Abstract
Plants from the Casuarinaceae family enter symbiosis with the actinomycete Frankia leading to the formation of nitrogen-fixing root nodules. We observed that application of the auxin influx inhibitor 1-naphtoxyacetic acid perturbs actinorhizal nodule formation. This suggests a potential role for auxin influx carriers in the infection process. We therefore isolated and characterized homologs of the auxin influx carrier (AUX1-LAX) genes in Casuarina glauca. Two members of this family were found to share high levels of deduced protein sequence identity with Arabidopsis (Arabidopsis thaliana) AUX-LAX proteins. Complementation of the Arabidopsis aux1 mutant revealed that one of them is functionally equivalent to AUX1 and was named CgAUX1. The spatial and temporal expression pattern of CgAUX1 promoter:β-glucuronidase reporter was analyzed in Casuarinaceae. We observed that CgAUX1 was expressed in plant cells infected by Frankia throughout the course of actinorhizal nodule formation. Our data suggest that auxin plays an important role during plant cell infection in actinorhizal symbioses.
Actinorhizal plants, which belong to eight families of angiosperms, can form nitrogen-fixing nodules in symbiosis with the soil actinomycete Frankia (Benson and Silvester, 1993). The symbiotic interaction starts, under conditions of nitrogen deprivation, by an exchange of signals between the plant roots and bacteria. The chemical nature of Frankia nodulation factors is unknown, but data suggest that it has different biochemical properties from that of Rhizobium (Cérémonie et al., 1999). During intracellular infection, Frankia signals lead to root hair deformation, some of which become infected. At the same time, limited cell divisions are triggered in the cortex, creating a so-called prenodule. Prenodule function is not known, but it is an obligatory step of intracellular infection (Laplaze et al., 2000). Concomitantly, cell divisions occur in the pericycle in front of the xylem pole leading to the formation of a nodule lobe primordium. The growing nodule lobe is infected by Frankia hyphae coming from the prenodule. The structure of the new organ formed upon infection largely differs from legume nodules even if the infection mechanisms share common features (Pawlowski and Bisseling, 1996). Actinorhizal nodules are considered as modified lateral roots because (1) they originate from divisions in the pericycle in front of a xylem pole; (2) they have a lateral root-like structure with a central vasculature, infected cells in the cortex, and an apical meristem; and (3) in some species (e.g. Casuarina sp.) a so-called nodule root is produced at the apex (Obertello et al., 2003). Little is known about the mechanisms of actinorhizal nodule development.
The plant hormone auxin is involved in many developmental processes (Tanaka et al., 2006) and is the key signal controlling lateral root development (Casimiro et al., 2003). Auxin transport across the plant is polarized and perturbations of polar auxin transport using inhibitors such as naphthylphthalamic acid or mutants result in dramatic alteration of the plant developmental pattern (Reed et al., 1998). The existence of auxin transporters has been predicted for a long time to account for polar auxin transport (Goldsmith, 1977). Characterization of Arabidopsis (Arabidopsis thaliana) mutants perturbed in auxin transport or sensitivity led to the identification of auxin efflux and influx facilitators encoded by the PIN and AUX-LAX genes, respectively (Kramer and Bennett, 2006). The latter are encoded by a small gene family (four genes) in Arabidopsis (Parry et al., 2001b). Only one member of the AUX-LAX family has been characterized to date: AUX1 is involved in gravitropism (Bennett et al., 1996) and lateral root initiation (Marchant et al., 2002). AUX1 has recently been shown to encode a high-affinity auxin influx transporter by heterologous expression in Xenopus oocytes (Yang et al., 2006). The mechanism of transport remains to be elucidated, but is predicted to occur by proton symport (Kerr and Bennett, 2007).
Auxin transport is also thought to be involved in the establishment of legume symbiosis. Local auxin transport inhibition is triggered by spot inoculation of rhizobia, leading to subsequent accumulation of auxin at the site of infection as shown by the use of the GH3:gusA auxin response marker in white clover (Trifolium repens; Mathesius et al., 1998) and Lotus japonicus (Pacios-Bras et al., 2003). In legumes forming indeterminate nodules, flavonoids are produced as a response to bacterial lipochitin oligosaccharides (Mathesius et al., 2000) and act as inhibitors of auxin efflux transport (Brown et al., 2001), leading to local accumulation of auxin necessary for cell division and subsequent nodule primordium formation (Wasson et al., 2006). Moreover, the expression of auxin influx transporters in Medicago is associated with nodule primordium development and vasculature differentiation (de Billy et al., 2001).
A role of auxin during actinorhizal symbiont dialogue has also been suggested because some Frankia strains can produce different forms of auxin in culture (Gordons et al., 1988; Hammad et al., 2003). However, no link has been made between the production of hormones by Frankia and establishment of symbiosis. The symbiotic bacteria Rhizobium produce auxins that were proposed to be involved in establishing symbiosis with legume plants (Badenoch-Jones et al., 1983). Indeed, a Bradyrhizobium japonicum mutant producing 30-fold more indole-3-acetic acid (IAA) than the wild type has higher nodulation efficiency (Kaneshiro and Kwolek, 1985). Altogether, up to 80% of rhizobacteria are considered to produce auxins (Patten and Glick, 1996). However, nothing is known about the precise role of bacterial auxin during the processes of infection and symbiosis or how and when the plant cell perceives it.
In this study, we show that application of the auxin influx inhibitor 1-naphtoxyacetic acid (1-NOA) perturbs nodule formation. We therefore isolated a small family of AUX-LAX gene homologs in the actinorhizal plant Casuarina glauca. Among this family of genes, we identified CgAUX1, a homolog of AtAUX1, which carries an auxin carrier function as shown by functional complementation of the Arabidopsis aux1 mutant. Expression of CgAUX1 is found in all Frankia-infected cells from the root hair to nodule nitrogen-fixing cells. We also bring evidence of differences between the genetic programs of lateral root and actinorhizal nodule primordium based on different patterns of CgAUX1 expression. Altogether, our results shed light on the role of auxin influx transport during actinorhizal nodule formation.
RESULTS
Inhibition of Auxin Influx Transport Using 1-NOA Perturbs Nodule Formation
We analyzed the effect of 1-NOA, a competitive inhibitor of auxin influx, on the C. glauca-Frankia interaction. 1-NOA is known to specifically inhibit AtAUX1 (Yang et al., 2006) and to mimic the aux1 mutant phenotype in Arabidopsis (Parry et al., 2001a). C. glauca plants were inoculated and grown in hydroponics in the presence of 25 μm 1-NOA. The number of nodulated plants (i.e. plants bearing prenodules or nodules) was checked every day after 10 d (Fig. 1A). We found in three independent experiments that 1-NOA treatments caused a 2-d delay in nodule appearance. The same effect was observed if the growth medium was changed every 3 d with fresh 1-NOA to prevent potential 1-NOA degradation (data not shown). Moreover, 24 d after inoculation, plants treated with 1-NOA mainly showed prenodules, whereas control plants showed nodules (Fig. 1, B and C). This 1-NOA effect on nodulation was not due to a more general effect on root growth because we found no significant differences in shoot or root weight in treated or nontreated plants (Student's t test; P < 0.1; no treatment [NT] roots, m = 0.059 g; NOA-treated roots, m = 0.062 g; NT shoots, m = 0.136 g; NOA-treated shoots, m = 0.137 g; dry weights; n = 20). Moreover, we also verified that addition of 25 μm 1-NOA had no deleterious effects on Frankia growth (Fig. 1D). We therefore conclude that inhibition of auxin influx transport using 1-NOA partially perturbs actinorhizal nodule formation in C. glauca.
Figure 1.
1-NOA treatment perturbs the nodulation process by specifically inhibiting auxin influx transport. A, Percentage of plants bearing symbiotic structures (prenodules or nodules) after inoculation by Frankia in absence (NT, black squares) or presence of 25 μm 1-NOA (NOA, white squares). The data presented correspond to one representative of three experiments with similar results (20 plants/treatment). B, 24-d-old roots of plants bearing nodules (arrowheads) upon NT. C, 24-d-old roots of plants bearing big prenodules (arrowheads) upon NOA treatment. D, Frankia exponential growth indicated as total protein content upon NT or 25 μm 1-NOA treatment (NOA). Bars = 1 cm (B and C). [See online article for color version of this figure.]
Identification of a Small Family of Auxin Influx Carrier Genes in C. glauca
Our data suggest a role for auxin influx carriers encoded by AUX1 homologs during actinorhizal nodule development. AUX-LAX gene homologs were therefore isolated from C. glauca by amplifying genomic DNA with different sets of degenerate primers (Table I) designed in conserved regions of AUX-LAX proteins of Arabidopsis, Medicago truncatula, and poplar (Populus spp.). Seven different PCR products were produced, sequenced, and found to correspond to two different genes. The corresponding cDNAs (1,440 and 1,395 bp) were obtained by RACE-PCR. They were named CgAUX1 and CgLAX3 according to the sequence identity of the predicted proteins to Arabidopsis proteins, 85% for AUX1 and 87% for LAX3, respectively.
Table I.
Degenerate primers designed in the most conserved regions of the AUX-LAX genes and used for C. glauca genomic DNA amplification
| Primer Name | Sequence (5′-3′) | Direction-Position |
|---|---|---|
| AD1 | ATYCARCTHATWGCYTGYGC | Forward-exon 3 |
| AD2 | GACAARAGRACWTGGACWTA | Forward-exon 4 |
| AD3 | CACAT6GCRTGCATDATYTC | Reverse-exon 6 |
| AD4 | CCRAA6GCCCARTADAS6GC | Reverse-exon 6 |
| AF2 | CCACAT6GCRTGCATDATYTC | Forward-exon 3 |
| AF3 | TGGAC6TAYATHTTYGG6GC6TGY | Reverse-exon 5 |
The genomic sequences corresponding to CgAUX1 and CgLAX3 were amplified by PCR and found to be 2,942 and 2,224 bp long, respectively, from start to stop codon. Intron positions were conserved between Arabidopsis (Parry et al., 2001b) and C. glauca AUX-LAX genes. CgAUX1 and CgLAX3 have the same structure as AtAUX1 (Fig. 2A), whereas AtLAX3 (Fig. 2A) has one intron less in the C-terminal part of the gene, respectively. The same gene structure was found for all of the M. truncatula AUX-LAX genes (five genes; Schnabel and Frugoli, 2004), indicating common evolutionary origin. Interestingly, AUX-LAX gene structure is slightly different in rice (Oryza sativa sp. japonica ‘Nipponbare,’ five genes in the annotated genome; Tyagi et al., 2004), where three genes have seven exons like AtAUX1 and the two others have five and two exons, respectively. However, the position of the conserved introns is similar (data not shown), suggesting that this gene structure preceded the divergence of monocots and dicots and that a loss of introns is responsible for the observed differences in intron-exon number.
Figure 2.
C. glauca AUX-LAX genes family. A, Exon-intron structure of CgAUX1 and CgLAX3 compared to AtAUX1 and AtLAX3. Exons are shown as gray boxes and introns as black lines. B, Southern-blot experiments suggest that there are only two AUX-LAX genes in the C. glauca genome. Digested genomic DNA (with BamHI, EcoRI, or HindIII) was hybridized with a probe designed in a conserved region (lane 1), a CgAUX1-specific probe (lane 2), or a CgLAX3-specific probe (lane 3).
To estimate the number of AUX-LAX genes in the C. glauca genome, we conducted Southern-blot experiments using three different probes: a nonspecific probe designed in one of the most conserved regions of AUX-LAX genes (exon VII) and two gene-specific probes designed in CgAUX1 and CgLAX3 3′-untranslated regions. The conserved probe hybridized with a limited number of genomic DNA fragments in nonstringent conditions that could be assigned to either CgAUX1 or CgLAX3 using gene-specific hybridizations (Fig. 2B). This, together with the fact that we did not recover any other gene by PCR or in a C. glauca EST library (Hocher et al., 2006), suggests that auxin influx carriers are encoded by a small gene family (possibly only two genes) in C. glauca.
CgAUX1 Encodes an Auxin Influx Carrier Functionally Equivalent to AtAUX1
Arabidopsis and C. glauca AUX-LAX deduced protein sequences were compared with a representative member of each class of the amino acid transporter family. A phylogenetic tree was generated using a neighbor-joining distance algorithm showing that AUX-LAX proteins belong to the amino acid and auxin permease (Young et al., 1999) family (Fig. 3A). Among the AUX-LAX proteins, two subclasses could be defined containing AtAUX1, CgAUX1, and AtLAX1 for the first subclass and AtLAX2, AtLAX3, and CgLAX3 for the second subclass (Fig. 3A). Comparison of protein sequences (Fig. 3B) shows that the N and C terminus sequences are the most divergent, whereas the central sequence is highly conserved. Out of 13 amino acids that have been shown to be important for AtAUX1 activity (Swarup et al., 2004), all are conserved in CgLAX3 and all but one in CgAUX1 (Fig. 3B).
Figure 3.
Sequence analysis. A, Arabidopsis and Casuarina AUX-LAX deduced protein sequences were aligned with a representative member of each subclass of the amino acid and auxin permease family: a Lys His transporter (AtLHT1, At5g40780), an amino acid permease (AtAAP1, At1g58360), and an aromatic and neutral amino acid transporter (AtANT1, At3g11900). The tree was elaborated using a neighbor-joining algorithm and rooted with the Suc transporter protein sequence (AtSUC1, At1g71880). Bootstrap analysis is shown for each branch (n = 100). B, Alignment of AtAUX1, CgAUX1, and CgLAX3 predicted protein sequences using ClustalW (Thompson et al., 1994). Amino acids known to be important for the activity of auxin influx carriers (Swarup et al., 2004) that are conserved are in bold and marked by a white arrowhead. The only amino acid that is not conserved in CgAUX1 is marked by a black arrowhead.
We tested whether CgAUX1 and CgLAX3 encode functional auxin influx carrier proteins equivalent to Arabidopsis AUX1 by carrying out a complementation analysis. CgAUX1 and CgLAX3 open reading frames were inserted between AtAUX1 promoter (ProAtAUX1) and terminator sequences in a binary vector and transformed into null aux1-22 mutants. We then analyzed whether that was sufficient to restore gravitropic phenotype in T1 plants 8 d after germination. aux1-22 plants transformed with an empty vector containing the AtAUX1 promoter and terminator sequences are agravitropic (Fig. 4A). In contrast, transformation with a vector expressing the AtAUX1 coding sequence under its own promoter and terminator rescued a wild-type gravitropic phenotype (Fig. 4A). In the same conditions, CgAUX1 was able to rescue a gravitropic phenotype to aux1 (Fig. 4A). However, expressing CgLAX3 under the control of the AtAUX1 promoter and terminator in aux1-22 mutant background could not restore a wild-type phenotype (Fig. 4A) even if CgLAX3 transcripts were detected in transgenic plants (Supplemental Fig. S1). We conclude that CgAUX1 is functionally equivalent to AtAUX1, whereas CgLAX3 is not. The inability of CgLAX3 to complement the aux1-22 mutant in the same conditions suggests that either CgLAX3 is not a functional auxin influx carrier or it is regulated differently at the translational or posttranslational level. The phylogenetic tree shows that LAX3 and AUX1 proteins belong to different subgroups, thus suggesting that LAX3 and AUX1 proteins might have diverged and have different functions and/or modes of regulation. This is further confirmed by the fact that AtLAX3 cannot complement the aux1-22 mutant when expressed under the AtAUX1 promoter and terminator (R. Swarup and M. Bennett, personal communication).
Figure 4.
Gravitropic response of aux1-22 Arabidopsis mutants complemented with Casuarina genes. CgAUX1 and CgLAX3 were expressed in aux1-22 mutants under the control of the AtAUX1 promoter and terminator sequences. A, Gravitropic response of T1 plants was assayed 24 h after a 90° gravistimulus, plants were grouped into eight classes, depending on the angle of the root apex. aux1-22 plants transformed with an empty vector (AtAUX1 promoter and terminator)-agravitropic phenotype. aux1-22 plants expressing AtAUX1 under the control of its own promoter-gravitropic phenotype. aux1-22 plants complemented with CgAUX1-gravitropic phenotype. aux1-22 plants complemented with CgLAX3-agravitropic phenotype. B, Gravitropic response of aux1-22 mutants complemented with CgAUX1 (homozygous T3 line), Col-0 plants, or aux1-22 mutants upon NT or 25 μm 1-NOA treatment (NOA). The percentage of plants in each group is shown as oriented white bars and plant number (n) is indicated in the circle.
We also checked whether CgAUX1 was sensitive to 1-NOA by attempting to disrupt the complementation of aux1 root gravitropism by CgAUX1. Treatment with 25 μm 1-NOA leads to a reversion to the mutant agravitropic phenotype (Fig. 4B) as in wild-type plants. This result indicates that CgAUX1, like AtAUX1 (Parry et al., 2001a), is sensitive to the auxin influx inhibitor 1-NOA.
CgAUX1 Expression Is Associated with Plant Cell Infection by Frankia, But Is Excluded from Nodule Primordia
Expression of CgAUX1 and CgLAX3 was analyzed in different C. glauca organs. Reverse transcription (RT)-PCR experiments detected CgAUX1 and CgLAX3 transcripts in all the organs tested (Fig. 5), showing that both genes are expressed throughout the plant.
Figure 5.
CgAUX1 and CgLAX3 are expressed in Casuarina root, shoot, and nodule. Nonquantitative RT-PCR analysis in mature nodule, shoot, and root using tubulin (CgTUB) as a control. A control without cDNA and a genomic DNA control were also included. The extra band in the CgLAX3 shoot RT-PCR most probably indicates the presence of some genomic DNA in our RNA sample.
We then focused our expression analysis on CgAUX1 because it encodes a functional auxin influx transporter. We cloned a 1.7-kb promoter fragment and fused it to the GUS reporter gene sequence in a binary vector, thus creating the ProCgAUX1:GUS construct. This construct was introduced into C. glauca and its close relative Allocasuarina verticillata by Agrobacterium tumefaciens-mediated genetic transformation (Franche et al., 1997). Similar patterns of expression were obtained in these two species. CgAUX1 is expressed in root tips (Fig. 6A) and in lateral root primordia (Fig. 6, B and C). Expression was also observed in the root (Fig. 6B) and shoot vasculature (data not shown). This expression pattern is very similar to the AtAUX1 expression pattern in Arabidopsis (Marchant et al., 2002). This, together with the complementation results, suggest that CgAUX1 is orthologous to AtAUX1 and is involved in the same biological processes (gravitropism and lateral root development) as AtAUX1.
Figure 6.
CgAUX1 nonsymbiotic expression pattern in A. verticillata. CgAUX1 expression is detected in root apex (A), mature root vasculature (B), and lateral root primordia (B and C). Bars = 50 μm (A and B) and 15 μm (C).
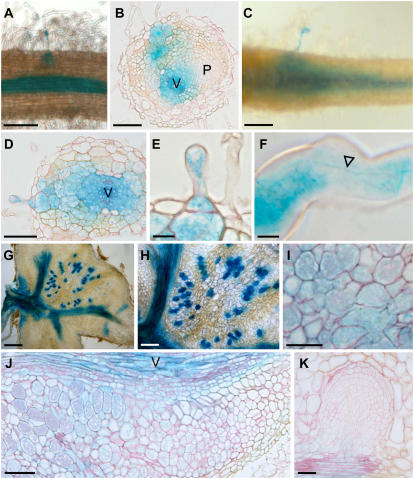
We then analyzed CgAUX1 expression during symbiotic interaction with Frankia. ProCgAUX1:GUS expression was studied 2, 7, 10, 14, and 21 d after inoculation (eight transgenic C. glauca plants/time point). All of the plants showed the same expression pattern. CgAUX1 expression was detected very early in very few root hairs from 10 d postinoculation (Fig. 7, A and C–F). Infecting Frankia hyphae were found in CgAUX1-expressing root hairs (Fig. 7F). At the same time, a higher expression level is clearly visible in the vasculature at the site of infection (Fig. 7, A–D). At later stages, CgAUX1 expression is associated with the infection process. Nodule sections showing strong staining in cortical cells that are infected and no staining in noninfected cells further confirm this pattern of expression (Fig. 7, G–J). Surprisingly, CgAUX1 is not expressed in the nodule primordium (Fig. 7B). This lack of expression in nodule primordia is confirmed by the analysis of nodule ramifications (Fig. 7K). We therefore found that CgAUX1 expression was associated with Frankia infection from the first stage of infection, but was excluded from nodule primordia.
Figure 7.
CgAUX1 expression pattern upon Frankia infection in C. glauca. CgAUX1 expression is observed in a few root hairs 10 d postinoculation (A and C–F). The presence of Frankia hyphae in a root hair expressing CgAUX1 is shown by an arrowhead (F). An increase in expression level is detected in the vasculature at sites of infection (A–D). CgAUX1 expression is associated with infection in nodules (G–J). No expression of CgAUX1 can be seen in primary nodule primordium (B) or nodule primordium ramification (K). V, Vasculature; P, nodule primordium. Bars = 5 μm (F), 10 μm (E), 25 μm (B and D), 50 μm (A, C, I–K), 125 μm (H), or 250 μm (G).
Frankia has been reported to synthesize different auxins (IAA and phenylacetic acid [PAA]). These bacterial auxins could be involved in the regulation of symbiotic genes in infected plant cells. Cg12 encodes a subtilisin-like protease specifically expressed in Frankia-infected cells (Svistoonoff et al., 2003). We therefore tested the effect of these auxins on the expression of CgAUX1 and Cg12. Plants were treated with different concentrations of IAA, naphthalene acetic acid, or PAA; roots were harvested and used to extract RNA. Quantification of gene expression by real-time RT-PCR experiments did not reveal any significant changes in CgAUX1 or Cg12 gene expression in response to any type of auxin (Supplemental Fig. S2). We also found no effect of plant nitrogen status on CgAUX1 expression in response to auxin (Supplemental Fig. S2).
DISCUSSION
The results presented here suggest that auxin influx activity is important for symbiotic interaction between C. glauca roots and the soil actinomycete Frankia. We first show that competitive inhibition of auxin influx using 1-NOA delays nodulation and confirms the involvement of auxin carriers in the process. This led us to isolate two members of a small family of auxin influx carrier genes in C. glauca. We found that CgAUX1 can complement the Arabidopsis aux1 mutant, whereas CgLAX3 could not. AtAUX1 was demonstrated to encode an auxin influx carrier in the Xenopus oocyte (Yang et al., 2006). We therefore conclude that CgAUX1 also encodes for an auxin influx carrier equivalent to AtAUX1.
The actinorhizal nodule is classically regarded as a modified lateral root (Pawlowski and Bisseling, 1996; Obertello et al., 2003). However, we observed that CgAUX1 is expressed in lateral root primordia, but not in nodule lobe primordia. These results suggest that these two organs have, at least in part, divergent development programs. This is in agreement with previous observations showing that some heterologous promoters used as molecular markers, such as 35S and AtUBQ1, drive different expression patterns in lateral root and nodule primordia in Casuarinaceae plants (Obertello et al., 2005). Nevertheless, because our analysis is only based on a promoter-GUS fusion, we cannot completely rule out that CgAUX1 is expressed in nodule primordia. We cannot exclude either that another AUX-LAX gene, such as CgLAX3, is involved in actinorhizal nodule primordium formation. Further studies will be needed to understand how much of the lateral root developmental program has been recycled during evolution to create the actinorhizal nodule developmental program. By comparison, in situ hybridization experiments suggest that AUX-LAX genes are expressed in vascular tissues and the nodule primordia during nodulation in the model legume M. truncatula (de Billy et al., 2001).
Interestingly, we found that CgAUX1 expression is closely associated with Frankia infection of plant cells during nodulation (summarized in Fig. 8A). We observed CgAUX1 expression already in Frankia-infected root hairs 10 d after infection. CgAUX1 was later expressed in all Frankia-infected cells in the prenodule and in the nodule regardless of their development stage (infection, nitrogen fixation, etc.). CgAUX1 expression was also detected in the vascular tissues in noninfected and infected roots and nodules. As a comparison, no expression of AUX-LAX genes was detected by in situ hybridization in Rhizobium-infected cells in the model legume M. truncatula (de Billy et al., 2001). To our knowledge, this is the first report of an auxin influx activity linked to plant cell infection by a soil microorganism. The signal responsible for the infection-specific expression of CgAUX1 is not known. We showed that auxin alone (IAA, naphthalene acetic acid, or PAA) cannot play this role. CgAUX1 expression may be induced by a symbiotic signal produced by Frankia. The expression of an auxin influx carrier in Frankia-infected cells would make them more permeable to auxin. Interestingly, some Frankia strains have been shown to produce different forms of auxin in culture, including IAA and PAA (Wheeler et al., 1984; Hammad et al., 2003). This could explain why actinorhizal nodules have been reported to contain more auxin than noninfected roots (Wheeler et al., 1979). We therefore speculate that CgAUX1 expression allows the entry and perception of Frankia-produced auxin and restricts it to infected plant cells (Fig. 8B). Auxin alone or in synergy with a symbiotic signal could induce changes in gene expression, cell metabolism, etc., in infected cells to allow the establishment of intracellular symbiosis (Fig. 8B). For example, the infection process is associated with remodeling of the cell wall to create an infection thread (Berg, 1999a). Many cell wall remodeling genes have been found to be auxin inducible in Arabidopsis (Neuteboom et al., 1999; Overvoorde et al., 2005; Esmon et al., 2006; Osato et al., 2006). Auxin could therefore induce genes encoding cell wall remodeling enzymes necessary for infection by Frankia. Moreover, infected cells are hypertrophied (Berg, 1999b), a phenotype that has been classically associated with auxin response (Teale et al., 2006). Further experiments will be needed to understand the interaction between cell wall remodeling and auxin transport during nodule formation.
Figure 8.
Putative CgAUX1 function during actinorhizal nodule formation. A, Summary of CgAUX1 expression pattern (blue color) at different steps of C. glauca-Frankia interaction. 1, Signal exchanges between the actinorhizal plant and Frankia lead to root hair infection. 2, Frankia penetrates a deformed root hair showing CgAUX1 expression and triggers cortical cell divisions. 3, Dividing cortical cells are infected by Frankia hyphae and hypertrophy, thus leading to the formation of a prenodule. At the same time, pericycle cell divisions occur in front of a xylem pole to form a nodule primordium. 4, Frankia hyphae coming from the prenodule invade the cortex of the nodule primordium. 5, In mature nodules, CgAUX1 expression is observed in infected cells and the vascular tissues. B, Proposed model of CgAUX1 role during the infection process. We propose that two signals occur in synergy during actinorhizal symbiosis. A specific signal that remains unknown is produced by the bacteria and triggers the production of the auxin influx carrier CgAUX1. A nonspecific signal, auxin, is also produced by the bacteria and acts in synergy with the specific signal to trigger the infection-related program by the plant.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Casuarina glauca seeds (purchased from Carter Seeds) were grown and inoculated by Frankia CcI3 strain as previously described (Franche et al., 1997). Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) and aux1-22 (Col-0 background) mutant seeds were obtained from the Nottingham Arabidopsis Stock Center. Plants were grown as previously described (Laplaze et al., 2005). Gravitropism assays were performed as previously described (Swarup et al., 2005).
Identification of CgAUX1 and CgLAX3 cDNA and Genomic Sequences
C. glauca genomic DNA was isolated from a young shoot apex using a MATAB extraction method (Ky et al., 2000). Amplification of AUX1 homologs was performed on genomic DNA using different sets of degenerate primers (Table I). Amplified fragments were cloned into pGEM-T easy (Promega) and sequenced. Total RNA was extracted on the whole root system by ultracentrifugation (Chirgwin et al., 1979). Polyadenylated RNA was purified using the Amersham mRNA purification kit. Full-length cDNA sequences were obtained by performing RACE-PCR on a root cDNA library (Marathon cDNA amplification kit; CLONTECH). cDNA was amplified using primers 5′-ATAGCATTATTTTGTCTGTGGGTTG-3′ and 5′-CAACCCACAGACAAAATAATGCTAT-3′ for CgAUX1 5′ and 3′ RACE, respectively, and primers 5′-TCACTGGGGCTACCAACATTCTCTA-3′ and 5′-TAGAGAATGTTGGTAGCCCCAGTGA-3′ for CgLAX3 5′ and 3′ RACE, respectively.
Full-length cDNA and genomic DNA were amplified using AUX1F, 5′-GCAGATCAGCCGGAATTTAG-3′; AUX1R, 5′-TGCTTTGGAAGCAAAGGAAT-3′; LAX3F, 5′-ACAATGGCTTCCGAGAAGGT-3′; LAX3R, 5′-GGCTAAATTCAATCCCACCGTA-3′, cloned into pGEM-T and sequenced.
Genomic DNA-Blot Analysis
Ten micrograms of DNA were digested with BamHI, EcoRI, and HindIII (New England Biolabs). DNA fragments were separated on a 1% agarose gel and capillary blotted onto a Hybond N+ membrane (Amersham). A 175-bp CgAUX1-specific probe was synthesized using primers 5′-AGCTAACACACCCCATAGTTTG-3′ and 5′-AATAATAAGCCTATGCTTTGGAAG-3′, a 234-bp CgLAX3-specific probe was synthesized using primers 5′-GCGTGTAAAGAGATTGGCATTT-3′ and 5′-TGAGCAAACACTACAACGGCTAA-3′ and a 174-bp AUX-LAX conserved probe was synthesized using primers 5′-CGTTTGGATTCGCCTGCACTCC-3′ and 5′-GAGATCCAACAGTTGAGTTGA-3′. Probes were labeled with α-32P dCTP by random priming. Hybridization was carried out in high-stringency conditions for CgAUX1- and CgLAX3-specific probes (65°C; 10-min washes with 2× SSC, 0.1% SDS; 1× SSC, 0.1% SDS; 0.5× SSC, 0.1% SDS) and low-stringency conditions for the AUX-LAX conserved probe (56°C; 10-min washes with 2× SSC, 0.1% SDS; 1× SSC, 0.1% SDS). Hybridization patterns were visualized using a Typhoon 8600 variable mode imager (Molecular Dynamics). DNA extractions and hybridization were repeated at least twice.
Nonquantitative and Quantitative RT-PCR
Total RNA was extracted on the whole-root system, shoot, or mature nodules by ultracentrifugation (Chirgwin et al., 1979). Poly(dT) cDNA was prepared out of 1 μg total RNA using the RT system (Promega) and three independent RT reactions were pooled for quantitative analysis. PCR reactions were carried out at 94°C for 5 min, followed by 40 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 60°C, and extension for 90 s at 72°C. Target amplifications were performed with CgAUX1- or CgLAX3-specific primer pairs designed on each side of the last intron (CgAUX1, 5′-GTTCTTCGGGCCCATAAACT-3′ and 5′-TGCTTTGGAAGCAAAGGAAT-3′; CgLAX3, 5′-ATTCCTGCCCTAGCACACAT-3′ and 5′-CCCACCGTAAAGAGATACCG-3′). Tubulin gene (CgTUB) expression was used as a control (CgTUB, 5′-CGCGGCCGCTGGAGAGGCGTC-3′ and 5′-GCAAGCTTTCGGATGCGATCC-3′). Quantitative PCR was performed on a Stratagene Mx3005P apparatus with the FullVelocity SYBR Green QPCR master mix (Stratagene) upon recommendation of the manufacturer. PCR was carried out in 96-well optical reaction plates heated for 5 min to 95°C, followed by 40 cycles of denaturation for 10 s at 95°C, and annealing extension for 30 s at 60°C. Target quantifications were performed with specific primer pairs designed using Beacon Designer 4.0 (Premier Biosoft International; CgAUX1, 5′-ACCAGGAGCAACCGGAAGAC-3′ and 5′-AGCACTTGCGCAACTTGATTG-3′; CgLAX3, 5′-CACTTGCGCGACCTGGTTAG-3′ and 5′-AAGAAGGCGATCCCAAGACG-3′; Cg12, see Hocher et al., 2006). Expression levels were normalized to ubiquitin (CgUBI; Hocher et al., 2006). All RT-PCR experiments were performed in triplicate and the presented values represent means ± sd.
Constructs and Generation of Transgenic Plants
For promoter studies, 1.7-kb genomic DNA fragments upstream of the CgAUX1 and CgLAX3 start codon (ATG) were amplified using the Universal GenomeWalker kit (CLONTECH) and cloned upstream of the GUS reporter gene in pBI101.3 binary vector (CLONTECH). For functional complementation, full-length CgAUX1 and CgLAX3 cDNA were fused with Arabidopsis AtAUX1 promoter (1.7 kb) and terminator (0.3 kb) in a pMOG402 binary vector (MOGEN International). Vectors were introduced into Agrobacterium tumefaciens C58C1 pGV3101 by electroporation. Transformation of Arabidopsis (Col-0 and aux1-22) was performed as previously described (Clough and Bent, 1998). Transformation of C. glauca and Allocasuarina verticillata plants was performed as previously described (Franche et al., 1997).
Microscopy and Root Sections
GUS assays were performed as previously described (Svistoonoff et al., 2003). Tissues were cleared in 70% ethanol for 2 d and then immersed in 50% (v/v) ethanol/10% (v/v) glycerol for 2 h, 30% (v/v) ethanol/30% (v/v) glycerol for 2 h, and in 50% (v/v) glycerol for 2 h. Seedlings were then mounted in 50% (v/v) glycerol and visualized on a Leitz DMRB microscope. For thin root sections, samples were fixed (Svistoonoff et al., 2003) and cleared in 70% ethanol for 2 d. Ethanol dehydration was performed (90%, 100% twice) at room temperature (15 min/step). Samples were then embedded in Technovit 7100 resin (Heraeus Kulzer) according to the manufacturer's instructions. Thin sections (5 μm) were cut with a Microm HM355S microtome. Sections were stained for 2 min in aqueous 0.05% ruthenium red solution and mounted in Clearium Mountant (Surgipath). For thick nodule sections, nodules were embedded in 3% agarose. Thick sections (55 μm) were cut with a Leica VT1000E vibratome.
Sequence data from this article have been deposited with the EMBL/GenBank libraries under accession numbers EF416279 and EF416280 for CgAUX1 gene and cDNA and EF416281 and EF416282 for CgLAX3 gene and cDNA.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR detection of CgLAX3 transcripts in complemented aux1 mutants.
Supplemental Figure S2. Quantitative expression levels of CgAUX1 and Cg12 in response to auxin.
Supplementary Material
Acknowledgments
We would like to thank our colleagues from Equipe Rhizogénèse and Dr. T. Tranbarger (Institut de Recherche pour le Développement Montpellier) for critical reading of the manuscript.
This work was supported by the Institut de Recherche pour le Développement and the British Council/Egide Alliance (grant no. 05752SM to L.L. and M.B.). B.P. was funded by the Ministère de l'Education Nationale, Enseignement Supérieur et Recherche.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Laurent Laplaze (laplaze@mpl.ird.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Badenoch-Jones J, Rolfe BG, Letham DS (1983) Phytohormones, rhizobium mutants, and nodulation in legumes. III. Auxin metabolism in effective and ineffective pea root nodules. Plant Physiol 73 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273 948–950 [DOI] [PubMed] [Google Scholar]
- Benson DR, Silvester WB (1993) Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev 57 293–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RH (1999. a) Frankia forms infection threads. Can J Bot 77 1327–1333 [Google Scholar]
- Berg RH (1999. b) Cytoplasmic bridge formation in the nodule apex of actinorhizal root nodules. Can J Bot 77 1351–1357 [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8 165–171 [DOI] [PubMed] [Google Scholar]
- Cérémonie H, Debellé F, Fernandez MP (1999) Structural and functional comparison of Frankia root hair deforming factor and rhizobia Nod factor. Can J Bot 77 1293–1301 [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ (1979) Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18 5294–5299 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- de Billy F, Grosjean C, May S, Bennett M, Cullimore JV (2001) Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact 14 267–277 [DOI] [PubMed] [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franche C, Diouf D, Le QV, Bogusz D, N'Diaye A, Gherbi H, Gobé C, Duhoux E (1997) Genetic transformation of the actinorhizal tree Allocasuarina verticillata by Agrobacterium tumefaciens. Plant J 11 897–904 [Google Scholar]
- Goldsmith MHM (1977) The polar transport of auxin. Annu Rev Plant Physiol 28 439–478 [Google Scholar]
- Gordons A, Stevens JR, Berry AM (1988) Cytokinin secretion by Frankia sp. HFPArI3 in defined medium. Plant Physiol 87 15–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad Y, Nalin R, Marechal J, Fiasson K, Pepin R, Berry AM, Normand P, Domenach AM (2003) A possible role for phenyl acetic acid (PAA) on Alnus glutinosa nodulation by Frankia. Plant Soil 254 193–205 [Google Scholar]
- Hocher V, Auguy F, Argout X, Laplaze L, Franche C, Bogusz D (2006) Expressed sequence-tag analysis in Casuarina glauca actinorhizal nodule and root. New Phytol 169 681–688 [DOI] [PubMed] [Google Scholar]
- Kaneshiro T, Kwolek WF (1985) Stimulated nodulation of soybeans by Rhizobium japonicum mutant (B-14075) that catabolizes the conversion of tryptophan to indol-3-acetic acid. Plant Sci 42 141–146 [Google Scholar]
- Kerr ID, Bennett MJ (2007) New insight into the biochemical mechanisms regulating auxin transport in plants. Biochem J 401 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Bennett MJ (2006) Auxin transport: a field in flux. Trends Plant Sci 11 382–386 [DOI] [PubMed] [Google Scholar]
- Ky CL, Barre P, Lorieux M, Trouslot P, Akaffou S, Louarn J, Charrier A, Hamon S, Noirot M (2000) Interspecific genetic linkage map, segregation distortion and genetic conversion in coffee (Coffea sp.). Theor Appl Genet 101 669–676 [Google Scholar]
- Laplaze L, Duhoux E, Franche C, Frutz T, Svistoonoff S, Bisseling T, Bogusz D, Pawlowski K (2000) Casuarina glauca prenodule cells display the same differentiation as the corresponding nodule cells. Mol Plant Microbe Interact 13 107–112 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56 2433–2442 [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HR, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14 23–34 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Weinman JJ, Rolfe BG, Djordjevic MA (2000) Rhizobia can induce nodules in white clover by “hijacking” mature cortical cells activated during lateral root development. Mol Plant Microbe Interact 13 170–182 [DOI] [PubMed] [Google Scholar]
- Neuteboom LW, Veth-Tello LM, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ (1999) A novel subtilisin-like protease gene from Arabidopsis thaliana is expressed at sites of lateral root emergence. DNA Res 6 13–19 [DOI] [PubMed] [Google Scholar]
- Obertello M, Santi C, Sy MO, Laplaze L, Auguy F, Bogusz D, Franche C (2005) Comparison of four constitutive promoters for the expression of transgenes in the tropical nitrogen-fixing tree Allocasuarina verticillata. Plant Cell Rep 24 540–548 [DOI] [PubMed] [Google Scholar]
- Obertello M, Sy MO, Laplaze L, Santi C, Svistoonoff S, Auguy F, Bogusz D, Franche C (2003) Actinorhizal nitrogen fixing nodules: infection process, molecular biology and genomics. Afr J Biotechnol 2 528–538 [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119 153–162 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C, Schlaman HR, Boot K, Admiraal P, Langerak JM, Stougaard J, Spaink HP (2003) Auxin distribution in Lotus japonicus during root nodule development. Plant Mol Biol 52 1169–1180 [DOI] [PubMed] [Google Scholar]
- Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ (2001. a) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J 25 399–406 [DOI] [PubMed] [Google Scholar]
- Parry G, Marchant A, May S, Swarup R, Swarup K, James N, Graham N, Allen T, Martucci T, Yemm A, et al (2001. b) Quick on the uptake: characterization of a family of plant auxin influx carriers. J Plant Growth Regul 20 217–225 [Google Scholar]
- Patten CL, Glick BR (1996) Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 42 207–220 [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Bisseling T (1996) Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell 8 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel EL, Frugoli J (2004) The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol Genet Genomics 272 420–432 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Laplaze L, Auguy F, Runions J, Duponnois R, Haseloff J, Franche C, Bogusz D (2003) cg12 expression is specifically linked to infection of root hairs and cortical cells during Casuarina glauca and Allocasuarina verticillata actinorhizal nodule development. Mol Plant Microbe Interact 16 600–607 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7 1057–1065 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J (2006) Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell Mol Life Sci 63 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7 847–859 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi AK, Khurana JP, Khurana P, Raghuvanshi S, Gaur A, Kapur A, Gupta V, Kumar D, Ravi V, Vij S, et al (2004) Structural and functional analysis of rice genome. J Genet 83 79–99 [DOI] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by Rhizobia. Plant Cell 18 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler CT, Crozier A, Sandberg G (1984) The biosynthesis of indole-3-acetic acid by Frankia. Plant Soil 78 99–104 [Google Scholar]
- Wheeler CT, Henson IE, MacLaughlin ME (1979) Hormones in plants bearing actinomycete nodules. Bot Gaz 140 52–57 [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 160 1123–1127 [DOI] [PubMed] [Google Scholar]
- Young GB, Jack DL, Smith DW, Saier MH (1999) The amino acid/auxin:proton symport permease family. Biochim Biophys Acta 1415 306–322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.