Glycosyl, or glycoside, hydrolases (GHs) comprise a structurally diverse group of enzymes that hydrolyze glycosidic bonds between carbohydrates, or between carbohydrates and other noncarbohydrate moieties, and that collectively exhibit a wide range of substrate specificities. GH enzymes from across the taxonomic spectrum were originally named based on substrate specificity, the corresponding International Union of Biochemistry and Molecular Biology (IUBMB) nomenclature system (EC 3.2.1.-), and the chronological order in which they were reported. However, as growing numbers of GH proteins, and later genes, were characterized, this strategy proved to be increasingly unsatisfactory and a complementary nomenclature was developed based on predicted protein sequence (Henrissat et al., 1998). This approach provides important insights into protein structure, evolutionary relationships, and an opportunity to infer mechanistic relationships. A regularly updated database, Carbohydrate-Active Enzymes (CAZY; www.cazy.org), currently lists 108 distinct GH families, a subset of which are further affiliated with 14 clans based on the presence of defined protein folds and conserved catalytic machineries.
This nomenclature initiative was developed largely in response to the rapidly growing numbers of reported microbial GHs, reflecting their numerous important industrial uses. Notable examples are endo-β-1,4-glucanases, or cellulases, which hydrolyze the β-1,4-glucosyl linkages of cellulose and several other plant cell wall polysaccharides, and are used in the generation of sugars from lignocellulosic biomass for ethanol production. Plant biologists are currently facing a similar nomenclature dilemma, since the sequencing of whole plant genomes has revealed many large GH families (Henrissat et al., 2001): a genome-scale assessment of Arabidopsis (Arabidopsis thaliana) in CAZY identified 393 GHs from 34 families (http://www.cazy.org/geno/3702.html), and equivalent families are emerging in many species as larger EST collections develop. GH activities from plants have long been studied in association with various aspects of growth, development, and cell wall metabolism, but plant GH enzymes have often been named with little consideration of their substrate specificity, molecular structure, or enzymatic reaction mechanism.
The current explosion of interest and new research opportunities in biofuels and bioenergy crops will inevitably result in renewed interest in identifying and annotating plant GHs, through their association with lignocellulosic biomass. Thus, this is an opportune time to adopt a new, rational, well-defined nomenclature. Importantly, it is apparent that many of these plant enzymes share similarities to previously classified families of microbial GHs, which is not clear from their original designations. While the IUBMB-recommended nomenclature (www.chem.qmul.ac.uk/iubmb), based on the substrates used and the reaction catalyzed, continues to provide critical information regarding enzyme function, we propose a complementary, standardized nomenclature for plant GHs, based on the rigorous scheme for naming their microbial counterparts, which is categorized by the catalytic domain of the enzyme (Henrissat et al., 1998).
We have selected plant endo-β-1,4-glucanases as a case study, since this family illustrates the problems with imprecise GH nomenclatures that have evolved over decades of research. In addition, this family of plant enzymes has a divergent subfamily structure that can usefully be incorporated into the development of a naming scheme. Early reports described the existence of plant cellulases (e.g. Hall, 1963) and cellulolytic activities have long been associated with both cell wall construction during cell expansion and the wall disassembly that accompanies processes such as fruit ripening and abscission (for review, see del Campillo, 1999; Rose and Bennett, 1999; Mølhøj et al., 2002) and cellulose biosynthesis (Nicol et al., 1998; Lane et al., 2001; Sato et al., 2001). Most of the plant “cellulases” studied to date are typical endoglucanases (EC 3.2.1.4) with low or no activity on crystalline cellulose, but clearly measurable activity on soluble cellulose derivatives, such as carboxymethyl cellulose, noncrystalline phosphoric acid swollen cellulose, and/or a variety of plant polysaccharide substrates, including xylans, 1,3-1,4-β-glucans, and glucomannans (Master et al., 2004; Yoshida and Komae, 2006; Urbanowicz et al., 2007). This is quite distinct from the case of microbial cellulases, whose modular structure and synergistic actions with a number of different enzymes allow effective degradation of crystalline cellulose. The sequencing of the first plant “cellulases”/endo-β-1,4-glucanases revealed that they belong to the GH9 family (Henrissat, 1991). This is one of the larger plant GH families (Henrissat et al., 2001), and studies of microbial GH9 proteins, including both endoglucanases (EC 3.2.1.4) and cellobiohydrolases (EC 3.2.1.91), have shown that they operate via an inverting mechanism to cleave the 1,4-β-glucosidic bond between two unsubstituted Glc units (Gebler, 1992). Analyses of complete plant genome sequences have now shown that this large multigene family can be subdivided into three distinct structural subclasses (Fig. 1), comprising members that are membrane anchored (with or without a cytosolic domain), secreted, or those that are secreted and have a carbohydrate binding module, CBM49 (Mølhøj et al., 2002; Libertini et al., 2004; Urbanowicz et al., 2007).
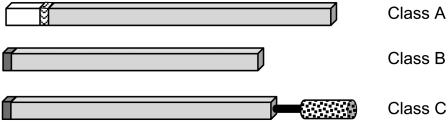
Figure 1.
Schematic representation of the modular plant GH9 family structure. Specific domains comprise the cytosolic domain (white), transmembrane domain (wavy lines), signal sequence (dark grey), GH9 catalytic domain (light grey), linker region (thick black line), and carbohydrate binding module (dots). Structural subclasses are represented by SlGHl9A1/TomCel3 (class A, U78526), SlGH9B1/TomCel1 (class B, U13054), and SlGH9C1/TomCel8 (class C, AF098292).
To standardize the nomenclature for these gene families, we suggest the following, using genes encoding GH9 enzymes as an example: an indication of the genus and species, followed by the designated the GH family (GH9). The letters A to C adjacent to the family number correspond to the domain structure, or subclass, of the corresponding protein (Fig. 1), which will provide additional information about potential function. We note that this nomenclature helps determine the structural subclass with which a particular GH9 gene is associated, rather than specifically suggesting an orthologous sequence. As an example of the naming scheme, we have applied the guidelines to rename the members of the GH9 family from tomato (Solanum lycopersicum), the plant species from which the greatest number of family members has been studied in detail. Historically, members of the tomato GH9 family have been referred to as TomCel1-8 and their new designations are shown in Table I. This nomenclature provides important information since, using SlGH9C1 gene as an example, the name indicates that this encodes the first described tomato (Sl, for S. lycopersicum) member of GH family 9 (GH9) with a class C domain structure, indicating that the protein contains a CBM49. This naming scheme has also been applied to the 25 member GH9 family in Arabidopsis (Table II), where a sequenced genome and ease of genetic manipulation is yielding information about the biological roles of many members of this family. Once the biochemical activity and substrate specificity of a particular protein are determined, the enzyme (as opposed to gene) can be given an appropriate EC number and a trivial name, followed by the GH family number, such as “Cel9” for a 1,4-β-endo-glucanase (EC 3.2.1.4) or “Xyn9” for an endo-1,4-β-xylanase (EC 3.2.1.8), based on the dominating activity of that protein. This approach has the advantage of allowing enzymologists to provide accurate and useful information when describing a protein that has been characterized in detail, but still enables facile gene annotation by nonexperts.
Table I.
GH9 proteins of tomato
| Current Designation | GenBank ID | New Designation | Featuresa |
|---|---|---|---|
| TomCel1 | U13054 | SlGH9B1 | SP, GH9 |
| TomCel2 | U13055 | SlGH9B2 | SP, GH9 |
| TomCel3 | U78526 | SlGH9A1 | CT, TM, GH9 |
| TomCel4 | U20590 | SlGH9B3 | SP, GH9 |
| TomCel5 | AF077339 | SlGH9B4 | SP, GH9 |
| TomCel6 | AAB46829b | SlGH9B5 | SP, GH9 |
| TomCel7 | Y11268 | SlGH9B6 | SP, GH9 |
| TomCel8 | AF098292 | SlGH9C1 | SP, GH9, CBM49 |
Features noted are cytosolic domain (CT), transmembrane domain (TM), signal peptide (SP), glycosyl hydrolase family 9 catalytic domain (GH9), and a family 49 carbohydrate binding module (CBM49).
Peptide fragment.
Table II.
GH9 proteins of Arabidopsis
| Chromosome Locus | GenBank ID | New Designation | Featuresa | Synonyms |
|---|---|---|---|---|
| At5g49720 | U37702 | AtGH9A1 | CT, TM, GH9 | Endo-1,4-β-glucanase 25, KORRIGANb, KOR1b |
| At1g65610 | AC001229 | AtGH9A2 | CT, TM, GH9 | Endo-1,4-β-glucanase 7, KOR2c |
| At4g24260 | AL078637 | AtGH9A3 | CT, TM, GH9 | Endo-1,4-β-glucanase 21, KOR3c |
| At3g43860 | AY072099 | AtGH9A4 | TM, GH9 | Endo-1,4-β-glucanase 16 |
| At1g70710 | AY048283 | AtGH9B1 | SP, GH9 | Endo-1,4-β-glucanase 8, cellulase 1d, AtCEL1d |
| At1g02800 | AF034573 | AtGH9B2 | SP, GH9 | Endo-1,4-β-glucanase 1, cellulase 2e, AtCEL2e |
| At1g71380 | U17888.1 | AtGH9B3 | SP, GH9 | Endo-1,4-β-glucanase 9, cellulase 3f, AtCEL3f |
| At1g22880 | AF000657 | AtGH9B4 | SP, GH9 | Endo-1,4-β-glucanase 3, cellulase 5f, AtCEL5f |
| At1g19940 | AC007797 | AtGH9B5 | SP, GH9 | Endo-1,4-β-glucanase 2 |
| At1g23210 | AC002311 | AtGH9B6 | SP, GH9 | Endo-1,4-β-glucanase 4 |
| At1g75680 | AC006434 | AtGH9B7 | SP, GH9 | Endo-1,4-β-glucanase 10 |
| At2g32990 | AC003033 | AtGH9B8 | SP, GH9 | Endo-1,4-β-glucanase 11 |
| At2g44540 | AC004521 | AtGH9B9 | SP, GH9 | Endo-1,4-β-glucanase 12 |
| At2g44550 | AC004521 | AtGH9B10 | SP, GH9 | Endo-1,4-β-glucanase 13 |
| At2g44560 | AC003672 | AtGH9B11 | SP, GH9 | Endo-1,4-β-glucanase 14 |
| At2g44570 | AC003672 | AtGH9B12 | SP, GH9 | Endo-1,4-β-glucanase 15 |
| AT4g02290 | AY079162 | AtGH9B13 | SP, GH9 | Endo-1,4-β-glucanase 17 |
| At4g09740 | AL161515 | AtGH9B14 | SP, GH9 | Endo-1,4-β-glucanase 18 |
| At4g23560 | AL035394 | AtGH9B15 | SP, GH9 | Endo-1,4-β-glucanase 20 |
| At4g38990 | AL035679 | AtGH9B16 | SP, GH9 | Endo-1,4-β-glucanase 22 |
| At4g39000 | AL035679, AK117850 | AtGH9B17 | SP, GH9 | Endo-1,4-β-glucanase 23 |
| At4g39010 | AY059825 | AtGH9B18 | SP, GH9 | Endo-1,4-β-glucanase 24 |
| At1g48930 | AC016041 | AtGH9C1 | SP, GH9, CBM49 | Endo-1,4-β-glucanase 5 |
| At1g64390 | AC066689 | AtGH9C2 | SP, GH9, CBM49 | Endo-1,4-β-glucanase 6 |
| At4g11050 | AF080120 | AtGH9C3 | SP, GH9, CBM49 | Endo-1,4-β-glucanase 19 |
Features noted are cytosolic domain (CT), transmembrane domain (TM), signal peptide (SP), glycosyl hydrolase family 9 catalytic domain (GH9), and a family 49 carbohydrate binding module (CBM49).
This nomenclature conforms to that used for bacterial endoglucanases with some slight modifications, since many of the families of plant GHs are much larger than those of bacteria (CAZY) and so the information designating the order in which they are reported is more easily presented by a numerical rather than an alphabetical system. For example, in Arabidopsis there are 49 members of family GH1, 33 of GH16, and 69 of GH28, all of which contain more members than there are letters in the alphabet (http://www.cazy.org/geno/3702.html).
The new nomenclature has been presented in the context of the plant GH9 family, but we further suggest that similar guidelines be adopted for naming members of other GH families and that future efforts to standardize nomenclature be coordinated in consultation with CAZY. Similarly, the use of the established naming schemes for carbohydrate binding modules (www.cazy.org; Boraston et al., 2004) will further help advance the study of analogous hydrolytic enzymes and other associated functional domains from different organisms, both within and between taxa.
References
- Boraston AB, Bolam HJ, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E (1999) Multiple endo-1,4-β-D-glucanase (cellulase) genes in Arabidopsis. Curr Top Dev Biol 46 39–61 [DOI] [PubMed] [Google Scholar]
- del Campillo E, Abdel-Aziz A, Crawford D, Patterson S (2004) Root cap specific expression of an endo-β-1,4-glucanase (cellulase): a new marker to study root development in Arabidopsis. Plant Mol Biol 56 309–323 [DOI] [PubMed] [Google Scholar]
- Gebler J, Gilkes NR, Claeyssens M, Wilson DB, Beguin P, Wakarchuk WW, Kilburn DG, Miller RC Jr, Warren RA, Withers SG (1992) Stereoselective hydrolysis catalyzed by related beta-1,4-glucanases and beta-1,4-xylanases. J Biol Chem 267 12559–12561 [PubMed] [Google Scholar]
- Hall CB (1963) Cellulase in tomato fruits. Nature 200 1010–1011 [DOI] [PubMed] [Google Scholar]
- Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PH, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47 55–72 [PubMed] [Google Scholar]
- Henrissat B, Teeri TT, Warren RAJ (1998) A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett 425 352–354 [DOI] [PubMed] [Google Scholar]
- Libertini E, Li Y, McQueen-Mason SJ (2004) Phylogenetic analysis of the plant endo-β-1,4-glucanase gene family. J Mol Evol 58 506–515 [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeijer A, Peng LC, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn J, et al (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master EM, Rudsander UJ, Zhou W, Henriksson H, Divne C, Denman S, Wilson DB, Teeri TT (2004) Recombinant expression and enzymatic characterization of PttCel9A, a KOR homologue from Populus tremula x tremuloides. Biochemistry 43 10080–10089 [DOI] [PubMed] [Google Scholar]
- Mølhøj M, Jorgensen B, Ulvskov P, Borkhardt B (2001) Two Arabidopsis thaliana genes KOR2 and KOR3, which encode membrane-anchored endo-1,4-β-D-glucanases, are differentially expressed in developing leaf trichomes and their support cells. Plant Mol Biol 46 263–275 [DOI] [PubMed] [Google Scholar]
- Mølhøj M, Pagant S, Höfte H (2002) Towards understanding the role of membrane-bound endo-β-1,4-lucanases in cellulose biosynthesis. Plant Cell Physiol 43 1399–1406 [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB (1999) Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci 4 176–183 [DOI] [PubMed] [Google Scholar]
- Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, Takabe K, Nishiyama Y, Kuga S, Sato S, et al (2001) Role of the putative membrane-bound putative endo-β-D-1,4-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis. Plant Cell Physiol 42 251–263 [DOI] [PubMed] [Google Scholar]
- Shani Z, Dekel M, Tsabary G, Shoseyov O (1997) Cloning and characterization of elongation specific endo-1,4-beta-glucanase (cel1) from Arabidopsis thaliana. Plant Mol Biol 34 837–842 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Catalá C, Irwin D, Wilson DB, Ripoll DR, Rose JKC (2007) A tomato endo-β-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49). J Biol Chem 282 12066–12074 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Komae K (2006) A rice family 9 glycoside hydrolase isozyme with broad substrate specificity for hemicelluloses in type II cell walls. Plant Cell Physiol 47 1541–1554 [DOI] [PubMed] [Google Scholar]
- Yung M, Schaffer R, Putterill J (1999) Identification of genes expressed during early Arabidopsis carpel development by mRNA differential display: characterization of ATCEL2, a novel endo-1,4-β-D-glucanase gene. Plant J 17 203–208 [DOI] [PubMed] [Google Scholar]