 apparently has been conserved in Physcomitrella could be that moss and fungi, unlike vascular plants, are able to undergo significant water loss and still be viable (Frank et al., 2005). Na+-ATPases could ensure that Na+ does not build up under these conditions. However, no up-regulation of PpENA1 was observed in Physcomitrella under severe drought imposed by placing protonemata on Whatman paper (data not shown).
The correlation between PpENA1 expression and salt tolerance raises the possibility that heterologous expression of PpENA1 may improve the salt tolerance of crop plants. Heterologous expression of AtNHX1 in diverse plants like Brassica, tomato (Solanum lycopersicum), and wheat (Triticum aestivum) allowed all three crops to grow under highly saline conditions and even produced a higher yield with similar properties to wild-type plants growing under nonstressful conditions (Zhang et al., 2001; Zhang and Blumwald, 2001; Xue et al., 2004). These results confirm that introducing a more efficient system for dealing with Na+ by changing a single trait is possible and can confer salt tolerance to crops not only in the greenhouse, but also in field trials (Xue et al., 2004). It will be of great interest to see whether the expression of PpENA1 in, for example, roots, where Na+ can be extruded to the large volume of growth medium, will be able to improve salt tolerance of vascular plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Physcomitrella patens (Hedw.) derived from a wild-type specimen collected in Gransden Wood in Huntingdonshire, UK (Ashton and Cove, 1977) was grown at 22°C on cellophane discs placed on solid minimal medium (Ashton et al., 1979), supplemented with ammonium tartrate when protonemata were required (0.5 g/L). Standard irradiance conditions were 16-h white light (fluorescent tubes; GRO-LUX; 100 μmol m−2 s−1) and 8-h darkness. Stress experiments were carried out on protonemata homogenized 10 d prior to the stress exposure or on 5-week-old gametophytes. Abiotic stress was induced by transferring the cellophane with Physcomitrella protonemata to medium or filter discs containing 100 μm methyl viologen (Sigma), 50 μm ABA (Sigma), 60 or 100 mm NaCl, 60 or 100 mm KCl, or 30 mm PEG 1,500 (Sigma). Supplemental CaCl2 was added to obtain the same level of free Ca2+ under all treatments. The amount of added CaCl2 required was calculated using MinTeq, version 2.30 (http://www.lwr.kth.se/English/OurSoftware/vminteq). The osmolarity of PEG 1,500 and NaCl was calculated according to the Merck Index (11th ed., 1989). Cold stress was imposed by placing the plates on ice. UV-B stress was imposed by placing Physcomitrella under a UVLMS-38 lamp fitted with an 8-W 302-nm tube (Ultra-Violet Products). The tissue was exposed to 0.23 mW cm−2 measured using an optical radiometer (MS-100; Ultra-Violet Products) fitted with a UV-B sensor (MP-131). Samples were frozen in liquid nitrogen upon harvesting and stored at −80°C until RNA was extracted.
The growth rate of wild-type and ena1 knockout lines under salt stress was determined by generating protoplasts (Schaefer and Zryd, 1997). After 2 weeks, the recovered protoplasts were transferred to standard medium, containing 100 or 200 mm of NaCl or KCl. Growth was followed for 4 to 8 weeks by measuring the diameter of 40 to 60 colonies per treatment.
RNA Extraction, cDNA Synthesis, and Real-Time qRT-PCR
Total RNA was extracted from protonemata or gametophytes using TRIzol (Invitrogen) according to the manufacturer's instructions. Purified RNA was treated with DNaseI using the DNA-free kit (Ambion). RNA integrity was checked on a 1% (w/v) agarose gel containing ethidium bromide. cDNA was synthesized using SuperScript III first-strand synthesis system (Invitrogen) and 2.5 μm oligo(dT) (18–20) primer according to the manufacturer's protocol. Four control genes—histone3 (BT003326), elongation factor 1α (P13905), actin2 (NP_188508), and tubulin β1 (NP_177706)—were used in each experiment, as described in Lunde et al. (2006). Primers specific to PpENA1 were designed, a forward primer (5′-AAGGCATTACCTGGGAGTGGAT) annealing to the coding region and a reverse primer (5′-ACAGCATGGGTGCGGATTCT) annealing to the 3′ UTR. Three splice forms of PpENA2 have been identified (Benito and Rodríguez-Navarro, 2003). PCR primers annealing to PpENA2 (5′-ATGGCGACCTTGGCAGTCAG, 5′-CAGGGTGGGAACTGGCACG) were designed to the coding region upstream of the splice sites to include all splice variants. Primers used for PpMdhar2 (monodehydroascorbate reductase) are described in Lunde et al. (2006). qRT-PCR analysis was performed essentially as described in Burton et al. (2004). For each primer pair, the specific PCR fragment was generated, purified, and quantified using a known DNA standard (pUC19/HpaII digested; Geneworks). The size in base pairs and identity were confirmed by sequencing. A dilution series of each PCR fragment was prepared covering seven orders of magnitude (107 − 101 copies of PCR fragment per microliter). Three replicates of each of the seven standard dilutions were included in each RT-PCR experiment to allow quantification of the corresponding cDNA using Rotor-Gene version 4.6 software (Corbett Research). qRT-PCR results were normalized to the four control genes described above to obtain the normalized mRNA copies per nanoliter cDNA. In the abiotic stress experiment, the relative changes in mRNA were obtained by dividing the qRT-PCR results (normalized mRNA copies per nanoliter cDNA) for the treated tissue with the qRT-PCR results for the corresponding untreated control tissue.
Genomic DNA Purification, Genomic Walking, Promoter Analyses, and BLAST Searches
Genomic DNA was purified as in Schlink and Reski (2002). The PpENA1 promoter sequence was obtained by genomic walking as described in Siebert et al. (1995). Genomic DNA (15 μg) was digested overnight with 100 units of DraI, NruI, HpaI, StuI, SmaI, EcoRI, HincII, or PvuII, phenol-chloroform purified and ligated to a 5′ adaptor to generate eight different templates. The 5′ sequences were obtained by nested PCR using gene-specific and adaptor-specific primers. The overlapping sequences obtained from the eight templates were aligned using Contig Express (Vector NTI 8). The promoter sequence was analyzed using NSITE-PL (www.softberry.com) and PLACE (http://www.dna.affrc.go.jp/PLACE). ESTs encoding PpENA1 and PpENA2 were obtained by BLAST searches in the two Physcomitrella databases (www.cosmoss.org; http://moss.nibb.ac.jp). Corresponding genomic sequences were obtained from BLAST searches of the draft release of the whole-genome shotgun sequences (http://moss.nibb.ac.jp).
Generating PpENA1 Knockouts and GUS-Promoter Fusions
To obtain stable knockouts of PpENA1, a cassette containing p35S-nptII-CamVter was excised from pMBL6 (http://www.biology.wustl.edu/moss/pmbl6.jpg) using ClaI and inserted into a unique ClaI site in the PpENA1 gene (Benito and Rodríguez-Navarro, 2003), generating pCL247. To increase the efficiency of homologous recombination, pCL247 was digested with EcoRI, which cuts twice in the vector backbone. Physcomitrella was transformed with 10 to 30 μg DNA and selected on geneticin (Gibco) according to Schaefer and Zryd (1997). To confirm integration, several independent PCR reactions were performed on genomic DNA purified from resistant colonies using primers oCL183 (5′-ATTGAACAAGATGGATTGCACGC) and oCL184 (5′-TCAGAAGAACTCGTCAAGAAGGCG) annealing to the selective cassette (p35S-nptII-CamVter) and primers oCL181 (5′-AACGGGTACGCTGACGGAGG) and oCL182 (5′-CGTCCCCAGAGATGCTATGT) annealing 5′ or 3′ of the ClaI site, respectively (Fig. 3). Genomic DNA was checked by PCR using primers annealing to the 5′ and 3′ end of PpENA2, respectively (5′-ATGGTCGACATCCGAGAGTTGA, 5′-CTACATAGTGTAAGGGGTGGTG), to ensure that PpENA2 was intact in the transformants.
The 1,448-bp DNA fragment directly upstream from the PpENA1 coding region was obtained by nested PCR using genomic DNA as template. The first PCR was done using oCL136 (CTTGTCCCCAGAGCCCTCC) and oCL138 (GCAGGTAGCTTCCAAATTCTCT), and the second PCR using oCL137 (CACCTTCGCAACTTCGCAAC) and oCL139 (CGTACACCAGCAGACATTAGTG). The fragment was cloned into PCR8/GW/TOPO (Invitrogen) and transferred to a destination vector containing the GUS reporter gene (pMDC162; Curtis and Grossniklaus, 2003) using Gateway technology according to the manufacturer's protocol (Invitrogen). The PpENA1 promoter-GUS construct, due to sequence-specific homologous recombination, is precisely targeted to the native PpENA1 gene. GUS staining was performed according to Jefferson (1989) and visualized using a stereomicroscope.
Intracellular Na+ and K+ Levels and Chlorophyll Content
Five-week-old gametophytes were transferred from standard medium to medium containing 100, 200, or 400 mm NaCl. In the standard medium, 1 mm CaCl2 was added and 1.3, 1.7, 2.0 mm CaCl2, respectively, were added to the NaCl-containing medium to keep the level of available Ca2+ constant (see above). After 1, 3, or 7 d of exposure, gametophytes were washed for 5 s in 10 mm CaCl2 and allowed to drain briefly on Whatman filter paper. The single short wash was selected to minimize leakage and loss of intracellular K+ and Na+ during washing. Preliminary experiments had shown that, for tissue exposed to 100 or 200 mm NaCl for 4 h, a 5-s wash led to a 75% and 87% reduction in NaCl compared to an unwashed control. A subsequent second washing step led to only a further 2% to 4% decrease in tissue Na+.
The water content of the gametophytes was determined by weighing the samples before and after drying overnight at 80°C. Na+ and K+ content was determined after digestion in 10 mm nitric acid overnight at 80°C and analyses using a flame photometer (Sherwood 420).
The total chlorophyll content of protonemata was determined by measuring the absorbance of acetone extracts at 652, 647, and 663 nm (Arnon, 1949; Frank et al., 2005). All P values for statistical significance were determined using a two-tailed, Student's t test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EF683141.
Acknowledgments
We acknowledge Dr. Jesse Machuka as part of the Physcomitrella EST Program at the University of Leeds and Washington University for supplying pMBL6. We also thank Neil J. Shirley for excellent technical assistance and Alonso Rodríguez-Navarro for kindly supplying the vector containing PpENA1.
This work was supported by the Australian Research Council and the State Government of South Australia (grants to D.P.D., A.K.J., M.T.) and by the Danish Research Council (grant to C.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christina Lunde (chlu@life.ku.dk).
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts—polyphenoloxidase in Beta vulgaris. Plant Physiol 24 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ (1977) Isolation and preliminary characterization of auxotrophic and analog resistant mutants of moss, Physcomitrella patens. Mol Gen Genet 154 87–95 [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss Physcomitrella patens using auxin and cytokinin resistant mutants. Planta 144 427–435 [DOI] [PubMed] [Google Scholar]
- Bañuelos MA, Quintero FJ, Rodríguez-Navarro A (1995) Functional expression of the ENA1(PMR2)-ATPase of Saccharomyces cerevisiae in Schizosaccharomyces pombe. Biochim Biophys Acta 1229 233–238 [DOI] [PubMed] [Google Scholar]
- Bañuelos MA, Sychrova H, Bleykasten-Grosshans C, Souciet JL, Potier S (1998) The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144 2749–2758 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Maldonado-Gama M, Pantoja O (1999) Abscisic acid induction of vacuolar H+-ATPase activity in Mesembryanthemum crystallinum is developmentally regulated. Plant Physiol 120 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito B, Garciadeblas B, Rodríguez-Navarro A (2002) Potassium- or sodium-efflux ATPase, a key enzyme in the evolution of fungi. An Microbiol (Rio J) 148 933–941 [DOI] [PubMed] [Google Scholar]
- Benito B, Rodríguez-Navarro A (2003) Molecular cloning and characterization of a sodium-pump ATPase of the moss Physcomitrella patens. Plant J 36 382–389 [DOI] [PubMed] [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley: quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45 437–448 [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Ratnadewi D, Reski R (2005) Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 220 384–394 [DOI] [PubMed] [Google Scholar]
- Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45 146–159 [DOI] [PubMed] [Google Scholar]
- Garciadeblas B, Rubio F, Quintero FJ, Bañuelos MA, Haro R, Rodríguez-Navarro A (1993) Differential expression of 2 genes encoding isoforms of the ATPase involved in sodium-efflux in Saccharomyces cerevisiae. Mol Gen Genet 236 363–368 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong DM, Guo Y, Schumaker KS, Zhu JK (2004) The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 134 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger KM, Jia ZP, Young PG (1996) Functional expression of the Schizosaccharomyces pombe Na+/H+ antiporter gene, sod2, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93 5031–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1989) The GUS reporter gene system. Nature 342 837–838 [DOI] [PubMed] [Google Scholar]
- Kamisugi Y, Cuming AC (2005) The evolution of the abscisic acid-response in land plants: comparative analysis of group 1 LEA gene expression in moss and cereals. Plant Mol Biol 59 723–737 [DOI] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T-halophila. Plant Cell Environ 29 1220–1234 [DOI] [PubMed] [Google Scholar]
- Kishor PBK, Hong ZL, Miao GH, Hu CAA, Verma DPS (1995) Overexpression of delta-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108 1387–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CD, Sehgal A, Atwal K, Wallace JC, Cove DJ, Coates D, Quatrano RS, Bahadur S, Stockley PG, Cuming AC (1995) Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell 7 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde C, Baumann U, Shirley NJ, Drew DP, Fincher GB (2006) Gene structure and expression pattern analysis of three monodehydroascorbate reductase (Mdhar) genes in Physcomitrella patens: implications for the evolution of the MDHAR family in plants. Plant Mol Biol 60 259–275 [DOI] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Munns R, Cramer R, Ball MC (1999) Interactions between rising CO2, soil salinity and plant growth. In Y Luo, HA Mooney, eds, Carbon Dioxide and Environmental Stress. Academic Press, London, pp 139–167
- Nishiyama T, Fujita T, Shin-I T, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K, et al (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implications for land plant evolution. Proc Natl Acad Sci USA 100 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo JM, Cubero B, Leidi EO, Quintero FJ (2006) Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot 57 1181–1199 [DOI] [PubMed] [Google Scholar]
- Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, et al (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135 2150–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279 207–215 [DOI] [PubMed] [Google Scholar]
- Reski R (1998) Development, genetics and molecular biology of mosses. Bot Acta 111 1–15 [Google Scholar]
- Rodríguez-Navarro A, Rubio F (2006) High-affinity potassium and sodium transport systems in plants. J Exp Bot 57 1149–1160 [DOI] [PubMed] [Google Scholar]
- Saavedra L, Svensson J, Carballo V, Izmendi D, Welin B, Vidal S (2006) A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. Plant J 45 237–249 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Murata N (2002) The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant Cell Environ 25 163–171 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zryd JP (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schlink K, Reski R (2002) Preparing high-quality DNA from moss. Plant Mol Biol Rep 20 423a–423f [Google Scholar]
- Shen JQ, Casaretto JA, Zhang P, Ho THD (2004) Functional definition of ABA-response complexes: the promoter units necessary and sufficient for ABA induction of gene expression in barley (Hordeum vulgare L.). Plant Mol Biol 54 111–124 [DOI] [PubMed] [Google Scholar]
- Shi HZ, Ishitani M, Kim CS, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HZ, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21 81–85 [DOI] [PubMed] [Google Scholar]
- Shi HZ, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HZ, Zhu JK (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol 50 543–550 [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarczynski MC, Jensen RG, Bohnert HJ (1993) Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 259 508–510 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DP, Duan XL, Wang BY, Hong BM, Ho THD, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM (2004) Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167 849–859 [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10 88–94 [DOI] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30 529–539 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]