Abstract
The crystal structure of Pterocarpus angolensis seed lectin is presented in complex with a series of high mannose (Man) oligosaccharides ranging from Man-5 to Man-9. Despite that several of the nine Man residues of Man-9 have the potential to bind in the monosaccharide-binding site, all oligomannoses are bound in the same unique way, employing the tetrasaccharide sequence Manα(1–2)Manα(1–6)[Manα(1–3)]Manα(1–. Isothermal titration calorimetry titration experiments using Man-5, Man-9, and the Man-9-containing glycoprotein soybean (Glycine max) agglutinin as ligands confirm the monovalence of Man-9 and show a 4-times higher affinity for Man-9 when it is presented to P. angolensis seed lectin in a glycoprotein context.
For already a long time, plants are known to express lectins in relatively large amounts in their storage organs (seeds, rhizomes) and in lower concentrations in their vegetative parts. The seeds from legume plants have traditionally been excellent sources for lectins of a variety of specificities. For several decades, the legume lectin family has served as the model system of choice for the study of protein-carbohydrate recognition (Sharon and Lis, 1990; Loris et al., 1998). Many principles have been discovered first for legume lectin family members and were later confirmed for other families of carbohydrate-recognizing proteins (Loris, 2002). The legume lectin family covers the widest possible range of carbohydrate specificities among all known lectin families. Variations of the lengths, sequences, and conformations of five loops that constitute the carbohydrate-binding site determine mono- and oligosaccharide specificity in a complex way (Sharma and Surolia, 1997; Loris et al., 1998). Avidity and higher order specificity are generated via a variety of quaternary structures leading to the formation of homogeneous cross-linked lattices (Bhattacharyya et al., 1988; Sacchettini et al., 2001). The past decade has provided a wealth of structural and thermodynamic data that provide insight on how oligosaccharide specificity, quaternary structure, and metal binding cooperate to generate a variety of biological effects.
Despite the wealth of data obtained from x-ray crystallography, relatively few structures of lectins in complex with large oligosaccharides are available. Only 12 out of the more than 500 structures of lectins and carbohydrate-binding domains currently present in the Protein Data Bank (Berman et al., 2002) have a bound ligand corresponding to at least a pentasaccharide. These can be placed into three groups. The first one concerns lectins in complex with a fucosylated or nonfucosylated complex-type biantennary oligosaccharide or with the pentasaccharide GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man derived thereof. They include the legume lectins Lathyrus ochrus lectin, concanavalin A (con A) and Pterocarpus angolensis seed lectin (PAL; Bourne et al., 1994b; Moothoo and Naismith, 1998; Buts et al., 2006), bovine galectin 1 (Bourne et al., 1994a), and DC-SIGN, a C-type lectin (Feinberg et al., 2001). The second group concerns sialylated pentasaccharides recognized by cholera toxin (Merritt et al., 1994) and influenza virus hemagglutinin (Ha et al., 2003; Gamblin et al., 2004). When looking at the high Man family of glycans, the only crystal structure of a lectin in complex with a complete Man-9 entity is that of the anti-HIV protein cyanovirin (Botos et al., 2002). A number of other structures of complexes with pentasaccharides derived from Man-9 are also available for rat Man-binding protein (Weis et al., 1992), cyanovirin-N (Botos et al., 2002), snowdrop lectin (Wright and Hester, 1996), and artocarpin (Jeyaprakash et al., 2004).
Detailed thermodynamic data for carbohydrate binding are equally limited to mono-, di-, or trisaccharides in most cases (Dam and Brewer, 2002). Exceptions are the interactions of high Man oligosaccharides with con A (Mandal et al., 1994), cyanovirin-N (Shenoy et al., 2002), and the human antibody 2G12 (Wang et al., 2004), of the pentasaccharide GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man with PAL, con A, and other Diocleinae lectins (Mandal et al., 1994; Dam et al., 1998; Buts et al., 2006) and of bivalent pentasaccharides and sialofetuin with different galectins (Ahmad et al., 2004; Dam et al., 2005). Only for con A, PAL, and cyanovirin-N is at least one corresponding crystal structure available.
The Man-binding lectin from the seeds of P. angolensis (PAL) has been previously studied in detail by x-ray crystallography and a variety of biophysical techniques. This resulted in a clear picture of how this lectin recognizes mono-, di-, and trisaccharides (Loris et al., 2004) as well as complex-type oligosaccharides (Buts et al., 2006) and how its structure, stability, and activity respond to demetalization (Garcia-Pino et al., 2006). Here we report the crystal structures of PAL in complex with a series of high Man ligands ranging from Man-5 to Man-9. To our knowledge, not only is this the first time that the structure of a full-length high Man ligand is determined in complex with a plant protein, it is only the second complex for any lectin and the first time the whole range of naturally occurring high Mans are probed systematically.
RESULTS AND DISCUSSION
Overall Structure
The crystal structures of PAL in complex with six high Man-type glycans were determined at high resolution (Table I). The crystals contain a dimer of PAL in their asymmetric unit, the two subunits of which will be termed A and B. The overall structure of the PAL dimer has previously been described in detail (Loris et al., 2003, 2004) and remains unaltered in the complexes presented in this article. These two subunits are chemically identical and show only minor conformational differences that are irrelevant for carbohydrate binding. Nevertheless, they present different crystal packing environments to the carbohydrate-binding site.
Table I.
X-ray data collection and refinement statistics
| Man-5 | Man-6 | Man-7D1 | Man-7D3 | Man-8D1D3 | Man-9 | |
|---|---|---|---|---|---|---|
| Beamline | X11 | ID14-1 | X13 | X11 | X11 | X11 |
| Wavelength (Å) | 0.81 | 0.934 | 0.81 | 0.81 | 0.81 | 0.81 |
| Detector | MAR CCD | ADSC Q4 CCD | MAR CCD | MAR CCD | MAR CCD | MAR CCD |
| Unit cell | ||||||
| a (Å) | 56.83 | 56.74 | 56.80 | 56.63 | 56.70 | 56.69 |
| b (Å) | 83.57 | 83.33 | 83.27 | 83.47 | 83.35 | 83.21 |
| c (Å) | 122.81 | 122.81 | 123.13 | 122.35 | 122.69 | 122.92 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Resolution limits (Å) | 15.0−1.8 | 15.0−2.1 | 15.0−1.9 | 15.0−2.1 | 15.0−2.2 | 15.0−1.8 |
| No. of measured reflections | 218,624 | 151,954 | 270,422 | 144,738 | 147,111 | 310,693 |
| No. of unique reflections | 54,225 | 34,604 | 46,128 | 34,386 | 29,897 | 54,041 |
| Rmerge | 0.047 | 0.095 | 0.076 | 0.094 | 0.057 | 0.080 |
| Completeness (%) | 99.0 | 100.0 | 98.4 | 99.9 | 99.2 | 98.8 |
| 〈I/σ(I)〉 | 17.9 | 12.4 | 15.0 | 11.7 | 12.2 | 14.4 |
| Rcryst | 0.178 | 0.180 | 0.177 | 0.184 | 0.174 | 0.175 |
| Rfree | 0.207 | 0.216 | 0.197 | 0.218 | 0.209 | 0.200 |
| Ramachandran profile | ||||||
| Core (%) | 88.0 | 86.1 | 87.4 | 87.4 | 86.4 | 88.4 |
| Add. allowed (%) | 12.0 | 13.9 | 12.6 | 12.6 | 13.6 | 11.6 |
| Disallowed (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Protein Data Bank entry | 2PHX | 2PHF | 2PHR | 2PHT | 2PHU | 2PHW |
The carbohydrate-binding site of subunit A is involved in crystal packing and therefore limits the conformations that are accessible to the bound carbohydrate. After the desoaking step (see “Materials and Methods”), a molecule of Manα(1–3)Man remains bound (Loris et al., 2005), but in a number of cases this can still be substituted by a different carbohydrate ligand if the concentration of the replacing ligand is sufficiently high (Loris et al., 2003, 2004; Buts et al., 2006). The interactions and carbohydrate conformations observed in this binding site then have to be interpreted as situations that are possible in solution, but not necessarily reflecting the dominant one.
The carbohydrate-binding site of subunit B on the other hand faces a large solvent channel and can easily accommodate large oligosaccharide ligands without having to disturb either the crystal packing or the ligand conformation. Interactions and ligand conformations observed in this binding site are therefore assumed to reflect the dominant situation in solution. Further interpretation of the results presented in this article will be based upon the situation observed in subunit B unless otherwise stated.
Recognition Sequence for the Lectin
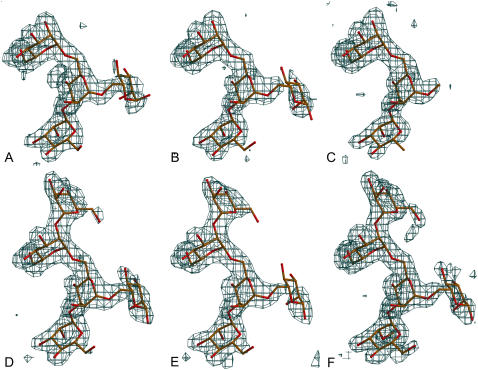
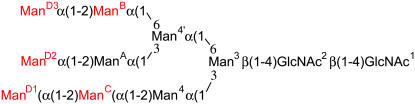
The chemical structures of the different high Man oligosaccharides used in this study and the fragments that could be built in the respective electron density maps are shown in Figure 1. The soaking experiments resulted in all cases in occupation of the carbohydrate-binding site of the B-subunit with four to five carbohydrate residues resulting from the high Man oligosaccharides. The carbohydrate sequence that is systematically observed is ManD3α(1–2)ManBα(1–6)[ManAα(1–3)]Man4′α(1–, corresponding essentially to the D3 arm and part of the D2 arm of the high Man chains (Fig. 2). ManB occupies the monosaccharide-binding site (nomenclature of individual monosaccharides according to Koles et al., 2004).
Figure 1.
Oligosaccharides used in this work. The Man residues that show clear electron density and interact with the lectin are highlighted in blue for each carbohydrate. The monosaccharide nomenclature used (Koles et al., 2004) is indicated for Man-9.
Figure 2.
Different electron density maps for all of the oligosaccharides used in this study as observed in the binding site of monomer B. All maps were calculated by removing the sugar residues from the final coordinates and applying one round of slow-cool refinement to remove potential bias of Man-9 and drawn at a level of 3 σ. The atomic model is superimposed. A, Man-5. B, Man-6. C, Man-7D1. D, Man-7D3. E, Man-8D1D3. F, Man-9. [See online article for color version of this figure.]
In all but one case (Man-7D3) the carbohydrate-binding site of subunit-A contains the disaccharide Manα(1–3)Man, which results from the crystallization (see “Materials and Methods”). In the Man-7D3 structure on the other hand, the sequence Manα(1–2)Manα(1– is observed in the binding site of subunit-A, indicating that again the carbohydrate binds with the D3 arm (the D1 and D2 arms both lacking a terminal α1–2 linked Man) despite that more extensive interactions are not possible due to crystal packing limitations.
Interactions between Carbohydrate and Protein
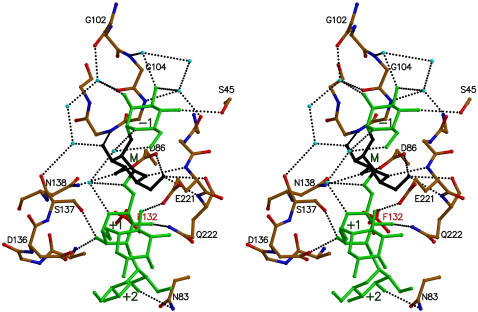
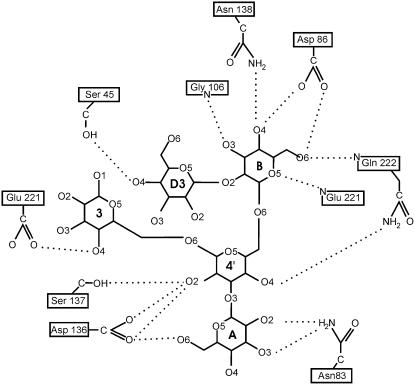
Manα(1–2)Manα(1–6)[Manα(1–3)]Manα(1– interacts with the protein via a network of van der Waals contacts and hydrogen bonds that is illustrated in Figures 3 and 4 and Table II. This interaction network builds on those observed earlier for the smaller constituents Manα(1–2)Man and Manα(1–3)[Manα(1–6)]Man. The monosaccharide-binding site dominates over all subsites in both the number of hydrogen bonds formed and the amount of surface buried upon complexation, a feature seen in the overall majority of protein:carbohydrate complexes.
Figure 3.
Details of interactions between Man-9 and PAL. Residues of the binding site of PAL are colored according to atom type and selected residues are labeled. The pentasaccharide moiety of Man-9 that is visible in the structure is shown in green, except for the Man residue in the monosaccharide-binding site that is shown in black. Water molecules are drawn as light blue spheres and hydrogen bonds are indicated by dotted lines.
Figure 4.
Two-dimensional schematic of the hydrogen bond network between Man-9 and PAL. Different Man residues are labeled according to the nomenclature of Koles et al. (2004).
Table II.
Protein:carbohydrate hydrogen bonds (Å)
nr, Not relevant as the corresponding sugar residue is not present in this particular polysaccharide; no, not observed in this complex.
| H-Bond | Man-5 | Man-6 | Man-7D1 | Man-7D3 | Man-8D1D3 | Man-9 | |
|---|---|---|---|---|---|---|---|
| ManD3 (site −1) | Ser-45(OG)Man(O4) | nr | nr | nr | 3.16 | 3.21 | 3.25 |
| ManB (site M) | Asp-86(OD1)Man(O4) | 2.67 | 2.59 | 2.54 | 2.56 | 2.52 | 2.57 |
| Asp-86(OD2)Man(O6) | 2.83 | 2.82 | 2.73 | 2.87 | 2.76 | 2.73 | |
| Gly-106(N)Man(O3) | 2.80 | 2.80 | 2.88 | 2.82 | 2.74 | 2.88 | |
| Asn-138(ND2)Man(O4) | 3.06 | 3.01 | 3.03 | 2.93 | 3.05 | 2.96 | |
| Glu-221(N)Man(O5) | 3.08 | 3.09 | 3.08 | 3.19 | 3.09 | 3.09 | |
| Gln-222(N)Man(O6) | 3.14 | 3.13 | 3.09 | 3.03 | 3.15 | 3.17 | |
| Man4′ (site +1) | Asp-136(OD1)Man(O2) | 2.98 | 2.87 | 2.63 | 2.76 | 2.44 | 2.86 |
| Asp-136(OD2)Man(O2) | 3.04 | 3.06 | no | no | no | 3.04 | |
| Ser-137(OG)Man(O2) | 2.70 | 2.68 | 2.76 | 2.67 | 2.83 | 2.71 | |
| Gln-222(NE2)Man(O4) | 2.49 | 2.52 | 2.57 | 2.46 | 2.49 | 2.51 | |
| ManA (site +2) | Asn-83(ND2)Man(O2) | 3.37 | 3.18 | 3.24 | 3.03 | 3.13 | no |
| Asn-83(ND2)Man(O3) | 3.13 | 3.07 | 3.00 | 3.09 | 3.07 | 3.29 | |
| Asp-136(OD1)Man(O6) | 2.98 | 3.47 | no | 3.41 | 2.65 | no | |
| Man3 | Glu-221(OE1)Man(O4) | 2.65 | 3.51 | no | no | 3.00 | 2.71 |
The contact surface between Man-9 and PAL is 130 Å2 (Table III), significantly lower than the 165 Å2 observed in the complex with the complex-type biantennary oligosaccharide NA2F and its pentasaccharide constituent GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man (Buts et al., 2006). This agrees with the lower affinity for Man-9 (see below). The major differences between these complexes are found in the −1 subsite where ManD3 has a smaller contact area with the protein than the corresponding GlcNAc in the NA2F complex and the absence of interactions between the protein and ManD2 (which is completely disordered in the Man-9 complex while the corresponding GlcNAc in the NA2F complex is well ordered and contributes to binding).
Table III.
Contact surfaces (listed for each sugar residue and each polysaccharide in Å2)
NA2F corresponds to the decasaccharide Galβ(1–4)GlcNAcβ(1–2)Manα(1–6)[Galβ(1–4)GlcNAcβ(1–2)Manα(1–3)]Manβ(1–4)GlcNAcβ(1–4)[Fucβ(1–6)]GlcNAc (Buts et al., 2006) and is included for comparison.
| Man-5 | Man-6 | Man-7d1 | Man-7d3 | Man-8d1d3 | Man-9 | Na2F | ||
|---|---|---|---|---|---|---|---|---|
| ManD3 (site −1) | All | – | – | – | 30.8 | 29.3 | 29.6 | 47.1 |
| Apolar | – | – | – | 13.9 | 12.2 | 12.7 | 36.8 | |
| Polar | – | – | – | 17 | 17.1 | 16.9 | 10.3 | |
| ManB (monosaccharide binding site) | All | 51.9 | 51.6 | 52 | 45.6 | 45.5 | 45.2 | 42.1 |
| Apolar | 21.2 | 21.2 | 21.6 | 20 | 19.9 | 19.6 | 17.8 | |
| Polar | 30.8 | 30.4 | 30.4 | 25.6 | 25.6 | 25.7 | 24.3 | |
| Man4′ (site +1) | All | 24.3 | 23.2 | 28 | 22.1 | 21.7 | 22.6 | 19.1 |
| Apolar | 9.5 | 8.9 | 13.1 | 7.3 | 6.8 | 7.3 | 6.3 | |
| 1 | Polar | 14.8 | 14.4 | 14.9 | 14.8 | 14.9 | 15.2 | 12.8 |
| ManA (site +2) | All | 24.2 | 24.3 | 24 | 24.7 | 24.2 | 22.6 | 18.2 |
| Apolar | 10 | 11.1 | 14.2 | 10.6 | 10.6 | 9.7 | 9.9 | |
| 2 | Polar | 14.2 | 13.3 | 9.9 | 14.1 | 13.6 | 13.6 | 8.3 |
| (Site +3) | All | – | – | – | – | – | – | 20.8 |
| (N-Acetylglucosamine) | Apolar | – | – | – | – | – | – | 13.3 |
| Polar | – | – | – | – | – | – | 7.4 | |
| (Site +4) | All | – | – | – | – | – | – | 4.4 |
| (Gal) | Apolar | – | – | – | – | – | – | 1.4 |
| 4 | Polar | – | – | – | – | – | – | 3 |
| Man3 | All | 6.9 | 4.6 | – | 4.5 | 5.1 | 5.2 | 13.2 |
| (N-Acetylglucosamine) | Apolar | 0.7 | 0.6 | – | 0.8 | 0.4 | 1.1 | 9.1 |
| Polar | 6.2 | 4 | – | 3.7 | 4.7 | 4 | 4 | |
| Total | All | 107.4 | 103.9 | 104.6 | 127.7 | 125.9 | 130.4 | 165.0 |
| Apolar | 41.5 | 41.0 | 49.2 | 52.5 | 49.9 | 55.7 | 94.8 | |
| Polar | 65.9 | 62.1 | 55.4 | 75.1 | 76.0 | 74.8 | 70.0 |
Alternative Binding Modes
A unique and identical binding mode is observed for all high Man oligosaccharides. This comes as a surprise as one would envisage several of the Man residues of Man-9 to be able to occupy the monosaccharide-binding site. Based upon the affinities for different mono-, di-, and trimannoses (Buts et al., 2006) one would expect a very poor selectivity resulting in sliding of the oligosaccharide over the binding surface of the lectin. To look for possible explanations for this apparent super-selectivity, we applied a modeling approach to identify possible factors hindering alternative binding modes.
All possible alternatives of Man residues that can be imagined to occupy the monosaccharide-binding site are summarized in Figure 5. The branched Mans Man3 and Man4′ are sterically excluded from the monosaccharide-binding site because of substitutions on O3 and O6 and in the case of Man3 also because of its β-linkage.
Figure 5.
Potential multivalency of Man-9. Each of the five Man residues that can theoretically bind in the monosaccharide-binding site of PAL are highlighted in red. In the crystal structures only ManB is observed to bind in the monosaccharide-binding site. Theoretically, each of the three arms could interact with a different lectin molecule via ManD1, ManD2, ManD3, and ManC. The other four Mans are excluded from the monosaccharide-binding site for steric reasons. Even with the binding mode observed via ManC as shown in Figures 1 to 3, the D3 arm remains free to interact with a second lectin molecule. Still, only a 1:1 stoichiometry is observed in solution and a unique binding mode in the crystal.
The terminal Mans ManD1, ManD2, and ManD3 as well as ManC on the other hand are not excluded from the monosaccharide-binding site in any trivial way. To get a more quantitative answer, we calculated the conformational energy maps for the disaccharide Manα(1–2)Man with the nonreducing Man in the monosaccharide-binding site. The optimal conformation in the lectin context is centered around Φ = 80°, Ψ = 160° (see “Materials and Methods” for torsion angle definitions), which also corresponds to the main low energy conformation in its isolated state, although the energy minimum seems to be more shallow than on the equivalent maps calculated for Manα(1–2)Man in the binding mode observed in the crystal structure. Thus we cannot provide for an explanation as to why PAL does not recognize Man-9 via ManD1, ManD2, ManD3, or ManC in its monosacchride-binding site.
Finally, ManA and Man4 remain as potential alternative candidates to occupy the monosaccharide-binding site. This would correspond to the binding of the trisaccharide Manα(1–3)[Manα(1–6)]Man in its reverse orientation, Manα(1–3)Man occupying the monosaccharide-binding site and the +1 subsite. The isolated disaccharide Manα(1–3)Man is known to bind to PAL in this manner (Loris et al., 2004). Therefore, we modeled Manα(1–3)[Manα(1–6)]Man in the PAL-binding site based upon the coordinates of the Manα(1–3)Man complex and allowed the α(1–6) linkage to vary. The α(1–6) linkage is highly flexible due to its additional Ω torsion angle. With Ω in the gauche-trans conformation (+60°), the third Man (ManB or Man4′, respectively) points directly toward the solvent, making any further substitution possible (although without generating additional specific contacts). However, steric hindrance is expected for the chitobiose stem, especially when Man4 is located in the monosaccharide-binding site because of the β-linkage in the Man3β(1–4)GlcNAc2 moiety. Therefore, ManA and Man4 are probably excluded from the monosaccharide-binding site.
In conclusion, four binding modes different from the one observed in our crystal structures (Fig. 5) as well as multivalent behavior of the oligosaccharides remain theoretically possible. Although we predict that the affinities for these alternative binding modes would be slightly lower (probably similar to that of a monosaccharide), the binding mode observed in the crystal is not expected to be all dominating. Our earlier work also showed that isolated Manα(1–3)[Manα(1–6)]Man and Manα(1–2)Man each bind in a single binding mode (Loris et al., 2004) while two are expected. For Manα(1–2)Man, the 5-times higher binding constant (compared to Man) could still explain the observation of an apparent single-binding mode in the crystal structure. For Manα(1–3)[Manα(1–6)]Man, this argument is already much weaker as this trisaccharide binds only twice as strong as Man (Buts et al., 2006). However, in the case of Man-9 with a total of at least five possible binding modes, it is expected that the binding mode observed in our crystal structures would constitute well below 50% of the population, creating a contradiction between experiment and theory.
In the case of Man-7D1, it is even more remarkable that ManB and not ManC is not found in the monosaccharide-binding site. This would result in the recognition of at least ManD1(α1–2)ManC though the −1 and M subsites. Assuming additivity of the subsite interactions, this binding mode should occur with an affinity of at least 5 times that of Man. The binding mode observed in the crystal with ManB occupying the monosaccharide-binding site and Man4′ and ManA on the +1 and +2 subsites corresponds to the situation of the trisaccharide Manα(1–3)[Manα(1–6)]Man that binds with only twice the affinity of Man. The selection of the binding mode of Man-7D1 remains therefore unexplained.
Thermodynamics of Oligomannose Binding
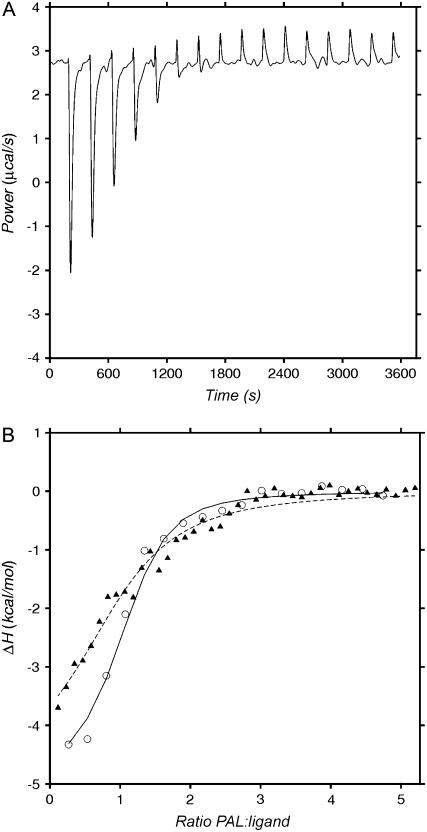
To confirm the results obtained by crystallography, the solution binding of PAL to Man-5, Man-9, and soybean (Glycine max) agglutinin (a homotetrameric protein containing one Man-9 high Man oligosaccharide chain on each subunit) were measured by isothermal titration calorimetry (ITC). Examples of the experimental data are shown in Figure 6 and results are summarized in Table IV. Of interest is that binding of PAL to Man-9 and soybean agglutinin (SBA) is not exclusively enthalpy driven, but also shows a favorable entropy term. This seems to be a key feature for the better carbohydrate ligands of PAL as it was previously also observed for GlcNAc(β1–2)Man (Buts et al., 2006). In the case of con A, a favorable entropy contribution to the binding of Manα(1–2)Man was attributed to a sliding mechanism (Brewer and Brown, 1979; Mandal et al., 1994), which was later confirmed by x-ray crystallography (Moothoo et al., 1999). The unique recognition mode observed in all our crystal structures refutes this explanation for PAL, as does the observed 1:1 stoichiometry in ITC experiments with GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man (Buts et al., 2006), Man-5, Man-9, and SBA (when concentration is expressed in SBA monomers). This 1:1 stoichiometry is also at odds with the potential multivalence of Man-9 (three terminal Mans that each could interact with a different molecule of PAL).
Figure 6.
A, Titration of 6.2 mm PAL into 0.14 mm SBA. The solid line represents the power required to maintain the calorimeter cell at constant temperature. Each dip in the line corresponds to the injection of a constant amount of PAL, resulting in heat production by the binding reaction and a temporary reduction in the power requirement. Integration of the surface area for each peak yields the total heat production corresponding to the formation of a known amount of complex. The positive final peaks correspond to the small heat of dilution of PAL into the saturated solution. B, Integrated and dilution-corrected heats of binding for the titrations of PAL into SBA (white circles, solid line) and Man-9 (black triangles, dotted line). The lines represent the fits for a 1:1 binding model.
Table IV.
Energetics of binding
| Carbohydrate | No. of Experiments | c Value | Stoichiometrya | Kass | ΔG0b | ΔH0b | TΔS0b |
|---|---|---|---|---|---|---|---|
| 103m−1 | kcal/mol | kcal/mol | kcal/mol | ||||
| Manc | 4 | 3.8 | 0.90 | 1.9 | −4.4 | −6.3 | −1.9 |
| ManαMec | 3 | 5.4 | 0.95 | 3.4 | −4.8 | −6.5 | −1.7 |
| Manα(1–2)Manc | 3 | 13.0 | 0.92 | 15.4 | −5.7 | −7.0 | −1.3 |
| Manα(1–3)[Man(α1–6)]Manc | 3 | 1.4 | 0.95 | 5.6 | −5.1 | −7.1 | −2.0 |
| Manα(1–3)[Manα(1–3)[Manα(1–6)]Manα(1–6)]Man (Man-5) | 3 | 2.3 | 0.95 | 6.6 | −5.2 | −8.7 | −3.5 |
| Man-9 | 2 | 2.8 | 1.0d | 23 | −5.8 | −4.8 | 1.1 |
| Soybean agglutinin | 2 | 11.2 | 0.93 | 80 | −6.7 | −4.8 | 1.9 |
Obtained from fitting the ITC data. The reported values for Kass, ΔG0, ΔH0, and TΔS0 are determined with n fixed at 1.0. These values do not differ significantly from those obtained when treating the number of binding sites as a variable.
The errors on ΔG0 and ΔH0 are of the order of magnitude of 0.1 kcal/mol while for TΔS0 they are 0.2 kcal/mol.
Taken from Buts et al. (2006) and included for comparison.
As the correct concentration of Man-9 could not be determined sufficiently accurately (due to availability of sufficient material) the number of binding sites was kept fixed at 1.0 in accordance with the data obtained from Man5 and soybean agglutinin.
The thermodynamic data can be compared with similar data obtained for con A (Mandal et al., 1994). In the case of con A, binding of Man-9 is much tighter (binding constant 1.1 106 m−1) and entirely enthalpy driven (Mandal et al., 1994). The binding stoichiometry in the case of con A is also 1:1 despite the potential of multivalency of the carbohydrate. Unfortunately, no crystal structure of con A in complex with a high Man oligosaccharide, other than the trimannose core Manα(1–6)[Manα(1–3)]Man (Naismith and Field, 1996), is available. Nevertheless, given the affinities and observed binding modes for Manα(1–2)Man and Manα(1–6)[Manα(1–3)]Man, con A recognizes probably the same epitope as PAL but with different energetics.
MATERIALS AND METHODS
Sugar and Protein
All carbohydrates were obtained from DEXTRA laboratories. The nomenclature used to identify the Man residues in high Man oligosaccharides is taken according to Koles et al. (2004). PAL was purified from mature seeds by affinity chromatography. Extracts from uncoated ground and defatted Pterocarpus angolensis seeds were fractionated with ammonium sulfate. The 30% to 60% ammonium sulfate fraction was suspended in 100 mm NaCl + 10 mm phosphate buffer pH 7.4 and then dialyzed extensively against the same buffer. After removal of all remaining insoluble material by centrifugation (30 min at 24000g), the resulting solution was applied to a fetuin-sepharose column. After extensive washing to remove all unbound protein, the lectin was eluted using 0.3 m Man in 100 mm NaCl + 10 mm phosphate buffer pH 7.4. The protein was subsequently extensively dialyzed against 50 mm phosphate pH 7.2 + 150 mm NaCl. Protein concentrations were determined from UV absorption spectroscopy at 280 nm using the specific absorption coefficient ɛ of 37,410 m−1 cm−1 as calculated according to Gill and von Hippel (1989).
Crystallization and Data Collection
Cocrystallization of PAL with the different oligomannoses is not possible due to the amounts of sugar needed. For each sugar, only 20 μg was available (except for Man-9 [1 mg] and Man-5 [5 mg]), which is sufficient for just one soaking experiment in a 2 μL drop. Therefore, the complexes were prepared starting from crystals of PAL in complex with Manα(1–3)Man, which were obtained as described previously (Loris et al., 2005).
To produce complexes with high Man oligosaccharides, these crystals were first desoaked by transferring them to artificial mother liquor devoid of sugar (100 mm Na-cacodylate pH 6.5, 200 mm Ca-acetate, 20% [w/v] PEG-8000) for at least 1 week and refreshing the soaking solution twice. This treatment leads to crystals in which the binding site of subunit-B becomes empty (Loris et al., 2005; Buts et al., 2006). Desoaked crystals were then transferred to 1 μL drops containing 20 μg of the desired sugar. After an overnight soak, the crystals were then mounted in thin-walled glass capillaries for data collection at room temperature.
X-ray data were collected on EMBL beamlines X11 and X13 (DESY) as well as on ESRF beamline ID14-1 (ESRF). The data were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). Data were integrated with DENZO, merged with SCALEPACK, and converted to structure factor amplitudes using the CCP4 program TRUNCATE (CCP4, 1994). The statistics of the data collections are given in Table I.
Structure Determination
The crystal structure of the PAL:Manα(1–3)Man complex, stripped of water molecules, metal ions, and carbohydrate ligands, was used as the starting model in refinement with CNS 1.0 (Brünger et al., 1998). After an initial rigid body refinement, a slow cool stage was used to uncouple R and Rfree. From then on restrained positional and B-factor refinements were alternated with manual fitting in electron density maps using TURBO (Roussel and Cambillau, 1989). The refinement statistics are given in Table I. Superpositions of crystal structures were done using TURBO. Contact surfaces between carbohydrate and protein were calculated using NACCESS (S.J. Hubbard and J. Thornthon, University College, London, 1993). All figures were produced using MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997).
ITC
The heat accompanying the binding of PAL to oligosaccharides or SBA was measured using an Omega isothermal titration calorimeter (MicroCal) at a temperature of 25°C. Binding of Man-5 was measured by a direct titration with a PAL solution (0.97 mm in 50 mm phosphate pH 7.5, 150 mm NaCl) in the calorimeter cell and an oligosaccharide solution of 14.8 mm in the same buffer being injected from a 250 μL syringe (32 injections of 10 μL at 240 s intervals).
Because of limitations in material availability, binding of Man-9 and SBA was assessed via reverse titrations by placing the oligosaccharide (0.12 mm) or glycoprotein (0.14 mm) ligand in the calorimeter cell and titrating with PAL (3.1 mm or 6.3 mm). After the titration, the mixed solution was retrieved, stored at 4 degrees for 24 h, and centrifuged at 20,000g for 15 min. The PAL:Man-5 and PAL:Man-9 mixtures remained completely free of precipitation, whereas a large pellet was observed for the PAL:SBA cross-linked complex.
The cell and syringe concentrations [C]0 and [S]0 were chosen to ensure that the product c = n × Ka × [C]0 was between 1 and 10 and the ratio [S]0/n × [C]0 was 12.5 for the anticipated number of binding sites n. Each injection generated a heat burst, with the area under the curve being proportional to the heat of interaction. The heat effects accompanying ligand dilution were measured by titration into pure buffer and subtracted. After comparing different fitting models, all data were finally fitted with a Wiseman isotherm for a 1:1 binding model using a nonlinear regression procedure (Wiseman et al., 1989).
Modeling Calculations
Energy maps are calculated using the TRIPOS force field (Clark et al., 1989) together with the PIM parameterization (Imberty et al., 1999) developed for carbohydrates as a function of the Φ and Ψ dihedral angles defined as: Φ = O5 − C1 − O1 − Cx′ and Ψ = C1 − O1 − Cx′ − C(x + 1)′ for the β(1–2). For α(1–6) linkages, three dihedral angles Φ = O5 − C1 − O1 − C6′, Ψ = C1 − O1 − C6′ − C5′, and ω = O1 − C6′ − C5′ − O5′ were used.
Atomic coordinates and structure factor data were submitted to the Protein Data Bank and are available as entries 2PHX, 2PHF, 2PHR, 2PHT, 2PHU, and 2PHW.
Acknowledgments
The authors acknowledge the use of beamtime at the EMBL beamlines at the DESY (Hamburg, Germany) and ESRF (Grenoble, France) synchrotrons.
This work was supported by the Vlaams Interuniversitair Instituut voor Biotechnologie, the Onderzoeksraad of the Vrije Universiteit Brussel, and the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (post-doc fellowship to L.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Remy Loris (reloris@vub.ac.be).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Ahmad N, Gabius HJ, Sabesan S, Oscarson S, Brewer CF (2004) Thermodynamic binding studies of bivalent oligosaccharides to galectin-1, galectin-3 and the carbohydrate recognition domain of galactin-3. Glycobiology 14 817–825 [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al (2002) The Protein Data Bank. Acta Crystallogr D58 899–907 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya L, Khan MI, Brewer CF (1988) Interactions of concanavalin A with asparagine-linked glycopeptides: formation of homogeneous cross-linked lattices in mixed precipitation systems. Biochemistry 27 8762–8767 [DOI] [PubMed] [Google Scholar]
- Botos I, O'Keefe BR, Shenoy SR, Cartner LK, Ratner DM, Seeberger PH, Boyd MR, Wlodawer A (2002) Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J Biol Chem 277 34336–34342 [DOI] [PubMed] [Google Scholar]
- Bourne Y, Bolgiano B, Liao DI, Strecker G, Cantau P, Herzberg O, Feizi T, Cambillau C (1994. a) Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat Struct Biol 1 863–870 [DOI] [PubMed] [Google Scholar]
- Bourne Y, Mazurier J, Legrand D, Rouge P, Montreuil J, Spik G, Cambillau C (1994. b) Structures of a legume lectin complexed with the human lactotransferrin N2 fragment, and with an isolated biantennary glycopeptide: role of the fucose moiety. Structure 2 209–219 [DOI] [PubMed] [Google Scholar]
- Brewer CF, Brown RD III (1979) Mechanism of binding of mono- and oligosaccharides to concanavalin A: a solvent proton magnetic relaxation dispersion study. Biochemistry 18 2555–2562 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D54 905–921 [DOI] [PubMed] [Google Scholar]
- Buts L, Garcia-Pino A, Imberty A, Amiot N, Boons GJ, Beeckmans S, Versées W, Wyns L, Loris R (2006) Structural basis for the recognition of complex-type biantennary oligosaccharides by Pterocarpus angolensis lectin. FEBS J 273 2407–2420 [DOI] [PubMed] [Google Scholar]
- Clark M, Cramer RDI, van den Opdenbosch N (1989) Validation of the general purpose Tripos 5.2 force field. J Comput Chem 10 982–1012 [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D50 760–763 [DOI] [PubMed] [Google Scholar]
- Dam TK, Brewer CF (2002) Thermodynamic studies of lectin-carbohydrate interactions by isothermal titration calorimetry. Chem Rev 102 387–429 [DOI] [PubMed] [Google Scholar]
- Dam TK, Cavada BS, Grangeiro TB, Santos CF, de Sousa FA, Oscarson S, Brewer CF (1998) Diocleinae lectins are a group of proteins with conserved binding sites for the core trimannoside of asparagine-linked oligosaccharides and differential specificities for complex carbohydrates. J Biol Chem 273 12082–12088 [DOI] [PubMed] [Google Scholar]
- Dam TK, Gabius HJ, Andre S, Kaltner H, Lensch M, Brewer CF (2005) Galectins bind to the multivalent glycoprotein asialofetuin with enhanced affinities and a gradient of decreasing binding constants. Biochemistry 44 12564–12571 [DOI] [PubMed] [Google Scholar]
- Feinberg H, Mitchell DA, Drickamer K, Weis WI (2001) Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294 2163–2166 [DOI] [PubMed] [Google Scholar]
- Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, et al (2004) The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303 1838–1842 [DOI] [PubMed] [Google Scholar]
- Garcia-Pino A, Buts L, Wyns L, Loris R (2006) Interplay between metal binding and cis/trans isomerization in legume lectins: structural and thermodynamic study of P. angolensis lectin. J Mol Biol 361 153–167 [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182 319–326; erratum Gill SC, von Hippel PH (1989) Anal Biochem 189: 283 [DOI] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC (2003) X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 309 209–218 [DOI] [PubMed] [Google Scholar]
- Imberty A, Bettler E, Karababa M, Mazeau K, Petrova P, Pérez S (1999) Building sugars: the sweet part of structural biology. In M Vijayan, N Yathindra, AS Kolaskar, eds, Perspectives in Structural Biology. Indian Academy of Sciences and Universities Press, Hyderabad, India, pp 392–409
- Jeyaprakash AA, Srivastav A, Surolia A, Vijayan M (2004) Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity. J Mol Biol 338 757–770 [DOI] [PubMed] [Google Scholar]
- Koles K, van Berkel PHC, Pieper FR, Nuijens JH, Mannesse MLM, Vliegenthart JFG, Kamerling JP (2004) N- and O-glycans of recombinant human C1 inhibitor expressed in the milk of transgenic rabbits. Glycobiology 14 51–64 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24 946–950 [Google Scholar]
- Loris R (2002) Principles of structures of animal and plant lectins. Biochim Biophys Acta 1572 198–208 [DOI] [PubMed] [Google Scholar]
- Loris R, Garcia-Pino A, Buts L, Bouckaert J, Beeckmans S, De Greve H, Wyns L (2005) Crystallisation and crystal manipulation of the Pterocarpus angolensis seed lectin. Acta Crystallogr D61 685–689 [DOI] [PubMed] [Google Scholar]
- Loris R, Hamelryck T, Bouckaert J, Wyns L (1998) Legume lectin structure. Biochim Biophys Acta 1383 9–36 [DOI] [PubMed] [Google Scholar]
- Loris R, Imberty A, Beeckmans S, Van Driessche E, Read JS, Bouckaert J, De Greve H, Buts L, Wyns L (2003) Crystal structure of Pterocarpus angolensis lectin in complex with glucose, sucrose and turanose. J Biol Chem 278 16297–16303 [DOI] [PubMed] [Google Scholar]
- Loris R, Van Walle I, De Greve H, Beeckmans S, Deboeck F, Wyns L, Bouckaert J (2004) Structural basis of oligomannose recognition by the Pterocarpus angolensis seed lectin. J Mol Biol 335 1227–1240 [DOI] [PubMed] [Google Scholar]
- Mandal DK, Kishore N, Brewer CF (1994) Thermodynamics of lectin-carbohydrate interactions: titration microcalorimetry measurements of he binding of N-linked carbohydrates and ovalbumin to concanavalin A. Biochemistry 33 1149–1156 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol 277 505–524 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Sarfaty S, van den Akker F, L'Hoir C, Martial JA, Hol WG (1994) Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci 3 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moothoo DN, Canan B, Field RA, Naismith JH (1999) Manα(1-2)Manα-OMe-concanavalin A complex reveals a balance of forces involved in carbohydrate recognition. Glycobiology 9 539–545 [DOI] [PubMed] [Google Scholar]
- Moothoo DN, Naismith JH (1998) Concanavalin A distorts the b-GlcNac-(1-2)-Man linkage of b-GlcNac-(1-2)-a-Man-(1-3)-[b-GlcNac-(1-2)-a-Man-(1-6)] upon binding. Glycobiology 8 173–181 [DOI] [PubMed] [Google Scholar]
- Naismith JH, Field RA (1996) Structural basis of trimannoside recognition by concanavalin A. J Biol Chem 271 972–976 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276 307–326 [DOI] [PubMed] [Google Scholar]
- Roussel A, Cambillau C (1989) TURBO-FRODO. In Silicon Graphic Geometry Partner Directory. Silicon Graphics, Mountain View, CA, pp 71–78
- Sacchettini JC, Baum LG, Brewer CF (2001) Multivalent protein-carbohydrate interactions: a new paradigm for supermolecular assembly and signal transduction. Biochemistry 40 3009–301511258914 [Google Scholar]
- Sharma V, Surolia A (1997) Analyses of carbohydrate recognition by legume lectins: size of the combining site loops and their primary specificity. J Mol Biol 267 433–445 [DOI] [PubMed] [Google Scholar]
- Sharon N, Lis H (1990) Legume lectins—a large family of homologous proteins. FASEB J 4 3198–3208 [DOI] [PubMed] [Google Scholar]
- Shenoy SR, Barrientos LG, Ratner DM, O'Keefe BR, Seeberger PH, Gronenborn AM, Boyd MR (2002) Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: calorimetric and NMR characterization. Chem Biol 9 1109–1118 [DOI] [PubMed] [Google Scholar]
- Wang LX, Ni J, Singh S, Li H (2004) Binding of high-mannose-type oligosaccharides and synthetic oligomannose clusters to human antibody 2G12: implications for HIV-1 vaccine desing. Chem Biol 11 127–134 [DOI] [PubMed] [Google Scholar]
- Weis WI, Drickamer K, Hendrickson WA (1992) Structure of a C-type mannose-binding protein complexed with an oligosaccharide:. Nature 360 127–134 [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lung-Nan L (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 197 131–137 [DOI] [PubMed] [Google Scholar]
- Wright CS, Hester G (1996) The 2.0 A structure of a cross-linked complex between snowdrop lectin and a branched mannopentaose: evidence for two unique binding modes. Structure 4 1339–1352 [DOI] [PubMed] [Google Scholar]