Abstract
The Arabidopsis (Arabidopsis thaliana) genome encodes 51 proteins annotated as serine carboxypeptidase-like (SCPL) enzymes. Nineteen of these SCPL proteins are highly similar to one another, and represent a clade that appears to be unique to plants. Two of the most divergent proteins within this group have been characterized to date, sinapoyl-glucose (Glc):malate sinapoyltransferase and sinapoyl-Glc:choline sinapoyltransferase. The fact that two of the least related proteins within this clade are acyltransferases rather than true serine carboxypeptidases suggests that some or all of the remaining members of this group may have similar activities. The gene that encodes sinapoyl-Glc:malate sinapoyltransferase (sinapoyl-Glc accumulator1 [SNG1]: At2g22990) is one of five SCPL genes arranged in a cluster on chromosome 2. In this study, an analysis of deletion mutant lines lacking one or more genes in this SCPL gene cluster reveals that three of these genes also encode sinapoyl-Glc-dependent acyltransferases. At2g23000 encodes sinapoyl-Glc:anthocyanin acyltransferase, an enzyme that is required for the synthesis of the sinapoylated anthocyanins in Arabidopsis. At2g23010 encodes an enzyme capable of synthesizing 1,2-disinapoyl-Glc from two molecules of sinapoyl-Glc, an activity shared by SNG1 and At2g22980. Sequence analysis of these SCPL proteins reveals pairwise percent identities that range from 71% to 78%, suggesting that their differing specificities for acyl acceptor substrates are due to changes in a relatively small subset of amino acids. The study of these SCPL proteins provides an opportunity to examine enzyme structure-function relationships and may shed light on the role of evolution of hydroxycinnamate ester metabolism and the SCPL gene family in Arabidopsis and other flowering plants.
Plant secondary metabolites account for a great amount of the biochemical diversity that exists in nature. These compounds have been estimated to number between 100,000 to 200,000, with the majority of them yet to be studied in detail (Wink, 1988). As a class of molecules, plant secondary metabolites include compounds that possess many important properties: they function as antifeedents, phytoanticipins and phytoalexins, signaling molecules, UV protectants, and in a host of other physiologically important roles (Li et al., 1993; Wajant et al., 1994; Kuć, 1995; Kliebenstein et al., 2005; Taylor and Grotewold, 2005). Aside from their functions within the plant kingdom, they have also proven to be invaluable to humanity, playing long-standing roles in medicine and agriculture. Secondary metabolites are therefore not only crucial to our understanding of plant physiology, but are of great importance to our own health and survival as well.
A common metabolic step in the production of numerous secondary metabolites involves the attachment of an acyl group via an activated donor molecule (Croteau and Hooper, 1978; Dahlbender and Strack, 1986; Villegas and Kojima, 1986; Villegas et al., 1987; Kesy and Bandurski, 1990; Suzuki et al., 1994; Lenz and Zenk, 1995; Rabot et al., 1995; Yang et al., 1997; Dudareva et al., 1998; Fujiwara et al., 1998; Li et al., 1999; Walker et al., 1999; Lehfeldt et al., 2000; Shirley et al. 2001; Fröhlich et al., 2002; Nakayama et al., 2003). Although it is well documented that CoA thioesters often provide the energy necessary for transacylation reactions in secondary metabolism (St Pierre and De Luca, 2000), it is now clear that 1-O-Glc esters can serve as activated donor molecules as well due to their high free energy of hydrolysis (Dahlbender and Strack, 1984; Mock and Strack, 1993). The end products of these Glc ester-dependent reactions include many important compounds such as indoleacetyl myo-inositol, oak (Quercus spp.) gallotannins, isobutyryl Glc esters, certain acylated anthocyanins, and the phenylpropanoid-derived sinapate esters found in Arabidopsis (Arabidopsis thaliana) and other members of the Brassicaceae (Fig. 1; Kesy and Bandurski, 1990; Gläßgen and Seitz, 1992; Li et al., 1999; Lehfeldt et al., 2000; Shirley et al., 2001; Fröhlich et al., 2002).
Figure 1.
The pathway of sinapate ester biosynthesis in Arabidopsis. Multiple steps are required for the conversion of Phe to sinapic acid via p-coumaroyl-CoA. The enzymes required for the synthesis of sinapate esters in developing seeds are SGT and SCT. Upon germination, sinapoylcholinesterase (SCE) catalyzes the hydrolysis of seed reserves of sinapoylcholine, and SGT and SMT convert the released sinapate into sinapoylmalate. SST catalyzes the conversion of two molecules of sinapoyl-Glc to 1,2-disinapoyl-Glc and compound 1, a biosynthetic route that is predominant in etiolated seedlings. Dashed arrows indicate one possible route to the biosynthesis of compound 1, a reaction dependent upon SST. The action of SAT is required for the sinapoylation of Arabidopsis anthocyanins. Double arrows proceeding from p-coumaroyl-CoA indicate the biosynthesis of the cyanidin derivative via flavonoid metabolism and the subsequent acylation of the anthocyanin, presumably by a p-coumaroyl-CoA-dependent acyltransferase. This biosynthetic pathway yields one possible substrate for SAT, 3-O-(6-O-p-coumaroyl-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin (A5; nomenclature from Tohge et al., 2005), sinapoylation of which would result in the production of 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin (A11).
The enzymes responsible for the final step in the synthesis of isobutyryl Glc esters in Solanum berthaultii and Lycopersicon pennellii, as well as sinapoylcholine and sinapoylmalate in Arabidopsis (sinapoyl-Glc:choline sinapoyltransferase [SCT] and sinapoyl-Glc:malate sinapoyltransferase [SMT], respectively) are Ser carboxypeptidase-like (SCPL) proteins (Lehfeldt et al., 2000; Li and Steffens, 2000; Shirley et al., 2001). As a class of enzymes, SCPL proteins bear the trademark Ser-Asp-His catalytic triad along with other sequence features that are characteristic of known Ser carboxypeptidases (SCPs) such as carboxypeptidase-Y from yeast (Saccharomyces cerevisiae). In this article we report the characterization of three additional Arabidopsis SCPL proteins that are sinapoyl-Glc-dependent sinapoyltransferases. The functions of these proteins were identified through the phenotypic characterization of fast neutron-induced sinapoyl-Glc accumulator1 (sng1) deletion alleles in which the deletions affect not only the SNG1 gene, but also one or more of the SCPL genes that flank it. We report here that At2g23000 encodes a protein that catalyzes the sinapoyl-Glc-dependent sinapoylation of anthocyanins in Arabidopsis, demonstrating that this activity is not dependent upon a BAHD acyltransferase as has been suggested previously (Luo et al., 2007). Furthermore, we show that the SCPL protein encoded by At2g23010 catalyzes the disproportionation of two molecules of sinapoy-Glc to generate 1,2-disinapoyl-Glc (Fig. 1) and an additional, as yet unidentified compound. We have therefore designated the SCPL proteins encoded by At2g23000 and At2g23010 as sinapoyl-Glc:anthocyanin sinapoyltransferase (SAT) and sinapoyl-Glc:sinapoyl-Glc sinapoyltransferase (SST), respectively. Finally, we show that both SMT and the protein encoded by At2g22980 are capable of catalyzing the formation of 1,2-disinapoyl-Glc, although this activity is clearly not the primary function of SMT.
Like SCT and SMT, SST and SAT belong to a family of 51 SCPL proteins encoded by the Arabidopsis genome, and are members of a clade that includes 15 other closely related SCPL proteins (Fraser et al., 2005). The fact that SMT, SST, SAT, and the At2g22980 protein are between 71% and 78% identical to one another (Fraser et al., 2005) suggests that their analysis may permit the elucidation of structure-function relationships within this subclass of SCPL proteins. Thus, the SCPL sinapoyl-Glc acyltransferases (SGAs) represent additional members of an emerging class of enzymes that catalyze acyltransferase reactions in plant secondary metabolism.
RESULTS
sng1-5 and sng1-6 Harbor Deletions of an SCPL Gene Cluster on Chromosome 2
The SNG1 gene is one of five SCPL genes that are arranged in tandem on chromosome 2 (Fig. 2). The proteins encoded by these SCPL genes are highly similar, with any two of them being between 71% and 78% identical (Fraser et al., 2005). To identify mutations that affect the genes surrounding SNG1, a population of fast neutron-mutagenized plants was screened using the UV-phenotype characteristic of the sng1 mutant (Lehfeldt et al., 2000), and their biochemical sng1 phenotypes were confirmed by HPLC and genetic complementation tests. The genomic regions deleted from each mutant were initially estimated via Southern analysis (Lehfeldt et al., 2000). Additional PCR analysis of the SCPL gene cluster using both gene-specific and intergenic primers indicated that SNG1 (At2g22990) and the gene immediately downstream (At2g23000) were deleted in the sng1-5 mutant. Sequencing of genomic PCR products revealed that the sng1-6 mutant is missing a region of genomic DNA spanning At2g22980 through At2g23010 and four additional non-SCPL genes downstream, none of which encodes a SCPL protein (Fig. 2). In addition, a 170 bp fragment translocated from gene At4g14980 on chromosome 4 (Blastn E value = 1e −72) is inserted between the 5′ and 3′ sng1-6 deletion borders.
Figure 2.
The sng1 genotypes and the region surrounding the SNG1 locus. The SNG1 gene encodes SMT, and is one of five SCPL genes (white boxes) in a tandem cluster on chromosome 2. The deletions in the sng1-5 and sng1-6 mutants are designated by double-headed arrows, and include a number of non-SCPL genes (gray boxes) in sng1-6. Of the two SCPL genes uniquely deleted in sng1-6, At2g22980, and At2g23010, the latter encodes SST and its deletion is responsible for the lack of compound 1 accumulation in sng1-6. The combined absence of At2g22980, SMT, and At2g23010 explains the absence of 1,2-disinapoyl-Glc in the mutant. The deletion of At2g23000 in sng1-5 and sng1-6 results in the inability of these mutants to accumulate sinapoylated anthocyanins. Triangles indicate the sites of the EMS mutation in sng1-1 and the site of the T-DNA insertion in the SALK_133207 mutant.
Etiolated sng1-6 Seedlings Lack a Major Sinapate Ester Found in Etiolated Wild-Type, sng1-1, and sng1-5 Seedlings
The high sequence similarity shared by SMT and the SCPL proteins encoded by At2g22980, At2g23000, and At2g23010 suggested that one or more of the latter might also be sinapoyl-Glc-dependent sinapoyltransferases. To test this hypothesis, extracts from a variety of tissues from the sng1-5 and sng1-6 mutants were analyzed via HPLC to determine if any putative sinapate esters present in wild-type and sng1-1 plants were absent from one or both of the mutants. In these experiments, two compounds not previously identified in Arabidopsis with absorption spectra characteristic of sinapate esters were found to be present in wild-type, sng1-1, and sng1-5 seedling extracts, but were absent from sng1-6 extracts (Fig. 3A). Given that At2g22980 and At2g23010 are the only two SCPL genes uniquely deleted from the sng1-6 mutant relative to the sng1-5 mutant, the absence of these compounds (hereafter referred to as compounds 1 and 2) in the sng1-6 seedlings suggested that either At2g22980 or At2g23010 or both are required for their synthesis. Subsequent complementation analyses (see below) confirmed that this is indeed the case. Preliminary liquid chromatography/mass spectrometry (MS) analysis suggested that both compounds were disinapoylated monosaccharides (data not shown), suggesting that one of these two compounds might be1,2-disinapoyl-Glc, which has been shown to accumulate in the cotyledons of radish (Raphanus sativus) seedlings (Strack et al., 1984).
Figure 3.
Analysis of the sinapate esters in the sng1, SALK_133207, and transformed sng1-6 mutants. Extracts were prepared from 7-d-old, etiolated dark-grown seedlings and analyzed by HPLC with UV detection at 335 nm. A, Extracts from Columbia wild-type, sng1-1, sng1-5, and sng1-6 mutants as well as the sng1-6 mutant transformed with the At2g23010 genomic construct (pCC579). B, Extracts from the T-DNA insertional mutant of At2g23010 (SALK_133207), and sng1-6 transformed with either the At2g22980 genomic construct (pCC881) or the At2g22990/SMT construct (pCC398). The sng1 mutants are defective in SMT and accumulate sinapoyl-Glc (SG) in their leaves instead of accumulating the sinapoylmalate (SM) that is found in wild-type plants. Compounds 1 (a disinapoylated monosaccharide) and 1,2-disinapoyl-Glc do not accumulate in the sng1-6 mutant, both compounds accumulate in the sng1-6/pCC579 mutant, but only 1,2-disinapoyl-Glc accumulates in SALK_133207, sng1-6/pCC881, and sng1-6/pCC398.
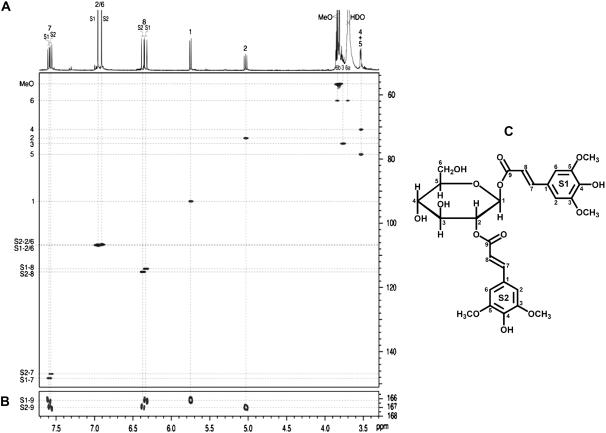
NMR Analysis Identifies Compound 2 as 1,2-Disinapoylglucose
NMR spectra of compound 2 not only established its identity as 1,2-disinapoyl-Glc but provided the full NMR data and its complete assignment. The key experiment was the long-range 13C–1H (HMBC) experiment, which delineated which sinapoyl unit was attached to the 1 versus 2 position (Fig. 4B). Then, with the coupling networks from each unit revealed in the normal way by COSY and HMBC correlations, all of the protons from each sinapoyl unit (S1 and S2) could be unambiguously assigned, with the possible exception of the methoxyl protons. Short-range 13C–1H (HSQC) correlations unambiguously established the assignments for the protonated carbons (Fig. 4A) and HMBC spectra identified all of the nonprotonated carbons. As seen from Figure 4A, some of the protons are poorly resolved in the one-dimensional (1D) proton spectrum, but the proton data are readily extracted from the HSQC spectrum (as well as by COSY; data not shown). The proton and carbon data presented in Tables I and II (in two solvents, CD3OD, and 9:1 acetone d6:D2O) are therefore unambiguously authenticated, again, with the possible exception of the methoxyl protons. Data presented in Tables I and II include previously obtained data (Strack et al., 1984), for comparison. NMR data have not yet been obtained to ascertain the structure of compound 1 due to its very low abundance in extracts.
Figure 4.
NMR analysis of 1,2-disinapoyl-Glc isolated from Arabidopsis. Spectra from isolated 1,2-disinapoyl-Glc in 9:1 acetone-d6:D2O. A, HSQC spectrum (with the 1D proton spectrum as the projection on top) showing all fully resolved short-range (1-bond) 13C–1H correlations. B, Small section of the HMBC (120 ms long-range coupling delay) highlighting each diagnostic ester carbonyl (C-9) correlation with its glucosyl 1 or 2 proton, delineating the two independent sinapoyl units attached to the 1 (S1) versus 2 position (S2) of Glc. C, The corresponding structure of 1,2-disinapoyl-Glc, compound 2.
Table I.
13C-NMR data for disinapoyl Glc derived from the usual array of 1D and 2D experiments, and comparison (assigned here) with prior data (Strack et al., 1984)
Ac, Acetone-D6; *, data from Strack et al. (1984).
| δ Carbon | δ MeOD | δ Ac/D2O | δ* MeOD |
|---|---|---|---|
| 1 | 94.2 | 93.2 | 94.1 |
| 2 | 74.4 | 73.5 | 74.4 |
| 3 | 76.2 | 75.2 | 76.1 |
| 4 | 71.4 | 70.8 | 71.4 |
| 5 | 79.3 | 78.5 | 79.2 |
| 6 | 62.4 | 61.8 | 62.4 |
| S1 sinapoyl unit | |||
| 1 | 126.3 | 125.3 | 126.4 |
| 2/6 | 107.3 | 106.9 | 107.3 |
| 3/5 | 149.6 | 148.5 | 149.5 |
| 4 | 140.3 | 139.7 | 140.2 |
| 7 | 149.1 | 148.3 | 149.0 |
| 8 | 114.7 | 114.2 | 114.7 |
| 9 | 167.4 | 166.1 | 167.1 |
| 3/5-OMe | 56.9 | 56.5 | 56.9 |
| S2 sinapoyl unit | |||
| 1 | 126.7 | 125.7 | 126.6 |
| 2/6 | 107.1 | 106.6 | 107.1 |
| 3/5 | 149.6 | 148.7 | 149.5 |
| 4 | 139.8 | 139.3 | 139.9 |
| 7 | 148.0 | 146.9 | 147.8 |
| 8 | 115.7 | 115.2 | 115.6 |
| 9 | 168.6 | 166.9 | 168.3 |
| 3/5-OMe | 56.9 | 56.5 | 56.9 |
Table II.
1H-NMR data for disinapoyl Glc derived from the usual array of 1D and 2D experiments, and comparison (assigned here) with prior data (Strack et al., 1984)
*, Data from Strack et al. (1984). We use the central CHD2OD peak as reference δ = 3.31 ppm; our data differ by approximately 0.04 ppm from Strack's. **, Assignments may be interchanged. s, Singlet; d, doublet; dd, doublet of doublets; m, multiplet.
| Proton | δ (m, J) MeOD | δ (m, J) Ac/D2O | δ* MeOD |
|---|---|---|---|
| 1 | 5.80 (d, 8.4) | 5.75 (d, 8.4) | 5.85 |
| 2 | 5.07 (dd, [9.5, 8.4]) | 5.03 (dd, 9.7, 8.4) | 5.12 |
| 3 | 3.73 (m) | 3.77 (m) | 3.77 |
| 4 | 3.518 (–) | 3.53 (–) | – |
| 5 | 3.515 (–) | 3.53 (–) | – |
| 6a | 3.76 (–) | 3.70 (–) | – |
| 6b | 3.89 (–) | 3.84 (–) | 3.95 |
| S1 sinapoyl unit | |||
| 2/6 | 6.89 (s) | 6.96 (s) | 6.91 |
| 7 | 7.64 (d, 16.0) | 7.60 (d, 15.9) | 7.67 |
| 8 | 6.32 (d, 16.0) | 6.33 (d, 15.9) | 6.36 |
| 3/5-OMe | 3.86** (s) | 3.83** (s) | 3.89** |
| S2 sinapoyl unit | |||
| 2/6 | 6.85 (s) | 6.91 (s) | 6.88 |
| 7 | 7.63 (d, 16.0) | 7.58 (d, 15.9) | 7.66 |
| 8 | 6.41 (d, 16.0) | 6.37 (d, 15.9) | 6.45 |
| 3/5-OMe | 3.84** (s) | 3.81** (s) | 3.87** |
Table III.
Identity and function of characterized SCPL acyltransferases in Arabidopsis
SCPL number designation refers to the nomenclature developed by Fraser et al. (2005). *, SST is the only known activity for At2g22980. Considering that future experiments may reveal that its prime role is in the synthesis of a compound other than 1,2-disinapoyl-Glc, SST is listed as a minor activity for this protein.
| Gene | SCPL No. | Major Activity | Minor Activity |
|---|---|---|---|
| At2g22980 | 13 | – | SST* |
| At2g22990 | 8 | SMT | SST |
| At2g23000 | 10 | SAT | – |
| At2g23010 | 9 | SST | – |
| At5g09640 | 19 | SCT | – |
At2g23010 Complements the sng1-6 Phenotype
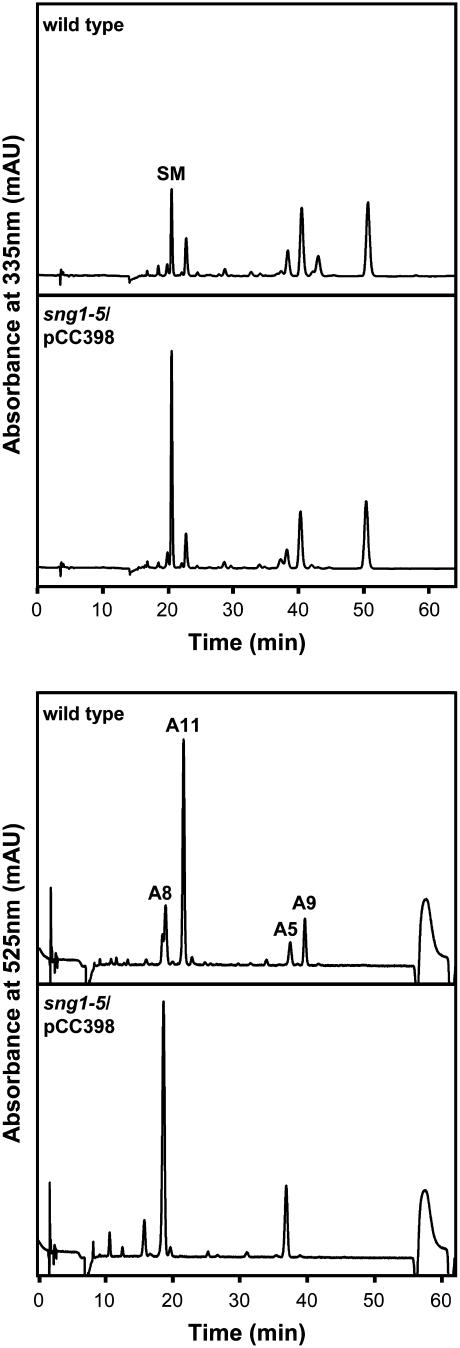
To test the hypothesis that At2g23010 is required for the synthesis of compound 1 and 1,2-disinapoyl-Glc, the sng1-6 mutant was transformed with pCC579, a vector containing the genomic upstream, downstream, and coding regions of the gene. HPLC analysis of extracts from etiolated, 6-d-old sng1-6/pCC579 seedlings showed that both compound 1 and 1,2-disinapoyl-Glc are present at wild-type levels in the transformed mutant seedlings, indicating that At2g23010 complements this aspect of the sng1-6 phenotype (Fig. 3A), and that this gene thus encodes SST.
To determine if the protein encoded by At2g23010 is uniquely responsible for the synthesis of 1,2-disinapoyl-Glc in etiolated seedlings, we obtained a SALK Institute T-DNA line (SALK_133207) harboring an insertion in At2g23010. Homozygosity for the T-DNA insertion was initially determined by screening for kanamycin resistance and by PCR genotyping, and Southern analysis was then used to confirm these results (data not shown). Although compound 1 is absent from extracts of etiolated SALK_133207 seedlings, 1,2-disinapoyl-Glc is still accumulated in the mutants seedlings (Fig. 3B), albeit at lower levels than in the wild type. These data suggested that one or more of At2g22980, SNG1, or At2g23000 are capable of synthesizing 1,2-disinapoyl-Glc.
At2g22980 and SNG1 May Contribute to 1,2-Disinapoylglucose Accumulation in Vivo
To test the hypothesis that one or more of the SCPL genes clustered near At2g23010 also have SST activity, two additional sng1-6 transgenic lines were generated: one harboring SNG1 (pCC398) and one harboring At2g22980 (pCC881) under the control of their native regulatory elements. Extracts from 6-d-old etiolated seedlings of each transgenic line contained 1,2-disinapoyl-Glc but not compound 1 (Fig. 3B), thus exhibiting a phenotype similar to that of the SALK At2g23010 knockout line. These data show that in addition to the At2g23010 protein, both of these enzymes have the capacity to synthesize 1,2-disinapoy-Glc.
In addition to the observed redundancy with respect to the genes missing from the sng1-6 mutants, results from enzyme assays suggest that another Arabidopsis protein, likely an SCPL enzyme, exhibits SST activity in vitro. We carried out enzyme assays using crude protein extracts from 6-d-old, etiolated wild-type and sng1-6 seedlings, and found roughly equivalent SST activity for both (data not shown). Given that at least five other clade 1 SCPL genes are expressed in seedlings (Fraser et al., 2005) and that the proteins encoded by these genes are similar to SST, it is reasonable to expect that one of these SCPL proteins might be able to catalyze the disproportionation reaction leading to the production of 1,2-disinapoyl-Glc in vitro. On the other hand, the lack of 1,2-disinapoyl-Glc in the sng1-6 mutant suggests that the corresponding enzyme(s) may not be expressed in cells in which sinapoy-Glc is available as a substrate.
1,2-Disinapoylglucose and Compound 1 Accumulate in sng1-1 and sng1-5 Leaves But Not in Wild-Type or sng1-6 Leaves
Although At2g23010 is expressed in wild-type leaves (Fraser et al., 2005), HPLC analysis shows that 1,2-disinapoyl-Glc is not present in wild-type leaf extracts (Fig. 5), suggesting that the expression of SST may be rendered moot by higher levels of SMT activity. In contrast, 1,2-disinapoyl-Glc is present in the leaves of the sng1-1 and sng1-5 mutants, suggesting that the cryptic levels of SST expression can be revealed when sinapoyl-Glc levels are not depleted by SMT as occurs in a wild-type background. Consistent with the hypothesis that At2g23010 encodes SST, 1,2-disinapoyl-Glc is not found in sng1-6 leaves (Fig. 5).
Figure 5.
Analysis of sinapate esters in mature leaves. Extracts were prepared from leaves of 5-week-old wild-type, sng1-1, sng1-5, and sng1-6 plants and were analyzed by HPLC using UV detection at 335 nm. SG, Sinapoyl-Glc; SM, sinapoylmalate; 1,2-DSG, 1,2-disinapoyl-Glc. 1,2-Disinapoyl-Glc accumulates in the leaves of only the sng1-1 and sng1-5 mutants.
In the course of investigating the bright trichomes1 (brt1) mutant, which is defective in a major sinapic acid:UDPG glucosyltransferase (Sinlapadech et al., 2007), we generated a brt1-1/sng1-1 double mutant. HPLC analysis of the double mutant showed that total sinapate ester content in the brt1-1/sng1-1 leaf extracts was not substantially different from that of the brt1-1 mutant; however, 1,2-disinapoyl-Glc levels were dramatically reduced compared to sng1-1 (Fig. 6). Taken together, these data suggest that SMT and SST are probably expressed in the same cell types where they compete with one another for substrate. Furthermore, they suggest that BRT1 is the major UDP-Glc:sinapic acid glucosyltransferase (SGT) expressed in cells that synthesize 1,2-disinapoyl-Glc, and conversely, that 1,2-disinapoyl-Glc is synthesized in only a subset of cells that accumulate sinapoyl-Glc.
Figure 6.
HPLC analysis of soluble secondary metabolites accumulated in the leaves of wild type, brt1-1, sng1-1, and the brt1-1/sng1-1 double mutant. Leaves were extracted with 50% MeOH and analyzed by HPLC. The elution of UV-absorbing compounds was monitored at 322 nm. SG, Sinapoyl-Glc; SM, sinapoylmalate; 1,2-DSG, 1,2-disinapoyl-Glc; Col, wild-type Columbia. 1,2-Disinapoyl-Glc accumulation is absolutely dependent upon the activity of the BRT1 glycosyltransferase.
The sng1-5 and sng1-6 Mutants Lack the Sinapoylated Form of the Major Anthocyanin in Arabidopsis
The most abundant anthocyanin in Arabidopsis is a sinapoylated cyanidin derivative, 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin (Fig. 1; Bloor and Abrahams, 2002; anthocyanin A11 according to the nomenclature of Tohge et al., 2005). Interestingly, visual inspection of 4-d-old light-grown seedlings revealed that whereas both wild-type and sng1-1 seedlings frequently show pigmentation in the upper portion of the hypocotyl and cotyledons, sng1-5 and sng1-6 plants never exhibit this coloration (data not shown). Reverse transcription-PCR expression analysis was also consistent with the hypothesis that At2g23000 might have a role in anthocyanin biosynthesis since the gene is expressed in senescing leaves (Fraser et al., 2005), a tissue that is frequently observed to accumulate anthocyanin pigments. The At2g23000 gene is also up-regulated in the pap1-D (production of anthocyanin pigment 1-Dominant) mutant, which accumulates anthocyanins to abnormally high levels (Tohge et al., 2005). To induce anthocyanin accumulation in a convenient and reproducible way, we stressed Murashige and Skoog-grown seedlings by exposure to 10% Suc for 5 d, which produced a distinct anthocyanin-less phenotype in the sng1-5 and sng1-6 mutants relative to wild type and sng1-1. HPLC analysis of acidic methanol extracts of these Suc-stressed wild-type and sng1-1 seedlings revealed the presence of a major anthocyanin that is absent from both sng1-5 and sng1-6 seedlings (Fig. 7). Analysis of this compound by matrix-assisted laser desorption ionization (MALDI) time of flight (TOF) yielded data that identified it as A11, the major anthocyanin in Arabidopsis (Bloor and Abrahams, 2002; Tohge et al., 2005). Furthermore, although wild-type Arabidopsis accumulates a number of sinapoylated anthocyanins (Bloor and Abrahams, 2002; Tohge et al., 2005), none of these compounds is present in the sng1-5 mutant, suggesting that sng1-5 lacks the enzyme required for the production of sinapoylated anthocyanins. Since the only difference between the sng1-1 and sng1-5 mutants is the absence of At2g23000 in sng1-5, these data suggest that At2g23000 encodes SAT.
Figure 7.
Analysis of anthocyanins in 11-d-old, Suc-stressed seedlings. Acidic methanol extracts were prepared from 11-d-old Columbia wild-type, sng1-1, sng1-5, and sng1-6 seedlings grown for 6 d in liquid Murashige and Skoog media containing 1% Suc, and for five additional days in Murashige and Skoog media containing 10% Suc. Extracts were analyzed by HPLC using detection at 525 nm. Peaks labeled A9 and A11 indicate the sinapoylated anthocyanins, 3-O-[6-O-p-coumaroyl-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin and 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)- β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin, respectively. A5 and A8 indicate nonsinapoylated anthocyanins 3-O-(6-O-p-coumaroyl-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin and 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin, respectively (Tohge et al., 2005).
At2g23000 Is Uniquely Responsible for Anthocyanin Sinapoylation in Vivo
The data above do not exclude the possibility that SMT is also an SAT, and that SMT and At2g23000 are redundant in function, at least with respect to anthocyanin sinapoylation. Since a T-DNA insertional mutant for At2g23000 is not currently available, the sng1-5 mutant was transformed with pCC398, the aforementioned vector harboring the SNG1 gene, to determine the effect of a loss of At2g23000 function alone. Twelve independent transformed lines from the T2 generation were obtained and screened via HPLC. Levels of sinapoylmalate for each of transformed lines varied, but in no case were sinapoylated anthocyanins detected (Fig. 8). The presence of sinapoylmalate in these plants indicate that SMT is expressed and catalytically active, and the lack of sinapoylated anthocyanin demonstrates that the enzyme is incapable of acting as a redundant SAT in vivo.
Figure 8.
Analysis of the sng1-5/SNG1 transformant. Acidic methanol extracts were prepared from 11-d-old, Suc-stressed Columbia wild type and sng1-5 transformed with the At2g22990/SMT genomic construct (pCC398) and analyzed by HPLC using detection at 335 and 525 nm. Both wild-type and sng1-5/pCC398 seedlings accumulate sinapoylmalate (SM) and nonsinapoylated anthocyanins (A5, A8) 3-O-(6-O-p-coumaroyl-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin and 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin, respectively, but only the wild type accumulates sinapoylated anthocyanins (A9 and A11) 3-O-[6-O-p-coumaroyl-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)- β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin and 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin, respectively.
DISCUSSION
SCPL proteins represent a class of proteins whose diversity of function has become apparent only recently. SCPL proteins have been designated as SCP like solely on the basis of their overall sequence similarity with respect to known SCPs but the enzymatic function of SCPL proteins implied by their annotation is entirely assumed. While the annotation is in some cases accurate, as with the Arabidopsis SCPL protein BRS1 (Zhou and Li, 2005), it is now clear that as a class, SCPL proteins are diverse in both the reactions they catalyze and the functions they serve. Indeed, the SCPL hydroxynitrile lyase from Sorghum bicolor (SbHNL; Wajant et al., 1994), the SCPL isobutyryl-Glc acyltransferases from L. pennellii (Li and Steffens, 2000), and the SCPL-SGAs from Arabidopsis and other members of the Brassicaceae all illustrate that SCPL proteins exhibit a range of catalytic activities (Lehfeldt et al., 2000; Shirley et al., 2001; Milkowski et al., 2004).
The Arabidopsis SCPL-SGA Gene Cluster Exemplifies a Mechanism Underpinning the Diversification of Plant Secondary Metabolism
Considering that the genes encoding SMT, SAT, SST, and the At2g22980 protein are arranged in a cluster, their divergence in substrate and product specificity serves as an excellent example of how gene duplication can contribute to metabolic diversity, and illustrates a means by which secondary metabolism in plants can evolve and diversify (Table III). Genes of secondary metabolism arranged in tandem often encode proteins that are similar in metabolic roles, substrate specificity, or catalytic activity (Frey et al., 1997; Hamberger and Hahlbrock, 2004; Bowles et al., 2005) but often exhibit distinct biochemical and biological functions. For example, Bx2 through Bx5, maize (Zea mays) genes involved in the biosynthesis of the natural pesticide 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one are clustered on the short arm of chromosome 4. These genes encode related cytochrome P450-dependent monooxygenases, each of which in turn metabolizes sequential 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one biosynthetic intermediates (Frey et al., 1997). Similarly, a number of Arabidopsis UDP-glucosyltransferases (UGTs) are encoded by tandemly arranged genes and retain the same catalytic activity and donor molecule specificity for UDP-Glc but differ in their acceptor substrate specificity (Bowles et al., 2005). For example, UGT73C6 has been identified as a UDP-Glc:flavonol-3-O-glycoside-7-O-glucosyltransferase (Jones et al., 2003); whereas, UGT73C5 has been identified as a deoxynivalenol glucosyltransferase (Poppenberger et al., 2003). The genes encoding these two enzymes are neighbors in a six-member gene cluster at the bottom of chromosome II. In contrast, UGT84A1 and UGT84A3, which are encoded by members of a gene cluster on chromosome IV, exhibit activity toward a broad range of hydroxycinnamic acids, with UGT84A1 and UGT84A3 showing the highest catalytic efficiencies for caffeic acid and ferulic acid, respectively (Lim et al., 2001). The SCPL-SGA gene cluster that includes SMT reflects aspects of both of these latter examples in that SST, SMT, and the protein encoded by At2g22980 are all sinapoyl-Glc-dependent SGAs capable of producing both 1,2-disinapoyl-Glc and distinct products as well.
The evolution of the SCPL-SGAs is likely to have taken place by an initial shift in catalytic activity from hydrolytic cleavage exhibited by SCPs to acyltransferase activity, followed by divergence of substrate specificity. The initial shift in catalytic activity may also represent the point of divergence in the function of the ancestral SCP from involvement in primary cellular processes (e.g. protein processing and degradation), providing the opportunity for members of the ancestral gene family to assume a role in plant secondary metabolism. Although a distinct function for At2g22980 has not yet been identified, and the principle role for SMT is clearly in sinapoylmalate biosynthesis, the partial redundancy of these proteins with SST indicates that the evolution of these genes has not yet resulted in enzymes with completely distinct acyl acceptor specificities. In contrast, the ability of SMT, SAT, and SST to function uniquely in the biosynthesis of sinapoylmalate, sinapoylated anthocyanins, and compound 1, respectively, appears to be shared by no other members of the protein family encoded by this gene cluster. In summary, this SCPL-SGA gene cluster appears to provide a glimpse of the gene evolution underlying plant secondary metabolism.
It is interesting to note that the SCPL-SGA gene cluster on chromosome II is one of seven tandem clusters found within the Arabidopsis SCPL gene family, with the three largest clusters all encoding clade IA SCPL proteins (Fraser et al., 2005). In fact, only two of 19 SCPL clade IA genes are found outside of these tandem gene clusters. In light of the results presented in this article, and the fact that the clade IA proteins are highly similar to one another, it is reasonable to expect that these other SCPL proteins not only function as SCPL-SGAs, but that they may overlap in acyl-donor and/or acceptor specificity as well. This is especially likely for the SCPL proteins encoded by the clade IA SCPL gene cluster located on the lower arm of chromosome I, given that they encode the most similar group of all SCPL proteins, with pairwise percent identities between 79% and 86% (Fraser et al., 2005). The presence of multiple gene clusters encoding highly similar SCPL proteins is also consistent with the hypothesis that clade IA SCPL proteins have evolved relatively recently and that they are, as a class, associated with plant secondary metabolism.
Sinapoylation by SCPL-SGAs Contributes to the Diversity of Plant Secondary Metabolites
Arabidopsis and other members of the Brassicaceae synthesize a wide array of sinapate esters in addition to those discussed here (Baumert et al., 2005). While the physiological roles of many of these compounds have yet to be determined, as with 1,2-disinapoyl-Glc (compound 2) and compound 1, their abundance and diversity make sinapate esters an interesting class of plant secondary metabolites, and it is likely many of these compounds are produced via SCPL-SGAs (Lehfeldt et al., 2000; Shirley et al., 2001; Baumert et al., 2005; Fraser et al., 2005). Anthocyanins, on the other hand, are widespread throughout the plant kingdom, and play significant roles in pollination, seed dispersal, and plant stress responses (Winkel-Shirley, 2001, 2002), so gaining an understanding of the steps involved in their biosynthesis, including acylation, is important to understanding plant biochemistry on a more general level. Anthocyanins vary widely in their patterns of acylation, with activated hydroxycinnamates commonly serving as acyl donors. It has been suggested that acyl groups may contribute to the stabilization of anthocyanins by allowing for molecular stacking, thereby preventing nucleophilic attack by water and preserving color (Brouillard, 1981, 1983; Hopp and Seitz, 1987). Acylation also affects the absorbance properties of anthocyanins, with different patterns of acylation resulting in observable differences in color (Honda et al., 2005), and it has been suggested that pH-dependent conformational changes brought about by the presence of acyl groups may be involved in localization of anthocyanins to the vacuole (Matern et al., 1986; Hopp and Seitz, 1987). Understanding the role that SCPL-SGAs may play in the acylation of anthocyanins may therefore provide important insights not only into plant secondary metabolism, but plant survival and adaptability as well.
Sinapoylated anthocyanins have been found in many different members of the Brassicaceae, including cabbage (Brassica capitata; Hrazdina et al., 1977; Wu and Prior, 2005), Matthiola incana (Saito et al., 1995, 1996), Sinapis alba (Takeda et al., 1988), and Orychophragonus violaceus (Honda et al., 2005). Sinapoylated anthocyanins are also found in some orchids (Tatsuzawa et al., 2004), and the major anthocyanin in wild carrot (Daucus carota) is sinapoylated (Harborne et al., 1983) where sinapoyl-Glc has been shown to serve as the activated acyl donor molecule (Gläßgen and Seitz, 1992). Based upon our findings, anthocyanin sinapoylation in some, or possibly all of these plants may be catalyzed by SCPL-SGAs and the acylation of anthocyanins with other hydroxycinnamates via SCPL-SGAs may serve as alternative catalysts to the more common BAHD acyltransferases (St Pierre and De Luca, 2000; Luo et al., 2007).
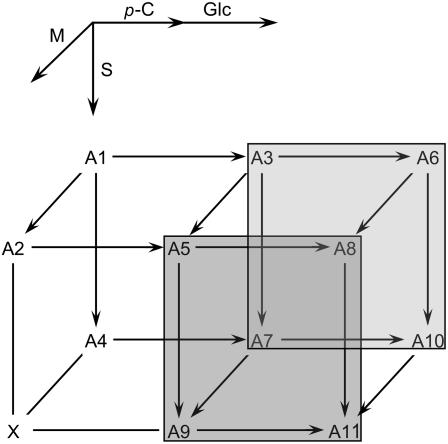
The metabolic route to sinapoylation still remains to be determined. While it is possible that SAT exhibits substrate specificity toward a single anthocyanin such as 3-O-(6-O-p-coumaroyl-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin (designated A5 in Tohge et al., 2005), an alternative is that the enzyme may be able to use multiple substrates as acyl acceptors (Fig. 9). For instance both A5 and A8 may act as acyl acceptors in the biosynthesis of A9 and A11, respectively (Fig. 9). A similar scenario appears to exist for the acyltransferases that add the malonyl and p-coumaroyl subunits to the glycosylated form of the core anthocyanin molecule, resulting in an anthocyanin biosynthetic metabolic grid (Fig. 9; Luo et al., 2007).
Figure 9.
Possible metabolic routes of anthocyanin acylation in Arabidopsis. Anthocyanins A1 to A11 have been identified in wild-type Arabidopsis and the pap1-D mutant (Bloor and Abrahams, 2002; Tohge et al., 2005). Arrows indicate potential acyltransferase reactions leading to the addition of malonyl (M), p-coumaroyl (p-C), and sinapoyl (S) groups, and glycosylation of the p-coumaroyl group (Glc). A1, 3-O-(2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-β-d-glucopyranosylcyanidin. A2, 3-O-(2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin. A3, 3-O-(6-O-p-coumaroyl-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-β-d-glucopyranosylcyanidin. A4, 3-O-[2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-β-d-glucopyranosylcyanidin. A5, 3-O-(6-O-p-coumaroyl-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl)-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin. A6, 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl]-5-O-β-d-glucopyranosylcyanidin. A7, 3-O-[6-O-p-coumaroyl-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-β-d-glucopyranosylcyanidin. A8, 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-β-d-xylopyranosyl-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin. A9, 3-O-[6-O-p-coumaroyl-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin. A10, 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-β-d-glucopyranosylcyanidin. A11, 3-O-[6-O-(4-O-β-d-glucopyranosyl-p-coumaroyl)-2-O-(2-O-sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin. 3-O-[2-O-(2-O-Sinapoyl-β-d-xylopyranosyl)-β-d-glucopyranosyl]-5-O-(6-O-malonyl-β-d-glucopyranosyl) cyanidin is designated as X since it has not been identified in Arabidopsis to date and, as a result, the lines that lead to and from X are not shown with arrowheads since it is not clear that it is a genuine biosynthetic intermediate in anthocyanin biosynthesis.
Substrate Specificity of the Arabidopsis SCPL-SGAs May Be Due to a Small Subset of Amino Acid Residues
SST, SAT, and the At2g22980 protein represent the latest additions to the class of SCPL-SGAs whose activities have been fully or partially characterized. As such, they not only expand the importance of this class of enzymes with respect to plant secondary metabolism, but they also provide an opportunity to study the relationship between the evolution of sequence and function. For example, the fact that SMT, SST, SAT, the protein encoded by At2g22980, and even SCT are relatively similar to one another would suggest that the relatively small number of amino acids by which these proteins differ is responsible for the differing substrate specificities exhibited by the Arabidopsis SCPL-SGAs, and that the most important amino acids affecting substrate specificity are likely to be an even smaller subset of residues clustered near the enzymes' active sites.
In conclusion, the SCPL-SGAs represent a growing class of enzymes important to plant secondary metabolism. Our research and the research of others suggests that in the plant kingdom, a significant number of SCPL proteins have been recruited to serve as Glc acyltransferases, utilizing 1-O-sinapoyl-Glc as an activated acyl donor molecule. The results of this article further suggest that the study of the SCPL-SGA gene cluster provides an opportunity to elucidate the relationship between gene sequence, enzyme function, and the evolution of metabolism.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown under a 16 h light/8 h dark photoperiod in soilless potting mix (Redi-Earth potting mixture, Scotts-Sierra Horticultural Products) at 23°C. For seedling plant material to be used in the analysis of sinapate ester accumulation, seeds were surface sterilized for 10 min in a 2:1 mixture of 0.1% Triton X-100 and household bleach. Seeds were rinsed thoroughly with sterile water and planted on modified Murashige and Skoog medium (Murashige and Skoog, 1962; ammonia-free medium to which 20.6 mm potassium nitrate was added in place of ammonium nitrate) containing 0.7% agar. For seedling plant material to be used for the isolation of 1,2-disinapoyl-Glc, nonsterilized seeds were sown on filter paper moistened with water. To induce anthocyanin production, seedlings were grown in liquid Murashige and Skoog media containing 1% Suc for 6 d, the media was then removed and replaced with 10% Suc Murashige and Skoog media and the seedlings were grown for an additional 5 d.
Analysis of Sinapate Esters and Anthocyanins
Sinapate esters were extracted from fresh plant tissue in 50% (v/v) methanol. Samples were incubated at 65°C for 30 min, centrifuged at 12,000g for 2 min, and analyzed by HPLC on a Microsorb-MV C18 column (Varian) using a gradient from 10% acetonitrile in 1.5% acetic acid to 35% acetonitrile in 1.5% acetic acid at a flow rate of 1 mL min−1. Sinapate esters were detected by their UV A335. Anthocyanins were extracted from fresh plant tissue in 4:46:50 acetic acid:water:methanol. Samples were incubated at 65°C for 30 min, centrifuged at 12,000g for 2 min, and analyzed by HPLC on an end-capped C18 column (Waters) using a gradient from 10% acetonitrile in 10% formic acid to 20% acetonitrile in 10% formic acid flow rate of 1 mL min−1. Anthocyanins were detected by their UV A525. Anthocyanin fractions corresponding to each peak were collected and analyzed by MALDI-TOF MS.
Isolation and Identification of 1,2-Disinapoylglucose (Compound 2)
For the identification of the sinapate esters not found in the sng1-6 mutant, sng1-1 seedlings were grown on moistened filter paper for 7 d in complete darkness and were extracted in 50% methanol. This extract was concentrated in vacuo, and analyzed using HPLC as described above. The pooled fractions containing 1,2-disinapoyl-Glc were again reduced in vacuo to 2 mL and analyzed by NMR.
A subsample (approximately 1 mg) of the putative 1(E),2(E)-di-O-sinapoyl-β-d-glucopyranoside (1,2-disinapoyl Glc) isolated by HPLC was dissolved in either CD3OD or 9:1 acetone-d6:D2O and transferred to a 5 mm OD NMR tube. The usual array of 1D (1H, 13C) and two-dimensional (2D; COSY, HSQC, HSQC-TOCSY, HMBC) experiments were run using standard Bruker pulse programs, from which unambiguous assignments were established. The spectra in perdeuteromethanol were acquired on a Bruker DRX-360 using an inverse (proton coils closest to the sample), 1H/broadband, gradient probe. The spectra in perdeutero-acetone:D2O were acquired on a Bruker DMX-500 equipped with an inverse, gradient, cryogenically cooled probe for enhanced sensitivity. HMBC spectra were acquired with both an 80 and 120 ms long-range coupling delay.
Construction of Transgenic Lines
To construct a plant transformation vector containing At2g23010 (pCC579) a cosmid carrying the corresponding genomic sequence (pCC301) was digested with NsiI to liberate a 7,500 bp fragment that contained the genomic coding sequence for At2g23010 as well as 2,200 bp upstream and 1,628 bp downstream of the At2g23010 open reading frame. This fragment was subsequently subcloned into the compatible PstI site of the binary vector, pCAMBIA 2300, generating pCC579. To construct the plasmid containing At2g22980 a cosmid carrying the corresponding genomic sequence (pCC305) was digested with HindIII and XbaI to liberate a 5,865 bp fragment that contained the genomic coding sequence of At2g22980, 1,066 bp upstream and 741 bp downstream of the At2g22980 open reading frame. The plasmid containing At2g22990 (SNG1) was constructed as described elsewhere (Lehfeldt et al., 2000).
Constructs for plant transformation were introduced into Agrobacterium tumefaciens C58 pGV3850 (Zambryski et al., 1983) by electroporation. Plant transformation was performed by the floral dip method (Clough and Bent, 1998) into sng1-6. Transformed seedlings (T1) identified by selection on Murashige and Skoog medium containing 50 mg L−1 kanamycin and 200 mg L−1 timentin were transformed to soil.
Matrix-Assisted Laser Desorption Ionization MS
Samples recovered after HPLC were spotted on a stainless steel MALDI plate with an equal volume of 5 mg/mL α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid and allowed to air dry. Mass spectra were acquired with an Applied Biosystems 4700 MALDI TOF/TOF instrument operating in positive reflectron mode. The instrument was initially calibrated with a series of peptide standards to a mass accuracy better than 50 ppm.
Acknowledgments
The authors would like to thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the Arabidopsis Biological Resource Center for the distribution of these materials.
This work was supported by the National Science Foundation (grant no. 0091419 to C.C.) and a graduate fellowship from Purdue University (to C.M.F.). J.R. was supported in part by funding through the Department of Energy Biosciences program (grant no. DE–AI02–00ER15067). NMR experiments on the Bruker DMX-500 cryoprobe system were carried out at the National Magnetic Resonance Facility at Madison with support from the National Institutes of Health Biomedical Technology Program (grant no. RR02301) and additional equipment funding from the University of Wisconsin National Science Foundation Academic Infrastructure Program (grant no. BIR–9214394), National Institutes of Health Shared Instrumentation Program (grant nos. RR02781 and RR08438), National Science Foundation Biological Instrumentation Program (grant no. DMB–8415048), and the U.S. Department of Agriculture. A portion of the mass spectrometry analysis was conducted at the Purdue University Metabolic Profiling Facility, supported by the National Science Foundation (grant no. DBI–0421102). This is journal number 2007–18146 from the Purdue University Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Clint Chapple (chapple@purdue.edu).
Open Access articles can be viewed online without a subscription.
References
- Baumert A, Milkowski C, Schmidt J, Nimtz M, Wray V, Strack D (2005) Formation of a complex pattern of sinapate esters in Brassica napus seeds, catalyzed by enzymes of a serine carboxypeptidase-like acyltransferase family? Phytochemistry 66 1334–1345 [DOI] [PubMed] [Google Scholar]
- Bloor SJ, Abrahams S (2002) The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59 343–346 [DOI] [PubMed] [Google Scholar]
- Bowles D, Isayenkova J, Lim EK, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8 254–263 [DOI] [PubMed] [Google Scholar]
- Brouillard R (1981) Origin of the exceptional colour stability of the Zebrina anthocyanin. Phytochemistry 20 143–145 [Google Scholar]
- Brouillard R (1983) The in vivo expression of anthocyanin colour in plants. Phytochemistry 22 1311–1323 [Google Scholar]
- Clough JS, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Croteau R, Hooper CL (1978) Metabolism of monoterpenes: acetylation of (-)-menthol by a soluble enzyme preparation from peppermint (Mentha piperita) leaves. Plant Physiol 61 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlbender B, Strack D (1984) Enzymatic synthesis of 1,2-di-dinapoylglucose from 1-sinapoylglucose by a protein preparation from cotyledons of Raphanus sativus grown in the dark. J Plant Physiol 116 375–379 [DOI] [PubMed] [Google Scholar]
- Dahlbender B, Strack D (1986) Purification and properties of 1-(hydroxycinnamoyl)-glucose: 1-(hydroxycinnamoyl)-glucose hydroxycinnamoyl-transferase from radish seedlings. Phytochemistry 25 1043–1046 [Google Scholar]
- Dudareva N, Dauria JC, Nam KH, Raguso RA, Pichersky E (1998) Acetyl-CoA-benzylalcohol acetyltransferase—an enzyme involved in floral scent production in clarkia breweri. Plant J 14 297–304 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Rider LW, Chapple C (2005) An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiol 138 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277 696–699 [DOI] [PubMed] [Google Scholar]
- Fröhlich B, Niemetz R, Gross GG (2002) Gallotannin biosynthesis: two new galloyltransferases from Rhus typhina leaves preferentially acylating hexa- and heptagalloylglucoses. Planta 216 168–172 [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Nakao M, Fukui Y, Yamaguchi M, Ashikari T, Kusumi T (1998) cDNA cloning, gene expression and subcellular localizationof anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J 16 421–431 [DOI] [PubMed] [Google Scholar]
- Gläßgen WE, Seitz HU (1992) Acylation of anthocyanins with hydroxycinnamic acids via 1-O-acylglucosides by protein preparations from cell cultures of Daucus carota L. Planta 186 582–585 [DOI] [PubMed] [Google Scholar]
- Hamberger B, Hahlbrock K (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci USA 101 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Mayer AM, Bar-Nun N (1983) Identification of the major anthocyanin of carrot cells in tissue culture as cyanidin 3-(sinapoylxylosylglucosylgalactoside). Z Naturforsch 38c 1055–1056 [Google Scholar]
- Honda T, Tatsuzawa F, Kobayashi N, Kasai H, Nagumo S, Shigihara A, Saito N (2005) Acylated anthocyanins from the violet-blue flowers of Orychophragonus violaceus. Phytochemistry 66 1844–1851 [DOI] [PubMed] [Google Scholar]
- Hopp W, Seitz HU (1987) The uptake of acylated anthocyanin into isolated vacuoles from a cell suspension culture of Daucus carota. Planta 170 74–85 [DOI] [PubMed] [Google Scholar]
- Hrazdina G, Iredala H, Mattick LR (1977) Anthocyanin composition of Brassica oleracea cv. Red Danish. Phytochemistry 16 297–299 [Google Scholar]
- Jones P, Messner B, Nakajima J, Schaffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278 43910–43918 [DOI] [PubMed] [Google Scholar]
- Kesy JM, Bandurski RS (1990) Partial purification and characterization of indol-3-ylacetylglucose: myo-inositol indol-3—ylacetyltransferase (indoleacetic acid-inositol synthase). Plant Physiol 94 1598–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Mitchell-Olds T (2005) The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr Opin Plant Biol 8 264–271 [DOI] [PubMed] [Google Scholar]
- Kuć J (1995) Phytoalexins, stress metabolism, and disease resistance in plants. Annu Rev Phytopathol 33 275–297 [DOI] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz R, Zenk MH (1995) Acetyl coenzyme A:salaturidinol-7-O-acetyltransferase from Papaver somniferum plant cell cultures: the enzyme catalyzing the formation of thebaine in morphine biosynthesis. J Biol Chem 270 31091–31096 [DOI] [PubMed] [Google Scholar]
- Li AX, Eannetta N, Ghangas GS, Steffens JC (1999) Glucose polyester biosynthesis: purification and characterization of a glucose acyltransferase. Plant Physiol 121 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AX, Steffens JC (2000) An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc Natl Acad Sci USA 97 6902–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson R, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276 4344–4349 [DOI] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, et al (2007) Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J 50 678–695 [DOI] [PubMed] [Google Scholar]
- Matern U, Reichenbach C, Heller W (1986) Efficient uptake of flavonoids into parsley (Petroselinum hortense) vacuoles requires acylated glycosides. Planta 167 183–189 [DOI] [PubMed] [Google Scholar]
- Milkowski C, Baumert A, Schmidt D, Nehlin L, Strack D (2004) Molecular regulation of sinapate ester metabolism in Brassica napus: expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant J 38 80–92 [DOI] [PubMed] [Google Scholar]
- Mock HP, Strack D (1993) Energetics of the uridine 5′-diphosphoglucose: hydroxycinnamic acid acyl-glucosyltransferase reaction. Phytochemistry 32 575–579 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15 473–497 [Google Scholar]
- Nakayama T, Suzuki H, Nishino T (2003) Anthocyanin acyltranferases: specificities, mechanism, phylogenetics, and applications. J Mol Catal B Enzym 23 117–132 [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glossl J, Luschnig C, Adam G (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278 47905–47914 [DOI] [PubMed] [Google Scholar]
- Rabot S, Peerless ACJ, Robins RJ (1995) Tigloyl-CoA-pseudotropine acyl transferase—an enzyme of tropane alkaloid biosynthesis. Phytochemistry 39 315–322 [Google Scholar]
- Saito N, Tatsuzawa F, Hongo A, Khin WW, Yokoi M, Shigihara A, Honda T (1996) Acylated pelargonidin 3-sambubioside-5-glucoside in Matthiola incana. Phytochemistry 41 1613–1620 [DOI] [PubMed] [Google Scholar]
- Saito N, Tatsuzawa F, Nishiyama A, Yokoi M, Shigihara A, Honda T (1995) Acylated cyanidin 3-sambubrioside-5-glucoside in Matthiola incana. Phytochemistry 38 1027–1032 [DOI] [PubMed] [Google Scholar]
- Shirley AM, McMichael CM, Chapple C (2001) The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J 28 83–94 [DOI] [PubMed] [Google Scholar]
- Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C (2007) The hyperfluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a gene encoding a sinapic acid: UDPG glucosyltransferase. Plant J 49 655–668 [DOI] [PubMed] [Google Scholar]
- St Pierre B, De Luca V (2000) Evolution of Acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In JT Romeo, R Ibrahim, L Varin, V De Luca, eds, Recent Advances in Phytochemistry, Volume 34: Evolution of Metabolic Pathways. Pergamon, Oxford, pp 285–315
- Strack D, Dahlbender B, Grotjahn L, Wray V (1984) 1,2-Disinapoylglucose accumulated in cotyledons of dark-grown Raphanus sativus seedlings. Phytochemistry 23 657–659 [Google Scholar]
- Suzuki H, Murakoshi I, Saito K (1994) A novel O-tigloyltransferase for alkaloid biosynthesis in plants: purification, characterization and distribution in Lupinus plants. J Biol Chem 269 15853–15860 [PubMed] [Google Scholar]
- Takeda K, Fischer D, Grisebach H (1988) Anthocyanin composition of Sinapis alba, light induction of enzymes and biosynthesis. Phytochemistry 27 1351–1353 [Google Scholar]
- Tatsuzawa F, Saito N, Seki H, Yokoi M, Yukawa T, Shinoda K, Honda T (2004) Acylated anthocyanins in the flowers of Vanda (Orchidaceae). Biochem Syst Ecol 32 651–664 [Google Scholar]
- Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8 317–323 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42 218–235 [DOI] [PubMed] [Google Scholar]
- Villegas RJA, Kojima M (1986) Purification and characterization of hydroxycinnamoyl D-glucose quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. J Biol Chem 261 8729–8733 [PubMed] [Google Scholar]
- Villegas RJA, Shimokawat T, Okuyama H, Kojima M (1987) Purification and characterization of chlorogenic acid: chlorogenate caffeoyl transferase in sweet potato roots. Phytochemistry 26 1577–1581 [Google Scholar]
- Wajant H, Mundry KW, Pfizenmaier K (1994) Molecular cloning of hydroxynitrile lyase from Sorghum bicolor (L.): homologies to serine carboxypeptidases. Plant Mol Biol 26 735–746 [DOI] [PubMed] [Google Scholar]
- Walker K, Ketchum REB, Hezari M, Gatfield DD, Goleniowski M, Barthol A, Croteau R (1999) Partial purification and characterization of acetyl coenzyme A: taxa-4(20),11(12)-dien-5 alpha-ol O-acetyl transferase that catalyzes the first acylation step of taxol biosynthesis. Arch Biochem Biophys 364 273–279 [DOI] [PubMed] [Google Scholar]
- Wink M (1988) Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor Appl Genet 75 225–233 [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5 218–223 [DOI] [PubMed] [Google Scholar]
- Wu X, Prior RL (2005) Identification and characterization of anthocyanins by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry in common foods in the United States: vegetables, nuts, and grains. J Agric Food Chem 53 3101–3113 [DOI] [PubMed] [Google Scholar]
- Yang Q, Reinhard K, Shiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA; anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35 777–789 [DOI] [PubMed] [Google Scholar]
- Zambryski P, Joos H, Gentello C, Leemans J, van Montagu M, Schell J (1983) Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J 2 2143–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Li J (2005) Arabidopsis BRS1 is a secreted and active serine carboxypeptidase. J Biol Chem 280 35554–35561 [DOI] [PubMed] [Google Scholar]