Abstract
Protein diversification is commonly driven by single amino acid changes at random positions followed by selection, but, in some cases, the structure of the gene itself favors the occurrence of particular kinds of mutations. Genes encoding hydroxyproline-rich glycoproteins (HRGPs) in green organisms, key protein constituents of the cell wall, carry short-repeat modules that are posited to specify proline hydroxylation and/or glycosylation events. We show here, in a comparison of two closely related Chlamydomonas species—Chlamydomonas reinhardtii (CC-621) and Chlamydomonas incerta (CC-1870/3871)—that these modules are prone to misalignment and hence to both insertion/deletion and endoduplication events, and that the dynamics of the rearrangements are constrained by purifying selection on the repeat patterns themselves, considered either as helical or as longitudinal face modules. We suggest that such dynamics may contribute to evolutionary diversification in cell wall architecture and physiology. Two of the HRGP genes analyzed (SAG1 and SAD1) encode the mating-type plus and minus sexual agglutinins, displayed only by gametes, and we document that these have undergone far more extensive divergence than two HRGP genes (GP1 and VSP3) that encode cell wall components—an example of the rapid evolution that characterizes sex-related proteins in numerous lineages. Strikingly, the central regions of the agglutinins of both mating types have diverged completely, by selective endoduplication of repeated motifs, since the two species last shared a common ancestor, suggesting that these events may have participated in the speciation process.
Hyp-rich glycoproteins (HRGPs) represent a family of proteins that self-assemble to form vital scaffolding in the cell walls of plants, where the relationship of this scaffolding to the abundant structural polysaccharides—cellulose, hemicellulose, and pectin—found in most types of cell wall is not well understood (Cassab, 1998; Showalter, 2001). HRGP families in higher plants include the extensins, with a characteristic Ser-(Hyp)4 repeat motif, and the arabinogalactan proteins (AGPs), rich in Ala-Hyp repeats interspersed with Ser. Chlamydomonas, a unicellular soil alga, produces a variety of HRGPs, which self-assemble to form a cell wall without additional polysaccharide components (for review, see Roberts et al., 1985). Because the chlorophyte algae share a deep common ancestor with higher plants (Lewis and McCourt, 2004), analysis of their HRGPs is expected to yield insight into higher plant extracellular matrix biology and evolution.
In the Chlamydomonas lineage, HRGPs not only self-assemble as cell walls, but also participate in sexual recognition between mating-type plus and minus gametes. Enormous HRGPs (>1,000 kD; approximately 230–240 nm in length; Adair et al., 1983; Goodenough et al., 1985), called Sag1 (plus) and Sad1 (minus) sexual agglutinins, are displayed on gametic flagellar membranes and mediate the initial sex-specific and species-specific adhesion events between interacting gametes that culminate in zygotic cell fusion (Goodenough et al., 1985, 1995, 2007; Goodenough and Heuser, 1999; Ferris et al., 2005).
All known HRGPs in Chlamydomonas are chimeric (Kieliszewski and Lamport, 1994), with an elongated Hyp-rich domain (hereafter called shaft) and one or two terminal globular domains (heads; Goodenough and Heuser, 1985, 1988a, 1988b; Goodenough et al., 1986; Goodenough, 1991; Hallmann, 2006). In each case, the shafts, like the nonchimeric extensins, carry short repetitive amino acid motifs (e.g. PPSPX, PS, PPX; Woessner and Goodenough, 1989, 1992; Waffenschmidt et al., 1993; Suzuki et al., 2000; Ferris et al., 2001, 2005; Hallmann, 2006), and they form polyproline II (PII) helices (Ferris et al., 2001, 2005).
The PII helix is a secondary structure, adopted by peptide domains with substantial (hydroxy) Pro content, in which 3.34 amino acids generate 1 nm of shaft length (van Holst and Varner, 1984; Stapley and Creamer, 1999; Rucker et al., 2003). Because there are approximately three amino acids per helical gyre, this generates three longitudinal faces along the long axis of a shaft, each face separated from the next by approximately 120° (Ferris et al., 2005). The highly elongated helix enables the formation of space-filling fibers, a property that has been independently exploited by animals (collagens) and plants (HRGPs) in constructing their extracellular matrices.
A recurrent theme within the HRGPs (as well as the collagens) is significant posttranslational modification. Encoded Pro residues are commonly hydroxylated in the endoplasmic reticulum (Harwood et al., 1974; Bolwell et al., 1985; Pihlajaniemi et al., 1991; Annunen et al., 1997, 1998; Yuasa et al., 2005), where, in collagen, the Hyp residues have been shown to stabilize the triple helix (for reviews, see Kivirikko and Pihlajaniemi, 1998; Myllyharju, 2003; Myllyharju and Kivirikko, 2004). In HRGPs, the Hyp and Ser residues are usually O-glycosylated, presumably in the Golgi (for review, see Colley, 1997), albeit a recent study suggests that some HRGP O-glycosylation may occur in the endoplasmic reticulum (Estévez et al., 2006). Glycosylation bestows several important attributes to biopolymers designed to self-assemble in extracellular spaces: increased solubility, increased resistance to proteolysis, rigidity for structural fidelity, malleability for matrix remodeling, and, presumably but not yet demonstrated, specificity of intermolecular interactions (for review, see Van den Steen et al., 1998). A glycosylated PII shaft essentially presents itself to the cell as a sequence—or, perhaps more aptly, a bottle brush—of specified sugar residues, much like animal proteoglycans.
It has been proposed (Kieliszewski and Lamport, 1994) that particular HRGP repeats signal particular hydroxylation/glycosylation events, in which case multiple forms of modification enzymes are expected to be involved. Prolyl 4-hydroxylases are encoded by a multigene family of at least six genes in Arabidopsis (Arabidopsis thaliana; Hieta and Myllyharju, 2002; Tiainen et al., 2005) and 10 genes in Chlamydomonas reinhardtii (Keskiaho et al., 2007) and, while a search for the genes encoding the relevant glycosyltransferases is only now under way (e.g. Egelund et al., 2007), these are presumed to be members of large families as well. Some progress has been made in identifying features of the HRGP repeats as glycosylation codes (Kieliszewski and Lamport, 1994; Shpak et al., 1999; Ferris et al. 2001, 2005; Zhao et al., 2002; Tan et al., 2003; Estévez et al., 2006), but much remains to be learned about which residues in the repeats and/or local conformations are important in guiding which prolyl 4-hydroxylases to particular Pros and directing glycosyltransferases to add particular sugar residues to particular positions (Estévez et al., 2006; Keskiaho et al., 2007).
The iterative nature of proteins with short repeats also renders the genes vulnerable to rearrangements: They are prone to undergo slipped-strand misalignment during replication and recombination, generating insertions and deletions (indels; Smith, 1976; Levinson and Gutman, 1987; Elder and Turner, 1995; Li et al., 2002; Lai and Sun, 2003), and they are poised to endoduplicate. If the HRGP repeats indeed function as signals for posttranslational modification, then rearrangements that generate noninterpretable information and hence a dysfunctional glycoprotein product are presumably subject to negative selection, whereas rearrangements that preserve or create interpretable information would either be neutral or fodder for positive selection. An intriguing possibility, developed in this study, is that such events have generated species-specific sex-recognition features of the sexual agglutinins of Chlamydomonas.
To study the evolution of HRGPs, we compared the shaft sequences of the sex-related Sag1 and Sad1 agglutinins and the sex-unrelated cell wall proteins Vsp3 and Gp1 in two species, C. reinhardtii (CC-621) and Chlamydomonas incerta (CC-1870/3871), estimated to have last shared a common ancestor <10 million years ago (Coleman and Mai, 1997; Liss et al., 1997; Popescu et al., 2006); analysis of the globular domains of these chimeric proteins will be presented in a separate report (J.-H. Lee, S. Waffenschmidt, and U.W. Goodenough, unpublished data). We found, for the cell wall proteins, that gene rearrangements have occurred and, in one case, have generated shafts of different lengths, but the shafts maintain the same overall subdomain structure and frame of repeating motifs; whether the rearranged proteins generate cell walls with novel properties awaits functional tests. For the agglutinins, gene rearrangements are far more extensive and portions of the shafts are totally different in sequence, providing another example of the rapid evolution of sex-related proteins (Civetta and Singh, 1998; Swanson et al., 2001a; Swanson and Vaquier, 2002; Clark et al., 2006). These differences may be relevant to species formation and/or isolation in the Chlamydomonas lineage.
RESULTS
Longitudinal Face versus Helical Modules
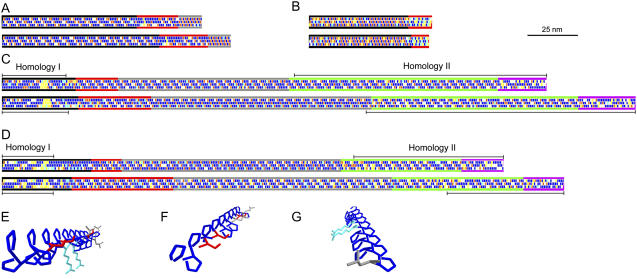
The iterative nature of the shaft sequences and the large size of the agglutinin shafts prompted us to devise a means to visualize shafts at the protein sequence level in the context of the PII helix. Such diagrams make it easier to compare a given shaft from two different species and they serve to emphasize the three longitudinal faces of a shaft sequence previously noted in Ferris et al. (2005). In the diagrams (Fig. 1, A–D), the primary amino acid sequence of each shaft is arranged in three separate rows such that residues (rectangles) in every third position appear in a single row. For example, three PPSPX repeats (PPSPXPPSPXPPSPX) in a primary sequence generate P-P-P-X-S-, P-X-S-P-P-, and S-P-P-P-X- rows, meaning that long iterations of PPSPX repeats create long iterations of PPPXS repeats when considered as longitudinal faces of the PII helix.
Figure 1.
Diagrammatic representations of chimeric HRGP shaft domains from Chlamydomonas. A, Gp1. B, Vsp3. C, Sag1. D, Sad1 longitudinal face diagrams. Amino acids are depicted as rectangles along the three faces of PII helices (Ferris et al., 2005). Blue, Pro; red, Ser; light blue, charged; white, other. Yellow denotes guest amino acids: Two or more sequential amino acids other than Pro that have the potential to disrupt the helical configuration (Creamer, 1998). Diagrams at the top derive from C. reinhardtii, at the bottom from C. incerta. Subdomains are indicated by colored longitudinal lines as follows: A, Black, main shaft; red, kink; gray, neck. B, Black, main shaft; red, P3X3 subdomain. C and D, Black, 2A; red, 2B; gray, 2C; green, 2D; purple, 2E (Ferris et al., 2005). Homology I and II regions are described in text. E to G, Three-dimensional reconstruction of a PII helix from the PPPPPSPPSPRPPRPPPLPPSPPPPLL sequence of the 2A subdomain of Sag1 C. reinhardtii, emphasizing the three longitudinal faces. The γ-carbon-to-γ-carbon spacing of Pro amino acid residues along a PII longitudinal face is calculated to be 0.934 nm; Pro spacing along a PII helical face is calculated to be 0.643 nm. E, View of all three longitudinal faces looking from the N terminus. F, View of the first and third faces from the N terminus. G, View of the first and second faces from the C terminus. Blue, Pro; red, Ser; light blue, Arg; gray, Leu. Backbone atoms are omitted for simplification. Images generated using DeepView, version 3.7 (http://www.expasy.org/spdbv).
It is generally assumed (Kieliszewski and Lamport, 1994) that posttranslational modification enzymes read the primary sequence of repeat modules as they are spirally displayed along the PII helix (helical modules, PPSPX in the above example), and this may prove to be the case in many instances. But, given that PII helices also carry three longitudinal faces that are linearly displayed along the long axis of a shaft (longitudinal face modules, PPPXS in the above example), and given that the two sets of modules provide distinctive potential readouts, the longitudinal face modules represent additional candidates for modification enzyme recognition and/or for participation in interaction with other proteins.
To illustrate the three-dimensional topology of the longitudinal faces, the N-terminal region of the Sag1 shaft from C. reinhardtii (Fig. 1C, top) is modeled in Figure 1, E to G. Particular residues display a polarized distribution: One face contains a contiguous Pro stretch, the second contains a mix of Ser and Pro, and the third exposes two positively charged Args. Evidence that selection may in some cases act to preserve such longitudinal face modules is presented in a later section.
Figure 1, A to D, depicts the shafts of Gp1, Vsp3, Sag1, and Sad1, the eight HRGP proteins under study, for each of the two Chlamydomonas species. Pros and Sers, candidates for hydroxylation and/or glycosylation, are shown in blue and red, illustrating the probable space-filling pattern of sugars along the main axis. Charged amino acids (light blue) may mediate ionic interactions within or between shafts; notably, the first subdomains of Gp1 are largely devoid of charged amino acids (Fig. 1A), whereas the third subdomains of Sag1 and Sad1 display numerous charges along the axis (Fig. 1, C and D). Yellow represents guest amino acids—two or more contiguous non-Pro residues in a Pro-rich domain that have the potential to destabilize the locally driven PII configuration and promote bending (Creamer, 1998)—that are, for example, conspicuous throughout the Vsp3 sequences (Fig. 1B).
Whether such faces prove to be recognized by posttranslational modification enzymes or interacting molecules awaits experimental analysis. Meanwhile, the diagrams of Figure 1 serve to summarize and emphasize the overall organization of the proteins under study. In particular, they illustrate the conservation of the subdomain organization of each shaft, suggesting that this organization is functionally relevant to both wall assembly and sexual recognition.
Between-Species Comparisons: Evolutionary Dynamics of the HRGP Shafts
Between-species alignments of short-repeat sequences are reliable only if carried out at the nucleotide level and it proved necessary to develop novel strategies to analyze the HRGP repeats wherein the most parsimonious alignment is identified by the highest Ser codon matches (see “Materials and Methods”). The resultant alignments reveal complex histories for each of the four pairs of C. reinhardtii/C. incerta shaft domains.
Gp1
Gp1, a chimeric protein with a C-terminal head, resides in the outermost (W6B) layer of the C. reinhardtii wall and forms a regular hexagonal weave in conjunction with an underlying crystalline lattice formed by the Gp2 and Gp3 proteins (Goodenough and Heuser, 1985, 1988a). The shaft of Gp1 from C. reinhardtii displays three subdomains (Ferris et al., 2001): Most of the sequence is in PPSPX/PPX motifs, but these are interrupted by a kink region containing a long block of contiguous Pro (poly-Pro), and by a neck carrying a PS repeat. This subdomain organization, readily detected in the longitudinal face diagrams of Gp1 (Fig. 1A), is conserved between C. reinhardtii and C. incerta.
In contrast to the conservation of subdomain structure, alignment of the Gp1 shaft-encoding sequences from C. reinhardtii and C. incerta (supported by 67/75 Ser codon matches) reveals numerous codon substitutions, indels, and two extensive endoduplication events (Fig. 2), as detailed below.
Figure 2.
GP1 shaft sequences from C. reinhardtii and C. incerta showing nucleotide-based alignment. Ser residues are shaded dark gray for UCX codons and light gray for AGX codons. Non-Ser amino acids are shaded light gray when they share two nucleotides with the aligned Ser codon. Conserved nucleotide positions are marked with asterisks. Two predicted endoduplications are highlighted in black, with nucleotide identities shown above duplicates. Tajima-Nei distance: 0.203 ± 0.018. Sequence numbering on the alignments starts at nucleotide no. 118 of both subdomains; complete coding sequences are deposited at GenBank.
Of the 16 indels in the Gp1 alignment, 14 add 54 residues to the C. incerta shaft and two add four residues to the C. reinhardtii shaft. As a consequence, the C. incerta shaft is predicted to be longer (386 residues or 115 nm) than the C. reinhardtii shaft (336 residues or 100 nm; Fig. 1A). In C. reinhardtii, 9/15 (60%) of the PPX units have been created by indels that truncate PPSPX units; in C. incerta, 4/10 (40%) of the PPX units have been created by such indels. The three-residue PPX unit has the effect of bringing the longitudinal faces back into frame after one iteration because the addition of a PPX unit between PPSPX units creates PPPPXS longitudinal modules on two faces from otherwise PPPXS modules as noted earlier. Therefore, the PPSPX/PPX helical repeat structure, and the (Pro)3 to 4 XS longitudinal faces, are largely maintained despite numerous indels/substitutions.
Yet another measure of repeat-sequence conservation pertains to conservation of Pro and Ser positions (indel events are not included in the following calculations): Of the 209 Pro residues in the C. incerta Gp1 sequence, 123 (59%) are specified by the same codon in the C. reinhardtii sequence and 83 (49%) by a synonymous Pro codon; only three (1%) are changed to a different amino acid. Of the 70 Ser residues in the C. incerta sequence, 51 (73%) are specified by the same codon in C. reinhardtii, 14 (20%) by a synonymous Ser codon, and 5 (7%) are changed. By contrast, of the 59 X positions, only 20 are identical, 16 are synonymous, and 23 (40%) are changed to a different amino acid.
Despite this large X variation, there is a bias in the amino acids found in the X positions: Of the 112 X amino acids in the two shafts (excluding kink and neck and including indels), 66 are Ala, 15 Pro, 13 Val, nine Ser, three Glu, two Thr, and one each of Gly, Ile, Lys, and Leu. That is, the X positions are not drifting freely: They are restricted in composition. Taken together, what seems to be of primary importance to the Gp1 shaft sequence is the maintenance of particular (hydroxy) Pro residues relative to Ser and a subset of spacer (X) residues.
Vsp3
The Vsp3 protein of C. reinhardtii (Woessner and Goodenough, 1992; Woessner et al., 1994) is assumed to be a component of the W2 layer of the cell wall (Goodenough and Heuser, 1985) and carries a single globular head at the N terminus. Its shaft displays two subdomains (Fig. 1B): The dominant (PS)n repeat gives way at the nonhead C terminus to a conserved tract of 36 amino acids carrying blocks of three to four contiguous Pro residues separated by tracts of three to five amino acids that lack Ser residues (hereafter called P3X3 modules). Similar P3X3 tracts are also found at the nonhead N termini of two other cell wall proteins, Vsp1 (Waffenschmidt et al., 1993) and Zsp1 (Woessner and Goodenough, 1989), suggesting that such motifs may function as interaction domains. Shaft termini of chimeric HRGPs are observed to interact with partner proteins in forming both the fishbone units in the W2 cell wall and the hammock mastigonemes on the flagellum (Goodenough and Heuser, 1985); hence, this may be a common mode of self-assembly.
The longitudinal face patterns and the two distinctive subdomains of the Vsp3 shafts are conserved between C. reinhardtii and C. incerta (Fig. 1B) despite the rearrangements illustrated in the Figure 3A alignments (supported by 56/58 Ser codon matches). The subdomains containing the core (PS)x repeats are organized into nine units (Fig. 3B). Each unit shows size variations in its PS content, the shortest containing two PS repeats and the longest containing 33, with indels creating tracts of different lengths. Each (PS)x unit terminates in KX (where X is usually Ala; Fig. 3B). The KX motifs introduce charged amino acids and, as guest sequences, represent putative loci for PII helix interruptions (Fig. 1B, yellow). Given the variable length of (PS)x in each unit, the displays of KX on the two Vsp3 shafts are quite distinctive.
Figure 3.
A, VSP3 shaft sequences from C. reinhardtii and C. incerta showing nucleotide-based alignment. Shading and symbols as in Figure 2. Tajima-Nei distance: 0.209 ± 0.024. Sequence numbering on the alignments starts at nucleotide no. 793 of reinVSP3 and no. 781 of incVSP3; complete coding sequences are deposited at GenBank. B, Subdomain organization is delineated by KX (AAG/GXX codons; shaded) conserved between reinVSP3 and incVSP sequences, which define nine units (numbered 1–9) in (PS)x domain. Exceptions from the AAG/GXX rule are italicized. Length of (PS) repeats and the total number of (PS) are shown in parentheses at the end of each unit. Ct, C-terminal domain. C, Guide to the frame shifts occurring in the predicted endoduplication underlined and italicized in A.
There are two exceptional cases: (1) an endoduplication of 20 codons (highlight) is found in the C. incerta sequence; and (2) the sequence CCUUCU (which encodes PS) is endoduplicated eight times in the C. reinhardtii sequence (highlight). Underlined and italicized are sequences flanking this second event that have the interesting feature of going out of and then back into frame, as detailed in Figure 3C. Despite such events, the shafts of the two species preserve the basic (PS)x repeat structure and remain similar in predicted length (62 versus 60 nm).
Agglutinin Shafts: Common Features
A study of the plus (Sag1) and minus (Sad1) agglutinin shafts of C. reinhardtii has been published previously (Ferris et al., 2005). The two sequences are completely different from one another. Nonetheless, they display homologous subdomain organization: Subdomain 2A (tail hook) is without PPSPX repeats, carries numerous PPX units, is enriched in basic amino acids, and carries block interruptions (sequences not predicted to adopt PII conformations) at homologous positions; each central 2C subdomain is a stretch of uniquely reiterated PPSPX modules, flanked by nonreiterated PPSPX units in subdomains 2B and 2D, and the 2E (head loop) subdomain contains a mixture of PPSPX, PPX, and nonrepeating sequences. We have interpreted this topological conservation to indicate that (1) the plus and minus C. reinhardtii agglutinin genes derive from an ancient common ancestral gene and (2) the conserved subdomain organization of the shafts is relevant to their function (Ferris et al., 2005).
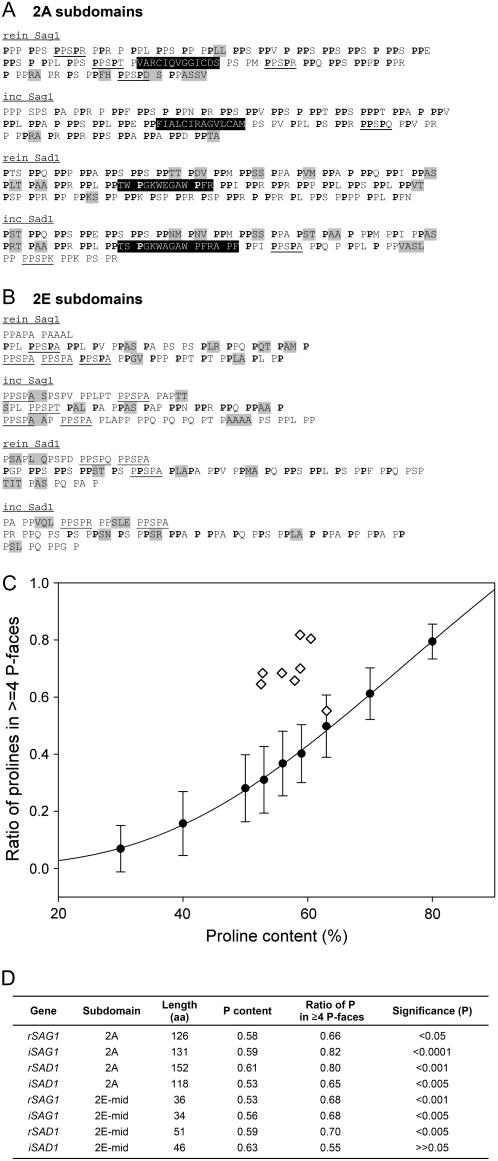
Here we report the sequences of the Sag1 (predicted length 323 nm) and Sad1 (289 nm) shafts of the sister species C. incerta. The proteins retain the same 2A to 2E topology as their C. reinhardtii counterparts (compare Fig. 4 with figure 7 in Ferris et al., 2005). Moreover, the 2C subdomains contain repeated iterations of a PPSPX sequence in both species: In C. incerta Sag1, this sequence reads (PPSPA, PPSPP, PPSPE)24; in C. incerta Sad1, it reads (PPSPA, PPSPE, PPSPT, PPSPQ, PPSPA, PALPT, PPSPV, PPSPA, PPSPE, PPSPF)7.
Figure 4.
Subdomain organization of the Sag1 (A) and Sad1 (B) agglutinin shafts from C. incerta. Repeating PPSPX motifs are in uppercase letters; PPX motifs are in uppercase in subdomains 2A and 2E. Nonrepeating residues and nonconforming positions in PPSPX units in 2B, 2D, and 2E are in lowercase. Repeats in 2C subdomains are aligned, with deviations from repeats in italics. Position judged to be missing is denoted with an asterisk (*). Block interruptions in subdomains 2A are indicated with angle brackets (< >). Format as in figure 7 of Ferris et al. (2005) for C. reinhardtii; alternative presentations of 2E subdomain sequences in Figure 5.
The four proteins share two additional common features. (1) Conservation of the PPSP tetrameric unit is observed throughout the 2B to 2D subdomains. Of the 551 PPSPX motifs in the four shafts, 90% preserve the PPSP sequence. In the 53 single-change variants, 3% are changed or deleted at the first Pro, 33% at the second Pro, 53% at the Ser, and 11% at the third Pro; of the 27 double-change variants, only one is changed or deleted at the first Pro, 89% at the second Pro, 93% at the Ser, and 15% at the third Pro. These gradients may indicate the relative importance of each position to proper hydroxylation/glycosylation/intermolecular recognition (Ferris et al., 2005), where it may be of significance that the highly conserved first and third Pro residues are adjacent to one other on the same longitudinal face (Fig. 1). (2) As noted earlier for Gp1, occupancy of the X position is again biased: Ala is by far the most common residue; Glu is abundant, followed by Gln (never used in Gp1), Val, and Thr; Pro is prominent in the two shafts with repeating PPSPP-containing units, but is not found elsewhere, and the remaining amino acids are of low abundance or absent altogether.
Agglutinin Shafts: Homology I and II Regions
When the between-species SAG1 and SAD1 sets are compared at the nucleotide level (Supplemental Fig. S1), plus-to-plus and minus-to-minus alignments are discernible, despite numerous endoduplications, indels, and substitutions, at the two shaft ends (supported by 7/14, 58/66, 6/6, and 30/36 Ser codon matches for Sag1 N and C termini and Sad1 N and C termini). Specifically, at the N-terminal end, alignment is evident from the start of 2A up to 18 amino acids before the start of 2B in both Sag1 sequences, and up to 65 (C. reinhardtii) or 31 (C. incerta) amino acids before the start of 2B in the two Sad1 sequences (Fig. 1, C and D, homology I). At the C-terminal end, virtually all of 2D and 2E are alignable in Sag1 comparisons; for Sad1, alignment picks up 186 (C. reinahrdtii) or 132 (C. incerta) amino acids into the 2D subdomain and continues to the termini (Fig. 1, C and D, homology II). By contrast, we have not found it possible to discern plus-to-plus and minus-to-minus alignments for portions of 2A and 2D (see above), 2B, or, by definition, the unique 2C repeats.
The aligned homology I and II regions offer the opportunity to identify features of the agglutinin shaft sequences that have been conserved not only since the recent C. reinhardtii/C. incerta divergence, but also since the ancient divergence of the SAG1 and SAD1 genes themselves. These features are likely to be important for posttranslational modification and/or for function. The analyses below focus on the 2A and 2E subdomains.
The 2A subdomains (Supplemental Fig. S1, A and C) are dominated by PPX motifs (Fig. 5A), where the X residue is often Arg or Lys. However, there is little conservation of X positions, even within plus or minus agglutinins, suggesting that selection is acting primarily on the retention of spaced PP dyads. Given that a PII sequence can be read as helical modules or as [n, n + 3] longitudinal face modules (Fig. 1), the PPX modules may be important for their helically displayed information and/or for generating the long (≥4 P) P faces that characterize the four 2A subdomains (Fig. 1, C and D, and as three-dimensional images in Fig. 1, E and G). These alternatives are considered more fully in the following section.
Figure 5.
Analysis of 2A and 2E shaft subdomains. A, 2A subdomains. PPX units are set off by spaces; light shading denotes guest sequences; dark shading denotes block interruptions; occasional PPSPX motifs are underlined. Boldfaced Pro residues participate in forming longitudinal face modules of ≥4 P. B, 2E subdomains, each displayed in three rows that correspond to within mating-type alignments (Supplemental Fig. S1). The first rows contain degenerate PPSPX residues; the third rows are also variable. The middle rows contain Pro residues (boldfaced) that participate forming longitudinal face modules of ≥4 P; these are considered in the simulation analysis. Other symbols as in Figure 5A. C and D, Simulation analysis. One hundred random protein domains, 100 amino acids in length, were generated with Pro content from 30% to 80%. Each sample was analyzed for its endowment of longitudinal faces containing ≥4 P; results plotted as circles in C. Diamonds in C show values for the actual sequences listed in D; P-value calculations (D) based on probability distribution of longitudinal Pro profiles in 100 randomly shuffled domains with the same Pro content.
Each 2A subdomain also carries a block-interruption sequence—a string of non-Pro or guest amino acids (highlighted in Fig. 5). The block interruptions are located in comparable positions in all four proteins and are of comparable length (13–15 amino acids). At the amino acid sequence level, the Sag1 and Sad1 block interruptions are completely different from one another. Within orthologous pairs, only five of the positions are conserved between the two Sag1 sequences, including two Cys positions, whereas 11 positions are conserved between the two Sad1 sequences. The 2A block interruption is posited to generate a bend in the distal end of the shaft (Ferris et al., 2005). It is not known whether it also functions in some aspect of adhesion and/or flagellar membrane association.
The boundaries between the 2E subdomains and their adjacent 2D subdomains are fuzzy and variable (Fig. 5B, first rows), after which the 2E subdomains resemble 2A in displaying numerous PPX modules and guest sequences (Fig. 5B) and in generating long (≥4 P) longitudinal P faces (Fig. 1, C and D).
Long P Faces in the 2A and 2E Subdomains of the Agglutinin Shafts
The four 2A subdomains and the four 2E subdomains of the agglutinin shafts share two common properties: (1) their sequences generate long (≥4 P) P faces (Fig. 1, E and G), and (2) they associate with the globular domains of the chimeric agglutinin protein (Ferris et al., 2005). This has prompted us to analyze the basis for generating long P faces given the possibility that such faces might play a role in shaft/globular-domain association.
Perfectly repeating helical PPX modules would by definition generate two longitudinal P faces. However, as illustrated in Figure 5, A and B, the PPX modules in the 2A and 2E subdomains are by no means perfectly repeated. Instead, the Pro residues that participate in generating longitudinal P faces (boldfaced) are recruited from diverse helical module contexts. Moreover, they persist despite the occurrence of numerous indels and substitutions within mating type, suggesting that the positions participating in longitudinal module generation may be under selection independently of any selection that may be operant to maintain helical modules.
The obvious objection to this proposal is that long P faces might simply be the random outcome of sequences having a high Pro content. To address this possibility, random sequences were generated with the same Pro content and length as the eight 2A and 2E subdomains. As detailed in Figure 5C and its legend, these proved to be significantly less likely to generate long P faces than seven of the actual sequences, the one exception being 2E Sad1 of C. incerta, which falls within the 95% expected line.
Nonhomologous Central Regions in the Agglutinin Shafts
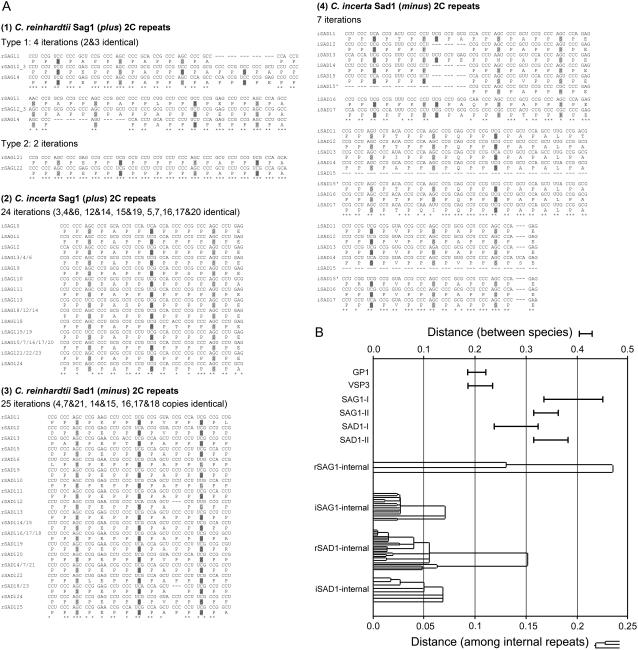
The homology I and II regions flank the middle portions of the shafts that we are unable to align, embedded in which are the repetitive 2C subdomains that are unique to each of the four proteins. Figure 6A presents a detailed analysis of each set of 2C internal repeats. Within each set, some repeats are found to be identical at the codon level, whereas others are increasingly divergent, allowing the construction of evolutionary trees for each subdomain. Evolutionary distances were estimated using the Tajima-Nei model (Tajima and Nei, 1984), which is applicable to sequences with unequal nucleotide composition.
Figure 6.
Evolutionary relationships of 2C internal endoduplications and of shaft sequences. A, Substitution patterns of aligned internal repeats in the four 2C subdomains of Sag1 and Sad1. Shading and symbols as in Figure 2. B, Tajima-Nei distances of alignable shaft sequences, where top scale is 2× the bottom scale. Top (I-bars), Between-species distances between the two cell wall sequences, Gp1 and Vsp3, and between Homology I and II regions of the Sag1 and Sad1 agglutinins. Bottom, Within-species endoduplicated repeats in the four 2C subdomains displayed as distance trees (neighbor-joining method).
These trees are plotted in Figure 6B, along with calculated between-species distances for the homology I and II regions and for the shafts of Gp1 and Vsp3. The between-species distances are greater for the two agglutinins than for the cell wall proteins, conforming to the well-documented tendency for sex-related genes to be more rapidly evolving than non-sex-related genes, as noted earlier. The within-species distances indicate that both sets of C. reinhardtii endoduplications initiated at a similar time, and that both sets of C. incerta endoduplications initiated at a similar, but more recent, time. These patterns are consistent with the possibility that the 2C endoduplications initiated at about the time that C. reinhardtii and C. incerta became genetically isolated.
A notable feature of the 2C subdomains is that the repeats generated in C. reinhardtii Sad1 (minus) and in C. incerta Sag1 (plus) both entail reiterations of the PPSPE PPSPA PPSPP motif, with the units being more degenerate in the older C. reinhardtii Sad1 sequence. Because we do not as yet know what role, if any, the 2C sequences play in sexual agglutination, we do not know whether this sequence convergence is biologically meaningful (e.g. whether it is necessary to adhesion that one shaft, from whichever mating type, carry a PPSPE PPSPA PPSPP repeat with its high density of negative charge) or whether it has occurred fortuitously.
DISCUSSION
Comparing the Evolution of Cell Wall and Sexual Agglutinin HRGP Shafts
Several mate recognition genes have been found to be endowed with repetitive modules (Gao and Garbers, 1998; Galindo et al., 2003; Kamei and Glabe, 2003). These have been posited to both encode multiple recognition sites and to generate variant sequences, via misalignment, that provide fodder for speciation events. The best-studied system in the California abalone involves the repetitive egg protein VERL and its partner sperm protein lysin. Species-specific dyads are apparently formed by a coevolutionary process wherein unequal crossing over between the long (153 amino acids) repeat units in VERL is accompanied by gene conversion to homogenize newly generated repeats, whereas positive selection for amino acid changes in lysin creates domains that bind to the novel VERL motifs (Swanson and Vacquier, 1998; Swanson et al., 2001a, 2001b; Galindo et al., 2003; Clark et al., 2006).
The HRGP shafts are also repetitive, and we document that they have been subject to major misalignment events since C. reinhardtii and C. incerta last shared a common ancestor <10 million years ago. Nevertheless, and unlike VERL, the repeated motifs that characterize particular shaft subdomains are strongly conserved in the surviving genes. We interpret such constraints to support the concept that the short (two- to six-residue) repeat units in these proteins generate glycomodules (Shpak et al., 1999) that are recognized by hydroxylation/glycosylation enzymes and that selection preserves these modules to guarantee production of a properly glycosylated shaft.
We have compared sex-related (agglutinin) and sex-unrelated (cell wall) shafts and find that, in several respects, they display similar evolutionary profiles: (1) Indels and nucleotide changes are tolerated only insofar as the overall pattern of repetitive motifs is not disrupted; (2) high rates of identical codons or synonymous substitutions are observed in Pro and Ser positions, suggesting that the placement of these amino acids is critical; and (3) only certain amino acids occupy the X positions of PPX and PPSPX units, presumably because these most comfortably accommodate the formation of the PII helix and the hydroxylation/glycosylation process (see also Ferris et al., 2005; “Discussion”).
A striking difference between the evolution of cell wall shafts and agglutinin shafts is that, whereas the cell wall sequences can be aligned without difficulty despite numerous codon changes, it is not possible to align approximately 50% of either of the plus-to-plus or the minus-to-minus agglutinin sequences. Not only do the agglutinins display unique central 2C endoduplications (see below), the sequences flanking one or both ends of these endoduplicated subdomains are also unique to each shaft even as they retain the signature motifs of their subdomains, suggesting that the events that generated the central 2C diversification have extended into, or originated from, the flanking regions. By contrast, the sequences at the proximal and distal ends of the shafts, while highly divergent, retain alignability.
Each 2C subdomain reiterates a particular sequence of repeat motifs, reminiscent of the 28 longer reiterations in VERL. Unlike the VERL reiterations, however, where a given module from one species may differ at a few amino acid positions from a second species, the plus 2C sequences from the two Chlamydomonas species are entirely different from one another (four reiterations of PPSPAPPA/LPPSPEPPSPAPPSPEPPSPAPPSPAPPSPAPPSPA versus 22 reiterations of PPSPEPPSPAPPSPP) and the minus 2C sequences are entirely different from one another (seven reiterations of PPSPAPPSPEPPSPTPPSPQPPSPAPALPTPPSPVPPSPAPPSPEPPSPF versus 24 reiterations of PPSPEPPSPAPPSPP).
Analysis of genetic distance among the repeats of the 2C subdomains, detailed in Figure 6, indicates that the 2C repeats diverged as endoduplication events, but their mode of origination is not easily explained. Not only is it the case that each 2C subdomain is a unique sequence, but also it is the case that its forerunner is not evident in the sequence of the other species. For example, if one posits that the ancestral Sad1 gene common to C. reinhardtii and C. incerta had a 2C sequence similar to the modern C. reinhardtii sequence, then the data indicate that, during C. incerta evolution, the 2C sequence was first eliminated entirely and then replaced by a second sequence that went on to undergo endoreduplication. The origin (as opposed to the propagation and diversification) of protein repeat motifs is itself obscure (Andrade et al., 2001). Nonetheless, the fact that such events took place in two independent genes in both mating types suggests that the process generating 2C divergence may play some role in generating the species specificity of gametic adhesion in this lineage.
Helical Modules versus Longitudinal Face Modules
The fact that the repeated modules of HRGP shafts generate information that guides posttranslational modification is widely accepted and supported by experimental studies (Shpak et al., 1999; Zhao et al., 2002; Tan et al., 2003; Estévez et al., 2006). Left unaddressed is whether the repeated modules are recognized in their PII helical context (helical modules) and/or whether information resides in the faces generated by the PII helix (longitudinal face modules; compare with Fig. 1, E and G). The γ-carbon-to-γ-carbon (on which hydroxylation occurs) spacing of Pro amino acid residues is calculated to be 0.643 nm along the helix and 0.934 nm along a face. Given that transcription factors recognize four to six nucleotide pairs on a DNA double helix (approximately 1–2 nm), it is plausible that modification enzymes could recognize three amino acid residues on a longitudinal face (approximately 2 nm). Indeed, sequence recognition scenarios are more restricted for helical faces than for longitudinal faces in that only two contiguous helical residues could be readily recognized at one time, the third residing on the opposite side of the helix.
Comparisons between closely related species can help guide this question because one can ask whether particular longitudinal module patterns are conserved between species even as helical modules diversify. Our analysis of the eight 2A and 2E subdomains of the agglutinin shafts indicates that maintenance of ≥4 P longitudinal face modules is under selection (Fig. 5): Pro residues at n and n + 3 positions generate long P faces despite many indels and substitutions that affect the sequence of helical modules. The 2E subdomain of C. reinhardtii Sag1 shafts has three distinctive structural features (Ferris et al., 2005): It is distinctly thinner in diameter than the rest of the shaft, suggesting a different endowment of sugar residues, it curves back on itself to form a loop, and it inserts into the globular head domain in the fashion of a lollipop stick. Similarly, the 2A subdomain interacts with a globular domain at the N terminus. Therefore, the longitudinal P blocks may carry distinctive hydroxylation/glycosylation information and/or may contribute to distinctive protein-protein associations.
Such proposals, it should be emphasized, in no way rule out roles for helical modules in HRGP biology. Indeed, we find most attractive the hypothesis that both modes of information will prove to be operant, either singly or collectively, in particular instances of HRGP hydroxylation/glycosylation, self-assembly, and interaction with other proteins.
Misalignment and Shaft Length Variation
Misalignment of repetitive HRGPs has generated shafts of varying lengths. Thus, the plus agglutinin shafts of C. reinhardtii and C. incerta are predicted to be 275 versus 323 nm, their minus shafts 258 versus 289 nm, and their Gp1 shafts 100 versus 115 nm. By contrast, their Vsp3 shafts are the same lengths (62 versus 60 nm) despite the occurrence of 12 indels adding/subtracting 42 amino acids to the approximately 205 amino acid sequences, suggesting that there may, in this case, be selection for length maintenance. The Vsp3 globular domains are far more strongly conserved in sequence than the heads of Gp1 and the agglutinins (J.-H. Lee, S. Waffenschmidt, and U.W. Goodenough, unpublished data) and, as noted below, share sequence homology with a head domain from Volvox carteri, suggesting a more stringent system for Vsp3 overall. In cell wall assemblies, shaft length variation would be expected to produce matrices with varying porosity and fiber density, and these would presumably be selectable traits.
Domain Swapping
The Vsp3 protein has had an interesting evolutionary history (Woessner et al., 1994): Its SPSPSPKA shaft repeat motif is found as well in a cell wall protein (WP6) from Chlamydomonas eugametos that has a very different head, and its head domain is 32% identical/19% strongly similar (our calculations) to the head of a cell wall protein (ISG) from V. carteri with a very different shaft [an irregular series of Ser-(Pro)3-7 units with many X residues]. The two head domains are also similar in predicted secondary structure, have Cys residues in identical positions, and share an intron position. C. reinhardtii and V. carteri are estimated to have last shared a common ancestor approximately 60 million years ago, whereas C. reinhardtii and C. eugametos diverged hundreds of millions of years ago (Pröschold et al., 2001). These observations suggest that, at some point during the ancient C. eugametos/C. reinhardtii radiation, the SPSPSPKA coding sequence came to associate with two different head domains and that, at some point during the C. reinhardtii/V. carterii radiation, a Vsp3-like head sequence came to associate with two different shaft domains.
Evidence for domain swapping has been reported as well for other HRGPs. (1) The VMP family of cell wall metalloproteinases in V. carteri (Hallmann et al., 2001) includes four genes, all induced by pheromone or wounding, that encode chimeric HRGPs with strongly similar globular head/enzyme domains, but shafts of different lengths and different P motifs. (2) In the large pherophorin family in Chlamydomonas and Volvox (Hallmann, 2006), two conserved globular domains are flanked by Pro-rich segments that are highly divergent in length, although similar in carrying homogeneous SP4 to SP7 sequences. (3) Baumberger et al. (2003) find evidence for domain swapping between members of the chimeric LRX extensin family during the common ancestry of Arabidopsis and rice (Oryza sativa).
Domain swapping is, of course, an important evolutionary dynamic, in general, but long Pro-rich repeats may well facilitate this process by enabling intra- and interchromosomal exchange. If chimeric HRGPs prove to be prone to such events, this would allow the generation of novel cell wall ideas that would promote matrix diversification (Baumberger et al., 2003).
Evolutionary Perspectives on HRGPs
This study represents, to our knowledge, the first comparison of HRGP shaft sequences between closely related species. We have shown that the divergence between C. reinhardtii and C. incerta can be explained by the occurrence of misalignments and, in the case of agglutinins, by a position-specific repeat generation mechanism that can replace its antecedent. These events occur in the context of purifying selection for particular modules, some of which may be recognized by their occurrence on the longitudinal faces of PII helices, and overall amino acid composition. Preserved misalignment events are more radical for the agglutinins than for the cell wall proteins, but the conservation of overall motifs is similarly stringent. Additional diversity may be generated by the occasional occurrence of domain swapping between heads and shafts.
Green algae assemble numerous kinds of cell walls (Hallmann, 2003), a trait that doubtless features in their spectacular radiation (Pickett-Heaps, 1975; Pröschold et al., 2001; Lewis and McCourt, 2004). Higher plant genomes carry numerous genes encoding HRGPs and a nonchimeric HRGP in Arabidopsis has been shown to play a specific role in establishing cell division planes in the embryo (Hall and Cannon, 2002), and AGPs have been shown to induce xylem differentiation in Zinnia (Motose et al., 2004) and to be required for female gametogenesis in Arabidopsis (Acosta-Garcia and Vielle-Calzada, 2004). Many of the plant wall genes have tissue-specific patterns of expression and are up-regulated by pathogenesis (Cassab, 1998; Baumberger et al., 2003; Estévez et al., 2006). Our studies indicate that plants have inherited from the algae a set of gene families that have the capacity to generate enormous matrix diversity. Because chimeric HRGPs are also abundant in sexual tissues of higher plants (Majewsk-Sawka and Nothnagel, 2000; Wu et al., 2001; Baumberger et al., 2003), these properties may also feature in higher plant speciation.
MATERIALS AND METHODS
Identification and Sequencing of Chlamydomonas incerta Genes
Orthologous Chlamydomonas incerta genes were identified by heterologous hybridization screening of the genomic library of C. incerta (CC-1870) generated as in Ferris et al. (1997). Probes were generated either from genomic or cDNA clones of Chlamydomonas reinhardtii genes. Hybridization screening and phage DNA purification were essentially as in Sambrook and Russell (2001). Genomic fragments were cloned and sequenced as in Ferris et al. (2005). cDNA sequences of C. incerta orthologs were predicted by comparing exon-intron structure of C. reinhardtii genes.
Nucleotide-Based Sequence Alignments
Nucleotide sequences for SAG1 and SAD1 pairs of agglutinin shaft domains from C. reinhardtii and C. incerta were initially aligned using ClustalW, version 1.7 (Thompson et al., 1994; located at http://npsa-pbil.ibcp.fr). However, it was difficult to assess the accuracy of these alignments given the numerous Pro positions and repetitive motifs. We therefore exploited the fact that Ser residues are specified by either UCX or AGX codons (shaded as indicated in figure legends). Because transition between UCX and AGX requires at least two nucleotide substitutions, ambiguous Clustal alignments were manually revised to maximize the frequency at which the same type of Ser codons was aligned. When a Ser failed to align with a Ser, the alignment was deemed acceptable if the corresponding codon differed from a Ser codon by a single nucleotide (shaded as indicated in figure legends). When indels were predicted, we looked for evidence of endoduplications, which presumably occurred following species isolation, meaning that their nucleotide divergence should be less than the average nucleotide divergence; duplications whose codons were >75% identical were shaded as indicated in figure legends, with nucleotide identity values above the shading. Underlined and italicized are endoduplicated sequences that are shifted in frame.
Nucleotide-Based Distance Calculation and Distance Tree Construction
Tajima-Nei distances and ses were calculated by MEGA 3.1 software (Kumar et al., 2004). Tajima-Nei distances were also used to construct distance trees of 2C iterated segments by the neighbor-joining method using MEGA 3.1 software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers. GP1 from C. reinhardtii and C. incerta: AF309494 and EF057410, VSP3 from C. reinhardtii and C. incerta: L29029 and AY795084, SAG1 from C. reinhardtii and C. incerta: AY450930 and AY937239, SAD1 from C. reinhardtii and C. incerta: AY450929 and AY858986.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure S1. Nucleotide-based alignment of shaft regions in SAG1 and SAD1 sexual agglutinin genes.
Supplementary Material
Acknowledgments
We thank Dr. Patrick Ferris for his important intellectual and technical contributions to this project.
This work was supported by the National Institutes of Health (grant no. GM–26150), the National Science Foundation (grant no. MCB 0326829), and the Deutsche Forschungsgemeinschaft (grant no. Wa 659/8–1 to S.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ursula Goodenough (ursula@biology.wustl.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Acosta-Garcia G, Vielle-Calzada J (2004) A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair WS, Hwang C, Goodenough UW (1983) Identification and visualization of the sexual agglutinin from mating-type plus flagellar membranes of Chlamydomonas. Cell 33 183–193 [DOI] [PubMed] [Google Scholar]
- Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134 117–131 [DOI] [PubMed] [Google Scholar]
- Annunen P, Autio-Harmainen H, Kivirikko KI (1998) The novel type II prolyl 4-hydroxylase is the main enzyme form in chondrocytes and capillary endothelial cells, whereas the type I enzyme predominates in most cells. J Biol Chem 273 5989–5992 [DOI] [PubMed] [Google Scholar]
- Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko KI (1997) Cloning of the human prolyl 4-hydroxylase α subunit isoform α (II) and characterization of the type II enzyme tetramer. J Biol Chem 272 17342–17348 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, Smmons MP, Bedinger P, Goff SA, Ringli C, et al (2003) Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice: a conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol 131 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Robbins MP, Dixon RA (1985) Elicitor-induced prolyl hydroxylase from French bean (Phaseolus vulgaris). Biochem J 229 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab GI (1998) Plant cell wall proteins. Ann Rev Plant Physiol Plant Mol Biol 49 281–309 [DOI] [PubMed] [Google Scholar]
- Civetta A, Singh RS (1998) Sex-related genes, directional sexual selection, and speciation. Mol Biol Evol 15 901–909 [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ (2006) Evolution of reproductive proteins from animals and plants. Reproduction 131 11–22 [DOI] [PubMed] [Google Scholar]
- Coleman AW, Mai JC (1997) Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J Mol Evol 45 168–177 [DOI] [PubMed] [Google Scholar]
- Colley KJ (1997) Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 7 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer TP (1998) Left-handed polyproline II helix formation is (very) locally driven. Proteins 33 218–226 [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64 439–451 [DOI] [PubMed] [Google Scholar]
- Elder JF, Turner BJ (1995) Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol 70 297–320 [DOI] [PubMed] [Google Scholar]
- Estévez JM, Kieliszewski MJ, Khitrov N, Somerville C (2006) Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol 142 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Pavlovic C, Fabry S, Goodenough UW (1997) Rapid evolution of sex-related genes in Chlamydomonas. Proc Natl Acad Sci USA 94 8634–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Waffenschmidt S, Umen JG, Lin H, Lee JH, Ishida K, Kubo T, Lau J, Goodenough UW (2005) Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17 597–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Drees J, Goodenough UW (2001) Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40 2978–298711258910 [Google Scholar]
- Galindo BE, Vacquier VD, Swanson WJ (2003) Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci USA 100 4639–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Garbers DL (1998) Species diversity in the structure of zonadhesin, a sperm-specific membrane protein containing multiple cell adhesion molecule-like domains. J Biol Chem 273 3415–3421 [DOI] [PubMed] [Google Scholar]
- Goodenough U, Lin H, Lee J-H (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol (in press) [DOI] [PubMed]
- Goodenough UW (1991) Chlamydomonas mating interactions. In M Dworkin, ed, Microbial Cell-Cell Interactions. American Society for Microbiology, Washington, DC, pp 71–112
- Goodenough UW, Adair WS, Collin-Osdoby P, Heuser JE (1985) Structure of Chlamydomonas agglutinin and related flagellar surface proteins in situ and in vitro. J Cell Biol 101 924–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Armbrust EV, Campbell AM, Ferris PJ (1995) Molecular genetics of sexuality in Chlamydomonas. Plant Mol Biol 46 21–44 [Google Scholar]
- Goodenough UW, Gebhart B, Mecham R, Heuser JE (1986) Crystals of the Chlamydomonas reinhardtii cell wall: polymerization, depolymerization, and purification of glycoprotein monomers. J Cell Biol 103 408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE (1985) The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze deep-etch technique. J Cell Biol 101 1550–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE (1988. a) Molecular organization of cell-wall crystals from Chlamydomonas reinhardtii and Volvox carteri. J Cell Sci 90 717–733 [Google Scholar]
- Goodenough UW, Heuser JE (1988. b) Molecular organization of the cell wall and cell-wall crystals from Chlamydomonas eugametos. J Cell Sci 90 735–750 [Google Scholar]
- Goodenough UW, Heuser JE (1999) Deep-etch analysis of adhesion complexes between gametic flagellar membranes of Chlamydomonas reinhardtii (Chlorophyceae). J Phycol 35 756–767 [Google Scholar]
- Hall Q, Cannon MC (2002) The cell-wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A (2003) Extracellular matrix and sex-inducing pheromone in Volvox. Int Rev Cytol 227 131–182 [DOI] [PubMed] [Google Scholar]
- Hallmann A (2006) The pherophorins: common, versatile building blocks in the evolution of extracellular matrix architecture in Volvocales. Plant J 45 292–307 [DOI] [PubMed] [Google Scholar]
- Hallmann A, Amon P, Godl K, Heitzer M, Sumper M (2001) Transcriptional activation by the sexual pheromone and wounding: a new gene family from Volvox encoding the modular proteins with (hydroxy)proline-rich and metalloproteinase homology domains. Plant J 26 583–593 [DOI] [PubMed] [Google Scholar]
- Harwood R, Grant ME, Jackson DS (1974) Collagen biosynthesis: characterization of subcellular fractions from embyonic chick fibroblasts and the intracellular localization of protocollagen prolyl and protocollagen lysyl hydroxylases. Biochem J 144 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieta R, Myllyharju J (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana: effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J Biol Chem 277 23965–23971 [DOI] [PubMed] [Google Scholar]
- Kamei N, Glabe CG (2003) The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev 17 2502–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskiaho K, Hieta R, Sormunen R, Myllyharju J (2007) Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA (1994) Extensin: repetitive motifs, functional sites, posttranslational codes and phylogeny. Plant J 5 157–172 [DOI] [PubMed] [Google Scholar]
- Kivirikko KI, Pihlajaniemi T (1998) Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol 72 325–398 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- Lai Y, Sun F (2003) The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol Biol Evol 20 2123–2131 [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4 203–221 [DOI] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM (2004) Green algae and the origin of land plants. Am J Bot 91 1535–1556 [DOI] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11 2453–2465 [DOI] [PubMed] [Google Scholar]
- Liss M, Kirk DL, Beyser K, Fabry S (1997) Intron sequences provide a tool for high-resolution phylogenetic analysis of volvocine algae. Curr Genet 31 214–227 [DOI] [PubMed] [Google Scholar]
- Majewsk-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429 873–878 [DOI] [PubMed] [Google Scholar]
- Myllyharju J (2003) Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol 22 15–24 [DOI] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI (2004) Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 20 33–43 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD (1975) Green Algae: Structure, Reproduction and Evolution in Selected Genera. Sinauer, Sunderland, MA
- Pihlajaniemi T, Myllyla R, Kivirikko KI (1991) Prolyl 4-hydroxylase and its role in collagen synthesis. J Hepatol 13 S2–7 [DOI] [PubMed] [Google Scholar]
- Popescu CE, Borza T, Bielawski JP, Lee RW (2006) Evolutionary rates and expression level in Chlamydomonas. Genetics 172 1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Marin B, Schlosser UG, Melkonian M (2001) Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Bobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152 265–300 [DOI] [PubMed] [Google Scholar]
- Roberts K, Grief C, Hills FJ, Shaw PJ (1985) Cell wall glycoproteins: structure and function. J Cell Sci Suppl 2 105–127 [DOI] [PubMed] [Google Scholar]
- Rucker AL, Pager CT, Campbell MN, Qualls JE, Creamer TP (2003) Host-guest scale of left-handed polyproline II helix formation. Proteins 53 68–75 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Showalter AM (2001) Introduction: plant cell wall proteins. Cell Mol Life Sci 58 1361–1362 [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation sites. Proc Natl Acad Sci USA 96 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP (1976) Evolution of repeated DNA sequences by unequal crossover. Science 191 528–535 [DOI] [PubMed] [Google Scholar]
- Stapley BJ, Creamer TP (1999) A survey of left-handed polyproline II helices. Protein Sci 8 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki L, Woessner JP, Uchida H, Kuroiwa H, Yuasa Y, Waffenschmidt S, Goodenough U, Kuroiwa T (2000) A zygote-specific protein with hydroxyproline-rich glycoprotein domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii (Chlorophyta). J Phycol 36 571–583 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Aquadro CF, Vacquier VD (2001. a) Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol Biol Evol 18 376–383 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD (1998) Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281 710–712 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD (2002) Rapid evolution of reproductive proteins. Nat Rev Genet 3 137–144 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Yang Z, Wolfner MF, Aquadro CF (2001. b) Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci USA 98 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, Nei M (1984) Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 1 269–285 [DOI] [PubMed] [Google Scholar]
- Tan L, Leykam JF, Kieliszewski MJ (2003) Glycosylation motifs that direct arabinogalactan addition to arabinogalactan proteins. Plant Physiol 132 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen P, Myllyharju J, Koivunen P (2005) Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J Biol Chem 280 1142–1148 [DOI] [PubMed] [Google Scholar]
- Van den Steen P, Rudd PM, Dwek RA, Opdenakker G (1998) Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol 33 151–208 [DOI] [PubMed] [Google Scholar]
- van Holst GJ, Varner JE (1984) Reinforced polyproline II conformation in a hydroxyproline-rich cell wall glycoprotein from carrot root. Plant Physiol 74 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt S, Woessner JP, Beer K, Goodenough UW (1993) Evidence that isodityrosine crosslinking mediates the insolubilization of cell-wall HRGPs in Chlamydomonas. Plant Cell 5 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW (1989) Molecular characterization of a zygote wall protein: an extensin-like molecule in Chlamydomonas reinhardtii. Plant Cell 1 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW (1992) Zygote and vegetative cell wall proteins in Chlamydomonas reinhardtii share a common epitope, (Ser Pro)x. Plant Sci 83 65–76 [Google Scholar]
- Woessner JP, Molendijk AJ, van Egmond P, Klis FM, Goodenough UW, Haring MA (1994) Domain conservation in several volvocalean cell wall proteins. Plant Mol Biol 26 947–960 [DOI] [PubMed] [Google Scholar]
- Wu H, de Graaf B, Mariani C, Cheung AY (2001) Hydroxyproline-rich glycoproteins in plant reproductive tissues: structure, functions and regulation. Cell Mol Life Sci 58 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J 41 81–94 [DOI] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DTA, Kieliszewski MJ (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31 431–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.