Abstract
Ethylene is the major effector of ripening in many fleshy fruits. In apples (Malus x domestica) the addition of ethylene causes a climacteric burst of respiration, an increase in aroma, and softening of the flesh. We have generated a transgenic line of ‘Royal Gala’ apple that produces no detectable levels of ethylene using antisense ACC OXIDASE, resulting in apples with no ethylene-induced ripening attributes. In response to external ethylene these antisense fruits undergo a normal climacteric burst and produced increasing concentrations of ester, polypropanoid, and terpene volatile compounds over an 8-d period. A total of 186 candidate genes that might be involved in the production of these compounds were mined from expressed sequence tags databases and full sequence obtained. Expression patterns of 179 of these were assessed using a 15,720 oligonucleotide apple microarray. Based on sequence similarity and gene expression patterns we identified 17 candidate genes that are likely to be ethylene control points for aroma production in apple. While many of the biosynthetic steps in these pathways were represented by gene families containing two or more genes, expression patterns revealed that only a single member is typically regulated by ethylene. Only certain points within the aroma biosynthesis pathways were regulated by ethylene. Often the first step, and in all pathways the last steps, contained enzymes that were ethylene regulated. This analysis suggests that the initial and final enzymatic steps with the biosynthetic pathways are important transcriptional regulation points for aroma production in apple.
Apples (Malus x domestica Borkh. also known as Malus pumila) produce a blend of volatile compounds upon ripening (Dimick and Hoskin, 1983; Young et al., 2004), including alcohols, aldehydes, ketones, sesquiterpenes, polypropanoids, and esters. These aroma compounds are produced from primary metabolites via at least four pathways. Straight chain esters are synthesized from lipids that are broken down initially through β-oxidation, then by lipoxygenase activity (Rowan et al., 1999; Dixon and Hewett, 2000). The resulting hydroperoxides are then converted first to aldehydes, then to alcohols, and finally to esters. Branched chain esters, on the other hand, are produced from the breakdown of Ile (Rowan et al., 1996). Only a handful of genes have been described relating to the production of esters in apple, including an alcohol acyl transferase (AAT) and an alcohol dehydrogenase (ADH; Defilippi et al., 2005b; Souleyre et al., 2005). Apples also produce E,E and Z,E isomers of the sesquiterpene α-farnesene, via the mevalonate pathway (Ju and Curry, 2000), with the final step catalyzed by a terpene synthase (Pechous and Whitaker, 2004). Estragole has been proposed to be synthesized from the phenylpropanoid pathway (Gang et al., 2001).
In apple, ethylene is central to ripening, inducing significant changes in gene expression (Lay-Yee et al., 1990). In commercial cool stores ethylene is routinely scrubbed from the atmosphere to prevent ripening. Good storing varieties that ripen more slowly, for example, have been linked to alleles of ACC SYNTHASE (MdACS1) that synthesize less of this ripening hormone (Sunako et al., 1999). Ripening can also be delayed through the application of the ripening inhibitor, 1-methylcyclopropene, which binds to the ethylene receptor (Sisler and Serek, 2003) or by transgenic silencing approaches that reduce 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase and/or ACC synthase concentrations (Dandekar et al., 2004). ‘Greensleeves’ apples with reduced concentrations of ACC oxidase, through antisense suppression, have reduced autocatalytic ethylene production, as well as reduced ripening, showing a reduction in fruit softening, sugars, availability of aroma precursors, and synthesis of esters and α-farnesene, while volatile aldehydes and alcohols were only marginally repressed and the sugar/acid balance was unaffected by the reduction of ethylene, implying that ethylene does not regulate all aspects of flavor production (Dandekar et al., 2004).
Here we describe a microarray approach to identify the ethylene-regulated transcriptional control points of aroma production in ripening apple fruit. Such an approach has proven successful in the study of ripening-associated processes in other fruit including strawberry (Fragaria x ananassa; Aharoni and O'Connell, 2002), pear (Pyrus communis; Fonseca et al., 2004), and tomato (Solanum lycopersicum; Alba et al., 2005). Other genomic studies of aroma biosynthesis in apple have included EST annotation of putative biosynthetic genes (Newcomb et al., 2006) and statistical and reverse transcription (RT)-PCR analysis or their tissue and developmental expression (Park et al., 2006). We have developed a ‘Royal Gala’ line of apple containing an antisense ACC oxidase gene (Ross et al., 1992) that has lost its ability to synthesize ethylene. This allows ripening to be synchronized by the simultaneous application of ethylene to fruit and the subsequent analysis of traits associated with ripening, such as increased volatile production, to be analyzed. When these antisense ACC oxidase lines were induced to ripen through the application of exogenous ethylene, volatiles were measured and microarrays containing 15,720 oligonucleotides derived from a nonredundant set of apple ESTs (Newcomb et al., 2006) were used to detect changes in the expression of individual genes. We identify members of multigene families encoding candidate aroma biosynthetic genes that were ethylene regulated, especially in key steps at the beginning and end of each biosynthetic pathway.
RESULTS
Generation of ACC Oxidase Mutants That Produce No Detectable Ethylene
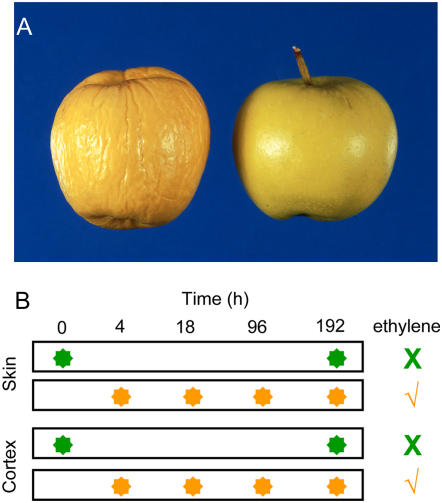
We generated eight lines of transgenic ‘Royal Gala’ apple targeting the ACC oxidase gene that is predominantly expressed in fruit tissue (ACO1; Ross et al., 1992) in an antisense orientation to a 35S promoter. In contrast to the previously reported ACC oxidase antisense lines in ‘Greensleeves’ that produce reduced concentrations of ethylene (less than 90% of untransformed lines; Dandekar et al., 2004), we identified one of the eight lines of transgenic ‘Royal Gala’ (AO3) that produced no detectable ethylene (Table I). The vegetative habit of these AO3 antisense plants were similar to untransformed plants, they set fruit with a similar frequency, and their fruit had a similar appearance. In contrast to the untransformed plants the AO3 antisense fruit had no obvious aroma and did not soften even when left for 3 months at room temperature (Fig. 1). More detailed storage analysis focused on a comparison of AO3 fruit with control untransformed fruit. Following 80 d storage at −1°C, AO3 and control fruit were brought to 22°C to stimulate the climacteric response. Under these conditions, ethylene production in AO3 fruit peaked at less than 2% of that in control fruit. AO3 fruit also showed a much lower rate of CO2 production (Table II). Application of external ethylene did not stimulate internal ethylene production in the AO3 fruit but did increase CO2 production to a concentration similar to that observed in the control fruit (Table II), suggesting that exogenous ethylene could be applied to induce a ripening response. Non-ethylene-treated AO3 fruit were much firmer and contained slightly higher concentrations of sugar than the control fruit at the final stages of this experiment (Table II). There were no obvious changes in color in response to ethylene over the treatment period (data not shown).
Table I.
Analysis of antisense MdACO1 lines in apple
| Transgenic Line | No. Fruit | Days to Ethylene Climacteric | Ethylene Production |
|---|---|---|---|
| nL g−1h−1 | |||
| Control | 9 | 29.1 ± 1.6a | 137.6 ± 7.6 |
| AO1 | 4 | 24.8 ± 1.2 | 157.8 ± 10.0 |
| AO2 | 10 | 19.6 ± 1.8 | 72.9 ± 4.3 |
| AO3 | 10 | Not detectable | Not detectable |
| AO4 | 3 | 30.0 ± 4.0 | 81.52 ± 9.3 |
| AO5 | 10 | 23.8 ± 0.7 | 133.5 ± 14.2 |
| AO7 | 10 | 23.2 ± 3.5 | 57.2 ± 11.4 |
| AO8 | 9 | 21.8 ± 1.7 | 107.8 ± 5.5 |
Values are given as means ± se.
Figure 1.
Phenotype of AO3 apples and microarray experimental design. A, Fruit from untransformed ‘Royal Gala’ (left) and ACO antisense line AO3 (right), after storage at 22°C for 85 d. B, Experimental design for the volatile and gene expression analysis. Non-ethylene-treated AO3 apples (green circles) were harvested at 0 and 192 h. Ethylene-treated apples (orange circles) were exposed to a continuous stream of 120 mg/m3 ethylene and harvested at 4, 18, 96, and 192 h. Volatiles were measured for each of the time points, and skin and cortex tissue was collected for microarray analysis.
Table II.
Ripening characteristic of wild type and AO3 ‘Royal Gala’ apple fruit
| Genotype | Treatment | No. Fruit | Ethylene Production at Climacteric Peak | CO2 Production at Climacteric Peak | Firmness | Sugar |
|---|---|---|---|---|---|---|
| nL g−1h−1 | μL g−1h−1 | N | °Brix | |||
| ‘Royal Gala’ | Air | 9a | 157.3 ± 18.4b | 15.6 ± 1.6 | 66.6 ± 2.6 | 14.4 ± 0.3 |
| AO3 | Air | 15 | 3.0 ± 1.3 | 4.4 ± 2.1 | 104.1 ± 3.1 | 16.2 ± 0.3 |
| AO3 | Ethylene | 16 | 2.5 ± 0.8 | 14.8 ± 1.1 | 93.6 ± 3.6 | 16.0 ± 0.3 |
Fruit from AO3 transgenic apples and untransformed ‘Royal Gala’ apples were stored at −1°C for 80 d followed by holding at 22°C for 14 d.
Values are given as means ± se.
Ethylene-Induced Volatiles
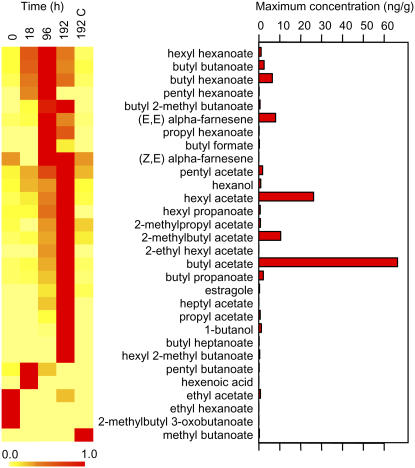
Apple fruit from A03 plants were induced to ripen by continuous streaming of 120 mg/m3 exogenous ethylene. Two samples of three fruit were selected at 0, 4, 18, 96 (4 d), and 192 h (8 d; Fig. 1), the volatiles measured, and the abundance of RNA transcripts from the skin and cortex determined using a 15,720 oligonucleotide apple microarray. For this experiment, apples from AO3 lines that were not treated with ethylene were sampled 192 h after the start of the experiment as a no-ethylene control. The total volatile concentrations produced by the apples that had not been exposed to ethylene (0 h) were approximately 10 ng/g fresh weight. Eight days after exposure to ethylene the total volatile concentrations had increased 12-fold, with approximately 80% of the volatiles at maximum concentration at either 96 or 192 h after exposure to ethylene. The control apples that had not been exposed to ethylene had a total volatile concentration at 192 h that was similar to that of the initial time point (approximately 12 ng/g fresh weight). In total, 30 volatile compounds were identified from the headspace above the fruit (Fig. 2). Over three-quarters (25 out of 30) of these compounds were esters, with the remaining compounds coming from the compound classes: alcohols, terpenes, alkenoic acids, and aromatics. At the initial time point, 19 of the total 30 compounds were detectable, though at very low concentrations (with the detectable compounds at a concentration of less than 5% of their maximum concentration).
Figure 2.
Volatiles produced by AO3 fruit after exposure to ethylene. Figure on the left represents the time of maximum volatile production (red). The relative volatile concentrations were clustered by the timing of peak of production; 192 C represents the 192-h control with no added ethylene. The graph on the right shows the maximum concentration of each volatile produced.
The esters butyl and hexyl acetate were the major compounds at all time points, including in the control apples that were not exposed to ethylene. The concentrations of these compounds at all time periods accounted for approximately 44% of the total volatiles by mass. Both compounds increased linearly, with butyl acetate increasing 16-fold and hexyl acetate 8-fold at the 192-h time point. The two alcohol precursors for these compounds, butanol and hexanol, were also detected among the volatiles, reaching their maximum concentration at 192 h after ethylene exposure. 2-Methylbutyl acetate was the third most abundant ester and showed a similar proportional increase over this time to butyl and hexyl acetate. As no 2-methylbutanol was observed in the profiles, it was assumed that this intermediate was rapidly utilized by alcohol AT(s).
The sesquiterpene, α-farnesene, like the dominant acetates, was detected at time zero and increased rapidly with the application of ethylene. α-Farnesene was present as both the (E,E) and (Z,E) isomers, but the (Z,E) isomer accounted for less than 1% of the total α-farnesene present in the ripening apples, with a maximum concentration measured 96 h after the application of ethylene. The phenylpropanoid, estragole, was not detected until the 96- and 192-h time points.
Clustering the volatiles by the timing of maximal production revealed some putative groupings of compounds (Fig. 2). The concentration of the dominant acetates continued to rise throughout the sampling period, while some of the lower abundance esters such as the formates, hexanoates, and butanoates peaked at 96 h, implying that the pools of these acid precursors might be limited.
Ethylene-Induced Gene Expression Changes
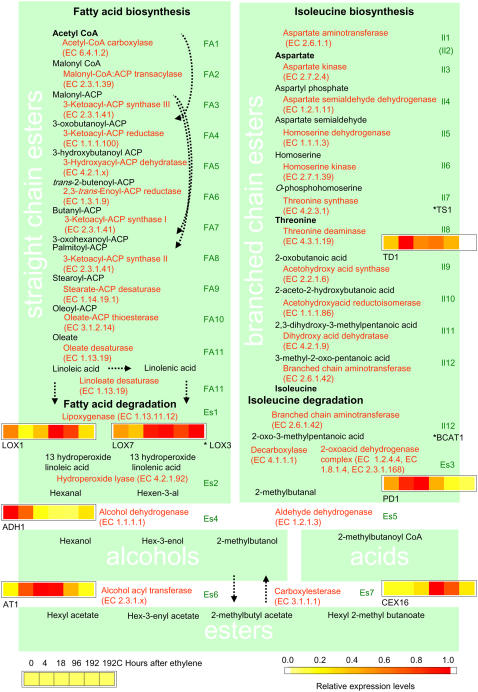
In apple, at least four pathways are responsible for the production of the volatiles observed during fruit ripening (Figs. 3–5). Straight chain esters in apple are derived from fatty acids such as linoleic and linolenic acid (Rowan et al., 1999), which in turn are synthesized from acetyl-CoA (Ohlrogge and Jaworski, 1997). In apple, branched chain esters such as 2-methylbutyl acetate are derived from Ile (Rowan et al., 1996), which is synthesized from Asp via Thr (Azevedo et al., 1997; Fig. 3). The sesquiterpene α-farnesene is produced via the mevalonate pathway (Fig. 4). Volatile compounds from the phenylpropanoid pathway, e.g. estragole (and eugenol), are likely to be produced from Phe (Gang et al., 2001; Fig. 5). We conducted a systematic survey of the ‘Royal Gala’ EST collection for candidate genes that are likely to encode enzymes involved in these four pathways. From the nonredundant set of ‘Royal Gala’ EST sequences (Newcomb et al., 2006), we initially identified candidate genes that represent each step of a flavor biosynthesis pathway using tBLASTn cutoff of e < 10−0.05. Sequences of the longest available cDNA representing the gene together with other available plant sequences from the gene family were then further assessed using phylogenetic analysis to verify assignments to a gene family and differentiate allelic variants from paralogous genes (data not shown). In total, 186 genes were identified as potential aroma-related genes (Table III). Many of the steps in each pathway are encoded by multigene families and therefore there is the potential that multiple enzymes could be involved in each step, acting singly or together. The number of family members for each enzymatic step ranged from 1 to 16, with the carboxylesterases containing most members. Each unique enzyme was assigned a gene name (Supplemental Table S1). A 15,720 oligonucleotide apple microarray was used to measure the abundance in RNA extracted from fruit skin and cortex tissue for each gene. As aroma develops predominantly in the skin (Guadagni et al., 1971), we initially focused our analysis on this tissue. ANOVA was used to identify genes that changed their expression in skin tissue after the induction of ripening with ethylene. To reduce the chance of selecting genes that change over time without the induction of ethylene, our ANOVA modeled the 192-h control (no ethylene treatment) as a zero time point. Using this analysis method, 537 oligonucleotides were selected that were significantly altered in expression levels over time in skin using a false discovery rate (FDR) threshold of 0.05. When these features were clustered using hierarchical clustering, three main expression patterns were observed. A total of 235 features showed a rapid induction with ethylene (4–18 h after ethylene exposure), 248 showed a slower ethylene response (96–192 h after exposure), and 54 were inhibited by the addition of ethylene (Supplemental Table S2). Of the 186 candidate aroma genes 179 (96%) were represented by 270 oligonucleotides on the microarray. When the 270 oligos were compared with the list of 537 selected genes, 17 genes were identified as having a significantly altered expression upon ethylene exposure in skin. The expression patterns of these 17 genes were mapped onto the predicted biosynthetic pathways (Figs. 3–5). The microarray expression patterns of all the 186 candidate aroma genes were plotted. In addition to the 17 statistically significantly genes, a further nine genes that visually showed an induction with ethylene were also identified. Quantitative RT-PCR (qPCR) was used to verify the changes in expression of all 26 genes. In all cases primers were designed to span the region of the gene selected for the microarray oligonucleotide. Eighty-four percent of the statistically selected and 33% of the visually selected genes showed patterns in the qPCR similar to those obtained in skin using microarrays (Supplemental Fig. S1). This analysis resulted in 17 genes that we are confident to assign as aroma-related genes that are regulated by ethylene in skin. Henceforth, unless mentioned otherwise, all positive assertions regarding ethylene regulation have been verified by qPCR.
Figure 3.
Schematic of the two pathways that make precursors for ester biosynthesis. The fatty acid pathway contributes to the straight chain esters while the Ile biosynthesis pathway contributes to the branched chain pathway. Colored sections show the relative expression patterns of genes that were differentially expressed in skin in response to ethylene treatment, as detected by microarray and confirmed by qPCR. Red depicts the level of highest expression and white the lowest. Sections from left to right are 0, 4, 18, 96, and 192 h after ethylene addition and 192 h no-ethylene control. Genes that showed changes in expression in skin on the microarray but were not confirmed by qPCR are indicated with an asterisk.
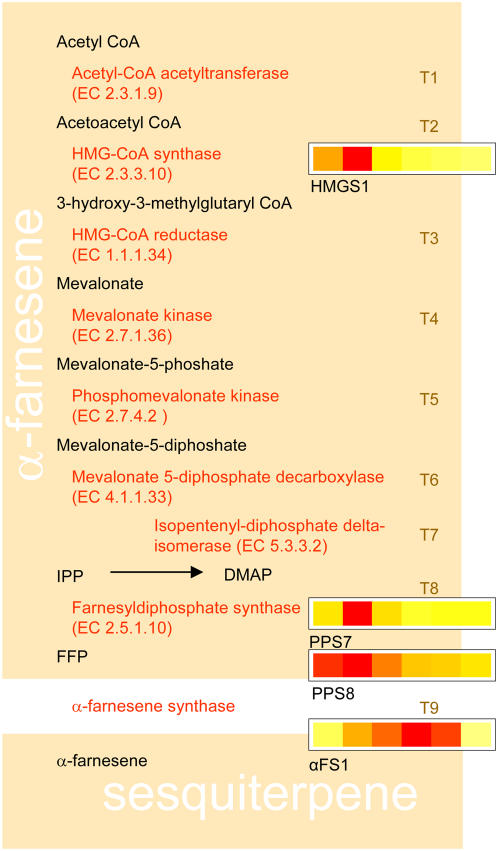
Figure 4.
Schematic of the α-farnesene synthesis pathway with sections as described for Figure 3.
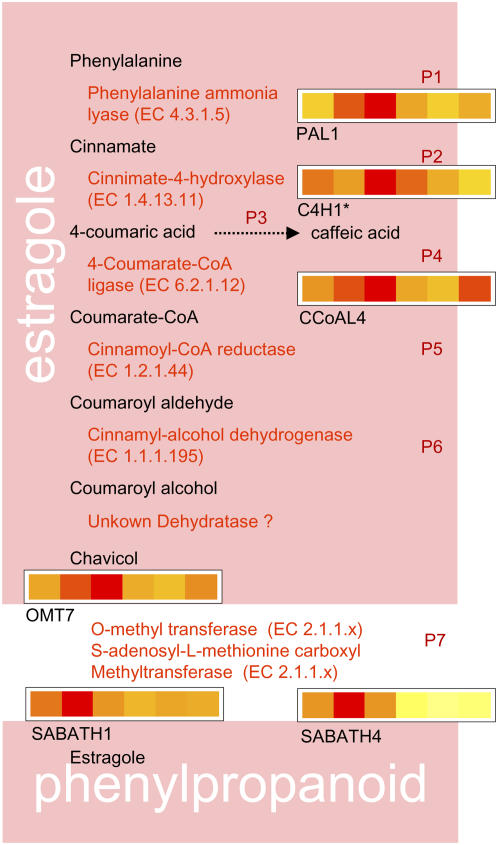
Figure 5.
Possible biosynthetic pathway for estragole. Schematic of candidate genes in the phenylpropanoid pathway with sections as described for Figure 3.
Table III.
Candidate genes involved in aroma biosynthesis in apple
| Pathway | Enzyme | Step | Enzyme Name | EC No. | No. Genes | No. Oligos | Genes with Oligos |
|---|---|---|---|---|---|---|---|
| Fatty acid biosynthesis | ACC | FA1 | Acetyl-CoA carboxylase | 6.4.1.2 | 3 | 3 | 3 |
| MAT | FA2 | Malonyl-CoA ACP transacylase | 2.3.1.39 | 2 | 3 | 2 | |
| KASIII | FA3 | 3-Ketoacyl ACP synthase III | 2.3.1.41 | 1 | 0 | 0 | |
| KAR | FA4 | 3-Ketoacyl ACP reductase | 1.1.1.100 | 1 | 1 | 1 | |
| DH | FA5 | 3-Hydroxyacyl ACP dehydratase | 4.2.1.x | 2 | 2 | 2 | |
| ER | FA6 | 2,3-Trans-enoyl ACP reductase | 1.3.1.9/10 | 1 | 2 | 1 | |
| KASI | FA7 | 3-Ketoacyl ACP synthase I | 2.3.1.41 | 1 | 1 | 1 | |
| KASII | FA8 | 3-Ketoacyl ACP synthase II | 2.3.1.41 | 1 | 1 | 1 | |
| SAD | FA9 | Stereate ACP desaturase | 1.14.19.1 | 2 | 3 | 2 | |
| OTE | FA10 | Oleate-ACP thioesterase | 3.1.2.14 | 1 | 2 | 1 | |
| FAD | FA11 | Oleate and linoleate desaturase | 1.13.19.x | 3 | 3 | 2 | |
| Total | 18 | 21 | 16 | ||||
| Isoleucine biosynthesis | AAT | IL1 | Aspartate aminotransferase | 2.6.1.1 | 4 | 5 | 4 |
| AATL | IL2 | Aspartate aminotransferase like | ? | 4 | 7 | 4 | |
| AK | IL3 | Aspartate kinase | 2.7.2.4 | 1 | 1 | 1 | |
| ASADH | IL4 | Aspartate semialdehyde dehydrogenase | 1.2.1.11 | 0 | 0 | 0 | |
| HSD | IL5 | Homoserine dehydrogenase | 1.1.1.3 | 1 | 1 | 1 | |
| HSK | IL6 | Homoserine kinase | 2.7.1.39 | 1 | 1 | 1 | |
| TS | IL7 | Threonine synthase | 4.2.3.1 | 2 | 3 | 2 | |
| TD | IL8 | Threonine deaminase | 4.3.1.19 | 1 | 3 | 1 | |
| ALS | IL9 | Acetolactic synthetase | 2.2.1.6 | 4 | 6 | 4 | |
| AHIR | IL10 | Acetohydroacid isomeroreductase | 1.1.1.86 | 1 | 1 | 1 | |
| DHAD | IL11 | Dihydroxy acid dehydratase | 4.2.1.9 | 1 | 3 | 1 | |
| BCAT | IL12 | Branched chain aminotransferase | 2.6.1.42 | 5 | 11 | 5 | |
| Total | 25 | 42 | 25 | ||||
| β-Oxidation | ACD | Box1 | Acyl-CoA dehydrogenase | 1.3.99.3 | 1 | 4 | 1 |
| MFP | Box2 | Multifunctional protein | 4.2.1.17 | 12 | 12 | 10 | |
| 3KT | Box3 | 3-Ketoacyl-CoA thiolase | 2.3.1.16 | 3 | 2 | 2 | |
| Total | 16 | 18 | 13 | ||||
| Ester biosynthesis | LOX | Es1 | Lipoxygenase | 1.13.11.12 | 9 | 15 | 9 |
| HPL | Es2 | Hydroperoxide lyase | 4.2.1.92 | 2 | 3 | 2 | |
| BCKDC E1-a | Es3a | Branched chain dehydrogenase | 1.2.4.4 | 3 | 4 | 3 | |
| BCKDC E1-b | Es3b | Branched chain dehydrogenase | 1.2.4.4 | 2 | 2 | 2 | |
| BCKDC E2 | Es3c | Branched chain dehydrogenase complex | 2.3.1.168 | 2 | 3 | 2 | |
| BCKDC E3 | Es3d | Dihydrolipoyl dehydrogenase | 1.8.1.4 | 2 | 3 | 2 | |
| PD | Es3e | Pyruvate decarboxylase | 4.1.1.1 | 2 | 2 | 1 | |
| ADH | Es4 | Alcohol dehydrogenase | 1.1.1.1 | 9 | 10 | 9 | |
| ALDH | Es5 | Aldehyde dehydrogenase | 1.2.1.3 | 8 | 14 | 8 | |
| AT | Es6 | Acyl transferase | 2.3.1.x | 15 | 26 | 15 | |
| CXE | Es7 | Carboxylesterase | 3.1.1.1 | 16 | 19 | 16 | |
| Total | 70 | 101 | 69 | ||||
| Terpene biosynthesis | ACOAAT | T1 | Acetyl-CoA acetyltransferase | 2.3.1.9 | 3 | 9 | 3 |
| HMGS | T2 | HMG-CoA synthase | 2.3.3.10 | 1 | 1 | 1 | |
| HMGR | T3 | HMG-CoA reductase | 1.1.1.34 | 2 | 4 | 2 | |
| MK | T4 | Melalonate kinase | 2.7.1.36 | 1 | 2 | 1 | |
| PMK | T5 | Phosphomevalonate kinase | 2.7.4.2 | 1 | 1 | 1 | |
| M5DD | T6 | Mevalonate 5-diphosphate decarboxylase | 4.1.1.33 | 1 | 2 | 1 | |
| IDDI | T7 | Isopentyl-diphosphate Δ-isomerase | 5.3.3.2 | 2 | 3 | 2 | |
| PPS | T8 | Polyprenyl synthase | 2.5.1.1/10/29 | 9 | 18 | 9 | |
| AFS | T9 | α-Farnesene synthase | None | 1 | 1 | 1 | |
| Total | 21 | 41 | 21 | ||||
| Phenylpropanoid biosynthesis | PAL | P1 | Phenyl-alanine ammonia lyase | 4.3.1.5 | 2 | 1 | 1 |
| C4H | P2 | Cinnimate 4-hydroxylase | 1.14.13.11 | 1 | 4 | 1 | |
| C3H | P3 | 4-Coumaric acid-3-hydroxylase | 1.14.18.1 | 2 | 5 | 2 | |
| CCoAL | P4 | 4-Coumarate-CoA ligase | 6.2.1.12 | 12 | 15 | 12 | |
| CCoAR | P5 | Cinnamoyl-CoA reductase | 1.2.1.44 | 3 | 4 | 3 | |
| CADH | P6 | Cinnamyl-ADH | 1.1.1.195 | 3 | 3 | 3 | |
| SABATH | P7 | S-adenosyl-l-Met carboxyl methyltransferase | 2.1.1.x | 6 | 6 | 6 | |
| OMT | P7 | O-methyl transferase | 2.1.1.x | 7 | 9 | 7 | |
| Total | 36 | 47 | 35 | ||||
| Grand total | 186 | 270 | 179 |
Expression Changes in Aroma-Related Genes
Within the fatty acid biosynthesis pathway (Fig. 3) we identified 18 potential genes that contribute to this pathway. All but two steps were represented on the microarray (Table III); these two steps were the 3-ketoACP synthase III and a desaturase. Sixteen of the 18 genes were represented by 21 oligonucleotides on the array. None significantly changed in expression with ethylene application. β-Oxidation reduces the chain length of fatty acids by two carbons per round of oxidation. In apple, β-oxidation is likely involved in the production of four carbon esters such as butyl acetate. None of the genes predicted to encode enzymes involved in the three steps of β-oxidation changed in expression significantly in skin in response to ethylene on the microarray. There are two enzymatic steps involved in fatty acid degradation, the initial steps toward ester biosynthesis. Lipoxygenases catalyze the first committed step in the pathway of which there were seven genes. One of these genes (LOX3) was selected by the microarray analysis but not verified by qPCR. However, two visually selected genes (LOX1 and LOX7) are both induced by ethylene. Neither of the 2-s step enzymes (hydroperoxide lyases) is ethylene regulated.
The Ile biosynthesis pathway is represented by 12 enzymatic steps (Azevedo et al., 1997). Of the 25 genes that were identified that may play a role in this pathway, 25 were represented on the array with 42 oligos. All but one step (Asp semialdehyde dehydrogenase) are represented on the array. Two genes, a Thr Synthase1 and a Branched Chain Aminotransferase1 (BCAT1) were selected as changing significantly in skin expression after the addition of ethylene (Fig. 3), but neither were verified by qPCR (Supplemental Fig. S1). However, the visually selected Thr Deaminase1 (TD1), the first committed step in Ile biosynthesis, is ethylene induced according to qPCR results (Supplemental Fig. S1). The branched chain backbone can either be decarboxylated and then reduced to form the alcohol moiety (2-methylbutanol) of the ester or ligated to CoA to form ultimately the acid moiety (2-methylbutanoate). In bacteria, ligation onto CoA is performed by members of the branched chain dehydrogenase complex. None of the three genes on the array that are members of the complex was selected as responding to ethylene. This reaction could also be performed by pyruvate decarboxylase. When PD1 was examined, it was found to have a rapid increase in expression with ethylene. However, this enzyme is also involved in glycolysis and therefore may be playing a role in the climacteric respiratory burst.
The resulting aldehydes from the fatty acid and Ile degradation pathways can be reduced to alcohols by ADHs, of which there were 10 genes identified. One of these (ADH1) decreased in response to ethylene. This gene has previously been shown to decrease in expression with the addition of ethylene (Defilippi et al., 2005b). The final biosynthetic step in ester formation is catalyzed by acyl transferases (ATs). These enzymes are members of the BAHD superfamily of enzymes, of which at least two subfamilies have been found to encode transferases with AAT activity. Fifteen ATs were identified, including MpAAT1 (Souleyre et al., 2005; AT1), and the ortholog of SAAT (Beekwilder et al., 2004; AT6), both representing different subfamilies of AATs. Of these AT1 was found to change significantly, increasing in expression with the addition of ethylene as also found previously (Defilippi et al., 2005b). Carboxylesterases can hydrolyze esters and therefore potentially can return alcohols and acids to their respective pools within the fruit. Members of the CXE carboxylesterase family from fruit can catalyze this reaction (Ileperuma et al., 2007). Sixteen CXE carboxylesterases have oligonucleotides on the array, one of which showed a significant increase in expression with application of ethylene (CXE16).
The sesquiterpene biosynthetic pathway is represented by nine enzymatic steps; 21 genes encode enzymes that may be involved in this pathway, all of which were represented at least once by 41 individual oligos on the microarray. Four of the genes showed a change in expression in skin due to the application of ethylene (Fig. 4). The second step corresponding to 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase was rapidly induced by ethylene (HMGS1). None of the other enzymatic steps had candidates that showed an increase in expression until the second to last step that is represented by the polyprenyl synthetases, which includes the geranyl diphosphate, farnesyl diphosphate, and geranylgeranyl diphosphate synthases. There are nine representatives of these genes on the microarray, two of which showed changes in gene expression. One increased rapidly in expression (PPS7) and a second showed a small increase followed by a decrease in expression (PPS8). The final step in sesquiterpene biosynthesis in apple is catalyzed by α-Farnesene Synthase1 (AFS1) and (Pechous and Whitaker, 2004; Green et al., 2007) was highly induced by ethylene.
Although it has been proposed that estragole is synthesized from the phenylpropanoid pathway, some of the enzymatic steps are poorly understood. While the beginning of this pathway is well known as it feeds into the lignin and color pathways, the second half was mined for theoretical components except for a proposed dehydratase step for which mining was not conducted. Combining the known and unknown sections, there are 36 potential candidate genes identified in this pathway with 47 oligonucleotides representing 35 of these genes on the microarray. Genes involved in the first steps of this pathway are ethylene responsive; Phe Ammonia Lyase1 (PAL1) showed a rapid increase of expression (Fig. 5). The second step, cinnimate 4-hydrolase, has only one predicted gene (C4H1) with four oligos. Two of these oligos changed significantly, and qPCR showed an up-regulation of this gene by ethylene (Fig. 5; Supplemental Table S3). The fourth step, p-coumarat-CoA ligase (CCoAL4), showed a rapid induction with ethylene. It is of interest that these early steps are shared with the general flavanoid biosynthetic pathway that branches off at this point. The final step in the estragole biosynthetic pathway is predicted to involve a methyl transferase class of enzyme. There are two families of enzyme that can add a methyl group, the O-methyltransferase (OMT) and the SABATH families. Both add the methyl group using Met as a donor provided by a SAM synthetase, the first step in the ethylene biosynthesis pathway. There were six members of the SABATH family, two of which (SABATH1 and 4) were represented by oligonucleotides that showed an increase of expression upon the addition of ethylene. Of the seven OMTs, OMT7 was ethylene induced. OMT7 was most similar to caffeic acid OMT (Gowri et al., 1991) and therefore could possibly be catalyzing the final step in estragole biosynthesis.
Analysis of Other Ethylene-Regulated Processes
Volatile production is only one aspect of ethylene-induced ripening with 17 aroma-related genes of the 537 oligos selected as changing in expression in skin tissue. To identify the function of other genes that are also associated with ethylene-induced ripening, the clusters of differentially expressed genes were examined further. The selected genes showed three main clusters of expression pattern, rapid induction, slow induction, and inhibition; the identity of these genes were further examined by categorizing them into functional groups that are associated with a ripening response (Table IV). The rapid induction cluster (peaking at 4–18 h) comprised 235 genes. Approximately 10% could be assigned a role in secondary metabolism, 8% in transcriptional regulation, 5.5% in primary metabolism, 5% in defense, 5% in oxygen-related genes including chloroplast genes, 5% in regulation of proteins such as kinases, and 3.5% in flavanoid, anthocyanin pathway, and 2.5% in ethylene-related response genes. The slow response cluster (peaking at 4–8 d) comprising 248 genes had a similar proportion of transcription factors (6%), protein regulation (5%), and secondary metabolism genes (8.5%), and lower proportions of defense genes (2.5%), ethylene response genes (0.4%), flavanoid, anthocyanin (0.4%), and primary metabolism genes (3%). In addition, there was an increased proportion of cell wall-related enzymes (4.5%) and receptors (2%). The ethylene-inhibited cluster (genes that decrease in expression because of ethylene) comprised 54 genes. This number is too small to warrant making generalized categories. Of interest, however, was that secondary metabolism genes comprised 10% of the genes that were down-regulated.
Table IV.
Functional categories of genes involved in ripening processes that are regulated by ethylene
| Category | Skin
|
Skin and Cortex
|
||||
|---|---|---|---|---|---|---|
| Early Response | Late Response | Inhibited by Ethylene | Ethylene Up-Regulated | Ethylene Down-Regulated | Tissue Differences | |
| Total genes | 235 | 248 | 54 | 728 | 244 | 941 |
| Ethylene | 6/2.6a | 1/0.4 | 0/0 | 9/1.2 | 1/0.4 | 5/0.5 |
| Primary metabolism | 13/5.5 | 8/3.2 | 2/3.7 | 32/4.4 | 12/5 | 42/4.5 |
| Oxygen | 12/5.1 | 0/0 | 3/5.6 | 9/1.2 | 9/3.6 | 15/1.6 |
| Defense | 12/5.1 | 6/2.4 | 0/0 | 24/3.3 | 9/3.6 | 28/3 |
| Sugars starch | 2/0.9 | 3/1.2 | 0/0 | 12/1.6 | 1/0.4 | 7/0.7 |
| Cell wall | 2/0.9 | 11/4.4 | 3/5.6 | 16/2.2 | 4/1.6 | 10/1.1 |
| Secondary metabolism | 23/9.8 | 21/8.5 | 5/9.3 | 80/11 | 26/10.7 | 114/12.1 |
| Color | 8/3.4 | 1/0.4 | 1/1.9 | 17/2.3 | 1/0.4 | 15/1.6 |
| Transcription regulators | 19/8.1 | 15/6 | 2/3.7 | 44/6 | 15/6.1 | 66/7 |
| Phosphatase kinase | 11/4.7 | 13/5.2 | 2/3.7 | 40/5.5 | 10/4.1 | 57/6 |
| Protein turnover | 3/1.3 | 4/1.6 | 0/0 | 12/1.7 | 4/1.6 | 23/2.4 |
| Unknown | 44/18.7 | 55/22.2 | 15/27.8 | 167/23 | 62/25.4 | 194/20.6 |
Actual number of genes followed by percentage of genes that change in response to ethylene.
To expand the study of gene expression analysis, a second ANOVA model incorporating the cortex samples was conducted to identify those genes that show a change in expression by tissue, time, and tissue by time. There were 941 genes selected that had a significantly different expression (with an FDR threshold of 0.05) pattern between the two tissues. When the genes selected by time and tissue by time were combined, as these could be classed as ethylene-responsive genes, there were 972 oligonucleotides that changed. These could be separated into those that had a maximum peak in either the 0- or 192-h control (244 ethylene repressed genes) and those that had a maximum expression either at 4, 8, 96, or 192 h after exposure to ethylene (728 ethylene-induced genes). A list of these genes is provided in Supplemental Table S3 and their predicted function is summarized in Table IV. In the ethylene up-regulated gene group, there was a large proportion of genes that were in the cell wall category, while there were similar proportions of genes that were involved in primary and secondary metabolism in both the up-regulated and down-regulated groups, implying that there are large metabolic changes occurring during the ripening process. There were similar proportions of transcription factors and protein regulatory proteins in all the categories.
Within these lists, there contained many other genes related to ripening traits. These included genes were involved in ethylene production and detection, such as SAM synthase (oligo 146858) and EIN4-like receptors (oligos 166801 and 16752). There are representatives of glycolysis and the Kreb's cycle, color-related genes, and those involved in sugar metabolism and cell wall-related genes such as polygalcturanase (oligo 315849; Atkinson et al., 1998). All these processes are of interest, are relevant to ripening, and can be found in the Supplemental Data, but are outside the scope of this article.
DISCUSSION
Ethylene-Induced Processes in Ripening Apples
From a molecular perspective ethylene-induced ripening is a transcriptionally controlled event, therefore many of the changes in physiology can be inferred from the changes seen in gene expression (Solano et al., 1998). We have employed a genomics approach to identify genes that may play a role in aroma production during ripening. We have identified genes that change in expression due to ethylene in skin and tissue-regulated genes. From a list of 186 potential aroma-related genes we have collated the gene expression data for 179 genes and within these identified enzymatic steps that are likely to be key transcriptional control points in the production of aroma compounds in apple (Supplemental Table S1).
Many of the enzymatic steps are represented by multigene families, often only a single member of these families show an ethylene-regulated change in expression. Following the hypothesis that the subset of the paralogs that are transcriptionally regulated in fruit by ethylene encode the enzymes involved in aroma biosynthesis, we have identified candidates for such genes, for example, the last step of the phenylpropaniod pathway involved in the transfer of a methyl group onto a polyphenyl backbone. This step can be synthesized by one of two multigenes, the SABATHs and the OMTs. Of the six SABATHs and the seven OMTs for which we have transcriptional data, only two SABATHs (SABATH1 and 4) and one OMT (OMT7) are ethylene regulated (Supplemental Table S1).
The enzymes in the pathways involved in the biosynthesis of aroma compounds are not coordinately regulated by ethylene. Only certain steps seem to be ethylene regulated. Of the genes that changed significantly upon ethylene exposure, in all pathways the last steps had candidates that were regulated by ethylene (Figs. 3–5). For example, AFS1 catalyzes the final step in sesquiterpene biosynthesis in apple, converting farnesyl diphosphate to α-farnesene (Pechous and Whitaker, 2004; Green et al., 2007). AFS1 is significantly up-regulated in response to ethylene; indeed, AFS1 is among the highest induced genes in this study. This suggests that the most important point of control involves the final step, which produces the volatile product from a nonvolatile set of precursors. The last step in ester biosynthesis is also ethylene regulated. From a total of 15 ATs only AT1 is ethylene induced. This gene (AT1; also known as MpAAT1) is expressed late in fruit ripening (Defilippi et al., 2005b; Souleyre et al., 2005) and from in situ hybridization analysis the protein product is found predominantly in fruit skin (Li et al., 2006). MpAAT1 is able to use a range of alcohols and CoA acids to make a range of esters found in apple fruit (Souleyre et al., 2005). Interestingly, a carboxylesterase (CXE16) is ethylene induced, increasing its expression in ripe fruit. Certainly increased carboxylesterase activity has been found in apple fruit tissues (Goodenough and Entwistle, 1982). It is possible that CXE16 may contribute to an increase in carboxylesterase activity and the hydrolysis of esters and the increase in alcohols seen in the volatile profile late in ethylene treatment of AO3 fruit.
A typical control point for biochemical pathways is often the first committed step in the process. There is evidence for regulation by ethylene at the beginning of many of the aroma pathways. For example, in branched chain ester biosynthesis there are two major pathways, the primary production of Ile from Thr and then the secondary breakdown of Ile to make branched chain esters. Upon ethylene induction, Ile shows a rapid buildup in concentrations in the skin of apple fruit (Defilippi et al., 2005a). Entrance into the pathway responsible for Ile biosynthesis is controlled by TD1. TD1 is regulated by ethylene in skin, as seen by qPCR, while other steps along the pathway do not appear to be ethylene regulated. A second example is the LOX genes, which catalyze the first step in the fatty acid breakdown pathway, where two members are induced by ethylene.
The LOX genes are an example where some members of the multigene family are ethylene regulated while others are not. From the set of 11 lipoxygenase- related genes two are up-regulated in response to ethylene (LOX1, 7). Three other LOX genes (LOX2, 4, 5) are expressed significantly more highly in skin than in cortex but are not ethylene regulated according to microarray analysis. Potentially all these LOX genes could be contributing to aroma production. In tomato, for instance, of the five lipoxygenase genes identified three are ethylene regulated, two positively so, and of these only one lipoxygenase (LOXC) has an impact on flavor profiles when expression levels are reduced using antisense technology (Chen et al., 2004). The ortholog of LOXC from apple (LOX5) is not regulated by ethylene. While we see an increase in expression of these genes, Defilippi et al. (2005a) have shown that there is no increase in LOX activity in the skin after ethylene-induced ripening, suggesting that increased quantities of LOX isozyme are not required for ester biosynthesis in apple fruit. While this hypothesis may hold for some enzymes, it is of interest that in the absence of ethylene there was a detectable aroma being produced in the ACC oxidase antisense lines, with the concentrations increasing rapidly with the application of exogenous ethylene. These observations may be explained by multiple levels of regulation of aroma production, by the many enzymatic paralogs for each biosynthetic step. The existence of multigene families also presents opportunity for organelle- and tissue-specific expression for different members. While many of the LOXs are certainly expressed in apple fruit skin, not all may be expressed in the same cellular compartments. Analysis using PSORT (Nakai and Horton, 1999) suggests that LOX1, 2, and 7 are likely to be cytoplasmic while LOX4 may be chloroplastic.
The Regulation of Aroma Biosynthetic Genes by Transcription Factors
Many of the central genes involved in ethylene perception and the downstream signal transduction pathway have been identified through genetics studies in tomato and Arabidopsis (Arabidopsis thaliana; Adams-Phillips et al., 2004). Ethylene signal transduction is effected by changes in transcription starting with the binding of EIN3 and EIL proteins to the promoters of EREBP genes (Solano et al., 1998). However, few transcription factors that directly act on aroma and other ripening-associated genes have been identified. From our analysis of apple genes, 36 transcription factors were identified as changing expression in skin and 59 in the skin and cortex. These included some genes that have already been associated with ripening. One of the first transcription factors in the Arabidopsis ethylene cascade is EIN3 (Chao et al., 1997). Upon the detection of ethylene there is an EIN-like gene that shows a decrease in expression levels. In Arabidopsis the EIN-like genes activate a class of genes called AP2/EREBP binding proteins. Two genes with homology to this class of transcription factor (oligos 149233, 186231) changed during ethylene induction. Oligonucleotide 186231 shows homology to ERF3, a transcription factor known to be induced by ethylene and involved in transcriptional repression (Fujimoto et al., 2000). A MADS-box gene (oligo 173646) that is related to the AP1 class of MADS-box transcription factors showed a decrease in expression. The apple gene (oligo 173646) has previously been described as MdMADS5 and shown to have AP1-like properties, promoting flowering in Arabidopsis (Kotoda et al., 2002). Also identified were two squamosa-binding proteins that change in expression upon the detection of ethylene (oligos 121375, 139438). Squamosa-binding proteins have also been implicated in ripening in tomato (Manning et al., 2006).
One approach to identifying candidate transcription factors that regulate pathways of interest is on the basis of shared patterns of gene expression across multiple treatments or time series. We have investigated transcription factors that might be involved in regulating aroma pathways by clustering aroma biosynthesis genes with the 36 transcription factors by their expression in our ethylene-induction array experiment. Within the cluster of genes that are rapidly induced by ethylene (Fig. 6), the final step in the volatile phenylpropanoid pathway and a NAM class transcription factor (oligo 229542) were coordinately induced. This NAM has homology to the AT1G01720 (ATAF1) homologs, which have been shown to be induced by biotic and abiotic stress, again suggesting a role in the defense response (Hegedus et al., 2003). There were four transcription factors that were rapidly activated upon the application of ethylene (BHLH oligo 188523, NAM oligo 229542, myb oligo 319213, HS oligo 298511), followed by a rapid decline. The BHLH showed the highest similarity to a jasmonic acid-insensitive BHLH and also to an enhancer of glabra 3 EGL3, a myc gene that coactivates the color pathway in Arabidopsis, and may play a role in the activation of the anthocyanin biosynthesis genes associated with ethylene induction. This analysis provides candidate transcription factors for further research on ethylene-induced control of ripening in apple.
Figure 6.
Grouping of potential aroma biosynthetic and transcription factor encoding genes by relative expression patterns using hierarchical clustering. Expression was normalized to a maximum level of 1 (red). Ester biosynthesis candidate genes, green; terpene synthesis genes, brown; phenylpropanoid genes, red, and are grouped with transcription factors (black). Circles indicate the last enzymatic steps in the each pathway.
In summary, we have identified many genes involved in the ripening process of apple fruit, with a focus on genes involved in the synthesis of aroma compounds. We have found that the genes involved in aroma biosynthesis are not coordinately regulated by ethylene but typically only the first and final steps are ethylene regulated.
MATERIALS AND METHODS
Generation of Transgenic Apple Lines
A binary vector was constructed using the pART7/pART27 (Gleave, 1992) system to contain the gene MdACO1 (Ross et al., 1992) in an antisense orientation to the 35S promoter. Transgenic apple (Malus x domestica ‘Royal Gala’) plants were produced as described by Yao et al. (1995). Eight independent transgenic lines were established in a containment greenhouse and grown as described by Yao et al. (1999). To generate fruit from these lines, flowers were pollinated manually with compatible ‘Granny Smith’ pollen.
Analyses of Ethylene, CO2, Sugar, Volatile Flavor Components, and Firmness
Fruit from the MdACO1 transgenic lines and a nontransgenic control were grown under identical conditions and harvested and weighed at maturity (when fruit on the ‘Royal Gala’ control plants had reached a skin background color level 4–5 as determined by the ‘ENZA Fruit Gala/Royal Gala’ background color charts). Ethylene concentrations were measured by placing fruit in 1,215 cm3 respiration containers for 30 min, 1 cm3 samples were then withdrawn from the container headspace and ethylene measured by flame ionization chromatography (PU 4500 Chromatograph, Phillips). Ambient ethylene concentrations in empty respiration jars were used as controls. The fruit was then stored at 22°C and ethylene production of individual fruit was monitored every 3 d for 60 d. For the AO3 transgenic line and nontransgenic control, fruit stored at −1°C for 80 d was transferred to 22°C for 14 d. A sample of AO3 fruit was treated with flowing air containing ethylene (120 mg/m3) and a second sample, along with the control fruit, was treated with air for 6 d. The ethylene production of individual fruit was monitored at days 1, 3, and 6 and thereafter every day until day 14. When ethylene production reached a maximum (at day 8), CO2 production was measured using gas chromatography. At day 14, fruits were destructively sampled for analysis of soluble solids concentration (SSC) using a digital refractometer (model PR-1, Atago) and fruit firmness using a Materials Testing Machine (model 4301, Instron).
Volatile Analysis
To increase the number of apple fruit, vegetative tissue from the A03 line was grafted onto four ‘M.9’ rootstocks. Two sets of two trees were treated as biological repeats, and treated in parallel. Apples were harvested at maturity and then stored at 4°C for a week. The fruits were warmed to room temperature for a day and exposed to 120 mg/m3 ethylene continuously until volatiles were measured. Two samples of three fruit per replicate were sampled after 1 d at room temperature, before exposure to ethylene and then at 4, 18, 96 (4 d), and 192 h (8 d) of ethylene exposure. A control set of transgenic fruits were stored without ethylene and sampled at 192 h. Each sample was weighed and placed into a 2 L sampling vessel. The headspace in the sealed flask was allowed to equilibrate for 1 h at 24°C prior to flushing. Dried air was introduced to sweep the headspace (25 mL min−1) for 1 h onto a volatile absorbent trap (100 mg Chromosorb 105). Where analysis was not immediate traps were stored at −20°C prior to drying. Traps were dried with a N2 flow at 10 psi, 35°C for 15 min before analysis by gas chromatography/mass spectromy.
The trapped headspace material was thermally desorbed at 175°C onto a 30 m × 0.32 mm i.d., 0.5 μm film DBWax gas chromatography column (J & W Scientific), the outlet of which was split between a flame ionization detector (for quantification) and a mass spectrometer, mass spectrometry for component identification. The oven temperature program was 30°C for 6 min, then ramped at a rate of 3°C min−1 to 102°C followed by 5°C min−1 to 210°C. This final temperature was maintained for 5 min. The carrier gas was He at 30 cm s−1 and both detectors were maintained at 220°C. Peaks were converted into mass using an average detector response factor based on a standard containing ethyl butanoate, butyl acetate, 2-methylbutyl acetate, butanol, methyl hexanoate, ethyl hexanoate, hexyl acetate, and hexanol in pentane. Component identification was based on calculation of retention indices, mass spectra of authentic standards, and comparison with library spectra (NIST 98, Wiley 7, and in house).
Microarray Analysis
Following volatile detection, apple fruits were peeled and skin tissues and cortex tissues (excluding the core) for each time point were snap frozen in liquid nitrogen before storage at −80°C. Total RNA was extracted using a method to extract RNA from pine (Pinus taeda) needles (Chang et al., 1993). RNA was cleaned using RNAeasy cleanup kit (Qiagen) according to the manufacturer's protocol. A 1 in 5 dilution of RNA was checked for quality using an Agilent 2100 Bioanalyzer, with a RNA cassette according to the manufacturer's protocols. RNA was labeled with either Cy3 or Cy5 flourescent dye (GE Healthcare) using an amino-allyl dye coupling reaction. RT of the RNA consisted of taking 50 μg of RNA was added to 3 μL Oligo dT 23mer with a dAGC anchor (100 mm) in a total volume of 19.5 μL. This was heated to 70°C for 10 min and cooled to 4°C on ice, 6 μL transcriptor buffer, 2 μL dithiothreitol (100 mm), and 2 μL dNTP mix (dA, GCTP 7.5 mm, dTTP and amino allyl dUTP 3.75 mm), and 10 units of Transcriptor was added (total reaction volume 30 μL) and incubated for 42°C for 30 min. The reaction was stopped by adding 1 μL 20 mm EDTA, the RNA degraded by adding 1 μL NaOH (500 mm), heating at 70°C for 10 min, and cooling on ice, the reaction was neutralized by adding 1 μL HCl (500 mm). cDNA was precipitated with ethanol and reasuspended in 5 μL Na2CO3 (100 mm pH 9.0). Cy dye NHS esters (GE Healthcare) were resuspended in 22 μL dimethyl sulfoxide, and 5 μL was added to the resuspended cDNA, incubated for 2 h in the dark at room temperature, and cleaned using a PCR purification column (Qiagen) as described in the manufacturer's protocols, eluting from the column with 52 μL of water.
Apple microarrays containing 15,723 45 to 55 mer oligonucleotides, representing 15,102 nonredundant Malus sequences, with a constant melting temperature designed to ‘Royal Gala’ ESTs from the database (Newcomb et al., 2006) were used to measure global gene expression patterns. Oligos were printed on epoxy slides (MWG) using a Biorobotics II robot. Each sampling point was represented with four microarrays, with each of the two biological samples repeated in a dye swap experiment. Each array was hybridized with sheared genomic DNA from apple ‘Royal Gala’ in one channel to allow direct comparison between arrays. A total of 2.5 μg of sheared genomic DNA was labeled using a radprime labeling kit (Invitrogen) as described in the manufacturer's protocol, except 2 μL of 3 mm dAGCTP and 1.5 mm dT and amino allyl dUTP was used instead of the kit supplied nucleotides. Amino allyl incorporated DNA was ethanol precipitated and Cy dyes (GE Healthcare) added as described in the cDNA labeling protocol. Labeled cDNA and gDNA were mixed, and put through a further PCR cleanup column, eluting in 52 μL water. A total of 33 μL 20× SSC, 8.8 μL 5% SDS, 13.5 μL Liquid Block (GE Healthcare), and 114.7 μL water were added. The mixed DNA was denatured at 95°C for 10 min then kept at 60°C for hybridization. Hybridizations were performed eight at a time in a Lucidea Hybridization machine using chambers 2 to 5 and 9 to 11 (GE Healthcare). Microarrays were prehybridized at 45°C with 220 μL hybridization mixture without any labeled nuceotides for 15 min, using the mix step. The microarrays were then washed with wash 1 (2× SSC 0.3% SDS) and flushed with air. Hybridization mixtures containing the labeled cDNA and gDNA were injected onto the slide and hybridized for a minimum of 16 h at 45°C again using the mix step. The microarrays were washed with wash 1 for 1.2 min and cooled to 30°C, the microarrays were washed again with wash 1 for 1.2 min, wash 1 for 2.4 min, wash 2 (0.5× SSC, 0.3% SDS) for 2.4 min (twice), and then once with wash 3 (0.5× SSC). The microarrays were then air dried and scanned using a Genepix 4000B scanner (Axon). Spots were aligned using Genepix 4 software (Axon).
Data Processing
All analysis was done in R- and S-Plus 6.1 (Insightful). Microarrays were normalized in R- and S-Plus using modules from the BioConductor limma package (Smyth and Speed, 2003) and an in-house normalization protocol that had the following steps: All RNA and DNA channels were normalized with global mean normalization, combined into two files (containing all gDNA and all cDNA channels), and the distribution of intensities were normalized using quantile normalization (Bioconductor). A ratio (M) of the Cy3 and Cy5 values for each slide were calculated and the M values of the dye swaps were then smoothed with loess smoothing to remove dye bias. An absolute value for each spot was achieved by multiplying each ratio with the median gDNA value for that spot. This method has been used to select differentially expressed genes for a number of experiments and has a 75% to 80% qPCR verified success rate (out of 100 independent qPCR validations; data not shown). Genes were selected by identifying changes in the absolute values for each feature: First, features that were not represented on at least 12 of the 24 arrays for skin and 20 of the 48 arrays from the global analysis (from being flagged bad or not found) were removed. 12386 oligos were analyzed in the skin samples and 11682 oligos were analyzed for the global analysis. Second, for ethylene-induced skin genes a one-way ANOVA model of tissue, y = time, was used. This model treated the 8-d control sample that had no ethylene treatment as a zero time point (to identify genes controlled by ethylene rather than by changes over time). For global analysis of changes in tissue and time, an ANOVA model, y = tissue × time, was used, again treating the 8-d control sample as a zero time point. The number of significant differentially expressed genes was examined using a 0.05 threshold using a nonadaptive FDR control (Benjamini and Hochberg, 1995).
Real-Time RT-PCR Analysis
cDNA was synthesized from 2 μg of total RNA in a total volume of 50 μL with Superscript III reverse transcriptase according to the manufacturer's instructions (Invitrogen). Controls with no Superscript III reverse transcriptase were used to assess for potential genomic DNA contamination. Real-time primers were designed where possible as close to the region represented by the microarray oligos. cDNA used for real-time RT-PCR was synthesized in triplicate and optical density was measured for each sample. Each sample was then recombined for each time point and diluted to 15 ng μL−1. A total of 20 μL real-time PCR reactions were repeated four times on an ABI Prism 7900HT (Applied Biosystems) using 75 ng of cDNA, 0.4 μm primers, 0.2 mm of each dNTP, and a 0.1× concentration of SYBR Green I (Molecular Probes) as a reporter dye. Cycling conditions included an initial hot start at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 30 s. Each real-time PCR was ended by the addition of a dissociation curve analysis of the amplified product. This involved denaturation at 95°C for 15 s, cooling to 55°C for 20 s, and then gradual heating at 0.01°C s−1 to a final temperature of 95°C. Real-time PCR products were checked to ensure only single products were amplified. Three reference genes, Malus actin, Malus GAPDH, and a gene selected on the microarray as not changing over these and other experiments (oligonucleotide 23701) were analyzed in each real-time RT-PCR to normalize the expression patterns. Normalization factors were calculated by taking the geometric mean of the two least variable reference genes as determined by geNorm v3.4 (Vandesompele et al., 2002). Raw cycle threshold scores were converted to quantities representing relative expression levels using a modified comparative cycle threshold method (Pfaffl, 2001) and with correction for different amplification efficiencies (Ramakers et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. qPCR validation of aroma genes on the microarray.
Supplemental Table S1. A list of the putative aroma volatile genes found on the microarray with calculated P values from the microarray expression patterns for ethylene regulation in the skin, and differential tissue expression between skin and cortex.
Supplemental Table S2. A list of all selected genes that change during ethylene induction in the skin, and the relative expression level of that gene.
Supplemental Table S3. A complete list of genes that change with ethylene and that are differentially expressed between skin and cortex using a FDR cutoff of 0.05.
Supplementary Material
Acknowledgments
We would like to thank Dave Billing and Doug Burmeister for help with the ethylene induction, Carmen Villatoro for help with qPCR, and Ross Atkinson, Daryl Rowan, Eric Walton, and Ian Ferguson for critical reading of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew P. Gleave (agleave@hortresearch.co.nz).
The online version of this article contains Web-only data.
References
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9 331–338 [DOI] [PubMed] [Google Scholar]
- Aharoni A, O'Connell AP (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53 2073–2087 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Bolitho KM, Wright MA, Iturriagagoitia-Bueno T, Reid SJ, Ross GS (1998) Apple ACC-oxidase and polygalacturonase: ripening-specific gene expression and promoter analysis in transgenic tomato. Plant Mol Biol 378 449–460 [DOI] [PubMed] [Google Scholar]
- Azevedo RA, Arruda P, Turner WL, Lea PJ (1997) The biosynthesis and metabolism of the aspartate derived amino acids in higher plants. Phytochemistry 46 395–419 [DOI] [PubMed] [Google Scholar]
- Beekwilder J, Alvarez-Huerta M, Neef E, Verstappen FWA, Bouwmeester HJ, Aharoni A (2004) Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol 135 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc [Ser A] 57 289–300 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar AM, Teo G, Defilippi BG, Uratsu SL, Passey AJ, Kader AA, Stow JR, Colgan RJ, James DJ (2004) Effect of down-regulation of ethylene biosynthesis on fruit flavor complex in apple fruit. Transgenic Res 13 373–384 [DOI] [PubMed] [Google Scholar]
- Defilippi BG, Dandekar AM, Kader AA (2005. a) Relationship of ethylene biosynthesis to volatile production, related enzymes, and precursor availability in apple peel and flesh tissues. J Agric Food Chem 53 3133–3141 [DOI] [PubMed] [Google Scholar]
- Defilippi BG, Kader AA, Dandekar AM (2005. b) Apple aroma: alcohol acyltransferase, a rate limiting step for ester biosynthesis, is regulated by ethylene. Plant Sci 168 1199–1210 [Google Scholar]
- Dimick PS, Hoskin JC (1983) Review of apple flavor—state of the art. CRC Crit Rev Food Sci Nutr 18 387–409 [DOI] [PubMed] [Google Scholar]
- Dixon J, Hewett EW (2000) Factors affecting apple aroma/flavour volatile concentration: a review. N Z J Crop Hortic Sci 28 155–173 [Google Scholar]
- Fonseca S, Hackler L Jr, Zvara A, Ferreira S, Balde A, Dudits D, Pais MS, Puskas LG (2004) Monitoring gene expression along pear fruit development, ripening and senescence using cDNA microarrays. Plant Sci 167 457–469 [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Wang JH, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organizational-structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goodenough PW, Entwistle TG (1982) The hydrodynamic properties and kinetic constants with natural substrates of the esterase from Malus-pumila fruit. Eur J Biochem 127 145–149 [DOI] [PubMed] [Google Scholar]
- Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA (1991) Stress responses in alfalfa (Medicago-sativa L). 10. Molecular-cloning and expression of S-adenosyl-L-methionine-caffeic acid 3-O-methyltransferase, a key enzyme of lignin biosynthesis. Plant Physiol 97 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadagni DG, Bomben JL, Hudson JS (1971) Factors influencing development of aroma in apple peels. J Sci Food Agric 22 110–115 [Google Scholar]
- Green S, Friel EN, Matich A, Beuning LL, Cooney JM, Rowan DD, MacRae E (2007) Unusual features of a recombinant apple alpha-farnesene synthase. Phytochemistry 68 176–188 [DOI] [PubMed] [Google Scholar]
- Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53 383–397 [DOI] [PubMed] [Google Scholar]
- Ileperuma NR, Marshall SDG, Squire CJ, Baker HM, Oakeshott JG, Russell RJ, Plummer KM, Newcomb RD, Baker EN (2007) High resolution crystal structure of the plant carboxylesterase CXE1, from Actinidia eriantha, and its complex with a high affinity inhibitor paraoxon. Biochemistry 46 1851–1859 [DOI] [PubMed] [Google Scholar]
- Ju ZG, Curry EA (2000) Evidence that alpha-farnesene biosynthesis during fruit ripening is mediated by ethylene regulated gene expression in apples. Postharvest Biol Technol 19 9–16 [Google Scholar]
- Kotoda N, Wada M, Kusaba S, Kano-Murakami Y, Masuda T, Soejima J (2002) Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci 162 679–687 [Google Scholar]
- Lay-Yee M, Dellapenna D, Ross GS (1990) Changes in messenger-RNA and protein during ripening in apple fruit (Malus-domestica Borkh cv Golden Delicious). Plant Physiol 94 850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DP, Xu YF, Xu GM, Gu LK, Li DQ, Shu HR (2006) Molecular cloning and expression of a gene encoding alcohol acyltransferase (MdAAT2) from apple (cv. Golden Delicious). Phytochemistry 67 658–667 [DOI] [PubMed] [Google Scholar]
- Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38 948–952 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci 24 34–35 [DOI] [PubMed] [Google Scholar]
- Newcomb RD, Crowhurst RN, Gleave AP, Rikkerink EHA, Allan AC, Beuning LL, Bowen JH, Gera E, Jamieson KR, Janssen BJ, et al (2006) Analyses of expressed sequence tags from apple. Plant Physiol 141 147–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48 109–136 [DOI] [PubMed] [Google Scholar]
- Park S, Sugimoto N, Larson MD, Beaudry R, van Nocker S (2006) Identification of genes with potential roles in apple fruit development and biochemistry through large-scale statistical analysis of expressed sequence tags. Plant Physiol 141 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous SW, Whitaker BD (2004) Cloning and functional expression of an (E, E)-alpha-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219 84–94 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 52 1–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Morrman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Ross GS, Knighton ML, Layyee M (1992) An ethylene-related Cdna from ripening apples. Plant Mol Biol 19 231–238 [DOI] [PubMed] [Google Scholar]
- Rowan DD, Allen JM, Fielder S, Hunt MB (1999) Biosynthesis of straight-chain ester volatiles in red delicious and granny smith apples using deuterium-labeled precusors. J Agric Food Chem 47 2553–2562 [DOI] [PubMed] [Google Scholar]
- Rowan DD, Lane HP, Allen JM, Fielder S, Hunt MB (1996) Biosynthesis of 2-methylbutyl, 2-methyl-2-butenyl and 2-methylbutanoate esters in Red Delicious and Granny Smith apples using deuterium-labeled substrates. J Agric Food Chem 44 3276–3285 [Google Scholar]
- Sisler EC, Serek M (2003) Compounds interacting with the ethylene receptor in plants. Plant Biol 5 473–480 [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31 265–273 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souleyre EJF, Greenwood DR, Friel EN, Karunairetnam S, Newcomb RD (2005) An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavor. FEBS J 272 3132–3144 [DOI] [PubMed] [Google Scholar]
- Sunako T, Sakuraba W, Senda M, Akada S, Ishikawa R, Niizeki M, Harada T (1999) An allele of the ripening-specific 1-aminocyclopropane-1-carboxylic acid synthase gene (ACS1) in apple fruit with a long storage life. Plant Physiol 119 1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JL, Cohen D, Atkinson R, Richardson K, Morris B (1995) Regeneration of transgenic plants from the commercial apple cultivar Royal-Gala. Plant Cell Rep 14 407–412 [DOI] [PubMed] [Google Scholar]
- Yao JL, Cohen D, van den Brink R, Morris B (1999) Assessment of expression and inheritance patterns of three transgenes with the aid of techniques for promoting rapid flowering of transgenic apple trees. Plant Cell Rep 18 727–732 [Google Scholar]
- Young JC, Chu CLG, Lu XW, Zhu HH (2004) Ester variability in apple varieties as determined by solid-phase microextraction and gas chromatography-mass spectrometry. J Agric Food Chem 52 8086–8093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.