Abstract
Bacteria indigenous to water distribution systems were used to grow multispecies biofilms within continuous-flow slide chambers. Six flow chambers were also inoculated with an Escherichia coli isolate obtained from potable water. The effect of disinfectants on bacterial populations was determined after exposure of established biofilms to 1 ppm of hypochlorous acid (ClOH) for 67 min or 4 ppm of monochloramine (NH2Cl) for 155 min. To test the ability of bacterial populations to initiate biofilm formation in the presence of disinfectants, we assessed the biofilms after 2 weeks of exposure to residual concentrations of 0.2 ppm of ClOH or 4 ppm of NH2Cl. Lastly, to determine the effect of recommended residual concentrations on newly established biofilms, we treated systems with 0.2 ppm of ClOH after 5 days of growth in the absence of disinfectant. Whole-cell in situ hybridizations using fluorescently tagged, 16S rRNA-targeted oligonucleotide probes performed on cryosectioned biofilms permitted the direct observation of metabolically active bacterial populations, including certain phylogenetic groups and species. The results of these studies confirmed the resistance of established bacterial biofilms to treatment with recommended levels of disinfectants. Specifically, Legionella pneumophila, E. coli, and β and δ proteobacteria were identified within biofilms both before and after treatment. Furthermore, although it was undetected using routine monitoring techniques, the observation of rRNA-containing E. coli within biofilms demonstrated not only survival but also metabolic activity of this organism within the model distribution systems. The persistence of diverse bacterial species within disinfectant-treated biofilms suggests that current testing practices underestimate the risk to immunocompromised individuals of contracting waterborne disease.
Assessment of the microbiological safety of drinking water is based largely on the routine monitoring of water supplies for the presence of total coliforms and Escherichia coli. Detection of E. coli is considered indicative of recent fecal contamination and of the potential presence of enteric pathogens, while the presence of total coliforms is indicative of poor water quality.
Whereas routine water quality measurements assess the presence of planktonic bacteria, the vast majority of bacteria indigenous to aquatic environments exist attached to solid particles or surfaces. Within water distribution systems, significant bacterial populations exist as complex, structurally heterogeneous biofilms attached to pipe surfaces. Residence within these complex matrices provides organisms with higher localized nutrient concentrations than are commonly found in drinking waters (10, 14, 18, 22), and recent studies have shown that attached bacteria are more metabolically active than are their free-living counterparts (26, 29). Furthermore, biofilms afford bacteria significant protection from disinfecting agents (3, 31, 35), including hypochlorous acid and monochloramine. Biofilms are dynamic in nature, and portions are frequently sloughed off pipe surfaces for a variety of reasons. They can then provide an effective inoculum for previously disinfected waters.
The ability of total coliforms and E. coli to survive in biofilms is of marked significance to the water treatment industry, not only because the detection of these organisms in distribution water gives an incorrect indication of recent fecal contamination but also because the persistence of these organisms and their release from biofilms may mask true breakthrough events in water treatment. Several lines of evidence suggest the long-term survival of total coliforms and/or E. coli in distribution system biofilms. Among these are the recovery of total coliforms from distribution systems in which water leaving the treatment plant tested negative for coliforms (20, 21); the observation that environmentally derived strains of total coliforms and E. coli can grow in unsupplemented distribution system water (7); and the recovery of increased numbers of E. coli organisms from drinking water distribution pilot plants after the calculated inoculum should have theoretically washed out (12). Other studies have shown that E. coli can attach and become incorporated into biofilms within model distribution systems (8, 11), as well as into biofilms derived from groundwater populations (2).
In recent years, the advent of molecular detection techniques has greatly facilitated our understanding of bacteria indigenous to the environment. Whole-cell in situ hybridizations using fluorescently labeled oligonucleotides targeted to regions of the 16S rRNA molecule have permitted the identification of environmental bacteria with little disturbance of their surroundings (16, 29, 30). Because the ribosomal content of bacteria can be correlated with the growth rate, metabolic activity in environmental bacteria can be assessed by measuring fluorescence after hybridization (32). Whole-cell in situ hybridizations have been used for the direct identification of β and γ proteobacteria in biofilms formed on glass slides in drinking water (26) and for the examination of biofilm structure using embedded biofilm sections (36, 37, 39).
The studies reported here used whole-cell in situ hybridization on biofilm sections obtained from model distribution systems to assess the survival and growth capabilities of E. coli within multispecies biofilms after exposure to chlorine disinfectants. The responses of other organisms indigenous to water distribution systems, including Legionella pneumophila, sulfate-reducing bacteria (SRB) belonging to the δ subgroup of the proteobacteria, and the β proteobacteria, were also determined.
MATERIALS AND METHODS
Model distribution systems.
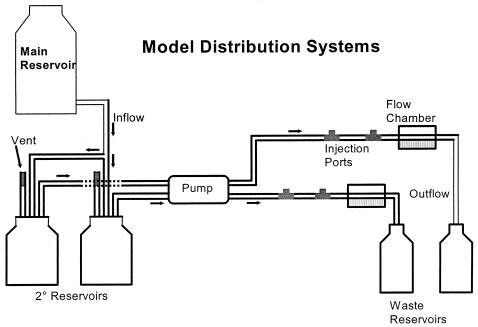
A 50-liter polypropylene carboy (Nalgene Nunc International, Rochester, N.Y.), acting as a primary reservoir, supplied irrigant to the model systems through a series of Tygon tubing, Y-connectors, and pipettes. Up to six 1-liter Wheaton bottles were used in parallel as secondary reservoirs. The system flow rate was controlled by a peristaltic pump (Masterflex L/S [Cole Parmer, Inc., Vernon Hills, Ill.] or Rabbit peristaltic pump [Rainin Instrument Co., Inc., Woburn, Mass.]) at 0.2 ml/min or a velocity of 1.2 mm/s. Two injection ports, located between the peristaltic pump and flow chamber, were constructed by filling the center opening of a T connector with silicone adhesive.
Flow chambers (5) comprised a glass slide (76 by 25 by 1.5 mm), two coverslips (50 by 22 mm, no. 1 thickness), and a ductile iron coupon (55 by 18 by 1.4 mm [BioSurface Technologies, Inc., Bozeman, Mont.]). Glass components were cleaned with acid-alcohol and ductile iron coupons were cleaned with 95% ethanol prior to assembly. The chambers were sealed using silicone adhesive and sterilized by baking at 160°C for at least 4 h. Silicone tubing (inner diameter, 1.02 mm) provided inflow and outflow to and from the chambers, which had an approximate volume of 0.2 ml (0.15 by 0.3 by 4.5 cm). Figure 1 presents a diagram of the model systems used in these studies.
FIG. 1.
Diagram of reservoirs and flow chambers assembled for the growth of biofilms formed by water distribution system populations.
Flow chambers were irrigated at room temperature (RT) with up to 30 liters of autoclaved tap water buffered with 0.54 mM phosphate (0.48 mM NaH2PO4, 0.06 mM Na2HPO4 [pH 7.0 to 7.5]; Sigma Chemical Co., St. Louis, Mo.) for 2 weeks on a flat surface in the dark. The city of Albany, N.Y., maintains distribution water at pH 8.5 for corrosion control. To increase the efficacy of chlorine disinfectant in a consistent manner, the pH within the systems was maintained between 7.0 and 7.5.
The initial sterility of each model system was ascertained by plating subsamples collected from the waste reservoirs onto R2A agar (Difco Laboratories, Detroit, Mich.) (34) and incubating them at RT for 10 days. Subsamples were also fixed with 3.7% formaldehyde and examined microscopically after fluorescent whole-cell hybridizations.
Inoculation of model systems.
To concentrate bacteria indigenous to water distribution systems, we filtered 5 liters of tap water onto sterile, prepackaged Millipore GS membrane filters (pore size, 0.2 μm [Millipore Corp., Bedford, Mass.]). Filters were placed in a sterile 50-ml disposable centrifuge tube containing 10 ml of tap water adjusted to pH 7 to 7.5 with 0.5 mM phosphate buffer and vortexed for 1 min to resuspend the particulate material. Flow chambers were slowly inoculated with 1 ml of tap water particulate by injection through surface-sterilized injection ports. Inoculation of flow chambers with distribution system concentrates was typically repeated on each of the first 5 to 7 days of each 2-week incubation period.
For experiments investigating the incorporation of E. coli into biofilms, strain 01571 was grown overnight in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) at 35°C. To acclimate the E. coli to the low-nutrient conditions within the flow chambers, 2 ml of log-phase culture was centrifuged at 11,750 × g for 5 min. The resulting pellet was resuspended in autoclaved tap water, inoculated into approximately 50 ml of autoclaved tap water, and incubated overnight at RT with shaking. On the following day, for each chamber, 1 ml of preconditioned E. coli was centrifuged at 11,750 × g for 5 min and resuspended in 1 ml of tap water containing the concentrated distribution system bacteria described above. A similarly prepared mixture of distribution system populations and E. coli was injected into the flow chambers through the injection port on four successive days.
Treatment with disinfectants.
Secondary reservoirs (Wheaton bottles) were filled with 1 liter of buffered tap water before the addition of either hypochlorous acid (ClOH; The Clorox Co., Oakland, Calif.) or monochloramine (NH2Cl). ClOH and NH2Cl concentrations in stock solutions were determined by the N,N-diethyl-p-phenylenediamine (DPD) ferrous titrimetric method (1) with one modification, namely, that HgCl2 was omitted from the phosphate buffer due to its toxicity (9). A scaled-down version of the titration method was used to test flow chamber discharge by addition of 5 ml of wastewater to a flask containing 250 μl each of phosphate buffer and DPD indicator solution. This mixture was titrated with a 1/20 dilution of ferrous ammonium sulfate titrant. Measurements were obtained immediately after collection of the samples.
NH2Cl was produced by reacting ammonium chloride (Sigma Chemical Co.) and hypochlorite ion (OCl−) in a 3:1 molar ratio at pH 10 (17). Concentrations in stock solutions were determined immediately prior to each experiment.
Enumeration of bacteria indigenous to tap water.
To estimate the number of culturable heterotrophic bacteria present in tap water concentrates, we serially diluted subsamples of each suspension 10-fold in buffered dilution water (1), spread plated them onto R2A agar, and incubated them for 10 days at RT before counting the colonies. For determination of the number of rRNA-containing organisms, a second subsample of each concentrate was fixed using 3.7% formaldehyde and stored at 4°C for subsequent use in the whole-cell hybridization procedure.
Tap water was analyzed for the presence of fecal coliforms using membrane filtration onto m-FC agar (1). On occasion, total coliforms and E. coli were assayed in the chamber outflows using the Colilert method in the Quanti-Tray format (IDEXX Laboratories, Inc., Westbrook, Maine).
Cryoembedding and sectioning.
Biofilms were embedded within flow chambers by using Tissue-Tek O.C.T. embedding compound (Sakura Finetek U.S.A. Inc., Torrance, Calif.), frozen on dry ice, and stored at −80°C (39). Before sectioning of the biofilms the flow chambers were snapped apart to release the embedded biofilm.
Cross-sections (6 μm thick) of embedded biofilm were obtained using a cryostat (Miles, Inc., Elkhart, Ind.). The sections were collected on baked Teflon-welled slides (14-mm-diameter wells [Erie Scientific, Portsmouth, N.H.]) that had been treated with 0.1% poly-l-lysine (Sigma) for 5 min at RT.
Oligonucleotide probe synthesis and labeling.
Oligodeoxynucleotide probes having a six-carbon linker containing a free amino terminus (Aminolink 2 [Applied Biosystems, Foster City, Calif.]) attached to the 5′ end were synthesized by the Molecular Genetics Core of the Wadsworth Center using a DNA synthesizer (model 8909; Perceptive Biosystems, Framingham, Mass.). Probes were deblocked and stored at −20°C until needed for labeling reactions. Methods for labeling probes with tetramethyl rhodamine-5 (and 6)-isothiocyanate (TRITC; Molecular Probes, Inc., Eugene, Oreg.) have been described previously (4). The probes used in these studies were as follows: primer 342, 5′-CTG CTG CSY CCC GTAG (38); Beta, 5′-TCA CTG CTA CAC GYG (positions 680 to 694); Delta, 5′-CGY GCG CCR CTY TACT (positions 90 to 105); Leg5, 5′-ACC GGA AAT TCC ACT ACC (positions 667-684); Leg8, 5′-GCT GCG CCA CTA ATT ATT T (positions 845 to 863); and Eco3, 5′-ACT TTA CTC CCT TCC TCC CCG (positions 443 to 463). Primer 342 is a universal probe, whereas Beta and Delta are specific for some members of the beta- and delta-proteobacteria. Leg5 and Leg8 are specific for Legionella spp., and Eco3 allowed identification of E. coli. Probe specificity was empirically determined using the Ribosomal Database Project (25).
Bacterial cultures and growth conditions.
The bacterial strains used in these studies included strains Acinetobacter calcoaceticus ATCC 23055, Agrobacterium radiobacter ATCC 19358, Comamonas testosteroni ATCC 11996, Salmonella enterica serovar Typhimurium ATCC 14028, Enterobacter aerogenes ATCC 33457 and 13048, Enterobacter cloacae ATCC 33457, Escherichia coli ATCC 43651, 25922 and 795, E. fergusonii ATCC 35469, E. vulneris ATCC 33821, E. blattae ATCC 29907, and E. hermannii ATCC 33650. Aureobacterium sp., Arthrobacter globiformis, Bacillus subtilis, Cytophaga pectinovorans, and Pseudomonas fluorescens were provided by S. Nierzwicki-Bauer, at Rensselaer Polytechnic Institute (Troy, N.Y.). Thames Water Utilities (Thames, United Kingdom) supplied 47 E. coli isolates, as well as 77 other coliforms, including several isolates each of Klebsiella terrigena, K. ornithinolytica, K. oxytoca, K. pneumoniae, K. ascorbata, Citrobacter freundii, Enterobacter aerogenes, E. sakazakii, E. intermedium, E. cloacae, E. agglomerans, E. amnigenus, E. taylorae, and Aeromonas schubertii. Desulfovibrio desulfuricans was provided by M. Wolin of the Wadsworth Center, Albany, N.Y. Legionella pneumophila isolates 1164, 1169, and 1818 were provided by D. Schoonmaker-Bopp, also of the Wadsworth Center. E. coli 01571 was isolated from potable water collected in Steuben County, N.Y., in July 1998. The majority of cultures were maintained on standard plate count agar at 4°C and transferred monthly. For whole-cell hybridizations, bacteria were grown in BHI broth at RT or 37°C. Log-phase cultures were fixed in 3.7% formaldehyde and prepared for the hybridization procedure. L. pneumophila isolates were grown on Legionella agar base medium (Difco Laboratories) supplemented with 0.2 g of l-cysteine-HCl and 0.125 g of ferric pyrophosphate per liter and adjusted to pH 7.1 to 7.2. Cultures were incubated at 37°C in a candle extinction jar (13). D. desulfuricans was grown anaerobically at 37°C in serum bottles containing LSTY broth reduced with 0.05% dithiothreitol (24). D. desulfuricans was transferred to new medium in serum bottles by using a sterile syringe and needle prereduced using 1.25% cysteine sulfide.
Fluorescent in situ hybridizations.
16S rRNA-targeted oligodeoxynucleotide probes were tested for specificity by hybridization to the laboratory cultures described above. Fixed bacterial cultures were washed and prepared on gelatin-coated slides as described previously (4). Since L. pneumophila isolates were grown on agar medium instead of broth, an inoculating loopful of organisms from an isolated colony was placed in 1 ml of phosphate-buffered saline (PBS) containing 3.7% formaldehyde and mixed well.
Well slides containing biofilm sections were held for 1 h at RT in Coplin jars containing PBS (pH 7.4) (Sigma Chemical Co.) with 3.7% formaldehyde and then dehydrated by serial exposure to 50, 80, and 100% ethanol for 3 min each (32). The slides were then treated for 1 min in 90:10 ethanol-formaldehyde and given a 5-min wash in filtered (0.2-μm pore size) and autoclaved distilled H2O.
Hybridizations using the universal 342, Beta, or Delta probes were carried out at 37°C as described previously (4), with either 5 μg of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes, Inc.) per ml or 2.5 μg of 7-diethylamino-3-4′-isothiocyanatophenyl)-4-methylcoumarin (CPI) (Molecular Probes, Inc.) per ml as a total-cell counterstain. The Eco3 probe, which has a Tm of 63°C, was resuspended in a mixture of 400 μl of formamide and 600 μl of hybridization mix for subsequent overnight hybridization at 37°C. For biofilm sections, the wash times were increased to three 30-min washes in 1× SET.
Planktonic distribution system populations or laboratory cultures underwent the same hybridization procedure, with the omission of the 1-h formaldehyde incubation and serial ethanol washes.
To confirm the presence of Legionella species in the biofilms, fluorescent-antibody staining was performed on biofilm cryosections by using fluorescein-labeled goat anti-L. pneumophila serogroup 1 to 14 immunoglobulin G and goat anti-Legionella species b to p immunoglobulin G (Monoclonal Technologies, Inc., Atlanta, Ga.) after hybridization with Leg5 and Leg8. After the third 30-min wash in 1× SET, the slides were air dried. The slide well was covered with antibody solution (Monoclonal Technologies, Inc.) containing 1% whole-goat serum (Organon Teknika Corp., West Chester, Pa.) and incubated at RT in the dark for 30 min in a box containing a moistened Kim Wipe. The slide was washed twice for 5 min each in a Coplin jar containing PBS.
Epifluorescence microscopy.
The results of hybridizations were examined using a Leitz DMR-B epifluorescence microscope (Leica Mikroskopie and System GmbH) equipped with a 100-W power supply, phase and filter sets appropriate for rhodamine (no. 513810 [Leica Inc., Deerfield, Ill.]), fluorescein (no. 513812), and DAPI (or CPI) (no. 513808) detection. Bacteria were viewed through a 100×/1.3 Plan Fluotar oil immersion objective. Photomicrographs were obtained using a Nikon UFX-DX automatic camera and Fujichrome 200-speed color slide film.
Effects of chlorine on whole-cell hybridization.
To determine the ability of the whole-cell hybridization procedure to detect the effects of chlorination on metabolically active (rRNA-containing) cells, chlorine was added to pure cultures of E. coli and bacteria obtained from distribution water concentrates and microscopically evaluated over time. E. coli (Thames Water Utilities isolate 159), grown to log phase in BHI broth and washed twice in sterile water, and bacteria obtained from distribution water concentrates were exposed to 1 ppm of ClOH. Subsamples were removed at 0, 10, and 30 min and immediately placed in sodium thiosulfate (Na2S2O3; final concentration 0.01%) before undergoing the hybridization procedure with the universal 342 probe. Control (untreated) samples of E. coli and distribution water bacteria were analyzed in parallel.
RESULTS
Specificity of oligonucleotide probes.
All E. coli strains, including ATCC cultures and environmental isolates, were detected by epifluorescence microscopy after hybridization with the Eco3 probe. Of the 77 other coliform strains used as negative controls, only two environmental strains of K. oxytoca hybridized with this probe. Biochemical identification of these strains was confirmed using API test strips (bioMérieux, Inc., Durham, N.C.). The Leg5, Leg8, Delta, and Beta probes hybridized with only the appropriate positive controls. The universal probe 342 hybridized with the 16S rRNA of all bacterial isolates and type strains tested.
Effects of chlorine on whole-cell hybridization.
The ability of the whole-cell hybridization procedure to measure the effects of chlorination on bacterial metabolic activity was tested on laboratory-grown and distribution system bacteria. As indicated in Table 1, the hybridization procedure has the sensitivity to measure the effect of 1 ppm of ClOH on bacteria within 10 min of exposure. Log-phase, laboratory-grown E. coli cells, which showed a 2-log-unit reduction in the numbers of bacteria detected, were more sensitive to chlorine treatment than were unamended bacterial populations freshly obtained from the distribution system.
TABLE 1.
Effects of 1 ppm of ClOH on the hybridization efficiency of Universal 342 probe to 16S rRNA in planktonic bacteriaa
| Time (min) | % of cells positive after hybridizationc
|
|||
|---|---|---|---|---|
| E. coli control | E. coli, 1 ppm of ClOH | TWb control | TW, 1 ppm of ClOH | |
| 0 | 108.5 ± 2.12 | 104.5 ± 2.12 | 20.7 ± 15.3 | 31.7 ± 7.14 |
| 10 | 103.5 ± 0.707 | 0.63 ± 0.891 | 25.8 ± 11.5 | 4.25 ± 4.60 |
| 30 | 98.2 ± 0.848 | 1.69 ± 2.39 | 39.9 ± 22.3 | 4.32 ± 2.66 |
The E. coli isolate, obtained from Thames Water Utilities, was grown to log phase in BHI broth and washed twice prior to treatment. The distribution system bacteria were treated and analyzed directly after they were sampled.
TW, bacteria present in tap water filter concentrates.
Two experiments were averaged, and the mean ± standard deviation is shown.
Metabolic activity within distribution system biofilms.
The results of fluorescent in situ hybridizations indicated that within distribution water concentrates, an average of 30.72% (n = 23, where n is the number of concentrated water samples examined, with a minimum of 200 bacteria enumerated per sample) of the bacterial cells staining with CPI also contained detectable amounts of rRNA. Percentages were not seasonal, ranging from a low of 7.94% in April 1999 to a high of 55.68% in April 1998. Each milliliter of distribution water concentrates, freshly prepared in an identical manner daily, contained an average of 6.7 × 106 cells, based on microscopic enumeration. Determining viability from CFU on R2A plates indicated that approximately 27.5% of total distribution water bacteria were culturable under the conditions used. In contrast, in situ hybridizations performed on cross-sections of distribution system biofilms obtained from flow chambers after a 2-week incubation period indicated that many bacteria contained detectable levels of rRNA after hybridization with the universal 342 probe (Fig. 2A and B). Biofilms were structurally heterogeneous, exhibiting pillars and interstitial channels. However, position within the biofilm, e.g., proximity to either the substrate coupon or water flow, had no apparent effect on the percentage of cells detected. Likewise, treatment with a variety of disinfectant regimens had no effect on the percentages of cells observed. Treatment regimens included exposure of biofilms to (i) 1 ppm of free chlorine for 67 min (1 CT), (ii) a residual of 0.2 ppm of free chlorine after the biofilms grew for 5 days without disinfectant, and (iii) 4 ppm of monochloramine for 155 min (Table 2). However, when flow chambers were treated continuously with a 2 ppm of NH2Cl, 4 ppm of NH2Cl, or 0.2 ppm of ClOH residual for 2 weeks, biofilm was not detected after embedding and sectioning.
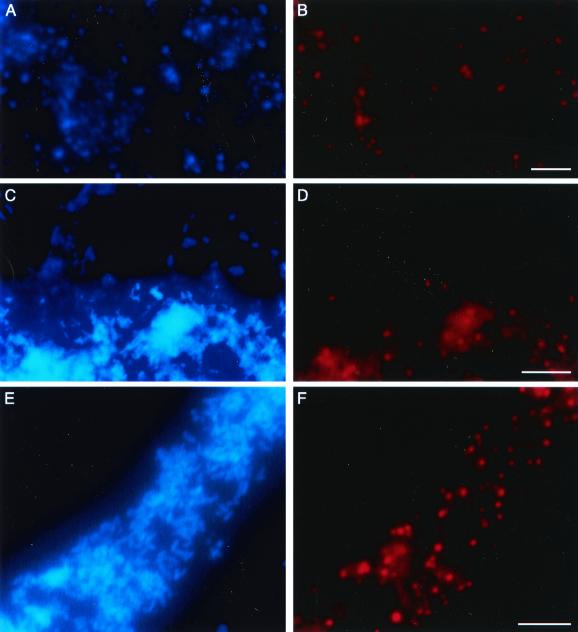
FIG. 2.
Micrographs of 2-week-old biofilms. (A, C, and E) Total bacteria were stained with CPI. (B, D, and F) Same fields; bacteria were detected after hybridization with a rhodamine-labeled 16S rRNA-targeted universal oligonucleotide probe (B), a probe specific for members of the δ proteobacteria (biofilm 1) (D), or a Legionella-specific probe (biofilm 10) (F). Bar, 5 μm.
TABLE 2.
Biofilm conditions under which the presence of E. coli and other populations were observeda
| Biofilm designation | Disinfectant concn and exposure time | No. of E. coli CFU inoculated |
|---|---|---|
| 1 | None | 0 |
| 2 | None | 0 |
| 3 | None | 0 |
| 4 | 1 CT ClOH (1 ppm for 67 min) | 0 |
| 5 | 0.2 ppm of residual ClOH starting on day 6 | 0 |
| 6 | 0.2 ppm of residual ClOH | 0 |
| 7 | None | 106 |
| 8 | 1 CT ClOH | 106 |
| 9 | 1 CT ClOH | 106 |
| 10 | 4 ppm of NH2Cl for 155 min | 106 |
| 11 | 2 ppm of residual NH2Cl | 106 |
| 12 | 4 ppm of residual NH2Cl | 106 |
Disinfectant type, concentration, and exposure time are summarized for 12 biofilms that were cryoembedded and sectioned for whole-cell hybridizations. E. coli 01571 was inoculated into biofilms 7 to 12.
Presence of SRB in biofilms.
Bacteria that were detected after hybridization with the Delta probe were considered to be SRB. SRB were present in significant quantities within each biofilm, even after exposure to free chlorine or monochloramine. In biofilm 1, the majority of SRB detected were large, short rods (Fig. 2D). In biofilms 4 and 7 to 10, the cells were smaller than those previously detected, although numerous SRB were observed. In most sections, SRB were scattered throughout the biofilm structure, including near the interface between the biofilm and water flow.
L. pneumophila in biofilms.
L. pneumophila and other Legionella species were detected in the treated and untreated biofilms 7 to 10 by using whole-cell in situ hybridizations (Fig. 2F) and fluorescent antibodies (data not shown). Because the Leg5 and Leg8 group probes hybridize with several Legionella species, the presence of L. pneumophila and nonpneumophila Legionella spp. was confirmed by using fluorescent antibodies. More bacteria stained with the antibodies than hybridized with the oligonucleotide probes. In these experiments, total bacteria were detected after staining with DAPI instead of CPI (Fig. 2E).
Survival and growth of E. coli in biofilms.
The ability of E. coli to survive in model distribution biofilms was examined using fluorescent in situ hybridizations performed on cross sections of biofilms obtained from flow chambers into which an environmental strain of E. coli had been inoculated. The results of whole-cell hybridizations with the universal 342 probe indicated that the total number of E. coli cells inoculated into each flow chamber was approximately 106, 70% of which were detected after hybridization and subsequently considered metabolically active. Mixtures of distribution water populations and E. coli injected into the flow chambers each day contained approximately 3% E. coli. As shown in Fig. 3, E. coli cells were detected within 2-week-old biofilms seeded with the environmental isolate. Similar to the results of studies using the universal probe, E. coli was detected in a seeded control (untreated) biofilm as well as in two seeded biofilms exposed to 1 CT free chlorine (Fig. 3B) and one biofilm exposed to 10 CT monochloramine (Fig. 3D). Since biofilms were embedded 10 days after the final inoculation of E. coli into the systems, E. coli is clearly able to survive for at least 10 days within a biofilm.
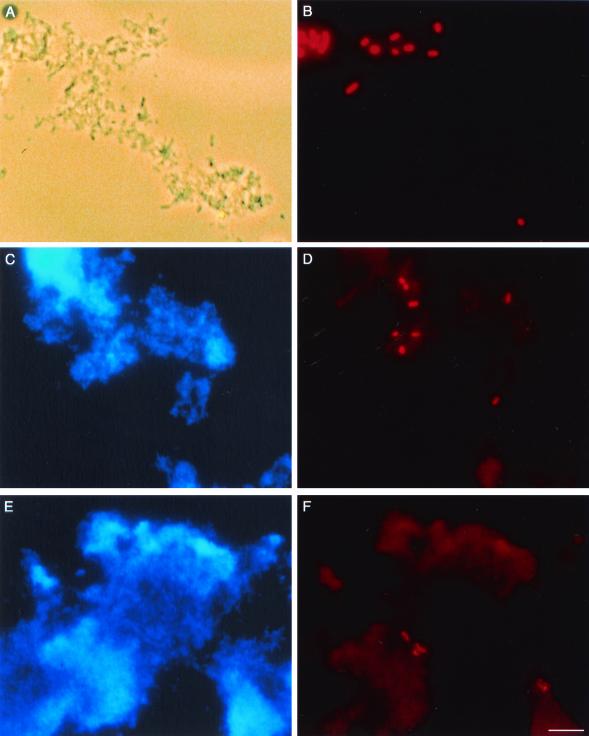
FIG. 3.
Micrographs of E. coli in 2-week-old biofilms. Total bacteria were visualized using phase-contrast microscopy (A) or CPI staining (C and E). E. coli was detected in the same fields after hybridization with the rhodamine-labeled 16S rRNA-targeted oligonucleotide Eco3 probe (B, D, and F). E. coli was inoculated into biofilms 8 and 10 and detected 10 days later, after exposure to 1 CT hypochlorous acid (A and B) or 4 ppm NH2Cl for 155 min (C and D). E. coli present in the Albany distribution system was detected in biofilm 4, exposed to 1 CT hypochlorous acid (E and F). Bar, 5 μm.
Fecal coliforms were not detected in 116 distribution system water samples (11.6 liters total tested, obtained between November 1997 and July 1999) using membrane filtration onto m-FC agar. However, a biofilm developed solely from distribution system populations contained bacteria that were detected after hybridization with the Eco3 probe (Fig. 3F) and confirmed by standard culturing methods. These indigenous E. coli cells were easily differentiated morphometrically from E. coli 01571 grown in the laboratory and seeded into biofilms in the experiments described above. The indigenous cells were less fluorescent and much smaller, suggesting that they had been exposed to adverse conditions for some time.
The ability of bacteriological tests used in the routine monitoring of potable water supplies to detect E. coli sloughing from biofilms was examined. Table 3 presents the occurrence of E. coli in chamber outflow samples using the Colilert Quanti-Tray method, beginning 1 day after the final inoculation of bacteria into flow chamber 3 and 7 days after final inoculation into flow chambers 7, 8, 10, 11, and 12. E. coli was detected in biofilm 3 on two occasions during the incubation period, although only bacterial concentrates obtained from distribution water were used as inocula. Incubation of subsamples of the E. coli-positive medium overnight on LES-Endo agar at 35°C and m-FC agar at 44.5°C gave rise to growth with a green metallic sheen and to blue colonies, respectively. In particular, the blue growth on m-FC agar at 44.5°C indicated the presence of fecal coliforms in the biofilm outflow. Confirmation of the presence of E. coli was completed using an API 20E test strip. The positive Quanti-Trays had been inoculated with flow chamber 3 outflow and 5 days after the final inoculation of distribution system population concentrates into the flow chamber system. The positive samples corresponded to days 8 and 10 of the 2-week incubation. Biofilm outflow samples from flow chambers 7, 8, and 10 also contained detectable numbers of culturable E. coli cells 7 to 10 days after the final inoculation of distribution system populations and E. coli, even after exposure to ClOH (chamber 8) or NH2Cl (chamber 10) on day 10 (Table 3). However, we were unable to culture E. coli from 100-ml samples of outflow from chambers 11 and 12, which contained residual NH2Cl for the entire 2-week incubation period.
TABLE 3.
E. coli detected in outflow from two flow chamber biofilm experiments by using Colilert Quanti-Tray MPNa
| Biofilm designation | Outflow sample day | Time (days) after final bacterial inoculation | Biofilm conditions | No. of E. coli or coliform cells/100 ml of outflow |
|---|---|---|---|---|
| 3 | 6 | 1 | Control (no E. coli added) | <1 |
| 3 | 7 | 2 | Control | <1 |
| 3 | 8 | 3 | Control | 3 |
| 3 | 9 | 4 | Control | 1 |
| 3 | 10 | 5 | Control | <1 |
| 8 | 11 | 6 | 1 CT ClOH (on day 14) | >201b |
| 11 | 2 ppm NH2Cl residual | <1 | ||
| 12 | 4 ppm NH2Cl residual | <1 | ||
| 7 | 12 | 7 | Control | 254 |
| 8 | 1 CT ClOH (on day 14) | 4,425 | ||
| 7 | 14 | 9c | Control | 750 |
| 8 | 1 CT ClOH (on day 14) | 504 | ||
| 10 | 4 ppm NH2Cl for 155 min (on day 14) | 4,725 |
Bacteria were inoculated once a day during the first 5 days of the 2-week incubation period.
The E. coli concentrations in the sample exceeded the upper detection limit of the Quanti-Tray. Subsequent samples were diluted before incubation with the Colilert medium.
Samples collected after chlorine and monochloramine exposures.
DISCUSSION
These investigations demonstrated that the mandated concentrations of the disinfectants chlorine and monochloramine had no measurable effect on the metabolic activity of specific phylogenetic groups of bacteria found in established biofilms. In particular, the results of whole-cell in situ hybridizations indicated the presence of metabolically active E. coli in biofilms both before and after disinfectant treatment. In addition, these organisms were demonstrated to occur in biofilms grown solely from bacterial concentrates obtained from distribution water that had been determined to be free of these organisms on the basis of routine monitoring techniques. These results are particularly important because the question of whether E. coli grows in drinking water biofilms or whether it merely survives for an undetermined maximum time in a stressed starvation state until reintroduced into a nutrient-rich environment suitable for growth has been a matter of debate (7, 28). Similar results showing the presence and possible growth of coliforms within distribution systems were obtained previously by using culturing techniques (20, 21). Long-term survival of E. coli cultures exposed to low-nutrient conditions has been documented previously (7, 12, 23). However, this is the first time that the presence of metabolically active E. coli indigenous to distribution system water has been shown within a biofilm by using direct microscopic techniques.
E. coli was detected by whole-cell hybridization with rhodamine-labeled Eco3 probe in a biofilm that had been exposed to 1 ppm of ClOH for 67 min after 2 weeks of incubation, even though E. coli had not been inoculated into the flow chamber. Because all 116 distribution system water samples tested negative for fecal coliforms (11.6 liters total tested, obtained between November 1997 and July 1999), using membrane filtration onto m-FC agar, the E. coli cells present in the distribution system were most probably injured, thereby escaping detection on a standard selective medium. Previous work has shown that injured coliforms can go undetected on selective media such as LES-Endo, although they can repair injury and subsequently grow (6, 27). During each of the first 5 to 7 days of each experiment, 100 ml of distribution water was tested for fecal coliforms, while bacterial concentrates from 500 ml were inoculated into each flow chamber. Since bacteria were inoculated into flow chambers from an initial volume that was fivefold the amount tested on m-FC agar, it is possible that fecal coliforms were simply absent from the volumes tested. However, since two flow chambers from separate experiments (chambers 3 and 4) contained E. coli, it seems likely that viable fecal coliforms were present in at least some of the original distribution water samples but were not culturable on m-FC agar due to injury or stress.
The smaller morphometry of indigenous E. coli obtained from the Albany distribution system, relative to laboratory-grown E. coli 01571, suggests that the indigenous E. coli had been in a low-nutrient environment for a significant length of time or had sustained injury. Although E. coli 01571 was originally isolated from potable water, it was grown in relatively rich culture medium while maintained in the laboratory. Before its inoculation into flow chambers, E. coli 01571 was incubated at RT for 24 h in the same autoclaved distribution water as was used for irrigation of the flow chambers, to reaccustom the bacteria to the relatively low-nutrient conditions of distribution systems. Even with that acclimation period and the 2-week incubation within flow chamber biofilms, E. coli 01571 cells detected by hybridization with the Eco3 probe were still larger than other bacteria within the biofilms (Fig. 3).
In contrast to the results of these investigations, McMath et al. (28) reported little to no detectable survival of E. coli or other coliforms in a model distribution system carrying chlorinated water. Since the presence of coliforms in the model distribution system outflows was evaluated using only Colilert medium, chlorine-injured or stressed coliforms may have escaped detection on that selective medium. Viable but noncultured coliforms might have been detected by using m-T7 agar (1) or by direct observation of coliform cells after hybridization with appropriate fluorescently labeled oligonucleotide probes targeted against 16S rRNA.
Other recent experiments have demonstrated the persistence of E. coli labeled with green fluorescent protein (GFP) in anaerobic biofilms derived from groundwater populations (2). While the requirement for creating GFP-labeled bacteria within the laboratory limits their usefulness in environmental studies, this confirms our work demonstrating the incorporation of E. coli into multispecies biofilms under low-nutrient conditions. Further work needs to be completed with GFP-labeled bacteria to determine how responsive the fluorescence in the cells is to changes in the physiological state of the bacteria. A second study examined E. coli in distribution water by using a fluorescently labeled peptide nucleic acid oligonucleotide probe targeting the V1 region of 16S rRNA (33). In this study, E. coli cells were exposed to 1.5 mg of ClOH per liter for 30 min and then stored at room temperature for 14 days before being detected by hybridization, although other measures of cell viability (plate counts, Colilert, and CTC reduction) showed no activity. This contrasts with our results demonstrating a change in the hybridization efficacy of our oligodeoxynucleotide (DNA) probe to E. coli after treatment with 1 mg of ClOH per liter for 10 min. The difference in results may be due to a difference in the target region for each probe. It is possible that the section of the 16S rRNA molecule targeted by the PNA probe, bases 71 to 86, is more strongly protected from chemical oxidation than is the target for our probe.
Demonstration of the persistence of E. coli within biofilms calls into question the utility of E. coli as an accurate indicator of recent fecal pollution. The determination of whether E. coli-positive distribution water samples indicate a true current contamination event or a release of E. coli residing in biofilms is a problem for public health officials, particularly when a decision on whether to order boiled-water alerts is necessary.
The study of SRB within biofilms is of interest because these organisms contribute to anaerobic corrosion of metal distribution pipes. As anaerobes, SRB might be expected to be limited to the most oxygen-deficient regions of biofilms. To facilitate their access to nutrients, they might also be expected to be in close proximity to the ductile iron coupon that comprised one side of the flow chamber. However, SRB were found evenly scattered throughout cross-sections of several biofilms, suggesting that sufficient dissolved oxygen within the distribution water was removed by surrounding aerobic heterotrophs to allow growth of SRB.
Members of the β proteobacteria were numerous in several biofilms exposed to the various disinfectant types and concentrations (data not shown). Several members of the β proteobacteria are typical constituents of drinking water supplies, confirming previous studies (26).
Legionella spp. are indigenous to both natural and man-made aquatic environments (15, 19). For example, they have been found in cooling towers and air conditioners, as well as in a wide variety of lakes. As such, Legionella spp. were used as control organisms in these studies. Both in situ hybridizations and fluorescent-antibody staining demonstrated the abundance of these organisms within distribution system biofilms.
Formation of distribution system biofilms resistant to disinfectants is a dynamic yet constant and reproducible process, as was observed by the persistence of a wide variety of bacterial species and groups within biofilms grown at different times of year and exposed to various chlorine disinfectant treatments.
Of three flow chambers treated with monochloramine residuals for their entire 2-week incubations, two were embedded and sectioned for subsequent examination using whole-cell hybridizations while the third was examined in a parallel study using scanning confocal laser microscopy (SCLM). Although biofilm was not observed on slides of 6-μm cryoembedded sections after whole-cell hybridizations, slight evidence of biofilm was observed by SCLM (data not shown). This suggests that small amounts of biofilm were probably lost during the embedding process, probably when the frozen O.C.T. block was removed from the flow chamber. However, since significant amounts of biofilm from all other flow chambers were successfully preserved for subsequent hybridizations, the biofilm loss was probably minimal. Also, because the entire chamber contents were not sectioned, it is possible that small amounts of biofilm were present but not detected.
In conclusion, these studies showed that noninjured E. coli cells survived for at least 10 days and maintained detectable amounts of RNA in distribution system biofilms. Within biofilms, they were protected from exposure to high levels of chlorine and monochloramine and remained viable. Injured E. coli and other coliforms routinely escaped detection and survived in a water supply that conformed to current water quality practices. Evidence indicated that injured fecal coliforms quickly repaired cellular damage once they were harbored in a biofilm and that they were subsequently culturable on standard selective media. Opportunistic pathogens such as L. pneumophila, as well as corrosion-causing SRB, were indigenous to water supply populations and were also protected from chlorine disinfectants within distribution system biofilms.
Although the current work has shown that the presence of E. coli in drinking water may represent either a true contamination event or a release of E. coli from a distribution system biofilm, E. coli is still important as an indicator of possible pubic health risk. Eradication of biofilm from distribution systems is not currently possible. Because the protection from disinfectants afforded to biofilm bacteria is not limited to any particular species, control of biofilm growth is needed to minimize the number and diversity of pathogens harbored within the biofilm matrix. Maintenance of the quality of the water entering the distribution system, as well as maintenance and repair of the distribution system itself, may go a long way toward prevention of risk of infection of sensitive individuals. However, the present studies indicate that recommendation of additional point-of-use water treatment for immunocompromised individuals may be prudent. In this manner, the higher cost of additional water treatment would be minimized by limiting the extra measures to the sector of the population at greatest risk of waterborne disease.
Acknowledgments
Some research materials were purchased with a grant awarded to M.M.W. by the Benevolent Foundation of the State University of New York at Albany.
We thank Terry Dixson and Tonia Carter for excellent technical assistance. The help of Yolanda Filippini and Diane Decker, who cryosectioned biofilms, is also gratefully acknowledged.
REFERENCES
- 1.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, D.C.
- 2.Banning, N., S. Toze, and B. J. Mee. 2003. Persistence of biofilm-associated Escherichia coli and Pseudomonas aeruginosa in groundwater and treated effluent in a laboratory model system. Microbiology 149:47-55. [DOI] [PubMed] [Google Scholar]
- 3.Boorman, G. A., V. Dellarco, J. K. Dunnick, R. E. Chapin, S. Hunter, F. Hauchman, H. Gardner, M. Cox, and R. C. Sills. 1999. Drinking water disinfection by-products: review and approach to toxicity evaluation. Environ. Health Perspect. 107(Suppl. 1):207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun-Howland, E. B., S. A. Danielsen, and S. A. Nierzwicki-Bauer. 1992. Development of a rapid method for detecting bacterial cells in situ using 16S rRNA-targeted probes. BioTechniques 13:928-933. [PubMed] [Google Scholar]
- 5.Caldwell, D. E., and J. R. Lawrence. 1988. Study of attached cells in continuous-flow slide culture, p. 117-138. In J. W. T. Wimpenny (ed.), CRC handbook of laboratory model systems for microbial ecosystems. CRC Press, Inc., Boca Raton, Fla.
- 6.Camper, A. K., and G. A. McFeters. 1979. Chlorine injury and the enumeration of waterborne coliform bacteria. Appl. Environ. Microbiol. 37:633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camper, A. K., G. A. McFeters, W. G. Characklis, and W. L. Jones. 1991. Growth kinetics of coliform bacteria under conditions relevant to drinking water distribution systems. Appl. Environ. Microbiol. 57:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camper, A., M. Burr, B. Ellis, P. Butterfield, and C. Abernathy. 1999. Development and structure of drinking water biofilms and techniques for their study. J. Appl. Microbiol. Symp. Suppl. 85:1S-12S. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson, K., L. Moberg, and B. Karlberg. 1999. The miniaturization of the standard method based on the N,N′-diethyl-p-phenylenediamine (DPD) reagent for the determination of free or combined chlorine. Water Res. 33:375-380. [Google Scholar]
- 10.Costerton, J. W., K.-J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and J. T. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 42:435-464. [DOI] [PubMed] [Google Scholar]
- 11.Daly, B., W. B. Betts, A. P. Brown, and J. G. O'Neill. 1998. Bacterial loss from biofilms exposed to free chlorine. Microbios 96:7-21. [PubMed] [Google Scholar]
- 12.Fass, S., M. L. Dincher, D. J. Reasoner, D. Gatel, and J.-C. Block. 1996. Fate of Escherichia coli experimentally injected in a drinking water distribution pilot system. Water Res. 30:2215-2221. [Google Scholar]
- 13.Feeley, J. C., G. W. Gorman, R. E. Weaver, D. C. Mackel, and H. W. Smith. 1978. Primary isolation medium for Legionnaires' disease bacteria. J. Clin. Microbiol. 8:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher, M., and K. C. Marshall. 1982. Are solid surfaces of ecological significance to aquatic bacteria? Adv. Microb. Ecol. 6:199-236. [Google Scholar]
- 15.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. Nature 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacangelo, J. G., V. P. Olivieri, and K. Kawata. 1991. Investigating the mechanism of inactivation of Escherichia coli B by monochloramine. J. Am. Water Works Assoc. 83:80-87. [Google Scholar]
- 18.Kefford, B., S. Kjelleberg, and K. C. Marshall. 1982. Bacterial scavenging: utilization of fatty acids localized at solid liquid interface. Arch. Microbiol. 133:257-260. [Google Scholar]
- 19.Kramer, M. H. J., and T. E. Ford. 1994. Legionellosis: ecological factors of an environmentally ‘new’ disease. Zentbl. Hyg. 195:470-482. [PubMed] [Google Scholar]
- 20.LeChevallier, M. W., N. J. Welch, and D. B. Smith. 1996. Full-scale studies of factors related to coliform regrowth in drinking water. Appl. Environ. Microbiol. 62:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeChevallier, M. W., T. M. Babcock, and R. G. Lee. 1987. Examination and characterization of distribution system biofilms. Appl. Environ. Microbiol. 53:2714-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeChevallier, M. W., W. Schulz, and R. G. Lee. 1991. Bacterial nutrients in drinking water. Appl. Environ. Microbiol. 57:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisle, J. T., S. C. Broadway, A. M. Prescott, B. H. Pyle, C. Fricker, and G. A. McFeters. 1998. Effects of starvation on physiological activity and chlorine disinfection resistance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maczulak, A. E., M. J. Wolin, and T. L. Miller. 1989. Increase in colonic methanogens and total anaerobes in aging rats. Appl. Environ. Microbiol. 55:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manz, W., U. Szewzyk, P. Ericksson, R. Amann, K.-H. Schleifer, and T.-A. Stenstrom. 1993. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl. Environ. Microbiol. 59:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFeters, G. A., J. S. Kippin, and M. W. LeChevallier. 1986. Injured coliforms in drinking water. Appl. Environ. Microbiol. 51:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMath, S. M., C. Sumpter, D. M. Holt, A. Delanoue, and A. H. L. Chamberlain. 1999. The fate of environmental coliforms in a model water distribution system. Lett. Appl. Microbiol. 28:93-97. [DOI] [PubMed] [Google Scholar]
- 29.Møller, S., C. S. Kristensen, L. Poulsen, J. M. Carstensen, and S. Molin. 1995. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl. Environ. Microbiol. 61:741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Møller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olivieri, V. P., A. E. Bakalian, K. W. Bossung, and E. D. Lowther. 1984. Recurrent coliforms in water ditribution systems in the presence of free residual chlorine, p. 651-666. In R. L. Jolley, R. J. Bull, W. P. Davis, W. Katz, M. H. Roberts, Jr., and V. A. Jacobs (ed.), Water chlorination: chemistry, environmental impact and health effects. Lewis, Chelsea, Mich.
- 32.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prescott, A. M., and C. R. Fricker. 1999. Use of PNA oligonucleotides for the in situ detection of Escherichia coli in water. Mol. Cell. Probes 13:261-268. [DOI] [PubMed] [Google Scholar]
- 34.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridgway, H. F., and B. H. Olson. 1982. Chlorine resistance patterns of bacteria from two drinking water distribution systems. Appl. Environ. Microbiol. 44:972-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K.-H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, P. S., R. Murga, R. Srinizasan, and D. de Beer. 1995. Biofilm structural heterogeneity visualized by three microscopic methods. Water Res. 29:2006-2009. [Google Scholar]
- 38.Vescio, P. A., and S. A. Nierzwicki-Bauer. 1995. Extraction and purification of PCR amplifiable DNA from lacustrine subsurface sediments. J. Microbiol. Methods 21:225-233. [Google Scholar]
- 39.Yu, F. P., and G. A. McFeters. 1994. Rapid in situ assessment of physiological activities in bacterial biofilms using fluorescent probes. J. Microbiol. Methods 20:1-10. [DOI] [PubMed] [Google Scholar]