Abstract
In angiosperms, the stigma provides initial nutrients and guidance cues for pollen grain germination and tube growth. However, little is known about the genes that regulate these processes in rice (Oryza sativa). Here, we generate rice stigma-specific or -preferential gene expression profiles through comparing genome-wide expression patterns of hand-dissected, unpollinated stigma at anthesis with seven tissues, including seedling shoot, seedling root, mature anther, ovary at anthesis, seeds 5 d after pollination, 10-d-old embryo, 10-d-old endosperm, and suspension-cultured cells by using both 57 K Affymetrix rice whole-genome array and 10 K rice cDNA microarray. A high reproducibility of the microarray results was detected between the two different technology platforms. In total, we identified 548 genes to be expressed specifically or predominantly in the stigma papillar cells of rice. Real-time quantitative reverse transcription-polymerase chain reaction analysis of 34 selected genes all confirmed their stigma-specific expression. The expression of five selected genes was further validated by RNA in situ hybridization. Gene Ontology analysis shows that several auxin-signaling components, transcription, and stress-related genes are significantly overrepresented in the rice stigma gene set. Interestingly, most of them also share several cis-regulatory elements with known stress-responsive genes, supporting the notion of an overlap of genetic programs regulating pollination and stress/defense responses. We also found that genes involved in cell wall metabolism and cellular communication appear to be conserved in the stigma between rice and Arabidopsis (Arabidopsis thaliana). Our results indicate that the stigmas appear to have conserved and novel molecular functions between rice and Arabidopsis.
In flowering plants, pollination is the first major reproduction process that results in the production of seeds. The process begins with the adhesion of pollen grains to the stigmatic tissue of style. Then, the highly desiccated pollen grains are rehydrated to reach a certain water content that allows them to regain active metabolisms. Once the pollen grain cell has established its internal polarity relative to an external signal, the pollen germinates and breaches the exine wall to emit a pollen tube. The newly formed pollen tube subsequently penetrates the stigma, grows down through the transmitting tissue of the style, and ultimately reaches an ovule, allowing fertilization to take place (Lord, 2003; Sanchez et al., 2004; Swanson et al., 2004; Boavida et al., 2005).
Successful pollination requires continued communication and coordination between the pollen and stigma and is essential to maximize the seed set. The stigma, the uppermost part of the pistil, is generally considered to be a passive structure for pollen grain capture and reception, germination, and initial growth of the pollen tubes. There are two major types of stigma, the dry and the wet types, which differ by the presence or absence of the secreting exudate compounds (Edlund et al., 2004). The adhesion and germination of pollen on the wet stigma is facilitated by the presence of the exudate, but its ingredients are highly variable among species (Edlund et al., 2004; Boavida et al., 2005). Tobacco (Nicotiana tabacum) is a wet stigma-type plant, and its stigma has three distinct zones: an epidermis with papillae, a subepidermal secretory zone, and a zone of parenchyma ground tissue (Kandasamy and Kristen, 1987). Elimination of the secretory zone involved in producing the exudates in tobacco by expressing a chimeric stigma-specific cytotoxic gene results in female sterility, but fertility could be restored upon application of the stigma exudate of wild-type tobacco plants (Goldman et al., 1994). In species with the dry stigma, including Arabidopsis (Arabidopsis thaliana), kale (Brassica oleracea), and rice (Oryza sativa), the epidermis is composed of large specialized papillae cells that interact directly with the surface of pollen, and the surface of each papilla is covered with a waxy cuticle overlaid with a distinct proteinaceous pellicle layer (Gaude and Dumas, 1986; Zinkl et al., 1999; Ciampolini et al., 2001). In Brassica, the stigmatic papillae cells completely lose their ability to capture the pollen grains when using acetone to strip off both the waxy cuticle and proteinaceous stigma pellicle (Heizmann et al., 2000). In addition, protease digestion of pellicles from kale also adversely affects pollen adhesion (Stead et al., 1980; Luu et al., 1997). These studies clearly demonstrate that the stigma plays crucial roles in pollination.
Nevertheless, the molecular roles of the stigma in pollination are largely unknown. Several stigma-specific expressed proteins have been identified, a few of which are found to be directly involved in the pollen-stigma interaction. SRK (S-locus receptor kinase) encoding a glycoprotein localizes to the stigmatic plasma membrane and interacts with the pollen coat S-locus Cys-rich protein (SCR) in the self-incompatibility response in Brassica (Nasrallah, 2000; Takayama and Isogai, 2005). Both SLG (S-locus glycoprotein) and SLR (S-locus-related protein) are expressed in stigma papillae cells, and their products localize to the stigma cell wall in Brassica (Kandasamy et al., 1989; Lalonde et al., 1989; Umbach et al., 1990). Although there is some controversy as to whether SLG and SLR are required for the self-incompatibility response in Brassica, both of them are shown to interact with several small pollen coat proteins, and these interactions are believed to generate adhesive forces of the pollen-stigma interaction (Luu et al., 1999; Takayama and Isogai, 2005). The pis63 gene encoding an unknown protein in B. napus is expressed in the stigma papillae cells, and a reduction in its transcript level results in reduced pollen germination and seed set (Kang and Nasrallah, 2001). Aquaporin-like proteins in the Brassica stigma may act as water channels in controlling water flow into the pollen grain from the stigma (Dixit et al., 2001). An ATP-binding cassette (ABC) transporter gene, designated as NtWBC1 (N. tabacum ABC transporter of the White-Brown Complex subfamily), is highly expressed in the stigmatic secretary zone, suggesting its important role in the reproductive process (Otsu et al., 2004). Recently, a stigma-specific peroxidase from the ragwort Senecio squalidus was identified, and its expression was confined to the specialized epidermal cells of the stigma (McInnis et al., 2005). The activity of stigma peroxidases has long been used to determine the pistil receptivity for pollen grains, and the high levels of the peroxidase activity show that the stigma is ripe for pollination (McInnis et al., 2006). But, the function of the peroxidase in pollination still is not clear.
Additional factors have been found to be involved in pollination in several species. Auxin has been proposed to operate as a pollination signal and promote the differentiation and development of ovules in flowers of orchid of the genus Phalaenopsis (O'Neill, 1997). In petunia (Petunia hybrida), the interaction of the male gametophyte with the stigmatic tissues is accompanied by a 3-fold increase in the ethylene production and a 1.5-fold increase in the auxin (indole-3-acetic acid [IAA]) content in the pollen-pistil system within 0 to 4 h after compatible pollination, and the growth of pollen tubes in the stylar tissues is accompanied by a further increase in IAA content and a decrease in the ethylene production by stigmatic tissues (Kovaleva et al., 2002). The ethylene/auxin status of the stigma has been suggested to control the processes of adhesion, hydration, and germination of pollen grains during pollination in petunia (Kovaleva et al., 2002). In our previous study, we found that an extensive set of genes is shared between abiotic stress responses and pollination/fertilization in rice, and nearly one-half of the genes expressed preferentially in the unpollinated pistils are responsive to dehydration; the majority of them are dehydration inducible, suggesting an overlap in the genetic programs controlling between these processes (Lan et al., 2005). Temperature is a major physical factor influencing the plant reproductive phase and limits geographical distributions of plant species. High temperature has been found to cause the loss of the sporophytic self-incompatibility response in Ipomoea fistulosa (Prabha et al., 1982). The stigmatic receptivity of peach (Prunus persica) loses the capacity to sustain the pollen tube penetration into the transmitting tissue, pollen germination, and adhesion as the temperature increases during pollination (Hedhly et al., 2005).
However, the molecular mechanisms of these events involved in pollination remain obscure. It is anticipated that, given recent rapid developments in functional genomics, genome-wide identification of stigma-specific genes could provide important initial steps in dissecting molecular control of pollination. Recently, two groups have carried out the stigma-specific gene profiling of Arabidopsis using whole-genome microarray analysis (Swanson et al., 2005; Tung et al., 2005). Tung et al. (2005) have identified 115 genes predicted to be expressed specifically in the stigma epidermis after comparing transcriptional profiles of the stigmas isolated from wild-type Arabidopsis and from the transgenic plants in which cells of the stigma epidermis were specifically ablated by expression of a cellular toxin. Swanson et al. (2005) have identified 317 genes that are highly expressed in the stigma after comparing wild-type stigma, ovary, and seedling transcriptional profiles, and the cDNA library subtractive hybridization has further confirmed the results. Both the stigma gene data sets are significantly enriched in those genes involved the cell wall metabolism, cellular communication, and signal transduction-related proteins. They are thought to play roles in the adhesion, hydration, and germination of pollen grain.
Cultivated rice is by far one of the most important food crops and is also considered the model monocot plant for molecular and genetic studies. Rice is primarily an autogamous, self-pollinating plant and has the typical dry stigma. But very little is known about stigma-specific genes that are required for ensuring successful pollination in rice. In this study, we generate rice stigma-specific or -preferential gene expression profiles by using both the commercially available 57 K Affymetrix rice whole-genome array and 10 K rice cDNA microarray (Lan et al., 2004). Gene Ontology (GO) analysis shows that several auxin-signaling components, transcription, and stress-related genes are significantly overrepresented in the rice stigma gene set. We also found that the genes involved in cell wall metabolism and cellular communication appear to be conserved in the stigmas between rice and Arabidopsis.
RESULTS
Gene Expression Profiles of Stigma versus Ovary by Two Independent Microarray Platforms
To generate stigma-specific and ovary-specific gene expression profiles, two independent microarrays were applied in this study. The stigma samples were hand collected by cutting the pistil just below the base of the plumose stigma, and the remainder of the pistil containing the style was used as the ovary samples. The important criterion for the dissection was that the stigma samples had the papillar cells but not the ovary samples.
To identify the genes preferentially expressed in the stigma, 57 K Affymetrix rice whole-genome array was used to generate the genome expression patterns across eight representative organs or tissues and suspension-cultured cells. They included seedling shoots (Sh), seedling roots (Rt), mature anthers (An), unpollinated stigmas at anthesis (St), ovaries at anthesis (Ov), seeds 5 d after pollination (5DAP), 10-d-old embryos (10EM), and 10-d-old endosperms (10EN). We performed three biological replicates for the stigma and ovary samples, respectively. The correlation coefficient value of each experiment was larger than 0.99. By using P value ≤ 0.05 and fold change ≥ 2 as cutoff to identify differentially expressed genes, 3,300 probe sets were up-regulated in the stigma compared to the ovary, and 4,125 probe sets were up-regulated in the ovary compared to the stigma (Supplemental Table S1). All microarray data are available online (http://www.ncbi.nlm.nih.gov/geo; http://plantbiol.genetics.ac.cn).
We also used the 10 K rice cDNA microarray constructed previously (Lan et al., 2004) to compare gene expression profiles of the stigma with the ovary. The gene expression profiles of the rest organs were generated previously (Lan et al., 2004, 2005). Three biological and one dye-swapped replicates were performed for the stigma versus ovary hybridizations. By using the same cutoff (fold change ≥ 2, P < 0.05), 453 cDNAs in total had more abundant expressions in the stigma compared to the ovary (Supplemental Table S2), and 257 cDNAs were identified as enriched in the ovary compared to the stigma (Supplemental Table S3).
The microarray results showed a high reproducibility using the two different platforms (cDNA microarray versus Affymetrix oligo array). For the 453 cDNAs preferentially expressed in the stigma identified using the 10 K rice cDNA microarray, 201 cDNAs' (using E value = 1e-20 and identity >80% as cutoff) expression patterns were confirmed by the Affymetrix GeneChip results (Supplemental Table S4), with about a 44.37% match. For the 257 cDNA clones highly expressed in the ovary, 109 cDNAs' expression patterns (using E value = 1e-20 and identity >80% as cutoff) were confirmed by the Affymetrix GeneChip results, with about a 42.41% match (Supplemental Table S5).
Identification and Functional Classification of the Stigma-Specific/-Preferential Genes
To identify rice stigma preferentially expressed genes, two statistical methods were conducted for data processing. First, Significance Analysis of Microarrays software package analysis was conducted for three biological samples replicates between the stigma and ovary. Using q value ≤ 0.05 and fold change ≥ 2 as cutoff, 3,300 probe sets were subsequently picked out, and their expression levels were significantly up-regulated in the stigma compared to the ovary. We also did a normal t test and added P value for these probe sets. Second, we used the Z-score transformation normalization method to compare the gene expression levels in stigma with other tested organs or tissues and suspension-cultured cells and directly calculated significant changes in the gene expression levels between them. For Z-score transformation normalization, we used average values of the stigma and ovary separately. In total, we identified 548 genes (665 probe sets) preferentially expressed in the stigma using Z score ≥ 2.32 and P value ≤ 0.01(Supplemental Table S6).
Among the identified 548 genes preferentially expressed in the stigma, 410 genes had putative functions, 103 genes were assigned as expressed proteins, 23 genes were assigned as hypothetical proteins, and 12 genes had no hits when blasted against The Institute for Genomic Research rice genome annotation database (http://www.tigr.org). The 410 genes were classified into 11 groups according to their annotations and one unclassified group: transcription, cell wall related, stress/defense, signal transduction, lipid metabolism, transport, hormone related, protein metabolism, carbohydrate and energy metabolism, nucleic acid metabolism, amino acid metabolism, and unclassified. The categories are listed in the order of gene numbers in each group (Supplemental Table S7). We list 149 genes derived from 173 probe sets with putative annotated functions and their ratios of the stigma/ovary of more than 10 (P < 0.05) in Table I. The largest category was transcription-related proteins, including two MADS-box proteins, four zinc-finger proteins, and six Myb- or Myb-like transcription factors. These proteins accounted for nearly 15% of the 410 putative functional genes. In the second category, including putative cell wall-localized enzymes (e.g. α-expansin, pectinesterase, and peroxidase), cellulose synthase, glycerol-3-P acyltransferase, and UDP-glucosyltransferase, the third category consisted of proteins that were potentially involved in stress/defense responses, such as glutathione S-transferase, phospholipase D, and wound-induced protein WI12. Many signal transduction- and transport-related proteins were enriched in the stigma. There was a large percentage of genes that may be related to lipid metabolism, including lipid binding protein, triacylglycerol lipase, and fatty acid elongase. The auxin signaling related proteins were identified to be highly expressed in the stigma, including auxin-responsive factor, five members of the auxin-responsive small auxin-up RNA (SAUR) gene family, and the highly expressed IAA-amido synthetase GH3.1 gene (see Table I; Supplemental Table S7).
Table I.
Candidate genes preferentially expressed in rice stigma
The total number of the preferentially expressed genes is 548 (665 probe sets). Expressed protein, hypothetical protein, and no hit genes are 103, 23, and 12, respectively. The table lists 149 (173 probe sets) of them with putative annotated functions, and their ratios of stigma/ovary are more than 10 (P < 0.05). ID, Identification.
| Probe Set IDa | Gene ID | Description | Stigma-Sb | Ovary-Sc | Ratiod | P Value |
|---|---|---|---|---|---|---|
| Transcription | ||||||
| Os.3386.1.S1_x_at | LOC_Os06g10350 | Anthocyanin regulatory C1 protein | 2,087.6 | 90.4 | 23.1 | 1.93E-04 |
| Os.2233.1.S1_at | LOC_Os04g47080 | Anthocyanin regulatory Lc protein | 7,912.47 | 75.93 | 104.2 | 1.98E-04 |
| Os.38982.1.S1_at | LOC_Os02g43820 | AP2 domain-containing protein | 1,498.93 | 72.97 | 20.5 | 1.67E-02 |
| Os.49746.1.S1_at | LOC_Os01g74020 | ARR1 protein like | 13,525.53 | 908.27 | 14.9 | 3.05E-04 |
| Os.31989.1.S1_at | LOC_Os03g02900 | B3 DNA-binding domain-containing protein | 3,014.57 | 260.47 | 11.6 | 2.72E-03 |
| OsAffx.27272.1.S1_at | LOC_Os05g41070 | bZIP transcription factor | 267.53 | 23.2 | 11.5 | 4.17E-02 |
| Os.46006.1.S1_at | LOC_Os07g47140 | CCT motif family protein | 10,524.93 | 313.83 | 33.5 | 8.37E-04 |
| Os.37618.1.S1_at | LOC_Os06g04850 | Homeobox-Leu zipper protein ATHB-4 | 1,608.2 | 93.23 | 17.2 | 2.78E-03 |
| Os.2365.1.S1_at | LOC_Os06g04870 | Homeobox-Leu zipper protein HAT1 | 9,315.13 | 352.67 | 26.4 | 1.31E-04 |
| Os.49761.1.S1_at | LOC_Os03g54170 | MADS-box transcription factor 34 | 3,492.13 | 276.37 | 12.6 | 6.24E-03 |
| OsAffx.21941.1.S1_s_at | LOC_Os01g19970 | myb-like transcription factor | 1,679.7 | 165.77 | 10.1 | 1.98E-04 |
| OsAffx.14605.1.S1_at | LOC_Os05g09020 | OsWRKY67 | 3,840.1 | 229.9 | 16.7 | 1.96E-03 |
| OsAffx.14605.1.S1_s_at | LOC_Os05g09020 | OsWRKY67 | 3,244.13 | 225.6 | 14.4 | 3.32E-07 |
| OsAffx.2905.1.S1_at | LOC_Os02g38320 | Protein-binding protein | 1,157.37 | 63.13 | 18.3 | 2.91E-04 |
| OsAffx.11799.1.S1_at | LOC_Os01g69200 | Regulatory protein | 2,144.53 | 2.9 | 739.5 | 2.25E-03 |
| Os.56298.1.S1_at | LOC_Os03g63810 | WRKY transcription factor 14 | 2,149.4 | 21.93 | 98 | 5.89E-04 |
| OsAffx.17417.1.S1_x_at | LOC_Os09g31140 | Zinc finger protein F15B8.140, Arabidopsis | 374.63 | 16.3 | 23 | 8.56E-03 |
| Os.49270.1.S1_at | LOC_Os06g40960 | Zinc finger protein | 5,652.4 | 172.57 | 32.8 | 1.37E-03 |
| Cell wall related | ||||||
| Os.53291.1.S1_at | LOC_Os03g64280 | 1-Aminocyclopropane-1-carboxylate oxidase | 861.17 | 85.33 | 10.1 | 7.57E-04 |
| Os.27155.1.S1_at | LOC_Os06g15170 | 3-Ketoacyl-CoA synthase | 2,845.73 | 76.87 | 37 | 1.19E-02 |
| Os.2367.1.S1_at | LOC_Os03g21820 | α-Expansin 10 precursor | 11,840.7 | 658.43 | 18 | 5.25E-04 |
| Os.31669.1.S1_at | LOC_Os01g53370 | Anthocyanidin 5,3-O-glucosyltransferase | 475.63 | 5.57 | 85.4 | 6.27E-03 |
| Os.31669.1.S1_x_at | LOC_Os01g53370 | Anthocyanidin 5,3-O-glucosyltransferase | 667.57 | 10.43 | 64 | 9.77E-03 |
| Os.8842.1.S1_at | LOC_Os07g32620 | Anthocyanidin 5,3-O-glucosyltransferase | 15,546.97 | 288.1 | 54 | 2.47E-04 |
| Os.57078.1.S1_at | LOC_Os05g45200 | Anthocyanidin 5,3-O-glucosyltransferase | 1,105.83 | 47.2 | 23.4 | 1.24E-02 |
| OsAffx.23995.2.S1_x_at | LOC_Os01g71094 | Basic 7S globulin 2 precursor | 349.03 | 4.27 | 81.8 | 3.36E-02 |
| Os.27968.1.S1_x_at | LOC_Os01g71094 | Basic 7S globulin 2 precursor | 2,661.17 | 184.47 | 14.4 | 1.16E-03 |
| Os.27968.1.S1_at | LOC_Os01g71094 | Basic 7S globulin 2 precursor | 3,755.6 | 271.67 | 13.8 | 5.83E-03 |
| Os.2075.1.S1_at | LOC_Os01g19220 | β-d-Xylosidase | 6,110.4 | 567.4 | 10.8 | 1.70E-04 |
| Os.14358.4.S1_x_at | LOC_Os02g12730 | β-Galactosidase precursor | 6,569.7 | 243.7 | 27 | 7.94E-04 |
| Os.14358.1.S1_at | LOC_Os02g12730 | β-Galactosidase precursor | 1,490.43 | 71.67 | 20.8 | 3.07E-04 |
| Os.2322.1.S1_at | LOC_Os10g33250 | CER1 | 28,284.37 | 469.33 | 60.3 | 4.07E-03 |
| Os.2322.2.S1_at | LOC_Os10g33250 | CER1 | 1,049.97 | 32.73 | 32.1 | 2.42E-02 |
| OsAffx.15853.1.S1_x_at | LOC_Os06g39970 | CESA11, cellulose synthase | 252.67 | 15 | 16.8 | 1.77E-02 |
| OsAffx.15853.1.S1_at | LOC_Os06g39970 | CESA11, cellulose synthase | 200.13 | 15.33 | 13.1 | 5.18E-03 |
| OsAffx.22999.1.S1_at | LOC_Os07g32710 | d-Genomic expressed protein | 19,698.87 | 292.03 | 67.5 | 4.54E-03 |
| OsAffx.4968.1.S1_at | LOC_Os06g30090 | DNA-binding protein | 325.67 | 2.43 | 133.8 | 4.58E-03 |
| Os.5804.1.S1_at | LOC_Os11g40210 | DNA-binding protein | 1,662.97 | 129.93 | 12.8 | 6.56E-04 |
| OsAffx.2049.2.S1_x_at | LOC_Os01g21310 | dTDP-Glc 4-6-dehydratase-like protein | 534.87 | 33.43 | 16 | 4.85E-02 |
| Os.26486.1.S1_at | LOC_Os09g25850 | gl1 protein | 19,971.97 | 353.67 | 56.5 | 2.77E-03 |
| OsAffx.30743.1.S1_at | LOC_Os01g14900 | Glycerol-3-P acyltransferase 1 | 135.2 | 5.87 | 23 | 5.01E-03 |
| Os.32123.1.S1_at | LOC_Os01g02930 | Glycosyltransferase | 7,300.53 | 480.7 | 15.2 | 1.54E-03 |
| Os.53164.1.S1_at | LOC_Os06g18010 | Hydroquinone glucosyltransferase | 7,375.97 | 115 | 64.1 | 6.97E-04 |
| Os.15570.1.S1_at | LOC_Os07g32630 | Hydroquinone glucosyltransferase | 10,598 | 395.8 | 26.8 | 1.99E-04 |
| OsAffx.31322.1.S1_at | LOC_Os11g36240 | Pectinesterase precursor | 6,228.2 | 1.53 | 4,061.9 | 9.86E-04 |
| Os.6192.1.S1_at | LOC_Os03g02460 | Retinol dehydrogenase 12 | 10,156.47 | 721.63 | 14.1 | 1.60E-06 |
| Stress/defense | ||||||
| Os.4377.1.S1_at | LOC_Os02g46970 | 4-Coumarate-CoA ligase 2 | 4,725.17 | 28.87 | 163.7 | 1.03E-03 |
| Os.46537.1.S1_at | LOC_Os10g04429 | Alcohol acyl transferase | 336.03 | 6.07 | 55.4 | 2.42E-03 |
| Os.23215.1.S1_at | LOC_Os06g24404 | Anther-specific Pro-rich protein APG precursor | 4,965.7 | 247.37 | 20.1 | 2.43E-03 |
| Os.22594.2.S1_at | LOC_Os01g03390 | Bowman-Birk-type bran trypsin inhibitor precursor | 206.1 | 9.63 | 21.4 | 1.29E-02 |
| Os.22594.1.S1_at | LOC_Os01g03390 | Bowman-Birk-type bran trypsin inhibitor precursor | 20,176.87 | 1,726.13 | 11.7 | 4.64E-03 |
| Os.3121.1.S1_at | LOC_Os10g28050 | Chitinase 2 | 11,909.67 | 287.67 | 41.4 | 7.30E-03 |
| Os.47136.1.A1_x_at | LOC_Os01g27360 | d-Genomic glutathione S-transferase GSTF1 | 1,140.33 | 36.13 | 31.6 | 2.45E-02 |
| Os.14539.1.S1_at | LOC_Os07g01600 | Disease resistance response protein 206 | 10,803.27 | 662.5 | 16.3 | 2.94E-03 |
| Os.12922.1.S1_at | LOC_Os09g36700 | Extracellular ribonuclease LE precursor | 9,753.43 | 146.17 | 66.7 | 1.47E-04 |
| Os.46551.1.S1_at | LOC_Os10g17260 | Flavonoid 3-monooxygenase | 3,769.07 | 16.27 | 231.7 | 6.04E-03 |
| Os.25651.1.S1_at | LOC_Os01g27360 | Glutathione S-transferase GSTF1 | 2,437.67 | 59.23 | 41.2 | 7.15E-03 |
| OsAffx.13105.1.S1_at | LOC_Os03g29250 | ids4-like protein | 134.93 | 9.43 | 14.3 | 6.16E-03 |
| Os.51747.1.S1_at | LOC_Os03g29250 | ids4-like protein | 1,188.83 | 94.13 | 12.6 | 2.91E-03 |
| OsAffx.14039.1.S1_s_at | LOC_Os04g30040 | Jacalin-like lectin domain containing protein | 348.73 | 5.33 | 65.4 | 7.62E-03 |
| OsAffx.25096.1.S1_at | LOC_Os03g15360 | Leucoanthocyanidin reductase | 4,015.07 | 294.57 | 13.6 | 9.87E-04 |
| Os.42490.1.S1_at | LOC_Os01g62070 | Metal tolerance protein C3 | 28,270.2 | 199.37 | 141.8 | 4.92E-03 |
| Os.42490.1.S1_x_at | LOC_Os01g62070 | Metal tolerance protein C3 | 27,456.03 | 206.4 | 133 | 3.13E-03 |
| OsAffx.10363.1.S1_at | LOC_Os06g03500 | NBS-LRR disease resistance protein | 225.27 | 17.27 | 13 | 2.59E-02 |
| Os.12851.1.S1_at | LOC_Os01g14670 | Nectarin-1 precursor | 6,357.33 | 141.37 | 45 | 6.85E-04 |
| Os.12761.1.S1_at | LOC_Os03g46060 | Osmotin-like protein OSML13 precursor | 19,564.23 | 702.03 | 27.9 | 2.60E-03 |
| Os.52536.1.S1_at | LOC_Os04g43800 | Phenyl-Ala ammonia-lyase | 6,313.3 | 20.2 | 312.5 | 2.92E-03 |
| Os.50455.1.S1_at | LOC_Os06g40170 | Phospholipase D-α-2 | 3,267.17 | 166.77 | 19.6 | 9.06E-05 |
| Os.11575.1.S1_a_at | LOC_Os03g46440 | Regulatory protein NPR1 | 1,400.97 | 120.6 | 11.6 | 9.44E-03 |
| OsAffx.13793.1.S1_at | LOC_Os04g12840 | Resistance protein | 1,537.77 | 142.13 | 10.8 | 4.86E-05 |
| Os.10600.1.S1_a_at | LOC_Os01g25820 | Respiratory burst oxidase 2 | 12,428.1 | 1,231.33 | 10.1 | 2.68E-03 |
| Os.57186.1.S1_at | LOC_Os05g27590 | Wound-induced protein WI12-containing protein | 1,239.17 | 45.27 | 27.4 | 3.73E-02 |
| Signal transduction | ||||||
| OsAffx.13408.1.S1_at | LOC_Os03g48560 | BGGP β-1-3-galactosyl-O-glycosyl-glycoprotein | 350.6 | 24.93 | 14.1 | 7.61E-03 |
| OsAffx.13408.1.S1_x_at | LOC_Os03g48560 | BGGP β-1-3-galactosyl-O-glycosyl-glycoprotein | 224.77 | 19.97 | 11.3 | 1.68E-03 |
| OsAffx.24314.1.S1_at | LOC_Os02g18930 | Calcineurin B-like protein 4 | 2,628.87 | 24.77 | 106.1 | 4.50E-03 |
| Os.15639.1.S1_at | LOC_Os06g40370 | CBL-interacting Ser/Thr-protein kinase 24 | 7,217.23 | 666.6 | 10.8 | 3.66E-03 |
| Os.48846.1.S1_at | LOC_Os09g24840 | GAST1 protein precursor | 3,067.53 | 136.57 | 22.5 | 1.28E-04 |
| Os.17201.1.S1_at | LOC_Os07g38810 | Lectin receptor-type protein kinase | 3,123.2 | 82.57 | 37.8 | 3.28E-03 |
| Os.6360.1.S1_at | LOC_Os08g02996 | Lectin-like receptor kinase 1 | 4,453.83 | 85.53 | 52.1 | 1.53E-03 |
| Os.6360.2.S1_x_at | LOC_Os08g02996 | Lectin-like receptor kinase 1 | 4,304.07 | 162.17 | 26.5 | 2.10E-03 |
| OsAffx.30716.1.S1_x_at | LOC_Os10g38960 | Lectin-like receptor kinase 7 | 3,306.8 | 244.23 | 13.5 | 3.84E-03 |
| Os.12660.1.S2_at | LOC_Os07g38800 | Lectin-like receptor kinase 7 | 2,745.87 | 27.73 | 99 | 2.88E-03 |
| Os.12660.1.S1_at | LOC_Os07g38800 | Lectin-like receptor kinase 7 | 938.63 | 16.2 | 57.9 | 1.14E-02 |
| Os.26943.1.S1_at | LOC_Os09g32840 | Nucleotide pyrophosphatase/phosphodiesterase | 3,333.23 | 205.1 | 16.3 | 1.96E-03 |
| OsAffx.29386.1.S1_x_at | LOC_Os08g27810 | OsWAK115 | 1,312.67 | 121.3 | 10.8 | 1.52E-02 |
| OsAffx.6309.1.S1_at | LOC_Os09g16980 | OsWAK86 | 2,528.73 | 9.33 | 270.9 | 2.87E-03 |
| Os.54625.1.S1_at | LOC_Os12g42020 | Protein kinase PVPK-1 | 7,055.77 | 441.67 | 16 | 2.73E-04 |
| Os.44814.1.S1_at | LOC_Os06g03610 | Protein kinase | 85.97 | 4.37 | 19.7 | 1.29E-02 |
| OsAffx.17892.1.S1_at | LOC_Os09g25540 | Receptor-like protein kinase precursor | 124.07 | 6.3 | 19.7 | 3.54E-02 |
| OsAffx.14892.1.S1_at | LOC_Os05g30740 | SRC2 | 823.27 | 54.3 | 15.2 | 6.54E-04 |
| Lipid metabolism | ||||||
| Os.5295.1.S1_at | LOC_Os08g10010 | Acyl-desaturase, chloroplast precursor | 12,530.77 | 158.63 | 79 | 4.74E-03 |
| Os.30537.1.S1_at | LOC_Os03g01820 | C-4 methylsterol oxidase | 7,759.1 | 306.77 | 25.3 | 1.48E-04 |
| Os.57035.1.S1_at | LOC_Os01g21300 | Catalytic/hydrolase | 605.6 | 45.2 | 13.4 | 1.21E-02 |
| Os.54454.1.S1_at | LOC_Os11g32650 | Chalcone synthase | 765.67 | 4.03 | 189.8 | 1.61E-02 |
| Os.57449.1.S1_x_at | LOC_Os11g32650 | Chalcone synthase | 10,288 | 150.9 | 68.2 | 3.56E-03 |
| Os.37295.1.S1_at | LOC_Os11g02440 | Chalcone-flavonone isomerase | 2,854.53 | 13.1 | 217.9 | 7.51E-03 |
| Os.6354.1.S1_s_at | LOC_Os12g02370 | Chalcone-flavonone isomerase | 13,196.17 | 228.1 | 57.9 | 1.56E-04 |
| Os.6354.1.S1_at | LOC_Os12g02370 | Chalcone-flavonone isomerase | 5,023.07 | 118.8 | 42.3 | 4.68E-04 |
| Os.53034.1.A1_at | LOC_Os11g18194 | Cycloartenol synthase | 10,955.6 | 23.7 | 462.3 | 4.83E-03 |
| Os.52235.1.S1_at | LOC_Os02g21810 | Cyt P450 | 2,250.9 | 4.37 | 515.5 | 6.50E-03 |
| Os.25621.2.S1_at | LOC_Os12g16720 | Cyt P450 | 16,932.33 | 546.27 | 31 | 2.06E-03 |
| Os.53717.1.S1_at | LOC_Os06g01250 | Cyt P450 | 13,610.93 | 88.73 | 153.4 | 2.18E-04 |
| Os.39637.1.A1_s_at | LOC_Os09g17000 | Glycerophosphoryl diester phosphodiesterase family protein | 6,529.73 | 75.93 | 86 | 3.88E-03 |
| Os.39637.1.A1_at | LOC_Os09g17000 | Glycerophosphoryl diester phosphodiesterase family protein | 4,584.1 | 73.8 | 62.1 | 2.93E-03 |
| Os.47906.1.A1_at | LOC_Os06g49770 | Lipid-binding protein | 7,864.5 | 210.5 | 37.4 | 3.39E-03 |
| OsAffx.32067.1.S1_x_at | LOC_Os12g37320 | Lipoxygenase 2.2, chloroplast precursor | 194.63 | 7.5 | 26 | 4.50E-03 |
| Os.41251.1.A1_at | LOC_Os01g43140 | Triacylglycerol lipase | 2,613.6 | 244.57 | 10.7 | 3.00E-04 |
| OsAffx.14502.1.S1_at | LOC_Os05g02350 | Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 | 393.47 | 12.6 | 31.2 | 1.50E-03 |
| Os.29290.1.S1_at | LOC_Os01g08780 | Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 | 4,496.2 | 65.27 | 68.9 | 3.62E-03 |
| Transport | ||||||
| Os.17440.1.S1_at | LOC_Os04g44610 | ABC transporter C05D10.3 in chromosome III | 9,093.1 | 674.9 | 13.5 | 5.79E-04 |
| Os.55221.1.S1_at | LOC_Os08g43120 | ABC transporter | 11,133.57 | 462.03 | 24.1 | 1.16E-02 |
| OsAffx.3427.1.S1_at | LOC_Os03g37960 | Acyl-CoA-binding protein | 782.93 | 58.17 | 13.5 | 4.37E-03 |
| Os.26802.2.S1_at | LOC_Os12g08090 | Amino acid transporter | 18,660.77 | 1,722.8 | 10.8 | 4.13E-04 |
| OsAffx.19991.1.S1_at | LOC_Os12g36660 | Antiporter/drug transporter/transporter | 1,091.07 | 41.33 | 26.4 | 5.61E-03 |
| OsAffx.24678.3.S1_at | LOC_Os02g41860 | Aquaporin PIP2.2 | 1,547.47 | 138.83 | 11.1 | 2.14E-03 |
| OsAffx.24678.2.S1_at | LOC_Os02g41860 | Aquaporin PIP2.2 | 790.03 | 73.83 | 10.7 | 8.10E-03 |
| Os.49307.1.S1_at | LOC_Os12g22284 | ABC subfamily G member 2 | 3,085.93 | 7.1 | 434.6 | 6.65E-04 |
| Os.52570.1.S1_at | LOC_Os12g44110 | ligA | 12,413.13 | 851.5 | 14.6 | 1.50E-02 |
| OsAffx.3105.1.S1_at | LOC_Os02g58530 | Major facilitator superfamily protein | 237.23 | 11.8 | 20.1 | 2.45E-02 |
| Os.27833.1.S1_at | LOC_Os12g32940 | Major myo-inositol transporter iolT | 2,600.73 | 98.1 | 26.5 | 5.41E-03 |
| OsAffx.20797.1.S1_at | LOC_Os10g02340 | Peptide transporter PTR2 | 275 | 12.63 | 21.8 | 4.68E-03 |
| Os.46447.1.S1_at | LOC_Os10g33210 | Peptide transporter PTR2 | 1,797.1 | 113.97 | 15.8 | 5.30E-03 |
| Os.11547.1.S1_s_at | LOC_Os06g48030 | Peroxidase 16 precursor | 9,654.5 | 622.3 | 15.5 | 5.94E-05 |
| Os.5843.1.S1_at | LOC_Os06g46799 | Peroxidase 39 precursor | 11,243.27 | 277.83 | 40.5 | 3.39E-03 |
| Os.53303.1.S1_at | LOC_Os06g46799 | Peroxidase 39 precursor | 1,034.43 | 56.8 | 18.2 | 5.73E-03 |
| Os.10754.1.S1_at | LOC_Os03g06520 | Sulfate transporter 3.1 | 7,211.23 | 668.47 | 10.8 | 3.14E-03 |
| Hormone related | ||||||
| Os.27822.2.S1_at | LOC_Os04g47520 | Auxin-independent growth promoter | 1,048.63 | 101.87 | 10.3 | 2.04E-02 |
| OsAffx.21458.1.S1_at | LOC_Os01g46030 | Brassinosteroid insensitive 1-associated receptor kinase 1 precursor | 278.63 | 17.87 | 15.6 | 6.22E-03 |
| OsAffx.28262.1.S1_at | LOC_Os07g04520 | Brassinosteroid insensitive 1-associated receptor kinase 1 precursor | 939.93 | 8.37 | 112.3 | 1.19E-02 |
| OsAffx.15447.1.S1_at | LOC_Os06g16000 | Cytokinin-O-glucosyltransferase 3 | 2,037.37 | 186.13 | 10.9 | 6.74E-04 |
| Os.36449.1.S1_at | LOC_Os01g57610 | IAA-amido synthetase GH3.1 | 10,154.47 | 533.07 | 19 | 4.14E-04 |
| OsAffx.32069.1.S1_at | LOC_Os12g37490 | N-Acetyltransferase | 405.6 | 22.13 | 18.3 | 8.10E-04 |
| Os.55720.1.S1_at | LOC_Os04g56690 | OsSAUR23 | 997.73 | 47.67 | 20.9 | 3.74E-03 |
| OsAffx.30169.1.S1_at | LOC_Os09g37430 | OsSAUR48 | 781.37 | 51.53 | 15.2 | 1.61E-03 |
| Os.51408.1.S1_at | LOC_Os09g37490 | OsSAUR54 | 1,076.63 | 64.17 | 16.8 | 1.47E-02 |
| Os.50785.1.S1_at | LOC_Os09g37500 | OsSAUR55 | 5,390.27 | 317.77 | 17 | 4.96E-03 |
| Protein metabolism | ||||||
| Os.54875.1.S1_at | LOC_Os05g04584 | 3-N-debenzoyl-2-deoxytaxol N-benzoyltransferase | 337.53 | 19.3 | 17.5 | 8.62E-04 |
| OsAffx.24010.1.S1_x_at | LOC_Os01g72020 | BOP2 | 806.33 | 30.57 | 26.4 | 2.00E-03 |
| Os.49276.1.S1_at | LOC_Os01g72020 | BOP2 | 1,908.9 | 162.2 | 11.8 | 1.03E-03 |
| Os.52793.1.S1_at | LOC_Os04g43840 | Carboxy-lyase | 5,289.97 | 30.97 | 170.8 | 2.02E-03 |
| Os.10714.1.S1_s_at | LOC_Os04g38450 | γ-Glutamyltranspeptidase 1 precursor | 1,0921.73 | 330.83 | 33 | 6.63E-04 |
| Os.10728.1.S1_at | LOC_Os02g46260 | Ser carboxypeptidase 1 precursor | 23,058.93 | 857.67 | 26.9 | 3.13E-03 |
| OsAffx.10889.1.S1_x_at | LOC_Os01g03170 | Seven in absentia protein family protein | 4,477.57 | 409.87 | 10.9 | 2.57E-04 |
| Os.3768.1.S1_at | LOC_Os02g44590 | Subtilisin-like protease precursor | 11,949.97 | 366.4 | 32.6 | 5.07E-03 |
| Os.8271.2.S1_x_at | LOC_Os01g67500 | Ubiquitin-protein ligase | 1,791.4 | 174.73 | 10.3 | 8.17E-03 |
| OsAffx.13747.1.S1_at | LOC_Os04g10360 | Xylem Ser proteinase 1 precursor | 4,292.53 | 12.23 | 350.9 | 6.60E-04 |
| Carbohydrate and energy metabolism | ||||||
| Os.53178.2.S1_at | LOC_Os05g47840 | ATIPT1 | 124.5 | 10.23 | 12.2 | 9.57E-03 |
| OsAffx.28287.1.S1_at | LOC_Os07g05380 | ATPase | 1,148.87 | 33.53 | 34.3 | 6.85E-03 |
| OsAffx.5436.1.S1_at | LOC_Os07g25540 | Disulfide oxidoreductase/monooxygenase/oxidoreductase | 197.67 | 8.37 | 23.6 | 1.74E-03 |
| OsAffx.31989.1.S1_at | LOC_Os12g32750 | Disulfide oxidoreductase/monooxygenase/oxidoreductase | 2,854.47 | 149.03 | 19.2 | 6.31E-04 |
| Os.18223.1.S1_s_at | LOC_Os12g32750 | Disulfide oxidoreductase/monooxygenase/oxidoreductase | 7,562.97 | 747.77 | 10.1 | 3.81E-04 |
| OsAffx.13016.1.S1_at | LOC_Os03g25150 | Flavonoid 3,5-hydroxylase 2 | 1,361.83 | 57.23 | 23.8 | 4.06E-02 |
| OsAffx.13016.1.S1_x_at | LOC_Os03g25150 | Flavonoid 3,5-hydroxylase 2 | 1,470.4 | 78.63 | 18.7 | 4.62E-02 |
| OsAffx.27142.1.S1_at | LOC_Os05g33644 | Hydrolase | 1,714.27 | 93.2 | 18.4 | 2.38E-02 |
| Os.46268.1.S1_at | LOC_Os10g02390 | NAD(P)H-dependent oxidoreductase | 7,951.73 | 3.63 | 2,188.6 | 6.30E-03 |
| Nucleic acid metabolism | ||||||
| Os.855.1.S1_at | LOC_Os01g03730 | Nuclease PA3 | 5,603.9 | 85.67 | 65.4 | 1.12E-02 |
| Os.55424.2.S1_at | LOC_Os02g50740 | Nucleotide binding protein | 2,223.23 | 164.1 | 13.5 | 8.77E-05 |
| Amino acid metabolism | ||||||
| Os.27876.1.S1_x_at | LOC_Os08g04540 | Aromatic-l-amino acid decarboxylase | 3,587.43 | 19.3 | 185.9 | 1.49E-02 |
| OsAffx.5702.1.S1_s_at | LOC_Os08g04560 | Aromatic-l-amino acid decarboxylase | 23,981.57 | 192.23 | 124.8 | 9.93E-04 |
| Unclassified | ||||||
| Os.48933.1.S1_at | LOC_Os05g37880 | axi 1-like protein | 12,167.7 | 1,092.03 | 11.1 | 1.32E-03 |
| OsAffx.3289.1.S1_at | LOC_Os03g21450 | Bromodomain-containing protein | 433.77 | 23.43 | 18.5 | 2.21E-02 |
| Os.28450.1.S1_at | LOC_Os01g70730 | FLP1 | 3,929.67 | 103.73 | 37.9 | 1.54E-04 |
| Os.6003.1.S1_x_at | LOC_Os09g26310 | Hypro1 | 3,628.77 | 109.37 | 33.2 | 4.80E-05 |
| Os.51638.1.S1_s_at | LOC_Os09g26310 | Hypro1 | 4,551.53 | 161 | 28.3 | 4.31E-04 |
| Os.49681.1.S1_at | LOC_Os03g07140 | Male sterility protein 2 | 2,714.93 | 42.37 | 64.1 | 4.57E-02 |
| Os.24786.1.S1_s_at | LOC_Os04g48870 | Nitrilase-associated protein | 4,117.03 | 126.8 | 32.5 | 2.12E-02 |
| OsAffx.26533.1.S1_at | LOC_Os04g48870 | Nitrilase-associated protein | 4,745.7 | 152.2 | 31.2 | 1.16E-02 |
| OsAffx.12915.1.S1_at | LOC_Os03g18300 | Pherophorin-like protein | 570.47 | 21.47 | 26.6 | 1.84E-03 |
| Os.46488.1.S1_at | LOC_Os10g05790 | Pro-rich protein | 28,988.2 | 512.93 | 56.5 | 1.96E-03 |
| OsAffx.16391.1.S1_at | LOC_Os07g24100 | Retrotransposon protein | 707.07 | 5.73 | 123.3 | 1.28E-04 |
| Os.28346.5.S1_x_at | LOC_Os03g63124 | Retrotransposon protein, unclassified | 242 | 23.33 | 10.4 | 1.57E-02 |
| Os.50793.1.S1_at | LOC_Os12g01900 | Retrotransposon protein, unclassified | 621.7 | 6.53 | 95.2 | 7.10E-03 |
| Os.55132.1.S1_at | LOC_Os03g26990 | VQ motif family protein | 222.07 | 20.03 | 11.1 | 1.21E-02 |
The gene probes of 57 K Affymetrix rice GeneChip. ID, Identity.
The gene's average signal intensity in stigma derived from three biological replicates of 57 K Affymetrix rice GeneChip. Stigma-S, Stigma signal.
The gene's average signal intensity in ovary derived from three biological replicates of 57 K Affymetrix rice GeneChip. Ovary-S, Ovary signal.
The ratios of stigma/ovary.
GO analysis of the rice stigma-preferential genes was performed, and the biological process term enrichment status and hierarchy are shown in Supplemental Figure S1. The results showed that the DNA-dependant transcription may play an important role in the stigma, and the biological processes that respond to hormone stimuli, especially to auxin, were overrepresented and seemed to be more significant in the rice stigma-specific or -preferential gene data set.
Comparison of the Stigma Preferentially Expressed Genes in Rice and Arabidopsis
We compared the stigma preferentially expressed genes in rice with that identified in Arabidopsis (Tung et al., 2005). We extracted the protein sequences for 115 Arabidopsis genes, blasted them against the rice Affymetrix consensus sequence database (https://www.affymetrix.com/analysis/netaffx/index.affx), and filtered 3,504 probe sets using E value ≤ 1e-4. We found 45 Arabidopsis genes hitting 92 rice probe sets with the stigma preferentially expression pattern (Supplemental Table S8). Meanwhile, we also blasted the sequences of the 665 rice probe sets against the Arabidopsis whole-genome protein sequence database (http://www.arabidopsis.org/Blast/index.jsp) and found that 90 rice probe sets (83 genes) were similar to 42 Arabidopsis stigma-specific/preferentially expressed genes (Supplemental Table S8). From both BLAST results, we found that they matched to each other well. The gene annotation showed that the majority of them encoded the cell wall-related and signal transduction-related proteins. We also listed the number of the stigma-preferential genes unique to rice (465) and Arabidopsis (70).
Validation of the Microarray Results
Real-time quantitative reverse transcription (qRT)-PCR analysis was employed to validate the candidate genes. In total, 34 of the identified genes preferentially expressed in stigma were selected, including 30 putative function genes belonging to different classification groups, two hypothetical proteins, one expressed protein, and one unknown function gene. The signal intensity range of the selected genes was from 107.8 to 2,8284.3 and the ratios (stigma versus ovary) from 2.6 to 2,188.5 (see Supplemental Table S1). To confirm whether these genes were stigma-specific by qRT-PCR analysis, the expression of the 34 candidate genes was compared between the stigma and the ovary, anther, seeding-shoot, or flag leaf (heading stage) samples (Table II). The results showed that their expression patterns detected by 57 K Affymetrix rice whole-genome array were in good correlation with those obtained by qRT-PCR.
Table II.
Verification of microarray results by real-time quantitative RT-PCR
ID, Identification.
| Probe Set IDa | Gene ID | Description | Stigma/Ovaryb | Stigma/Antherc | Stigma/Shootd | Stigma/Leafe |
|---|---|---|---|---|---|---|
| Os.12922.1.S1_at | LOC_Os08g10010 | Acyl-desaturase, chloroplast precursor | 182.34 ± 26.76 | 165.43 ± 23.822 | 2.29 ± 0.053 | 4,239.26 ± 1,003.77 |
| Os.2696.1.S1_at | LOC_Os04g15840 | α-Expansin 1 precursor | 1.46 ± 0.033 | 3.43 ± 0.25 | 10.18 ± 1.399 | 2.07 ± 0.089 |
| Os.31669.1.S1_at | LOC_Os01g53370 | Anthocyanidin 5,3-O-glucosyltransferase | 86.66 ± 1.7 | 21.05 ± 0.282 | 23.54 ± 0.327 | 482.61 ± 13.111 |
| OsAffx.5702.1.S1_s_at | LOC_Os08g04560 | Aromatic-l-amino acid decarboxylase | 2,087.29 ± 265.908 | 369.34 ± 36.352 | 68.17 ± 4.788 | 69.3 ± 4.887 |
| OsAffx.20044.1.S1_at | LOC_Os12g40080 | B3 DNA-binding domain containing protein | 12,823.5 ± 1,586.625 | 1,647.15 ± 159.426 | 1,204.98 ± 111.692 | 689.49 ± 58.868 |
| OsAffx.23995.2.S1_x_at | LOC_Os01g71094 | Basic 7S globulin 2 precursor | 32.59 ± 0.873 | 7.53 ± 0.117 | 25.4 ± 0.632 | 6.65 ± 0.097 |
| Os.22594.2.S1_at | LOC_Os01g03390 | Bowman-Birk-type bran trypsin inhibitor precursor | 24.59 ± 1.408 | 11.7 ± 0.514 | 14.42 ± 0.688 | 67.8 ± 5.111 |
| Os.2322.1.S1_at | LOC_Os10g33250 | CER1 | 319.88 ± 40.499 | 38.1 ± 3.04 | 1,362.35 ± 216.136 | 1,308.77 ± 206.473 |
| Os.27065.1.S1_a_at | LOC_Os01g59180 | Cyclin-like F box | 4.77 ± 0.059 | 14 ± 0.295 | 6.53 ± 0.098 | 25.4 ± 0.657 |
| Os.51106.1.S1_at | LOC_Os06g14540 | Endoglucanase 1 precursor | 252.24 ± 25.059 | 188.66 ± 17.754 | 130.53 ± 11.419 | 34.2 ± 2.167 |
| OsAffx.28068.1.S1_at | LOC_Os06g42730 | Esterase/lipase/thioesterase | 4.03 ± 0.053 | 2.26 ± 0.017 | 2.3 ± 0.018 | 6.33 ± 0.111 |
| OsAffx.21178.1.S1_x_at | LOC_Os01g68140 | Expressed protein | 97.87 ± 9.342 | 1,921.16 ± 303.246 | 4,341.75 ± 759.941 | 21,884.94 ± 4,580.042 |
| Os.5295.1.S1_at | LOC_Os09g36700 | Extracellular ribonuclease LE precursor | 1.34 ± 0.002 | 10.64 ± 0.161 | 1.53 ± 0.004 | 16.49 ± 0.296 |
| Os.29814.1.S1_at | LOC_Os03g62860 | Glucan endo-1,3-β-glucosidase A6 precursor | 119.47 ± 6.068 | 53.08 ± 2.238 | 134.61 ± 7.007 | 52.28 ± 2.196 |
| Os.50860.1.S1_at | LOC_Os09g27680 | Hypothetical protein | 19.23 ± 0.535 | 2.44 ± 0.02 | 9.97 ± 0.215 | 1.68 ± 0.008 |
| Os.55613.1.S1_at | LOC_Os01g65692 | Hypothetical protein | 96.45 ± 3.506 | 55.1 ± 1.758 | 55.83 ± 1.787 | 50.6 ± 1.58 |
| Os.12660.1.S2_at | LOC_Os07g38800 | Lectin-like receptor kinase 7 | 9.95 ± 0.143 | 6.95 ± 0.085 | 10.67 ± 0.158 | 3.35 ± 0.025 |
| Os.47906.1.A1_at | LOC_Os06g49770 | Lipid-binding protein | 117.27 ± 8.537 | 46.61 ± 2.735 | 16.09 ± 0.683 | 2.74 ± 0.042 |
| Os.31652.1.S1_at | LOC_Os01g06320 | myb-like DNA-binding domain protein | 37.96 ± 0.79 | 6.3 ± 0.066 | 148.69 ± 4.255 | 79.49 ± 1.99 |
| Os.46268.1.S1_at | LOC_Os10g02390 | NAD(P)H-dependent oxidoreductase | 51.29 ± 0.696 | 4.37 ± 0.022 | 5.15 ± 0.029 | 4.95 ± 0.027 |
| Os.10675.1.A1_at | LOC_Os03g46060 | Osmotin-like protein OSML13 precursor | 11.46 ± 0.017 | 26.72 ± 0.054 | 34.75 ± 0.076 | 52.39 ± 0.129 |
| OsAffx.30169.1.S1_at | LOC_Os09g37430 | OsSAUR48 | 15.39 ± 1.334 | 43.55 ± 5.216 | 19.91 ± 1.889 | 53.37 ± 6.738 |
| Os.51408.1.S1_at | LOC_Os09g37490 | OsSAUR54 | 105.91 ± 2.819 | 2.21 ± 0.01 | 2.79 ± 0.016 | 15.13 ± 0.235 |
| OsAffx.3726.1.S1_at | LOC_Os04g05050 | Pectate lyase 8 precursor | 14.62 ± 0.457 | 5.22 ± 0.101 | 6.46 ± 0.141 | 608.27 ± 45.45 |
| Os.53102.1.S1_s_at | LOC_Os09g21000 | Potassium transporter 12 | 38.62 ± 0.749 | 8.77 ± 0.101 | 12.95 ± 0.176 | 12.75 ± 0.172 |
| Os.10728.1.S1_at | LOC_Os02g46260 | Ser carboxypeptidase 1 precursor | 42.35 ± 0.681 | 43.97 ± 0.714 | 8.72 ± 0.081 | 49.59 ± 0.831 |
| Os.12761.1.S1_at | LOC_Os02g46260 | Ser carboxypeptidase 1 precursor | 125.09 ± 21.094 | 500.45 ± 108.954 | 71.65 ± 10.678 | 184.39 ± 33.617 |
| Os.41415.1.S1_at | LOC_Os01g40499 | Ser/Thr-protein kinase receptor precursor | 14.03 ± 1.286 | 5.34 ± 0.31 | 11.06 ± 0.921 | 4.3 ± 0.217 |
| OsAffx.14892.1.S1_at | LOC_Os05g30740 | SRC2 | 931.33 ± 5.402 | 3,901.73 ± 27.372 | 111.49 ± 0.446 | 126.66 ± 0.52 |
| Os.189.1.S1_at | LOC_Os08g43160 | TCP family transcription factor containing protein | 1.99 ± 0.006 | 1.87 ± 0.005 | 5.43 ± 0.042 | 2.79 ± 0.013 |
| Os.52592.1.S1_at | LOC_Os02g49480 | Transcription factor BIM1 | 8.67 ± 0.452 | 7.61 ± 0.373 | 6.02 ± 0.261 | 9.38 ± 0.507 |
| Os.8271.2.S1_x_at | LOC_Os01g67500 | Ubiquitin-protein ligase | 11.65 ± 0.081 | 25.74 ± 0.236 | 26.56 ± 0.246 | 45.2 ± 0.486 |
| Os.56298.1.S1_at | LOC_Os03g63810 | WRKY transcription factor 14 | 296.26 ± 15.558 | 95.22 ± 4.002 | 5.54 ± 0.087 | 5.51 ± 0.087 |
| Os.23805.1.S1_at | AK071040 | Unknown | 63.34 ± 1.016 | 77.33 ± 1.3 | 301.67 ± 6.661 | 1,212.9 ± 33.31 |
The gene probes of 57 K Affymetrix rice GeneChip.
Real-time RT-PCR results ± the sds for stigma/ovary.
Real-time RT-PCR results ± the sds for stigma/anther.
Real-time RT-PCR results ± the sds for stigma/shoot (seedling shoot).
Real-time RT-PCR results ± the sds for stigma/leaf (mature leaf).
Confirmation of the Candidate Stigma-Specific Genes by RNA in Situ Hybridization
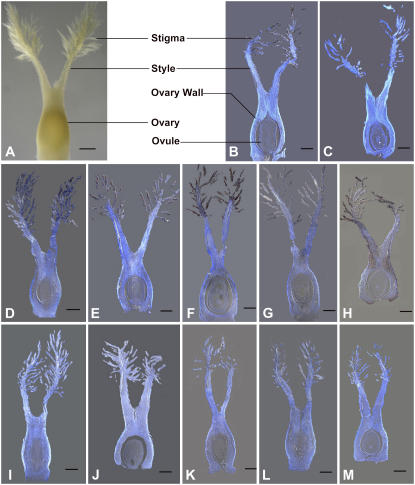
To further examine the expression of the candidate stigma-specific genes, we selected five genes to perform RNA in situ hybridization, including a Ser carboxypeptidase 1 precursor gene (LOC_Os02g46260), an extracellular ribonuclease LE precursor gene (LOC_Os09g36700), a putative CER1 gene (LOC_Os10g33250), a metal tolerance gene C3 (LOC_Os01g62070), and an unknown function gene (AK071040). They exhibited relatively high hybridization signals in the stigma microarray data sets, ensuring that their transcripts could be detected by the RNA in situ hybridization technology. A CCCH-type zinc finger protein gene (LOC_Os01g09620) expressed in the entire mature pistil according to the microarray results was used as a reference. Longitudinal sections through the center of pistil just before pollination were used for all the hybridizations. The results showed that the five selected genes exhibited unique expression patterns in the stigma papilla cells (Fig. 1) and that the hybridization signals of the examined genes were in good correlation with their intensities in 57 K Affymetrix rice GeneChip. Taken together, our results showed that the 548 genes preferentially expressed in the stigma thus identified represented good candidates for the stigma-specific genes in rice.
Figure 1.
Expression of five stigma preferential genes by RNA in situ hybridization. A, A rice pistil before pollination. B, D, E, F, G, and H, Hybridization by the antisense probes of LOC_Os02g46260, LOC_Os09g36700, LOC_Os10g33250, LOC_Os01g62070, AK071040, and LOC_Os01g09620, respectively. C, I, J, K, L, and M, Hybridization by their respective sense probes. Among them, the gene LOC_Os01g09620 (H and M) was used as a reference, because the microarray result showed that it was expressed in the entire mature pistil. The longitudinal section through the pistil before pollination was used for all the hybridizations. All of the slices were stained with Fluorescent Brightener 28 after hybridization. The hybridization signals are shown in dark purple. The structures of the pistil are illustrated in A and B. Bar = 100 μm.
cis-Acting Regulatory Element Analyses of the Candidate Stigma-Specific Genes
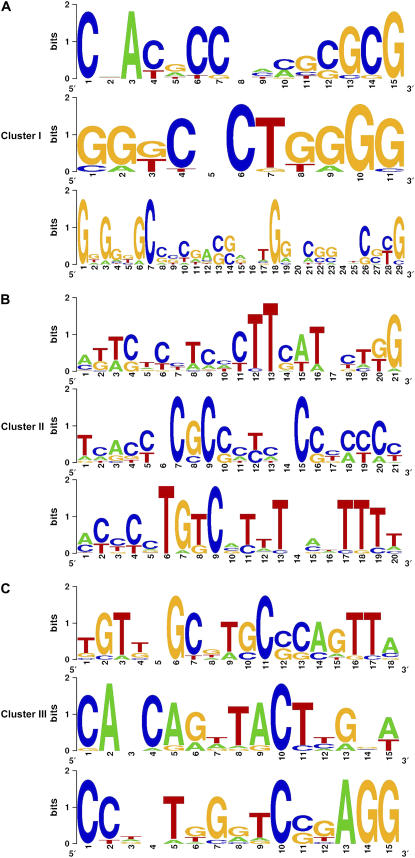
To identify possible cis-acting regulatory elements responsible for the transcription regulation of the candidate stigma-specific genes, we first did the hierarchical clustering analysis and found that nearly 35% of the identified genes (193) were mainly located in three clades (Supplemental Fig. S2). The common features were that all the genes in the three clades were specific or highly expressed in the stigma, and the correlation coefficients of the three clusters were 0.9277, 0.9389, and 0.8263, respectively. We named the three clades as Cluster I, II, and III, respectively. Then, 1,000-bp regions located in the upstream of the start codons of the genes from the three clusters were used for analysis. Several conserved motifs were subsequently identified using the MEME (Multiple Em for Motif Elicitation)/MAST (Motif Alignment and Search Tool) system (Bailey and Elkan, 1994; Bailey and Gribskov, 1998). Figure 2 shows the top three motifs identified for each cluster. The E values of the three motifs are: cluster I, 3.5e + 004, 3.3e + 006, and 7.3e + 001, respectively; cluster II, 1.1e + 006, 2.8e + 003, and 2.2e + 005, respectively; and cluster III, 7.5e + 005, 3.3e + 006, and 1.2e + 007, respectively. The common features of the cis-acting regulatory elements we identified from the conserved motifs using the PLACE database were mostly related to stress or defense responses (Prestridge, 1991; Higo et al., 1999). A large percentage of the genes in cluster I had the same motif named GCC box (Brown et al., 2003; Chakravarthy et al., 2003). Most of genes in cluster III had Myb-transcription factor recognition sites that are usually found in the promoters of the dehydration-responsive genes. One of the motifs (Fig. 2C, bottom) in cluster III appeared to have unknown function, indicating that they might represent new cis-acting elements in regulating the transcription of the stigma-specific genes.
Figure 2.
Putative cis-acting regulatory elements enriched in the stigma-specific or preferential genes. A to C, The top three candidate motifs of cluster I, cluster II, and cluster III, respectively. The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the heights of symbols within the stack reflect the relative frequencies of the corresponding nucleic acids at that position.
DISCUSSION
In this study, we have identified 548 genes expressed specifically or predominantly in the stigma papillar cells of rice by using 57 K Affymetrix rice whole-genome array and 10 K rice cDNA microarray. It is highly likely that they represent good candidates for the stigma-specific genes in rice. First, for the two different technological platforms we used, there existed good correlations, and the variables observed were reasonable according to several recent reports (Maruyama et al., 2004; Yauk et al., 2004; Hannah et al., 2005). Second, the qRT-PCR and RNA in situ hybridization confirmed the microarray results (Table II; Fig. 1). Third, the rice stigma gene data set is similar to that identified in Arabidopsis (Tung et al., 2005). Thus, the careful selection of the starting tissues is an effective approach to identify the stigma-specific genes using the whole-genome and cDNA microarrays.
The Stigma-Specific Genes Have Conservative Roles in Plants
The functional annotation of both rice and Arabidopsis genes specifically or preferentially expressed in stigma suggests that several groups of the genes appear to play conserved roles in the stigma. Forty-two Arabidopsis stigma-specific genes representing 36% of the gene set are highly similar to 83 genes representing about 15% of the rice stigma gene dataset (Supplemental Table S8). The majority of them belong to the cell wall-related and signal transduction groups, indicating that these two classes of genes have conserved functions in the stigma. Swanson et al. (2005) also found that these two functional categories were overrepresented in contrast to the distributions of the total stigma message RNA. The cell wall-related group represents the second largest group of the genes identified in the rice stigma gene data set, which include the genes encoding pectinesterase, peroxidase, and α-expansin. They are putative cell wall-localized enzymes and may be the most suitable candidates for the papilla cell wall loosening or expansion (Wu et al., 1996; Schopfer, 2001). The cellulose synthase, glycosyl transferase, and endoglucanase may be involved in the cell wall synthesis required for the pollen tube growth (Hong et al., 2001; Goubet et al., 2003; Aspeborg et al., 2005; Yokoyama and Nishitani, 2006). Consistent with the cell-cell communication in the pollen-pistil interaction, the rice stigma gene dataset also contained a significant proportion of signal transduction-related genes (8.9%). Protein kinases play crucial roles in a wide variety of cellular functions, including proliferation and differentiation, by adding phosphate groups to Thr, Tyr, and Ser residues of specific target proteins. The plant receptor-like kinases likely are transmembrane proteins that transduce external messages into the cell through their extracellular domains and intracellular kinase domains. Previously, this protein category was identified and implicated in the pollen-pistil interaction and pollen tube growth (Muschietti et al., 1998; Tang et al., 2002). In Brassica, the stigma-expressed SRK interacts with SCR and initiates a signal transduction cascade that inhibits the pollen rehydration and, consequently, the growth of self pollen, while allowing nonself pollen to grow (Kachroo et al., 2002; Takayama and Isogai, 2005). The putative protein kinases and putative protein receptor-like kinase identified in the rice stigma gene sets are likely involved in the pollen-stigma interaction and the early steps of pollination. The three genes confirmed by whole mount RNA in situ hybridizations in the Arabidopsis study all had the homologous genes in our stigma-specific gene list (Tung et al., 2005). At5g59810 is related to LOC_Os02g44590, encoding an expressed subtilisin-like protease precursor protein; At5g19880 is related to LOC_Os06g48030, encoding an expressed putative peroxidase 16 precursor; and At2g02850 is related to LOC_Os03g02400, encoding an expressed early nodulin-like protein 1 precursor protein. Thus, our results and those using Arabidopsis suggest that the stigma involves the genes with the highly conserved functions.
The Phytohormone Auxin and Stigma Function during Pollination
Auxin is the central growth regulator of a myriad of aspects of plant growth and developmental processes and appears to be actively transported throughout the plant to control cell division, extension, and differentiation (Benjamins et al., 2005). Several roles have been suggested for auxin in the pollination process. In orchid, it has been shown that the pollination and auxin regulate the ethylene production and ovary development, because when inhibitors of ethylene were used, the pollination- or auxin-induced ovary development was inhibited (Zhang and O'Neill, 1993). During Arabidopsis flower development, the concentration of free auxin increased gradually, starting at the floral organ tip visualized by immunolocalization with polyclonal antibodies against auxin and accumulated in pollen grains and stigma before fertilization (Aloni et al., 2006). In maize, the augmentation of auxin in the pistil tissues is important for egg cell differentiation (Mol et al., 2004).
However, it is unclear whether auxin is involved directly in the interaction of the pollen and stigma. In the hormone-related genes group from our data set, the auxin-related genes appear to be overrepresented (Table I; Supplemental Table S7), and the results of the GO analysis of the rice stigma also suggested that the auxin signaling plays a significant role in the rice stigma function (Supplemental Fig. S1). The five of the identified auxin-related genes are SAURs (Jain et al., 2006). They are LOC_Os04g56690, LOC_Os09g37430, LOC_Os09g37440, LOC_Os09g37490, and LOC_Os09g37500, namely OsSAUR23, OsSAUR48, OsSAUR49, OsSAUR54, and OsSAUR55, respectively. Interestingly, four of these belong to a big gene cluster that contains 17 OsSAURs contiguously arranged in chromosome 9 (Jain et al., 2006). The SAURs encode short-lived nuclear proteins that are induced within minutes after auxin application and may play a role in auxin-mediated cell elongation (McClure and Guilfoyle, 1989; Franco et al., 1990; Gee et al., 1991; Knauss et al., 2003). It was reported that a suitable amount of auxin could lead to loosening of the cell wall and promote cell growth through inducing H(+) secretion and activating cellulose synthesis (Fry et al., 1990). The auxin response factor seems to control the expression of the OsSAURs through the auxin-responsive elements present in their promoter regions (Ulmasov et al., 1997). These studies indicate that auxin may be involved in loosening the stigma papillar cell wall and promoting pollen tube growth through inducing the expression of OsSAURs. The IAA-amido synthetase GH3.1 gene (LOC_Os01g57610) is involved in the auxin homeostasis through the conjugation of IAA to amino acids (Terol et al., 2006). But it is unclear where the auxin comes from. Could it be synthesized in the stigma papillar cell or transported from other organs (see below)? Further analyses of these candidate genes will shed light on this aspect of auxin signaling in the stigma during pollination.
Transport Function in the Stigma
The transport-related proteins are mostly abundant in the rice stigma gene dataset and appear to be actively involved in the stigma function. The rice pollen is not fully dehydrated and metabolically active when shed from the anthers. They likely require the stigma to be fully developed to supply all the nutrition and metabolites for pollen germination and tube growth. The different kinds of transporters appear to be involved in the exchange of materials and information between pollen and stigma. The ABC transporter is one of the active transport systems of the cell, which is widespread in prokaryotes and eukaryotes. In tobacco, the gene NtWBC1 is developmentally regulated in the stigma/style, with mRNA accumulation increasing toward anthesis (Otsu et al., 2004). RNA in situ hybridization experiments demonstrated that NtWBC1 is predominately expressed in the stigmatic secretory zone. There are four ABC transporters expressed in the stigma papillar cells of rice (Table I; Supplemental Table S7). Two recent reports indicate that plant ABC transporters mediate the cellular and long-distance transport of the plant hormone auxin (Sidler et al., 1998; Multani et al., 2003). The functional study of the four ABC transporters may help to clarify the mechanism of the auxin signaling in the stigma during pollination. The potassium uptake gene expression was highly enriched in the stigma. Recently, it has been reported that potassium may regulate anther dehiscence, pollen imbibition, and papilla hydration (Rehman and Yun, 2006). In Brassica, aquaporin-like proteins in the stigma may act as water channels to regulate water flow into the pollen grain from the stigma (Dixit et al., 2001). There are 33 genes annotated for aquaporins in the rice genome that are predicted to enable the fast and controlled translocation of water across the membrane, and most of them have been identified and characterized recently (Sakurai et al., 2005; Guo et al., 2006). From our dataset, one aquaporin protein-encoding gene, PIP2.2 (LOC_Os02g41860), was identified to have a specific and enriched expression in the rice stigma papilla cells, suggesting that it may function in the stigma during pollination. However, it remains an important task to address their roles in pollination.
Possible Cross Talk of Stress Responses and Pollination
Previously, we detected an extensive overlap of the genes involved in pollination and abiotic stress responses (Lan et al., 2005). Consistent with this, many stigma-specific genes also were related to stress and defense genes, such as those encoding MLO-like proteins, heat shock protein-binding proteins, disease resistance proteins, Bowman-Birk-type bran trypsin inhibitor precursor proteins, and wound-induced protein WI12-containing protein (Table I; Supplemental Table S7). Defense-related compounds have been found to accumulate specifically in the stigma; the proteinase inhibitor proteins and chitinase are highly enriched in stigmas of Solanaceous plants (Leung, 1992; Atkinson et al., 1993). It has been shown that the pollination and wounding induced nearly identical flavonol kinetics in petunia and patterns of the accumulation in the outer cell layers and exudates of the stigma, suggesting that they share elements of a common signal transduction pathway (Vogt et al., 1994). In petunia flowers, both pollination and stigma wounding induced a transient increase in ethylene production and hastened corolla senescence (Woltering et al., 1997). Further, it has been reported that the SPP2 (Solanum pollinated pistil) dioxygenase from self-incompatible wild potato (Solanum chacoense) is predominantly expressed in the pistil and could be induced by wounding of the style as well as pollination/fertilization (Lantin et al., 1999). The SPP2 dioxygenase could be involved in the biosynthesis of deterrent alkaloids in reproductive tissues or in generating chemical signals involved in pollen tube guidance (Lantin et al., 1999). One lectin domain-encoding gene belonged to the stress-related group. A b-lectin receptor kinase gene has been shown to confer the rice blast resistance (Chen et al., 2006). Recently, Chu et al. (2006) have demonstrated that R gene xa13, a recessive allele conferring disease resistance against bacterial blight, plays a key role in both disease resistance and pollen development and a trade-off exists between fertility and pathogen defense, depending on the levels of expression of the xa13 gene.
Our analysis also revealed that the stigma-specific genes share some common cis-regulatory elements with the stress-responsive genes. A large percentage of genes in cluster I possess the same motif, named GCC box (Fig. 2). The GCC box has been found in many pathogen-responsive genes (Brown et al., 2003; Chakravarthy et al., 2003). GbERF belongs to the ethylene-responsive factor (ERF) family of transcription factors and regulates the GCC box containing pathogen-related genes (Qin et al., 2006). The GCC box is found not only in the pathogen-responsive genes but also existed in some abiotic stress-related genes. In Arabidopsis, AtERF7 binds to the GCC box, and overexpressing AtERF7 showed a reduced sensitivity to drought stress (Song et al., 2005). The rice OsBIERF3 could bind specifically to the GCC box sequence and was induced by salt, cold, drought, wounding, and the rice blast fungus (Cao et al., 2006). Further work is required to dissect the molecular basis of this possible cross talk.
In conclusion, we have identified a large set of genes that are specifically or highly enriched in the rice stigma. In addition to the conserved roles of the cell wall metabolism and cellular communication in the stigma, the identification of genes involved in the auxin signaling, transcription functions, and possible cross talk between pollination and stress/defense responses provides new insights into the molecular functions of the stigma in rice.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa) ‘Nipponbare’ seeds were germinated and grown in a growth container for 2 weeks (28°C, 16 h light, 8 h dark, and photo intensity of 240 μm photos m−2 s−1). Then, shoot and root were harvested for Sh and Rt, respectively. Other plant materials used in this study described below were harvested from rice plants grown under natural conditions in the field of the Institute of Genetics and Developmental Biology, Beijing, during May to October and Hainan, China, during December to April. An were collected at a stage 1 to 2 d before floret flowering; unpollinated St and Ov were dissected at a stage 0 to 1 d before floret flowering; pistils of 5DAP were dissected 5 d after anthesis, 10EM and 10EN were dissected from grains 10 d after flowering, respectively.
Suspension Cell Culture
The method used for suspension cell culture was described by Wang et al. (2005).
10 K cDNA Microarray Hybridization and Data Analysis
Total RNA was isolated using RNeasy kit (Qiagen). Double-strand cDNA was synthesized from 5 μg total RNA using cDNA Synthesis kit (TaKaRa). In vitro transcription from cDNA to cRNA was performed using T7 RiboMAX Express Large Scale RNA Production system (Promega). Then the cRNA was converted to DNA by using Superscript II RT kit (Invitrogen) and random primers. One microgram DNA product and random primers (9-mers) were annealed to denatured DNA template and extended by Klenow fragment (TaKaRa) in the presence of Cy5-dCTP/Cy3-dCTP (Amersham Pharmacia) for target preparation in microarray analysis. Hybridization and washing were performed as described in CyScribe Post-Labeling kit (Amersham Pharmacia) and CMTTM hybridization chamber (Corning) user manuals. Scanning and data acquisition were performed on a GenePix 4000B scanner using GENEPIX 4.0 software (Axon Instruments). GenePix Pro 4.0 output files were converted to The Institute for Genomic Research Multi Experiment Viewer file (.mev) by ExpressConverter V1.4 (http://www.tigr.org/software/tm4/utilities.html) and normalized (local lowess) by MicroArray Informatics Discovery System (http://www.tigr.org/software/tm4/). Then, spots flagged bad or not found by Genepix software were removed from further data analysis, and only those spots that showed fluorescent intensity levels in at least one channel above the background (local) + 2SD were used for further analysis. Those spots that exhibited a large difference between the duplicate experiments (dye swap) were regarded as outliers and removed manually. Hierarchical clustering was performed as described by Eisen et al. (1998).
Affymetrix GeneChip Hybridization and Data Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) and purified by using Qiagen RNeasy columns (Qiagen). For Affymetrix GeneChip (Affymetrix) analysis, 8 μg total RNA was used for making biotin-labeled cRNA targets. All the processes for cDNA and cRNA synthesis, cRNA fragmentation, hybridization, washing and staining, and scanning were conducted according to the GeneChip standard protocol (Eukaryotic Target Preparation, Affymetrix). Poly-A RNA Control kit and the One-Cycle cDNA Synthesis kit were used in this experiment as described in the Web site: http://www.affymetrix.com/products/arrays/specific/rice.affx. The information about GeneChip Rice Genome Array (MAS 5.0) could be accessed from the Affymetrix Web site: http://www.affymetrix.com/products/arrays/specific/rice.affx. Affymetrix GeneChip Operating software (GCOS) was used for data collection and normalization. The overall intensity of all probe sets of each array was scaled to 500 to guarantee that hybridization intensity of all arrays was equivalent, each probe set was assigned with P, A, and M and a P value from the algorithm in GCOS. To identify differentially expressed genes, the log2 transformed signal ratio of each gene was calculated using the GCOS baseline tool, and log2 (ratio) ≥ 1.33 (2.5-fold change) was used as cutoff.
Two statistical methods were used for data processing. First, analysis using the Significance Analysis of Microarrays software package was conducted for rice triplicate samples between stigma and ovary using q value ≤ 0.05 and fold-change ≥2-fold change as cutoff, and 7,425 probe sets were subsequently picked out and their expression levels were significantly different between stigma and ovary. We also did normal t test for each genes and added a P value for them. Second, we used the Z-score transformation normalization method to compare expression levels from stigma and other organs or tissues and suspension-cultured cells and directly calculated significant changes in gene expression between different samples. Z scores were calculated by taking the difference between the averages of stigma (Xi) and other test samples (μ, mean) and dividing by the sd of all the other samples using the following equation:
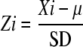 |
In total, we identified 665 probe sets preferentially expressed in stigma using Z score ≥ 2.32 and P value ≤ 0.01. For Z-score transformation normalization, we used the average value from stigma and ovary separately. The test samples included Sh, Rt, An, 5DAP, 10EM, 10EN, and suspension-cultured cell. In addition, we measured the relative ratio between stigma and other samples using the following equation:
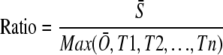 |
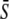 is the average of expression level in stigma tissue sample;
is the average of expression level in stigma tissue sample; 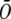 is the average of expression level in the ovary tissue sample; and T1, T2, …,Tn are the expression levels in other tissue samples. The ratios of all 665 probe sets were more than a 2-fold change.
is the average of expression level in the ovary tissue sample; and T1, T2, …,Tn are the expression levels in other tissue samples. The ratios of all 665 probe sets were more than a 2-fold change.
GO Analysis
We searched GO information for the 665 probe sets using EasyGO software (http://bioinformatics.cau.edu.cn/easygo/category_treeBrowse.html). Using the rice ‘Japonica’ gene database, we applied χ2 tests for the biological process search, and the cutoff for false discovery rate was adjusted and the P value was 0.0001.
Real-Time PCR
Total RNA preparation and real-time PCR were performed as previously described (Lan et al., 2004). In brief, 2 μg total RNA was used for cDNA synthesis with SuperScript III First-Strand Synthesis kit (Invitrogen). The cDNA samples were diluted to 8 and 2 ng/μL. Triplicate quantitative assays were performed on 1 μL of each cDNA dilution using the SYBR Green Master mix (Applied Biosystems) with an ABI 7900 sequence detection system according to the manufacturer's protocol (Applied Biosystems). Gene-specific primers were designed by using PRIMEREXPRESS software (Applied Biosystems). The relative quantification method (delta-delta threshold cycle) was used to evaluate quantitative variation between replicates examined. Amplification of 18S rRNA was used as an internal control to normalize all data.
RNA in Situ Hybridization
RNA in situ hybridization was performed as previously described (Lai et al., 2002). Mature rice pistils at the stage 0 to 1 d before floret flowering were fixed with formalin-acetic acid-alcohol fixative solution at 4°C overnight followed by dehydration steps and then embedded in paraffin (Paraplast Plus; Sigma). The tissues were sliced into 8-μm sections with a microtome (Leica RM2145; Leica Microsystems) and affixed to Poly-Prep slides (Sigma). The tissues were stained with Fluorescent Brightener 28 (Sigma) after hybridization. Images were observed under bright and fluorescence field at the same time through a microscope (Olympus BX51; Olympus Optical) and photographed using a Micro Color CCD camera (Apogee Instruments).
Promoter Analysis
The 1,000-bp regions located upstream of the start codons of genes of interest were used for analysis with the MEME/MAST system (http://meme.sdsc.edu/meme/meme.html) and PLACE (http://www.dna.affrc.go.jp/PLACE). Sequence logos of cis-acting regulatory elements were created by weblogo (http://weblogo.berkeley.edu/logo.cgi).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GO term enrichment status for the putative rice stigma-specific genes.
Supplemental Figure S2. Hierarchical clustering of the stigma-preferential genes from the three clusters with the eight tested organs or tissues and suspension culture cells.
Supplemental Table S1. Significantly expressed probe sets of stigma compared to ovary identified by 57 K Affymetrix rice whole-genome array.
Supplemental Table S2. Candidate cDNA clones highly expressed in stigma identified by 10 K cDNA microarray.
Supplemental Table S3. Candidate cDNA clones highly expressed in ovary identified by 10 K cDNA microarray.
Supplemental Table S4. Candidate genes highly expressed in stigma identified by both the two microarrays.
Supplemental Table S5. Candidate genes highly expressed in ovary identified by both the two microarrays.
Supplemental Table S6. Candidate probe sets highly expressed in stigma identified by 57 K Affymetrix rice whole-genome array.
Supplemental Table S7. Classifications of candidate genes highly expressed in stigma identified by 57 K Affymetrix rice whole-genome array.
Supplemental Table S8. Candidate stigma-specific genes conserved in rice and Arabidopsis.
Supplementary Material
Acknowledgments
We thank Kang Chong and Lingfeng Chen (Institute of Botany, Chinese Academy of Sciences) for technical assistance in microarray scanning and Zhen Su and Xin Zhou (China Agricultural University) for assisting on microarray analysis.
This work was supported by the Ministry of Science and Technology of China (grant no. 2005CB120800) and by the Chinese Academy of Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yongbiao Xue (ybxue@genetics.ac.cn).
The online version of this article contains Web-only data.
References
- Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of auxin in regulating Arabidopsis flower development. Planta 223 315–328 [DOI] [PubMed] [Google Scholar]
- Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, et al (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA (1993) Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14 48–54 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Malenica N, Luschnig C (2005) Regulating the regulator: the control of auxin transport. Bioessays 27 1246–1255 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Vieira AM, Becker JD, Feijo JA (2005) Gametophyte interaction and sexual reproduction: how plants make a zygote. Int J Dev Biol 49 615–632 [DOI] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM (2003) A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z (2006) Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol 163 1167–1178 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D'Ascenzo MD, Fobert PR, Despres C, Martin GB (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46 794–804 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampolini F, Shivanna KR, Cresti M (2001) Organization of the stigma and transmitting tissue of rice, Oryza sativa (L.)1. Plant Biol (Stuttg) 3 149–155 [Google Scholar]
- Dixit R, Rizzo C, Nasrallah M, Nasrallah J (2001) The brassica MIP-MOD gene encodes a functional water channel that is expressed in the stigma epidermis. Plant Mol Biol 45 51–62 [DOI] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16 S84–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AR, Gee MA, Guilfoyle TJ (1990) Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J Biol Chem 265 15845–15849 [PubMed] [Google Scholar]
- Fry SC, McDougall GJ, Lorences EP, Biggs KJ, Smith RC (1990) Oligosaccharins from xyloglucan and cellulose: modulators of the action of auxin and H+ on plant growth. Symp Soc Exp Biol 44 285–298 [PubMed] [Google Scholar]
- Gaude T, Dumas C (1986) Organization of stigma surface components in Brassica: a cytochemical study. J Cell Sci 82 203–216 [DOI] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ (1991) Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MH, Goldberg RB, Mariani C (1994) Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J 13 2976–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P (2003) AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Wang ZY, Lin H, Cui WE, Chen J, Liu M, Chen ZL, Qu LJ, Gu H (2006) Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res 16 277–286 [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedhly A, Hormaza JI, Herrero M (2005) The effect of temperature on pollen germination, pollen tube growth, and stigmatic receptivity in peach. Plant Biol (Stuttg) 7 476–483 [DOI] [PubMed] [Google Scholar]
- Heizmann P, Luu DT, Dumas C (2000) Pollen-stigma adhesion in the Brassicaceae. Ann Bot (Lond) 85 23–27 [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Verma DP (2001) A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88 360–371 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Nasrallah ME, Nasrallah JB (2002) Self-incompatibility in the Brassicaceae: receptor-ligand signaling and cell-to-cell communication. Plant Cell 14 S227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, Kristen U (1987) Developmental aspects of ultrastructure, histochemistry and receptivity of the stigma of Nicotiana sylvestris. Ann Bot (Lond) 60 427–437 [Google Scholar]
- Kandasamy MK, Paolillo DJ, Faraday CD, Nasrallah JB, Nasrallah ME (1989) The S-locus specific glycoproteins of Brassica accumulate in the cell wall of developing stigma papillae. Dev Biol 134 462–472 [DOI] [PubMed] [Google Scholar]
- Kang YR, Nasrallah JB (2001) Use of genetically ablated stigmas for the isolation of genes expressed specifically in the stigma epidermis. Sex Plant Reprod 14 85–94 [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L (2003) The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278 23936–23943 [DOI] [PubMed] [Google Scholar]
- Kovaleva LV, Zakharova EV, Skorobogatova IV, Karsunkina NP (2002) Gametophyte-sporophyte interactions in the pollen-pistil system. 3. Hormonal status at the progamic phase of fertilization. Russ J Plant Physiol 49 492–495 [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50 29–42 [DOI] [PubMed] [Google Scholar]
- Lalonde BA, Nasrallah ME, Dwyer KG, Chen CH, Barlow B, Nasrallah JB (1989) A highly conserved Brassica gene with homology to the S-locus-specific glycoprotein structural gene. Plant Cell 1 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Chen W, Lai Y, Suo J, Kong Z, Li C, Lu Y, Zhang Y, Zhao X, Zhang X, et al (2004) Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) with a 10K cDNA microarray. Plant Mol Biol 54 471–487 [DOI] [PubMed] [Google Scholar]
- Lan L, Li M, Lai Y, Xu W, Kong Z, Ying K, Han B, Xue Y (2005) Microarray analysis reveals similarities and variations in genetic programs controlling pollination/fertilization and stress responses in rice (Oryza sativa L.). Plant Mol Biol 59 151–164 [DOI] [PubMed] [Google Scholar]
- Lantin S, O'Brien M, Matton DP (1999) Pollination, wounding and jasmonate treatments induce the expression of a developmentally regulated pistil dioxygenase at a distance, in the ovary, in the wild potato Solanum chacoense Bitt. Plant Mol Biol 41 371–386 [DOI] [PubMed] [Google Scholar]
- Leung DWM (1992) Involvement of plant chitinase in sexual reproduction of higher plants. Phytochemistry 31 1899–1900 [Google Scholar]
- Lord EM (2003) Adhesion and guidance in compatible pollination. J Exp Bot 54 47–54 [DOI] [PubMed] [Google Scholar]
- Luu DT, Heizmann P, Dumas C (1997) Pollen-stigma adhesion in kale is not dependent on the self-(in) compatibility genotype. Plant Physiol 115 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu DT, Marty-Mazars D, Trick M, Dumas C, Heizmann P (1999) Pollen-stigma adhesion in Brassica spp involves SLG and SLR1 glycoproteins. Plant Cell 11 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38 982–993 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1989) Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243 91–93 [DOI] [PubMed] [Google Scholar]
- McInnis SM, Costa LM, Gutierrez-Marcos JF, Henderson CA, Hiscock SJ (2005) Isolation and characterization of a polymorphic stigma-specific class III peroxidase gene from Senecio squalidus L. (Asteraceae). Plant Mol Biol 57 659–677 [DOI] [PubMed] [Google Scholar]
- McInnis SM, Emery DC, Porter R, Desikan R, Hancock JT, Hiscock SJ (2006) The role of stigma peroxidases in flowering plants: insights from further characterization of a stigma-specific peroxidase (SSP) from Senecio squalidus (Asteraceae). J Exp Bot 57 1835–1846 [DOI] [PubMed] [Google Scholar]
- Mol R, Filek M, Machackova I, Matthys-Rochon E (2004) Ethylene synthesis and auxin augmentation in pistil tissues are important for egg cell differentiation after pollination in maize. Plant Cell Physiol 45 1396–1405 [DOI] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS (2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302 81–84 [DOI] [PubMed] [Google Scholar]
- Muschietti J, Eyal Y, McCormick S (1998) Pollen tube localization implies a role in pollen-pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell 10 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah JB (2000) Cell-cell signaling in the self-incompatibility response. Curr Opin Plant Biol 3 368–373 [DOI] [PubMed] [Google Scholar]
- O'Neill SD (1997) Pollination regulation of flower development. Annu Rev Plant Physiol Plant Mol Biol 48 547–574 [DOI] [PubMed] [Google Scholar]
- Otsu CT, daSilva I, de Molfetta JB, da Silva LR, de Almeida-Engler J, Engler G, Torraca PC, Goldman GH, Goldman MH (2004) NtWBC1, an ABC transporter gene specifically expressed in tobacco reproductive organs. J Exp Bot 55 1643–1654 [DOI] [PubMed] [Google Scholar]
- Prabha K, Sood R, Gupta SC (1982) High temperature-induced inactivation of sporophytic self-incompatibility in Ipomoea fistulosa. New Phytol 92 115–122 [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7 203–206 [DOI] [PubMed] [Google Scholar]
- Qin J, Zuo K, Zhao J, Ling H, Cao Y, Qiu C, Li F, Sun X, Tang K (2006) Overexpression of GbERF confers alteration of ethylene-responsive gene expression and enhanced resistance to Pseudomonas syringae in transgenic tobacco. J Biosci 31 255–263 [DOI] [PubMed] [Google Scholar]
- Rehman S, Yun SJ (2006) Developmental regulation of K accumulation in pollen, anthers, and papillae: are anther dehiscence, papillae hydration, and pollen swelling leading to pollination and fertilization in barley (Hordeum vulgare L.) regulated by changes in K concentration? J Exp Bot 57 1315–1321 [DOI] [PubMed] [Google Scholar]
- Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol 46 1568–1577 [DOI] [PubMed] [Google Scholar]
- Sanchez AM, Bosch M, Bots M, Nieuwland J, Feron R, Mariani C (2004) Pistil factors controlling pollination. Plant Cell 16 S98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P (2001) Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for the control of elongation growth. Plant J 28 679–688 [DOI] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R (1998) Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10 1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead AD, Roberts IN, Dickinson HG (1980) Pollen-stigma interaction in Brassica oleracea: the role of stigmatic proteins in pollen grain adhesion. J Cell Sci 42 417–423 [DOI] [PubMed] [Google Scholar]
- Swanson R, Clark T, Preuss D (2005) Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex Plant Reprod 18 163–171 [Google Scholar]
- Swanson R, Edlund AF, Preuss D (2004) Species specificity in pollen-pistil interactions. Annu Rev Genet 38 793–818 [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A (2005) Self-incompatibility in plants. Annu Rev Plant Biol 56 467–489 [DOI] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S (2002) A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Domingo C, Talon M (2006) The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371 279–290 [DOI] [PubMed] [Google Scholar]
- Tung CW, Dwyer KG, Nasrallah ME, Nasrallah JB (2005) Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol 138 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Lalonde BA, Kandasamy MK, Nasrallah JB, Nasrallah ME (1990) Immunodetection of protein glycoforms encoded by two independent genes of the self-incompatibility multigene family of Brassica. Plant Physiol 93 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Pollak P, Tarlyn N, Taylor LP (1994) Pollination- or wound-induced kaempferol accumulation in petunia stigmas enhances seed production. Plant Cell 6 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liang Y, Li C, Xu Y, Lan L, Zhao D, Chen C, Xu Z, Xue Y, Chong K (2005) Microarray analysis of gene expression involved in anther development in rice (Oryza sativa L.). Plant Mol Biol 58 721–737 [DOI] [PubMed] [Google Scholar]
- Woltering EJ, Vrije TD, Harren F, Hoekstra FA (1997) Pollination and stigma wounding: same response, different signal? J Exp Bot 48 1027–1033 [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Berndt ML, Williams A, Douglas GR (2004) Comprehensive comparison of six microarray technologies. Nucleic Acids Res 32 e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2006) Identification and characterization of Arabidopsis thaliana genes involved in xylem secondary cell walls. J Plant Res 119 189–194 [DOI] [PubMed] [Google Scholar]
- Zhang XS, O'Neill SD (1993) Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell 5 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126 5431–5440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.