Abstract
The tropical legume Sesbania rostrata provides its microsymbiont Azorhizobium caulinodans with versatile invasion strategies to allow nodule formation in temporarily flooded habitats. In aerated soils, the bacteria enter via the root hair curling mechanism. Submergence prevents this epidermal invasion by accumulation of inhibiting concentrations of ethylene and, under these conditions, the bacterial colonization occurs via intercellular cortical infection at lateral root bases. The transcriptome of both invasion ways was compared by cDNA-amplified fragment length polymorphism analysis. Clusters of gene tags were identified that were specific for either epidermal or cortical invasion or were shared by both. The data provide insight into mechanisms that control infection and illustrate that entry via the epidermis adds a layer of complexity to rhizobial invasion.
Legume plants can thrive in nitrogen-poor habitats because of their ability to establish a symbiotic interaction with soil bacteria, collectively called rhizobia. Bacterial Nod factors (NFs) are key signaling molecules for host specificity and they initiate the nodulation process in the plant (D'Haeze and Holsters, 2002; Geurts and Bisseling, 2002). Integration of plant developmental programs results in bacterial invasion and de novo cortical cell division, eventually leading to functional nodules with differentiated nitrogen-fixing bacteria.
Two invasion strategies have been studied in detail. The most common involves the curling of susceptible root hair cells in the elongation zone I of the root. Bacteria are entrapped in the curl and local plant cell wall hydrolysis and membrane invagination precede the formation of a tubular infection thread (IT) that guides the rhizobia through epidermis and cortex toward the incipient nodule. NF signaling is responsible for the induction of root hair curling (RHC) and IT formation and for the initiation of cell division. NFs activate an abundance of responses, including ion fluxes, membrane depolarization, phosholipase activity, phospholipid signaling, and calcium spiking (D'Haeze and Holsters, 2002; Oldroyd and Downie, 2004, 2006). A study of bacterial and plant mutants led to the hypothesis that NF perception triggers a signaling pathway, which prepares the plant for bacterial entry and cortical cell division, and an entry pathway for IT formation in the curled root hair (Ardourel et al., 1994). The latter response requires stringent NF recognition and has been proposed to be solely active within the epidermis that is also the site for nodulation checkpoints, such as inhibition by ethylene (Geurts et al., 1997; Oldroyd et al., 2001).
Direct cortical colonization (crack entry) takes place during lateral root base (LRB) nodulation on hydroponically grown roots of the semiaquatic legume Sesbania rostrata (Den Herder et al., 2006). In well-aerated soils, S. rostrata is invaded via the RHC mode and nodules appear in root zone I (Goormachtig et al., 2004a, 2004b). While RHC nodulation is inhibited by ethylene upon submergence, LRB nodulation makes active use of ethylene signaling to allow nodule formation under flooding conditions. No epidermal entry stages are apparent during this invasion. The bacteria gain direct access to the cortex via epidermal cracks at the sites of lateral root emergence (Den Herder et al., 2006). A few cortical cells are triggered by NFs to die, thus forming cavities that are colonized by the bacteria. These infection pockets serve as a launching point for ITs that grow toward the nodule primordium (Goormachtig et al., 2004a).
The occurrence of two different invasion strategies on the same plant allows us to compare the molecular mechanisms governing both processes. Hydroponic nodulation occurs without intervention of epidermal responses, in contrast to nodulation under aerated conditions where the epidermal root hairs are involved in the initial stages (Goormachtig et al., 2004b). Moreover, the structural requirements of NFs are more stringent for RHC than for crack-entry invasion (D'Haeze et al., 2000; Goormachtig et al., 2004b), supporting the hypothesis of an additional checkpoint for epidermal entry (Ardourel et al., 1994). These physiological and morphological dissimilarities indicate that each invasion pathway requires a specific set of genes and functions.
The advent of high-throughput transcript analysis tools has greatly improved our knowledge of the molecular mechanisms that regulate nodule initiation and development (Lievens et al., 2001; Fedorova et al., 2002; Poulsen and Pødenphant, 2002; Colebatch et al., 2004; El Yahyaoui et al., 2004; Küster et al., 2004; Manthey et al., 2004; Mitra and Long, 2004; Asamizu et al., 2005; Lohar et al., 2006; Starker et al., 2006). So far, all studies were focused on one invasion system. Here, we compared transcript profiles of RHC and LRB invasions in S. rostrata by using the PCR-based method of cDNA-amplified fragment length polymorphism (AFLP) because of its high sensitivity and reproducibility (Bachem et al., 1996; Breyne et al., 2003). Approximately 7,000 genes were screened, resulting in 627 differentially expressed tags. Statistical analysis of the data set revealed a core group of genes common to both invasion types as well as a large number of genes specific for one invasion mode.
RESULTS
Specific Sampling of Responsive Tissues
S. rostrata seedlings were planted in Leonard jars or in tubes with liquid medium to favor RHC invasion and LRB nodulation, respectively (see “Materials and Methods”; Goormachtig et al., 2004b). Approximately 900 plants in Leonard jars were inoculated with Azorhizobium caulinodans ORS571 (pBBR5-hem-gfp5-S65T) that constitutively expresses a GFP reporter (D'Haeze et al., 2004). The hydroponically grown roots were inoculated with wild-type ORS571 (pBBR-hem-gfp-S65T) or with the NF-deficient mutant ORS571-V44 (pBBR-hem-gfp-S65T) that contains a Tn5 insertion in the nodA gene (D'Haeze et al., 1998).
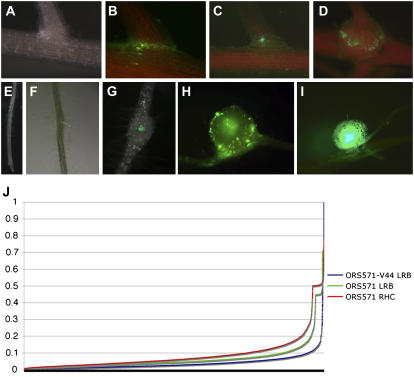
To enrich for responsive material, tissues in the process of being invaded by GFP-labeled rhizobia were harvested under fluorescent stereomicroscopy. For the hydroponic roots, time-based harvesting was possible because of the high synchronicity of LRB nodulation. Uninoculated LRBs and LRBs at 6, 12, 24, 48, and 72 h postinoculation (hpi) with ORS571 (pBBR-hem-gfp-S65T) were collected (Fig. 1, A–D). For the time course with A. caulinodans ORS571-V44 (pBBR-hem-gfp-S65T), uninoculated and LRBs at 24 and 48 hpi were taken.
Figure 1.
Tissue sampling and analysis of differentially expressed cDNA-AFLP tags. For LRB nodulation, plants were inoculated with either A. caulinodans ORS571-V44, a strain deficient in NF production, or A. caulinodans ORS571, both containing a GFP-expressing plasmid for visualization with fluorescence microscopy. A, As a negative control, LRBs of uninoculated plants were harvested. B to D, Representative stages of LRB nodulation at 6 (B), 24 (C), and 72 h (D) after inoculation with GFP-labeled A. caulinodans. E to I, A morphology-based approach used to collect tissue for RHC invasion. Representative stages are shown. As a negative control, 1.5-cm-long root segments without apical meristem were harvested (E). From inoculated plants, small root fragments with ITs were excised (F; rhc1, ±24–36 hpi). Later time points were root fragments with ITs and bumps indicative for cortical cell divisions (G; rhc2, ±48–60 hpi), primordia before the onset of nitrogen fixation (H; rhc3, ±72–100 hpi), and young fixing nodules (I; rhc4, ±140 hpi). J, Plot of log2(CV) values for all tags ordered along the abscissa. RHC samples contain more differentially expressed tags than LRB samples, which, in turn, have more samples than when inoculated with ORS571-V44.
Nodulation in Leonard jars via RHC invasion is not synchronized; hence, a morphology-based harvesting procedure was used (Fig. 1, E–I). From uninoculated roots, the zone directly behind the root apical meristem (designated rhc0) was collected (Fig. 1E). From plants inoculated with ORS571 (pBBR-hem-gfp-S65T), we excised small root fragments with IT-containing root hairs (rhc1, ±24–36 hpi; Fig. 1F), root fragments with ITs and small bumps, indicating cortical cell divisions (rhc2, ±48–60 hpi; Fig. 1G), young primordia before the onset of nitrogen fixation (rhc3, ±72–100 hpi; Fig. 1H), and young fixing nodules (rhc4, ±140 hpi; Fig. 1I).
cDNA-AFLP Transcript Profiling and Cluster Analysis
The samples were used for cDNA-AFLP transcript profiling to identify genes that were differentially regulated during nodule initiation in S. rostrata under hydroponic or aeroponic conditions (see “Materials and Methods”). The 128 primer combinations analyzed allowed visualization of some 7,000 transcript-derived tags. To determine what should be considered differential, approximately 3,000 tags were plotted on a graph, arranged by ascending log2 coefficient of variance (CV; see “Materials and Methods”). A distinct change in the curve was visible around log2(CV) = 0.1 (Fig. 1J). Hence, a tag was considered differential when its log2(CV) was 0.1 or higher. Of the 1,600 tags with a differential expression pattern in either the RHC or LRB nodulation systems or in both that were sequenced, 627 gave a significant BLAST hit (E value < 10−3) to sequences in public databases (Altschul et al., 1997) and were retained for further analysis.
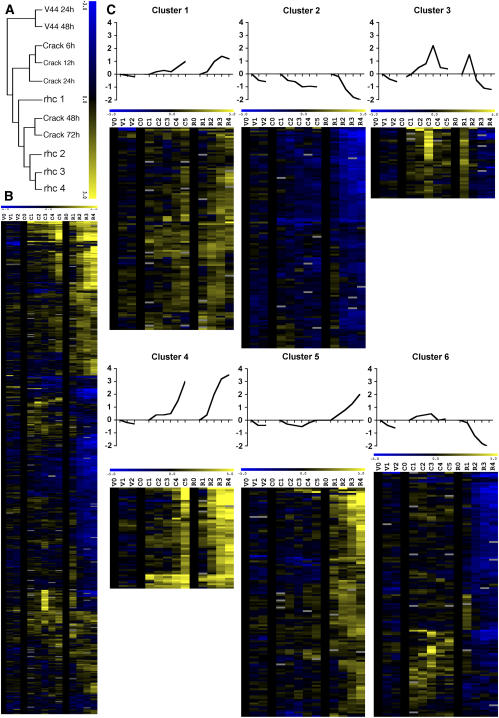
For a first insight into similarities between the transcript pools of each sample, the experiments were clustered hierarchically with the Pearson correlation as a statistical tool (see “Materials and Methods”; Fig. 2A). The base of the tree consisted of the V44 series; the inoculations with wild-type A. caulinodans split in three different clusters. The early LRB stages, crack 6, 12, and 24 h formed a subcluster (Fig. 2A), the rhc1 grouped separately, and the late time points of both the RHC and LRB samples, i.e. from 48 h after inoculation onward, clustered together (Fig. 2A).
Figure 2.
Cluster analysis. A, Hierarchical clustering of sample expression profiles visualizing the similarities in expression patterns at late time points. Differences between samples are more pronounced before 48 h. B, Hierarchical clustering of all 627 differentially expressed tags. Gene tags with a log2(CV) of at least 0.1 (corresponding to a 2-fold change in expression) were sequenced and expression profiles of the corresponding genes were clustered. Blue and yellow indicate down- and up-regulation, respectively. C, K means clustering of differentially expressed gene tags. Clusters 1 to 4 are considered common, cluster 5 harbors the RHC-specific genes, and cluster 6 contains tags specific for LRB invasion. From left to right: v, Hydroponically grown roots uninoculated (v0) or infected with A. caulinodans ORS571-V44 at 24 dpi (v1) and 48 dpi (v2); C, hydroponically grown roots, uninoculated (c0) and infected with A. caulinodans ORS571 at 6 (c1), 12 (c2), 24 (c3), 48 (c4), and 72 h (c5) postinoculation; r, aerated roots, uninoculated (r0) and corresponding to rhc1 (r1), rhc2 (r2), rhc3 (r3), and rhc4 (r4). The top sections depict expression patterns that are representative for most tags found in clusters 1 to 6.
Hierarchical cluster analysis revealed distinct expression profiles (Fig. 2B). The tags were separated into common and noncommon groups, the former referring to genes similarly regulated in both LRB and RHC nodulation and the latter to genes that were specifically expressed during LRB or RHC nodulation.
By K means analysis, used as a rough clustering method for the common and noncommon groups, six groups of tags were distinguished. The annotations, along with the data set are available online (Supplemental Table S1). Clusters 1 to 4 consisted of the 337 common gene tags. On average, cluster 1 contained tags that were gradually up-regulated in both invasion ways and reached a plateau at later stages; cluster 2 comprised mostly tags that were down-regulated in all series; tags corresponding to genes that were transiently up-regulated were grouped in cluster 3; and, finally, the common tags, whose transcript level did not reach a plateau at later time points, were found in cluster 4, with a subset of this cluster already expressed at the earliest time points. Cluster 5 displayed a predominantly RHC-specific pattern, whereas cluster 6 tags were specifically expressed during LRB nodulation, accompanied by some very early transient RHC tags.
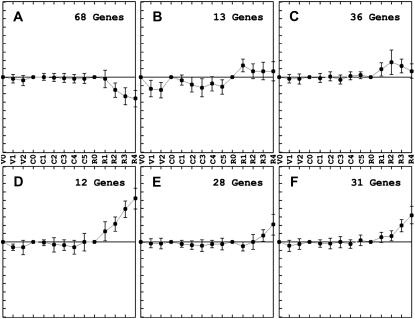
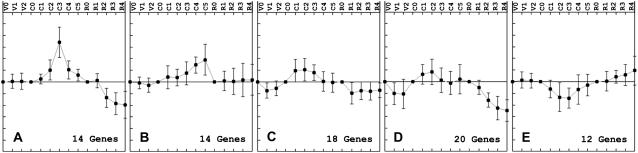
The RHC-specific tags (cluster 5) were further subclustered into six groups with distinct expression patterns (Fig. 3; Supplemental Fig. S1). Tags of subcluster 5A were down-regulated whereas those of subclusters 5B and 5C were transiently induced during RHC invasion. Subclusters 5D to 5F contained genes whose expression level gradually increased, starting at different time points. Subclustering of the LRB-specific tags (cluster 6) provided five groups, each represented by a specific expression pattern (Fig. 4; Supplemental Fig. S2). Subcluster 6A grouped tags that were transiently up-regulated and from which the expression level was highest at 24 hpi. Subcluster 6B contained tags from which the expression level steadily increased during the course of the experiment. Subgroups 6C and 6D also represented transiently expressed tags, but the expression did not reach the same level as that of subgroup 6A. Subgroups 6C and 6D differed with respect to time points at which the expression dropped again. Finally, tags that were transiently repressed during LRB invasion were grouped in subcluster 6E.
Figure 3.
RHC-specific gene tags. Average expression patterns of the six RHC-specific subclusters. Hierarchical clusters corresponding to these groups are available as Supplemental Figure S1. The same time points were used as those mentioned in Figure 2.
Figure 4.
LRB-specific gene tags. Average expression patterns of all five LRB-specific subclusters. Hierarchical clusters corresponding to these groups are available as Supplemental Figure S2. For the used time points, see Figure 2.
The large-scale profiling experiment was done only once because of the considerable work load and tedious harvesting procedures. Nevertheless, material from a biological repeat (see “Materials and Methods”) was analyzed by quantitative reverse transcription (qRT)-PCR for 10 tags, overall confirming the expression patterns obtained in the cDNA-AFLP (Supplemental Fig. S3).
Functional Classes in the Data Set
The differential tags were assigned to functional classes according to the 16 categories used in the Medicago EST Navigation System database (El Yahyaoui et al., 2004). Approximately 32% of all tags corresponded to putative genes or genes with unknown function; a putative function could be assigned to 68%. The relative contribution of the different functional classes in the RHC-specific, crack-entry-specific, and common clusters was very similar (Supplemental Fig. S4). However, different isoforms were often expressed in one or the other invasion way. For instance, several tags encoded peroxidase genes, of which some were common, but one was specific for crack entry, implying different roles and substrates. Similarly, of three NADPH oxidase-encoding tags, one was common, one was specific for RHC, and one for crack entry.
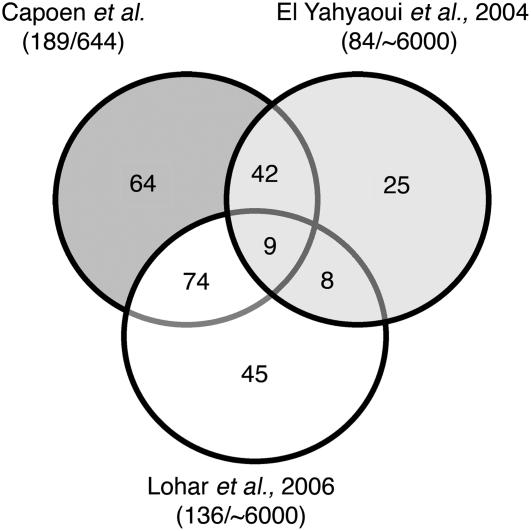
By means of BLASTN, we compared all S. rostrata tags with published expression profiling experiments in Medicago truncatula (El Yahyaoui et al., 2004; Lohar et al., 2006). Only tags with an E value under 10−3 were withheld for further analysis, resulting in 136 and 84 common tags with the data sets of Lohar et al. (2006) and El Yahyaoui et al. (2004), respectively (Fig. 5). Subsequently, expression patterns of these common tags were found to be similar in 61.0% (83 from 136) and 60.7% (51 from 84) of the cases, respectively. Nine tags had a similar expression pattern in the three different experiments.
Figure 5.
Data set comparison with existing nodulation transcript profiles. Graphical representation of similarities between this study and two similar array-based expression profiling experiments in M. truncatula (El-Yahyaoui et al., 2004; Lohar et al., 2006). The numbers of genes with similar expression patterns between the different data sets are shown in the overlaps within the Venn diagram.
SrLyr3, a LysM-Receptor-Like Kinase, Is Expressed during RHC and LRB Nodulation
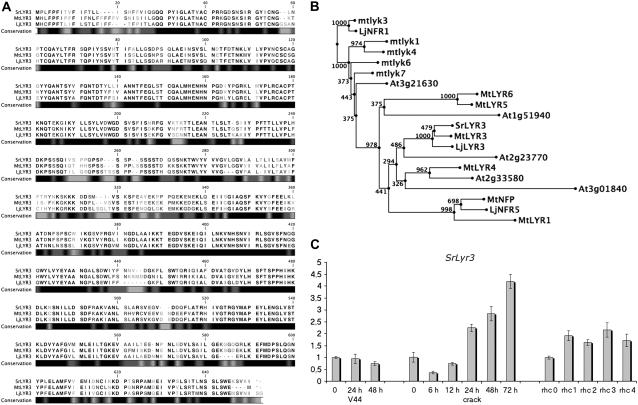
Many differential tags corresponded to putative kinases, receptor-like kinases (RLKs), and protein phosphatases, and might be important players in nodulation. One tag from a common up-regulated cluster was homologous to genes coding for the LysM domain-containing RLKs, the family to which the putative NF receptors belong (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Fig. 6A). The full-length cDNA clone was isolated with RACE (see “Materials and Methods”) and designated SrLyr3, because BLAST searches identified MtLyr3 of M. truncatula as the closest homolog (Arrighi et al., 2006). The corresponding amino acid sequence of SrLyr3 is 85% similar to that of MtLyr3 and 63% to the protein encoded by Arabidopsis (Arabidopsis thaliana) At2g23770. These LysM-RLKs belong to the same clade II as the NF receptor NFP, but they are located in a separate branch (Fig. 6B; Arrighi et al., 2006). The dissimilarity to NFP is demonstrated by differences in the kinase domain, such as the presence of an activation loop. However, the kinase domain might not be active because some important sequences, such as the P loop, the DFG motif, and the Ser/Thr of the activation loop are absent or have been substituted (Arrighi et al., 2006; Riely et al., 2006; Zhu et al., 2006).
Figure 6.
SrLyr3 sequence, expression profile, and phylogenetic analysis. A, Alignment of LYR3 orthologs in S. rostrata (SrLYR3), M. truncatula (MtLYR3), and L. japonicus (LjLYR3). B, Phylogenetic analysis of known M. truncatula, L. japonicus, S. rostrata, and Arabidopsis LysM domain-containing RLKs. The complete sequence was taken for comparison. C, qRT-PCR analysis of SrLyr3 during hydroponic (crack) and aeroponic (rhc) nodulation.
Confirmation that the gene is up-regulated in both invasion ways was obtained by qRT-PCR analysis on RNA from a biological repeat experiment. During LRB nodulation, SrLyr3 transcripts started to accumulate between 12 and 24 hpi and increased further at later time points (Fig. 6C). In the RHC series, the transcript level was already up-regulated at the stage of curled root hairs and remained more or less similar at later stages (Fig. 6C).
DISCUSSION
Commonalities and Differences
Both LRB and RHC nodulation on S. rostrata depend on perception of bacterial NFs and downstream events (D'Haeze et al., 2003; Goormachtig et al., 2004a, 2004b). The overall outcome of the processes is the same: the formation of functional nodules. The differences relate to the invasion mode, with NF perception linked to either cortical cell death or to inverted tip growth in a root hair, and to the local physiology of the nodulation zones. For RHC nodulation, as studied in M. truncatula and Lotus japonicus, several essential components of the NF perception, signal transduction, and response cascade have been identified. Examples are the M. trucatula genes coding for DOES NOT MAKE INFECTIONS1 (MtDMI1), MtDMI2, MtDMI3, the LysM-RLKs NFP and Lyk3 and their orthologs, and the transcription factors (TFs) NSP1 and NSP2 (Endre et al., 2002; Stracke et al., 2002; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Ané et al., 2004; Lévy et al., 2004; Mitra and Long, 2004; Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006). As no wide-scale genetic or genomic tools are available for S. rostrata, we have to resort to reverse genetics strategies to unravel the role of some of the key components in LRB nodulation (Capoen et al., 2005). Another approach to collect data on relevant functions for alternative nodulation processes in a nonmodel legume consists in a large-scale transcriptome analysis to hunt for differentially expressed genes (Goormachtig et al., 1995; Lievens et al., 2001; Schroeyers et al., 2004).
A comparative cDNA-AFLP transcriptome analysis of the two nodulation modes of S. rostrata allowed allocation of 627 tags in six clusters with differential expression profiles. The onset of each invasion strategy was characterized by specific gene expression. At later stages corresponding to primordium formation and nodule differentiation, many tags were common between the RHC and LRB nodulation series. The potential relevance of these differential tags will be discussed in the versatile S. rostrata symbiosis context and in the general context of legume nodulation.
Common Genes Related to Early Signaling Events
LRB and RHC nodulations depend both on NFs and genes involved in NF perception are expected to be up-regulated in both invasion modes from early stages on. Several tags with such an expression pattern are reminiscent of regulatory mechanisms, such as ubiquitin-dependent protein degradation (M12-180.8), transcriptional control (M12-288.3), chromosome reorganization (M21-459.1 and M33-349.4), and hormone perception. Tag M22-259.3 is related to MtRR4 (TC 103991), a response regulator involved in cytokinin signaling that is up-regulated during nodule development in M. truncatula (Gonzalez-Rizzo et al., 2006).
One of the earliest root hair responses to NFs is a rhythmic calcium oscillation that ensues within 10 min of NF addition (Oldroyd and Downie, 2006). Several tags related to calmodulin-like proteins as well as a Ca2+-dependent protein kinase, were up-regulated in the two invasion modes (Fig. 7A). Calcium spiking might also occur in cortical cells (Miwa et al., 2006), and inhibitors of Ca2+ spiking inhibit LRB nodulation (W. Capoen, W. D'Haeze, and M. Holsters, unpublished data). Hence, calcium signaling is presumably important in LRB as well as in RHC nodulations.
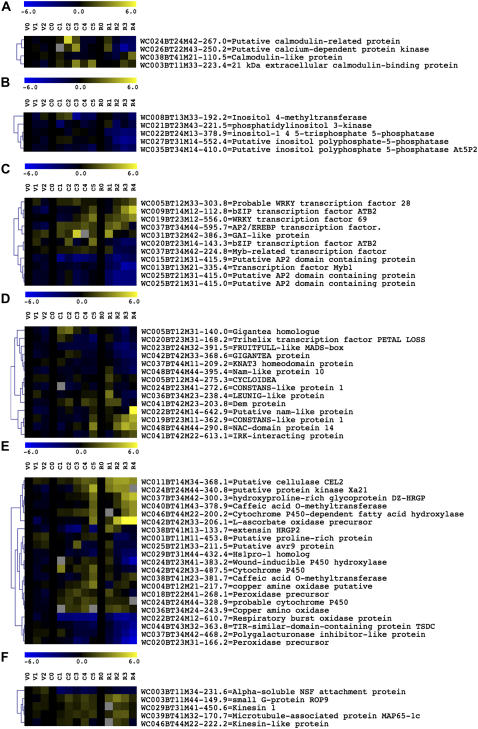
Figure 7.
Hierarchical cluster analysis of a selected group of tags according to the process in which they might be involved. A to F, Clustered tags involved in calcium signaling (A), phosphoinositol signaling (B), transcriptional regulation (C), meristem signaling (D), defense responses (E), and targeted vesicle trafficking (F).
One tag homologous to phosphatidyl inositol 3-kinase, three tags for putative inositol polyphosphate 5-phosphatases, and one tag similar to an inositol 4-methyltransferase were weakly and transiently up-regulated at early stages, followed by a down-regulation at later stages (Fig. 7B). Although phospholipids presumably play a role during early NF signaling, based on pharmacological evidence (den Hartog et al., 2003; Charron et al., 2004), our expression analysis suggests that transcripts for phospholipid signaling functions are repressed at later time points.
A common early up-regulated tag corresponded to SrLyr3, the S. rostrata ortholog of MtLyr3 (Arrighi et al., 2006), a recently described member of the M. truncatula LysM-RLK family. To this family belong MtLyk3 and MtNFP, the LysM domain proteins that play a key role in nodulation, presumably by binding NFs as ligands (Madsen et al., 2003; Radutoiu et al., 2003; Limpens et al., 2005; Mulder et al., 2006). SrLyr3 and MtLyr3 have a close homolog in Arabidopsis, a plant that is unable to establish symbioses (Fig. 6B). A preliminary expression analysis suggests that these LysM-RLKs are likely candidates to perceive endogenous signals involved in plant developmental programs or in pathogen defense (W. Capoen and M. Holsters, unpublished data).
Common Genes Related to Nodule Development
Plant development is controlled by specific TFs. Numerous TF tags were present in the data set, some up-regulated, some repressed (Fig. 7C). Among the latter were the GRAS protein GAI (a negative regulator of GA action), confirming the prediction for tight regulation of GA during nodule initiation (Lievens et al., 2005), two Myb1 like, two AP2 domain, and two WRKY TFs, related to the Arabidopsis orthologs WRKY28 and WRKY69 that are differentially up-regulated in biotic interactions (www.genevestigator.ethz.ch).
Remarkably, several differential tags were homologous to genes involved in growth and differentiation of shoots, flowers, and fruits (Fig. 7D), such as PETAL LOSS, the MADS-box protein FRUITFULL, Nam-like proteins 10 and 14, CONSTANS, the KNAT3 homeobox gene, GIGANTEA, CYCLOIDEA, LEUNIG, IRKI, an interactor of the inflorescence and root apices RLK (Hattan et al., 2004), and DEM, defective embryo and meristem (Irish, 1999; Ferrándiz et al., 2000; Kieffer and Davies, 2001; Komeda, 2004; Veit, 2004). Nodulation might have features in common with shoot apical meristem development, as suggested previously (Szczyglowski and Amyot, 2003). However, differentiation processes identified as shoot specific might also underlie root formation or have parallel programs for root development. Indeed, M. truncatula ESTs with closest homology to the S. rostrata tags were found in root tissues in the data set of The Institute for Genome Research. Moreover, in Arabidopsis, a CONSTANS-like 3 gene controlled lateral root development, a process that has much in common with nodule formation (Datta et al., 2006). Also KNAT3 expression occurred in young Arabidopsis root tissue and was specifically excluded from lateral root primordia (Truernit et al., 2006). Similarly, the KNAT3 gene was repressed during nodule formation.
Common Genes Related to Invasion
As both invasion ways make use of cortical ITs for bacterial penetration, tags that code for functions for IT growth might be shared. IT progression involves cell wall modifications and expression of specific matrix proteins (Brewin, 2004). Tags putatively coding for (hydroxy) Pro-rich proteins, a lignin biosynthesis enzyme (caffeic acid O-methyltransferase), and a cellulase are continuously up-regulated throughout nodulation. Six tags display homology to functions involved in reactive oxygen species (ROS) production and metabolism, such as two peroxidases, two distinct amine oxidases, a respiratory burst oxidase (NADPH oxidase), and an ascorbate oxidase (Fig. 7E; Supplemental Table S1). ROS components have been localized in ITs and hydrogen peroxide (H2O2)-driven cross-linking of root nodule extensins has been put forward as a driving force for IT progression (Brewin, 2004; Den Herder et al., 2007). Amine oxidases might provide a source of peroxide used by peroxidases to perform such cross-linking of glycoproteins (Brewin, 2004). The expression profiles of the two amine oxidases have been confirmed by RT-PCR and analysis of their function in nodulation is ongoing (J. Den Herder and M. Holsters, unpublished data).
ROS production and metabolism are also components of defense responses. Several tags corresponding to defense-related genes were up- or down-regulated during nodulation (Fig. 7E). Previous transcript profiling experiments in M. truncatula have revealed a similar behavior of defense-related genes (El Yahyaoui et al., 2004; Manthey et al., 2004). Rhizobial infection probably requires active repression of certain defenses (Mithöfer, 2002). Among the down-regulated genes were a TIR domain-containing protein TSDC, an Hs1pro1 homolog (nematode resistance), and a polygalacturonase inhibitor. To the up-regulated clusters belonged tags homologous to genes coding for a putative avr9 elicitor response protein, a protein with homology to a XA21-like receptor kinase, and several cytochrome P450 (Fig. 7E). Defense functions might restrict bacterial invasion because approximately 90% of all infection events abort prematurely in M. truncatula (Pauly et al., 2006), or they might protect the nodule that is rich in nutrients and an attractive target for pathogens.
Several tags correspond to functions involved in vesicle transport and vesicle targeting: two kinesins, a microtubule-associated protein MAP65-1c, α-soluble NSF attachment protein, and the small G protein ROP9 (Fig. 7F). ROPs have been implicated in tip growth and H2O2 production and might be involved in IT progression (Yang et al., 1994; Maunoury et al., 2007). In M. truncatula, a specific syntaxin MtSyP132 was localized to the plasma membrane surrounding ITs and infection droplets, illustrating the need for targeted exocytosis of IT development and growth (Catalano et al., 2007). The specific down-regulation of a SNAP protein in our data set (M34-231.6) might reflect a change in the nature of vesicles targeted to the site of infection (Fig. 7F).
RHC Nodulation-Specific Genes
Of the RHC-specific tags, 68 were strongly down-regulated and 122 up-regulated. Among the latter were tags corresponding to the nodulin genes MtN6 and MtN21 of M. truncatula. MtN6 expression precedes infection and has been proposed to play a role in the preparation of cells for IT passage (Mathis et al., 1999). Also LRB nodulation comes with IT formation, but differences in physiological conditions and hormone landscapes between LRBs and zone I might account for the specific involvement of this tag in RHC invasion.
A tag homologous to the auxin efflux carrier PIN2 is specific for RHC invasion. In M. truncatula, an orthologous tag is up-regulated during nodulation (Schnabel and Frugoli, 2004). A tag encoding an ADP-ribosylation factor has a similar expression pattern. ADP-ribosylation factors have been implicated in the correct targeting of both PIN2 and a ROP GTPase in growing root hair cells (Xu and Scheres, 2005), implying that these three proteins might polarize inverted tip growth in the root hair.
The number of RHC-specific tags is approximately 3-fold higher than that of crack-entry-specific tags, suggesting that the passage of the epidermis adds a layer of complexity and specificity to nodule initiation, as already illustrated by the more stringent NF structural requirements (D'Haeze et al., 2000; Goormachtig et al., 2004a). However, crossing the epidermis alone does not account for all the RHC nodulation-specific gene expression because the expression of more than 50% of the tags is modulated at time points when the ITs reach the primordium cells. Also, many RHC nodulation-specific tags are similar to genes coding for general metabolic functions, such as amino acid biosynthesis (Fig. 3; Supplemental Fig. S1). Thus, differences in tissue physiology between hydroponic LRBs and root zone I might contribute to the high number of RHC-specific genes. LRBs might, to a certain extent, be predisposed for nodule formation, for instance by the prevailing hormone concentrations. Indeed, in the root zone I of clover (Trifolium repens), NFs induced auxin landscapes that were similar to those preexisting at LRBs, suggesting a role for PIN2 in this process (Mathesius et al., 2000; van Noorden et al., 2006). Therefore, the RHC nodulation-specific expression pattern of an S. rostrata PIN2 tag surely needs further investigation.
LRB Nodulation-Specific Genes
LRB nodulation involves intercellular invasion and induction of local cell death for infection pocket formation. Ethylene, GA, and H2O2 are important players in the process (D'Haeze et al., 2003; Lievens et al., 2005). Tags in subclusters 6A, 6C, and 6D (Fig. 4; Supplemental Fig. S2) are good candidates to be involved in cell death execution because they are transiently induced and specific to crack entry. One tag (M34-318.5) was similar to superoxide dismutases involved in H2O2 generation. Moreover, tag M21-618.0 was homologous to a gene coding for a hexose transporter (Supplemental Fig. S2). A hexose transporter of Arabidopsis has been implicated in induction of programmed cell death (Nørholm et al., 2006). A papain-like Cys protease had a similar transient expression pattern (M33-163.9; Supplemental Fig. S2). Papain-like proteases are involved in cell death-related processes (Beers et al., 2000). Additionally, a GPI anchor transamidase, a type 2A protein phosphatase, a 7-transmembrane protein, and a His kinase were present in subcluster 6D (Supplemental Fig. S2). Tag M43-408.5 was similar to an Arabidopsis hydrolase (At2g14110) that was slightly differentially expressed upon osmotic stress and senescence-induced programmed cell death. Finally, tag M34-622.0 corresponded to a basic helix-loop-helix Arabidopsis TF (At4g37850) that was up-regulated upon Pseudomonas syringae infection, salt stress, and jasmonate treatment (www.genevestigator.ethz.ch). In comparison to the RHC clusters, very few of the LRB-specific tags were highly up-regulated or down-regulated nor did their expression level increase or decrease during the full course of nodulation (compare number of tags in subclusters 5A, 5D, 5E, and 5F [Supplemental Fig. S1] and in subcluster 6B [Supplemental Fig. S2]). The establishment of a nodule in the physiological context of the hydroponic LRBs seems simpler than that in the zone-I cortex of developing root hairs.
CONCLUSION
In summary, LRB nodulation shows less stringent NF structure requirements and fewer transcriptional changes than RHC nodulation in the same host. A number of plant functions have been identified that are potentially involved in preparing the root cortex for bacterial colonization. These tags will be helpful tools to further investigate the molecular characteristics of intercellular invasion in Sesbania and, by comparison, in other legume species that allow crack-entry invasion of symbiotic rhizobia.
MATERIALS AND METHODS
Plant Material and Bacterial Strains
Sesbania rostrata Brem seedlings were germinated and grown in tubes containing liquid medium or in Leonard jars as described (Goormachtig et al., 1995; Fernández-López et al., 1998). Plants were inoculated with Azorhizobium caulinodans ORS571 (pBBR5-hem-gfp5-S65T) or ORS571-V44 (pBBR5-hem-gfp5-S65T; Van den Eede et al., 1987; D'Haeze et al., 2004).
RNA Extraction and cDNA-AFLP Experiments
Samples were frozen in liquid nitrogen until RNA purification. RNA was extracted as described (Kiefer et al., 2000). The protocol for cDNA-AFLP was applied according to Breyne et al. (2002, 2003) with the most selective oligonucleotides (+2/+2), resulting in 128 primer combinations that maximally reduced template complexity, thus simplifying later analysis and band isolation. Gels were scored with the AFLP-QuantarTM-Pro (Keygene), analyzed with a Microsoft Access-based software application (ArrayAN; Vandenabeele et al., 2003), and normalized on the intensity levels of constitutive bands in each primer combination. A correction factor was introduced to account for differences in lane intensity because of loading differences and was calculated by dividing the sum of all individual band intensities within one lane by the average of all sums within the respective primer combination. Each band intensity value was divided by this correction factor. A CV was calculated as the sd on all values of the time course divided by the average expression over the time course.
Within each primer combination, 5% of the genes with the lowest CV value were marked as constitutively expressed. Per lane (time point), the intensities of these bands were summed and divided by the average of the sum, generating a second correction factor used to normalize the raw expression data generated by AFLP-QuantarTM-Pro. The CV was again calculated on these normalized data for each gene and used as a selection criterion for differential expression (Vandenabeele et al., 2003) with a cutoff value of log2(CV) = 0.1. Hierarchical clustering of the data was obtained with the Multiple Experiment Viewer (The Institute for Genome Research) software (Saeed et al., 2003). The K means clustering was done with the Euclidian distance. K means were calculated with a maximum of 50 iterations and the number of clusters mentioned; the resulting hierarchical clusters were made with the Euclidean distance and average linkage clustering.
Fragment Isolation, Sequencing, and Identification
Fragments were excised by superimposing the dried gel with an autoradiograph and eluted by incubation in 100 μL distilled water for 1 h. Of the eluate, 5 μL was used as a template for subsequent reamplification. After PCR reactions with the corresponding +2/+2 primer combinations, the resulting amplicons were sequenced directly. Low quality sequences were removed from the data set.
All sequences were compared to the nonredundant protein database at the European Molecular Biology Laboratory and to the M. truncatula Gene Index at The Institute for Genome Research with BLASTX and BLASTN algorithms, respectively (Altschul et al., 1997). Tags homologous (E value < 10−3) to accessions in either database were withheld in the final database.
qRT-PCR Analysis
Tissues obtained from a biological repeat were analyzed by qRT-PCR as described (Vlieghe et al., 2005). First-strand cDNA was synthesized with the Superscript RT II cDNA synthesis kit (Invitrogen). qRT-PCRs were run on a iCycler iQ (Bio-Rad) with a kit containing SYBR Green (Eurogentec). A ubiquitin gene was used as a constitutive control (for primer sequences, see Supplemental Table S2; Corich et al., 1998). All reactions were done in triplicate, averaged, and normalized with the 2−ΔΔCT method (Livak and Schmittgen, 2001).
In Silico Analysis
With the S. rostrata LYR3 tag as a query, both the genomic sequence (www.ncbi.nlm.nih.gov) and the Medicago EST (www.tigr.org) databases were searched with BLASTX and tBLASTN (Altschul et al., 1997). Of the sequences retrieved, redundant ones were removed by one-on-one alignments with ClustalW and the resulting nonredundant set was subjected to phylogenetic analysis. Sequences were aligned with the free workbench software (http://www.clcbio.com). Neighbor-joining trees were constructed and 1,000 iterations were done to test node significance for bootstrap analysis.
Gene expression of this experiment was compared with that of the microarray data from Lohar et al. (2006) and El Yahyaoui et al. (2004). To link the tags to the ESTs spotted on both arrays, we downloaded the EST sequences from the Gene Expression Omnibus (GEO GSE3441) of the National Center for Bioinformatics Information and received the corresponding sequences directly from El Yahyaoui et al. (2004). Through BLASTN, tags could be linked to ESTs from both experiments and general expression trends could be compared. No specific E-value threshold was used because the length of the tags varied a lot. Therefore, for correct assignment of the tags to ESTs, they were curated manually. The whole data sheet resuming the results for individual tags are also available for querying at http://www.psb.ugent.be/supplementary-data by clicking on the corresponding article.
Isolation of Full-Length Sequences
The full-length cDNA sequence of SrLyr3 was obtained by 5′ and 3′ RACE by means of the Smart RACE cDNA amplification kit (CLONTECH). The fragments were cloned in the pCRII-TOPO vector (Invitrogen). The full-length cDNA sequence of SrLyr3 could be amplified from S. rostrata cDNA with primers SrLYR3FULLS (5′-CCTTCCTGTGCATCTGCAAAAAC-3′) and SrLYR3FULLAS (5′-GGCTGGTATCTCATTCACAACCC-3′).
Sequence data for SrLyr3 from this article can be found in the GenBank/EMBL data libraries under accession number EF408056.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Hierarchical clustering of the six RHC-specific subgroups.
Supplemental Figure S2. Hierarchical clustering of the five crack-entry-specific subgroups.
Supplemental Figure S3. qRT-PCR verification of a subset of differential tags.
Supplemental Figure S4. Representation of all tags in functional classes.
Supplemental Table S1. Complete data set of identified tags.
Supplemental Table S2. List of qRT-PCR primers used.
Supplementary Material
Acknowledgments
The authors acknowledge Pascal Gamas and Jérôme Gouzy for providing the raw data from their published work, Christa Verplancke for skillful technical assistance, Tom Boonefaes for assistance with the computational analysis of the data, and Martine De Cock for help in preparing the manuscript.
This work was supported by the Research Foundation-Flanders (grants nos. G.0066.07 and G.0341.04 and predoctoral fellowship to J.D.H.) and the Institute for the Promotion of Innovation by Science and Technology in Flanders (predoctoral fellowship to W.C.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Sofie Goormachtig (sofie.goormachtig@psb.ugent.be) and Marcelle Holsters (marcelle.holsters@psb.ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané J-M, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GED, Ayax C, Lévy J, Debellé F, Baek J-M, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé J-C, Dénarié J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J-F, Barre A, Ben Amor B, Bersoult A, Campos Soriano L, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2005) Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol Plant-Microbe Interact 18 487–498 [DOI] [PubMed] [Google Scholar]
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J 9 745–753 [DOI] [PubMed] [Google Scholar]
- Beers EP, Woffenden BJ, Zhao C (2000) Plant proteolytic enzymes: possible roles during programmed cell death. Plant Mol Biol 44 399–415 [DOI] [PubMed] [Google Scholar]
- Brewin NJ (2004) Plant cell wall remodelling in the Rhizobium—legume symbiosis. CRC Crit Rev Plant Sci 23 293–316 [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M (2003) Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics 269 173–179 [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, et al (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M (2005) SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci USA 102 10369–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano CM, Czymmek KJ, Gann JG, Sherrier DJ (2007) Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225 541–550 [DOI] [PubMed] [Google Scholar]
- Charron D, Pingret J-L, Chabaud M, Journet E-P, Barker DG (2004) Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol 136 3582–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi M (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39 487–512 [DOI] [PubMed] [Google Scholar]
- Corich V, Goormachtig S, Lievens S, Van Montagu M, Holsters M (1998) Patterns of ENOD40 gene expression in stem-borne nodules of Sesbania rostrata. Plant Mol Biol 37 67–76 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, De Rycke R, Mathis R, Goormachtig S, Pagnotta S, Verplancke C, Capoen W, Holsters M (2003) Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc Natl Acad Sci USA 100 11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haeze W, Gao M, De Rycke R, Van Montagu M, Engler G, Holsters M (1998) Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol Plant-Microbe Interact 11 999–1008 [Google Scholar]
- D'Haeze W, Gao M, Holsters M (2004) A gfp reporter plasmid to visualize Azorhizobium caulinodans during nodulation of Sesbania rostrata. Plasmid 51 185–191 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, Holsters M (2002) Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12 79R–105R [DOI] [PubMed] [Google Scholar]
- D'Haeze W, Mergaert P, Promé J-C, Holsters M (2000) Nod factor requirements for efficient stem and root nodulation of the tropical legume Sesbania rostrata. J Biol Chem 275 15676–15684 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng X-W, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog M, Verhoef N, Munnik T (2003) Nod factor and elicitors activate different phospholipid signaling pathways in suspension-cultured alfalfa cells. Plant Physiol 132 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G, Schroeyers K, Holsters M, Goormachtig S (2006) Signaling and gene expression for water-tolerant legume nodulation. CRC Crit Rev Plant Sci 25 367–380 [Google Scholar]
- Den Herder J, Lievens S, Rombauts S, Holsters M, Goormachtig S (2007) A symbiotic plant peroxidase involved in bacterial invasion of the tropical legume Sesbania rostrata. Plant Physiol 144 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966 [DOI] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-López M, Goormachtig S, Gao M, D'Haeze W, Van Montagu M, Holsters M (1998) Ethylene-mediated phenotypic plasticity in root nodule development on Sesbania rostrata. Proc Natl Acad Sci USA 95 12724–12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 725–734 [DOI] [PubMed] [Google Scholar]
- Geurts R, Bisseling T (2002) Rhizobium Nod factor perception and signalling. Plant Cell (Suppl) 14 S239–S249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Heidstra R, Hadri A-E, Downie JA, Franssen H, van Kammen A, Bisseling T (1997) Sym2 of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol 115 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormachtig S, Capoen W, Holsters M (2004. a) Rhizobium infection: lessons from the versatile nodulation behaviour of water-tolerant legumes. Trends Plant Sci 9 518–522 [DOI] [PubMed] [Google Scholar]
- Goormachtig S, Capoen W, James EK, Holsters M (2004. b) Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc Natl Acad Sci USA 101 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormachtig S, Valerio-Lepiniec M, Szczyglowski K, Van Montagu M, Holsters M, de Bruijn FJ (1995) Use of differential display to identify novel Sesbania rostrata genes enhanced by Azorhizobium caulinodans infection. Mol Plant-Microbe Interact 8 816–824 [DOI] [PubMed] [Google Scholar]
- Hattan J, Kanamoto H, Takemura M, Yokoto A, Kohchi T (2004) Molecular characterization of the cytoplasmic interacting protein of the receptor kinase IRK expressed in the inflorescence and root apices of Arabidopsis. Biosci Biotechnol Biochem 68 2598–2606 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF (1999) Patterning the flower. Dev Biol 209 211–220 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kiefer E, Heller W, Ernst D (2000) A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol Biol Rep 18 33–39 [Google Scholar]
- Kieffer M, Davies B (2001) Developmental programmes in floral organ formation. Semin Cell Dev Biol 12 373–380 [DOI] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55 521–535 [DOI] [PubMed] [Google Scholar]
- Küster H, Hohnjec N, Krajinski F, El Yahyaoui F, Manthey K, Gouzy J, Dondrup M, Meyer F, Kalinowski J, Brechenmacher L, et al (2004) Construction and validation of cDNA-based Mt6k-RIT macro- and microarrays to explore root endosymbioses in the model legume Medicago truncatula. J Biotechnol 108 95–113 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet E-P, Ané J-M, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 [DOI] [PubMed] [Google Scholar]
- Lievens S, Goormachtig S, Den Herder J, Capoen W, Mathis R, Hedden P, Holsters M (2005) Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol 139 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens S, Goormachtig S, Holsters M (2001) A critical evaluation of differential display as a tool to identify genes involved in legume nodulation: looking back and looking forward. Nucleic Acids Res 17 3459–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Peñuela S, Samac D, Town C, Silverstein KAT, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception in rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick AM, Küster H (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant-Microbe Interact 17 1063–1077 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Weinman JJ, Rolfe BG, Djordjevic MA (2000) Rhizobia can induce nodules in white clover by “hijacking” mature cortical cells activated during lateral root development. Mol Plant-Microbe Interact 13 170–182 [DOI] [PubMed] [Google Scholar]
- Mathis R, Grosjean C, de Billy D, Huguet T, Gamas P (1999) The early nodulin gene MtN6 is a novel marker for events preceding infection of Medicago truncatula roots by Sinorhizobium meliloti. Mol Plant-Microbe Interact 12 544–555 [DOI] [PubMed] [Google Scholar]
- Maunoury N, Kondorosi A, Kondorosi E, Mergaert P (2007) Cell biology of nodule infection and development. In EK James, JI Sprent, MJ Dilworth, WE Newton, eds, Nitrogen-Fixing Leguminous Symbioses (Nitrogen Fixation: Origins, Applications, and Research Progress Series, Vol 7). Springer, Berlin (in press)
- Mithöfer A (2002) Suppression of plant defence in rhizobia-legume symbiosis. Trends Plant Sci 7 440–444 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Long SR (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol 134 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GED, Downie JA (2006) Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J 48 883–894 [DOI] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, Imberty A (2006) LysM domains of Medicago truncatula NFP protein involved in Nod factor perception: glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 16 801–809 [DOI] [PubMed] [Google Scholar]
- Nørholm MHH, Nour-Eldin HH, Brodersen P, Mundy J, Halkier BA (2006) Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett 580 2381–2387 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Engstrom EM, Long SR (2001) Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly N, Pucciariello C, Mandon K, Innocenti G, Jamet A, Baudouin E, Hérouart D, Frendo P, Puppo A (2006) Reactive oxygen and nitrogen species and glutathione: key players in the legume-Rhizobium symbiosis. J Exp Bot 57 1769–1776 [DOI] [PubMed] [Google Scholar]
- Poulsen C, Pødenphant L (2002) Expressed sequence tags from roots and nodule primordia of Lotus japonicus infected with Mesorhizobium loti. Mol Plant-Microbe Interact 15 376–379 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Riely BK, Mun J-H, Ané J-M (2006) Unravelling the molecular basis for symbiotic signal transduction in legumes. Mol Plant Pathol 7 197–207 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 374–378 [DOI] [PubMed] [Google Scholar]
- Schnabel EL, Frugoli J (2004) The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol Genet Genomics 272 420–432 [DOI] [PubMed] [Google Scholar]
- Schroeyers K, Chaparro C, Goormachtig S, Holsters M (2004) Nodulation-enhanced sequences from the water stress-tolerant tropical legume Sesbania rostrata. Plant Sci 167 207–216 [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Amyot L (2003) Symbiosis, inventiveness by recruitment? Plant Physiol 131 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Siemering KR, Hodge S, Grbic V, Haseloff J (2006) A map of KNAT gene expression in the Arabidopsis root. Plant Mol Biol 60 1–20 [DOI] [PubMed] [Google Scholar]
- Van den Eede G, Dreyfus B, Goethals K, Van Montagu M, Holsters M (1987) Identification and cloning of nodulation genes from the stem-nodulating bacterium ORS571. Mol Gen Genet 206 291–299 [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U (2006) Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol 140 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA 100 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit B (2004) Determination of cell fate in apical meristems. Curr Opin Plant Biol 7 57–64 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L (2005) The DP-E2F-like DEL1 gene controls the endocycle in Arabidopsis thaliana. Curr Biol 15 59–63 [DOI] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Cell polarity: ROPing the ends together. Curr Opin Plant Biol 8 613–618 [DOI] [PubMed] [Google Scholar]
- Yang W-C, de Blank C, Meskiene I, Hirt H, Bakker J, van Kammen A, Franssen H, Bisseling T (1994) Rhizobium Nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Riely BK, Burns NJ, Ané J-M (2006) Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.