Abstract
Natural genetic variation in fatty acid synthesis and modification pathways determine the composition of vegetable oils, which are major components of human diet and renewable products. Based on known pathways we combined diversity and genetic analysis of metabolites to infer the existence of enzymes encoded by distinct loci, and associated these with specific elongation steps or subpathways. A total of 107 lines representing different Brassica genepools revealed considerable variation for 18 seed fatty acid products. The effect of genetic variation within a single biochemical step on subsequent products was demonstrated using a correlation matrix of scatterplots, and by calculating relative step yields. Surprisingly, diploid Brassica oleracea segregating populations had a similar range of variation for individual fatty acids as across the whole genepool. This allowed identification of 22 quantitative trait loci (QTL) associated with activity in the plastid, early stages of synthesis, desaturation, and elongases. Four QTL were assigned to early stages of synthesis, seven to subpathway specific or general elongase activity, one to ketoacyl acyl-carrier protein synthetase, and two each to fatty acid desaturase and either desaturase or fatty acyl-carrier protein thioesterase. An additional 10 QTL had distinct effects but were not assigned specific functions. Where contrasting behavior in more than one subpathway was detected, we inferred QTL specificity for particular combinations of substrate and product. The assignment of enzyme function to QTL was consistent with the known position of some Brassicaeae candidate genes and collinear regions of the Arabidopsis (Arabidopsis thaliana) genome.
Plants are a vital source of renewable oils. Most vegetable oil currently produced meets the demand for human consumption, with as much as 25% of human calorific intake in developed countries derived from the constituent fatty acids (Broun et al., 1999). A third of plant oil harvested is already used for nonfood applications. Controlling the composition and maximizing the energy-efficient yield of oils within crop species have been recognized as major goals for plant breeders and the biotechnology industry. This requires an understanding of the genetic and biochemical basis of available variation.
Brassica rapeseed (Brassica napus), soybean (Glycine max), oil palm (Elaeis guinensis), and sunflower (Helianthus annuus) account for more than 65% of vegetable oil production worldwide (Gunstone, 2001). However, these represent a very limited sample of the diversity available within the plant kingdom, where at least 200 different fatty acids have so far been described, and it is thought that many more remain to be discovered (Van de Loo et al., 1993). The adaptive advantage or requirement for this diversity is unclear, although the ability of plants to tolerate high levels of unusual fatty acids is due to sequestration into oil bodies that appear to have no structural function (Millar et al., 2000).
The unique properties of many of the less abundant plant fatty acids have the potential for use in a range of industrial applications. To develop economically viable oilseed crops with modified fatty acid profiles, there is a requirement to manipulate the activity (or gene expression) of relevant key constituent steps in the synthetic or modification pathways, i.e. to carry out genetic metabolic engineering. This can be achieved either through up- or down-regulation of an introduced recombinant gene (transgenic), deletion of endogenous genes (mutagenesis), or by selection of appropriate combinations of the relevant naturally occurring alleles present in the gene pool. Evidence from natural plant populations suggests the latter is indeed a feasible approach (Millar et al., 2000).
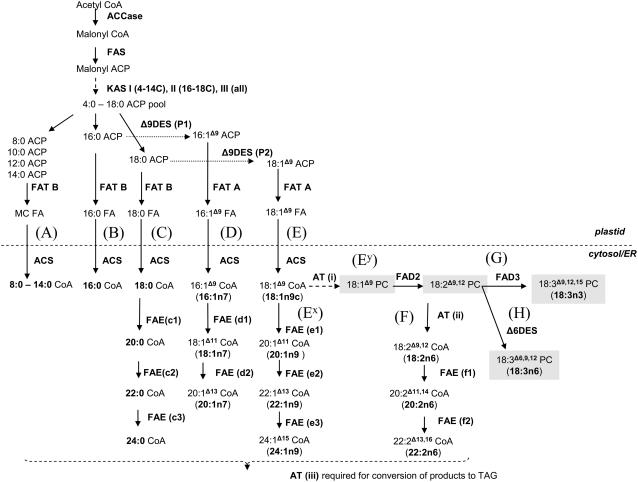
Figure 1 shows a simplified model of the enzymatic interactions involved in the synthesis of triacylglycerols within the developing seed and their division between the two cellular compartments (for review, see Ohlrogge et al., 1991; Slabas et al., 2001; Voelker and Kinney, 2001). The interplay of the various mechanisms described ultimately controls the final fatty acid composition, and together with consideration of the rate of de novo fatty acid synthesis, the yield of triacylglycerols formed from the condensation of diacylglycerol and acyl-CoA pools in the final step of the Kennedy pathway. Metabolic flux studies have revealed that over 50% of the carbon flux in developing B. napus seeds is directed toward triacylglycerol synthesis (Schwender et al., 2004).
Figure 1.
Current understanding of the pathways of fatty acid synthesis which are known to occur within the Brassicaceae. Dashed lines indicate multistep pathways, while shaded boxes indicate inferred steps. The capital letters in parentheses indicate the potential pathways by which triacylglycerols can be produced following ACP synthesis within the plastid. These letters are used to differentiate these pathways (A–H) as described within this article. Suffix annotations in parentheses after enzyme name have been added to simplify description of these enzymes within this article. ACCase, Acetyl-CoA carboxylase; FA, fatty acid; FAS, fatty acid synthase; FAT, fatty ACP thioesterase; DES, desaturase; ACS, acyl-CoA synthetase; MC, medium chain; AT, various acyl transferases; PC, phosphatidylcholine; TAG, triacylglycerol. Within the plastid, fatty acids are synthesized de novo from acetyl-CoA in a series of condensation and elongation reactions to form a pool of mostly 16:0- and 18:0-acyl chains linked to ACP, of which a portion are desaturated to monounsaturated acyl-ACPs. These acyl-ACPs are cleaved by specific thioesterases to free fatty acids and then converted to acyl-CoAs by acyl-CoA synthetases at or near the outer plastid envelope, so as to be available for passage to the cytoplasm (Pollard and Ohlrogge, 1999). Export to the cytoplasm, or entry to the eukaryotic pathway, diverts acyl chains away from membrane lipid synthesis in the plastid (the prokaryotic pathway) and makes them available for incorporation into triacylglycerols. Within the cytosol (primarily endoplasmic reticulum) further modification of the acyl chains occurs through the complex interaction of desaturation, elongation, and phospholipid/acyl-CoA exchange mechanisms. The extra carbon required for acyl chain elongation commonly observed in the Brassicaceae originates from a distinct cytosolic acetyl-CoA pool, probably derived from mitochondrial citrate metabolism (Fatland et al., 2005).
Economically, the Brassica genus is represented by oilseed, vegetable, fodder, and condiment crops, with the species B. napus (AC genome, n = 19), Brassica rapa (A genome, n = 10, syn. Brassica campestris), Brassica juncea (AB genome, n = 18), and Brassica carinata (BC genome, n = 17) providing about 12% of the worldwide edible vegetable oil supplies. The major fatty acids present within rapeseed (canola) are palmitic (16:0), oleic (18:1n9), linoleic (18:2n6), α-linolenic (18:3n3), eicosenoic (20:1n9), and erucic (22:1n9) acids. Of these, 18:1n9 is thermostable and thus valuable for cooking, while 18:2n6 is unsaturated with two double bonds providing nutritional benefits. The three double bonds within 18:3n3 lead to instability and rapid oxidation, thus reducing the shelf life of products, while 22:1n9 is poorly catabolized by the mammalian β-oxidation pathway (Sauer and Kramer, 1983). Conventional screening of parental germplasm and subsequent breeding selection has been successful in the development of the modern varieties of canola, which have been selected for low levels of both these fatty acids. However, complete elimination of 18:3n3 from the seed oil is probably impossible, since it plays an essential role in photosynthesis (Hugly et al., 1989), and furthermore it is critical for pollen development (McConn and Browse, 1996). Burns et al. (2003) have highlighted the value of identifying the quantitative trait loci (QTL) responsible for such traits as a valuable contribution to plant breeding via marker assisted selection.
Determining the genetic basis for the variation in plant metabolites has great potential for both modification of metabolic composition through classical breeding (Keurentjes et al., 2006) and for unraveling of metabolic, regulatory, and developmental pathways (Jansen and Nap, 2001). QTL analysis in segregating plant populations has been used to detect the presence of loci affecting metabolite profiles associated with particular synthesis and modification pathways. For example this approach has been applied to different classes of secondary metabolites, including flavonoids in maize (Zea mays) and poplar (Populus spp.; McMullen et al., 1998, 2001; Morreel et al., 2006), tetrahydrocannabinolic acid in hemp (Cannabis sativa; Pacifico et al., 2006), and glucosinolates in Arabidopsis (Arabidopsis thaliana) and Brassica (Kliebenstein et al., 2001; Li et al., 2001; Kroymann et al., 2003). However, it is recognized that in such analyses many loci may be involved (Kliebenstein et al., 2002).
Oil content and fatty acid composition are typically quantitative traits under polygenetic control, influenced by environmental conditions. It has been recognized that the identification and mapping of QTLs involved in the lipid biosynthesis pathway could provide information for improved breeding selection (Lionneton et al., 2002). Qiu et al. (2006) reported seven QTL and Ecke et al. (1995) reported three QTLs associated with oil content in B. napus. In both studies QTL showed close linkage to the 22:1n9 genes fatty acid elongase1.1 (FAE1.1) and FAE1.2, and a similar situation has been reported for B. juncea (Thormann et al., 1996; Cheung et al., 1998; Gupta et al., 2004). Genetic characterization of the loci involved in 18:1n9 content has been carried out in B. napus, B. rapa (Tanhuanpaa et al., 1998), and B. juncea (Sharma et al., 2002). Similar studies have also been carried out for low 18:3n3 (Tanhuanpaa et al., 1995; Jourdren et al., 1996) and for both linolenic and 22:1n9 (Thormann et al., 1996). Quantitative genetic studies of oil synthesis in the model dicotyledonous plant and closely related member of the Brassicaceae, Arabidopsis (Hobbs et al., 2004), has identified two major and two minor QTL that account for 43% of the variation in seed oil content, as well as other QTL affecting composition. This analysis used a recombinant inbred population derived from Landsberg erecta and Cape Verdi Islands ecotypes. Strong QTL identified for linoleic acid (18:2) and linolenic (18:3) acid content colocated with the FAD3 locus, while a QTL for 18:1n9 colocated with FAD2. Other less significant QTL were detected for 16:0, stearic (18:0), and 20:1n9 acids.
The fatty acid composition of Brassicaceae seed oils is thought to be genetically more variable than the composition of any other major vegetable oil (Sovero, 1993). This may reflect the position of Brassica crops as among the oldest cultivated plants known to man (Prakash, 1980; Yan, 1990). Diversity studies can demonstrate the presence of genes and allelic variation involved in the regulation of seed lipid traits (Mandal et al., 2002; O'Neill et al., 2003). Utilization of this natural allelic diversity would provide an alternative to a genetic modification approach. Classical breeding selection was responsible for the widespread adoption of canola and oilseed rape, as a result of significant changes in the fatty acid composition, as well of low glucosinolates in canola oilseed varieties. This involved the introgression of recessive alleles that reduced 22:1n9 levels from 60% to less than 2% (Morice, 1974). Extensive natural variation has been found in levels of seed oil content, very long chain fatty acids, and polyunsaturated fatty acids by surveying the Arabidopsis genepool (O'Neill et al., 2003), and a similar wide diversity of oil content and fatty acid profiles found within B. juncea (Mandal et al., 2002). The potential for the diploid Brassica C genome being exploited as a source of variation to modulate rapeseed oil yield and composition has to date not been comprehensively assessed. The development of structured core collections of ex situ genetic resource accessions representing domesticated vegetable Brassica oleracea, wild C genome species, and B. napus crop types (King et al., 2004) provides an opportunity to collate and interpret data from diversity screening in the context of the increasing volume of genetic, genomic, and metabolomic data.
The Brassica A and C diploid genomes are thought to have arisen from an ancient hexaploid ancestor in common with Arabidopsis, with a series of segmental chromosome duplications resulting in the presence of an average of three paralogous genes when compared to single gene loci within Arabidopsis (Lysak et al., 2005). The amphidiploid genomes B. napus, B. carinata, and B. juncea may typically contain six loci for each gene present within Arabidopsis, with a heterozygote line having the potential to contain 12 distinct alleles and sequence transcripts. We therefore surmised that by focusing our analysis on a single diploid crop genome and using homozygous doubled haploid (DH) populations, we would increase the likelihood of detecting significant genetic effects associated with fatty acid composition, and of deriving information about the key steps in the relevant complex metabolic pathways. We made use of two existing diverse reference mapping populations representing the Brassica C genome, and analyzed the results in the context of variation observed throughout the associated Brassica genepool.
This article demonstrates the power of combining analysis of natural variation in seed fatty acid composition with quantitative genetic analysis to provide novel information about the regulation of storage lipid biosynthetic pathways.
RESULTS
Fatty Acid Analysis
The seed weights of all the plants analyzed (Table I) ranged between 0.4 to 7.4 mg/seed. To assess the possible effect of this variation on the fatty acid composition, the correlation between micromoles per seed and micromoles per gram fresh weight values was calculated for each fatty acid. The values varied from 0.47 (20:0) to 0.96 (18:1n9), with a median correlation of 0.78. In addition we analyzed the percentage contribution of each fatty acid to the total content. We found that the combined results provided the most representative reflection of quantitative variation in synthesis and modification throughout the metabolic pathway. To determine any interaction with seed development, the possible contribution of QTL regulating seed weight was assessed in relation to effects on variation among individual fatty acids. Three QTL were identified on linkage group (LG) O4, O6, and O9 contributing to seed size variation. However, these did not correspond to any of the QTL accounting for differences in the total fatty acid content (G.J. King and G.C. Barker, unpublished data).
Table I.
Distribution of lines sampled from the Brassica genepool
Groups A to E represent lines contributing to the diversity survey, with material obtained from genetic resource collections. Groups F and G refer to the DH segregating mapping populations.
| Group | Species | Varietas or Subgroup | Crop Type | No. Lines |
|---|---|---|---|---|
| A | B. napus (AC) | oleifera | Modern oil crops | 20 |
| B | oleifera | Fodder rape (NZ) | 2 | |
| B | napobrassica | Swede | 5 | |
| B | biennis | Fodder rape | 3 | |
| C | B. oleracea (C) | acephala | Kales | 8 |
| C | capitata | Cabbages | 8 | |
| C | tronchuda | 3 | ||
| C | botrytis | Cauliflower | 15 | |
| C | italica | Broccoli | 10 | |
| C | alboglabra | Chinese kale | 2 | |
| C | gemmifera | Brussels sprouts | 6 | |
| C | gongylodes | Kohl rabi | 2 | |
| C | Others | Wild species | 3 | |
| D | B. rapa (A) | chinensis | Hybrid Chinese cabbage | 3 |
| D | pekinensis | Chinese cabbage | 4 | |
| D | Pak Choi group | Japanese greens, Neep greens Gp | 2 | |
| D | rapa | Turnip | 3 | |
| D | ruvo | Broccoletto | 2 | |
| D | trilocularis | Yellow sarson | 1 | |
| D | Others | Rapid cycling | 1 | |
| D | sylvestris | Wild species | 2 | |
| E | Brassica species (B) | carinata | Ethiopian mustard | 1 |
| E | “B” species | Wild varieties | 2 | |
| Total | 107 | |||
| F | B. oleracea (C) | AGDH population | Broccoli × Chinese kale segregants | 99 |
| G | B. oleracea (C) | NGDH population | Cauliflower × Brussels sprouts segregants | 69 |
| Total | 168 |
Sources of Variability
The variance components associated with the measurements of micromole per seed fatty acids were calculated (Table II). For most fatty acids, there was little variability between sample or occasions. For 10 of the 17 fatty acids most of the measured variation between lines was attributed to the genetic components of species, subspecies, and line. Between accession variance components differed between fatty acids, with a median of 27.6%. Although individual line means have a level of environmental variation, we are able to make inferences attributable to allelic variation across the genepool. The components contributing to the overall genetic variation varied between fatty acids. For most fatty acids, there are relatively small differences in relative amount between subspecies, with the exception of those that have been subject to recent selection in modern oilseed canola lines when these lines were included in the analysis (data not shown). A similar pattern of variance components was observed for the calculated step yields.
Table II.
Percentage of total variation accounted for by the different variance components in the genepool diversity assessment, for the different fatty acids
The modern oilseeds were excluded from this analysis. Significance of variance components other than between samples was assessed by comparing the change in deviance between models including the component and those excluding it to a χ2 distribution on the appropriate number of degrees of freedom. Significance levels are shown as follows: ***, <0.1%; **, <1%; *, <5%.
| Fatty Acid | Occasion | Species | Subspecies | Line | Accession | Sample |
|---|---|---|---|---|---|---|
| 14:0 | 1.9 | 0 | 26.8* | 4.3 | 56.9*** | 8.4 |
| 16:0 | 0 | 39.5*** | 2.6 | 25.4 | 27.6*** | 4.9 |
| 16:1n7 | 4.9 | 22.5* | 0 | 37.8* | 22.9*** | 11.9 |
| 18:0 | 0 | 27.0 | 5.9 | 19.2 | 36.5*** | 11.3 |
| 18:1n7c | 2.5 | 37.7*** | 1.7 | 31.8** | 17.2*** | 9.2 |
| 18:1n9c | 2.2 | 9.9 | 13.3 | 21.1 | 39.0*** | 14.4 |
| 18:2n6c | 2.9 | 19.7 | 9.0 | 15.1 | 46.5** | 6.7 |
| 18:3n3 | 0 | 19.6 | 16.8 | 31.6** | 22.1*** | 9.8 |
| 20:0 | 2.7 | 15.8 | 17.8 | 22.4 | 34.5*** | 6.7 |
| 20:1n7 | 1.7 | 10.2 | 20.2* | 50.6** | 6.4 | 10.9 |
| 20:1n9 | 2.2 | 37.7 | 4.5 | 30.4* | 18.7*** | 6.6 |
| 20:2 | 0 | 23.6 | 15.9 | 45.4** | 8.8*** | 6.3 |
| 22:0 | 5.2 | 29.2 | 33.0** | 8.6 | 18.8*** | 5.2 |
| 22:1n9 | 2.8 | 0 | 23.1* | 36.4 | 24.8*** | 12.8 |
| 22:2 | 0.5 | 0 | 18.2 | 27.1 | 41.0*** | 13.1 |
| 24:0 | 0 | 8.9 | 4.0 | 25.7 | 44.9*** | 16.5 |
| 24:1 | 0 | 11.1 | 0 | 36.2* | 40.6*** | 12.1 |
Diversity of Fatty Acid Amounts
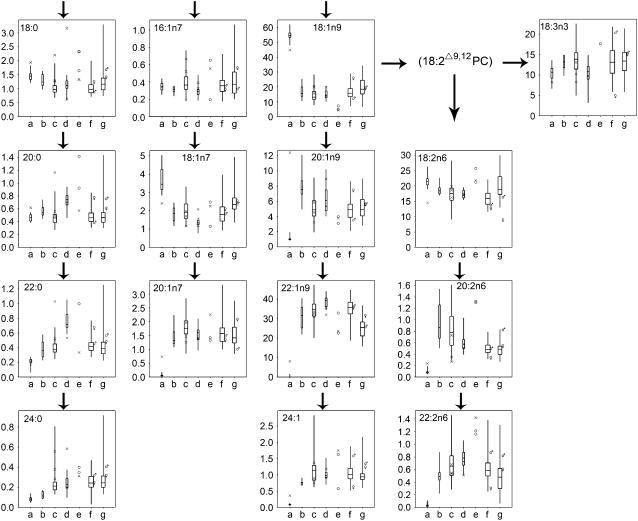
Both the distribution of micromole per seed and the percentage contribution of each fatty acid to the total content of fatty acid components across taxa at species level were plotted. Only the latter is shown in Figure 2 as the trends and relationships were found to be similar using both methods. The predominant fatty acids are 18:1n9, 18:2n6, 22:1n9, and 18:3n3. The levels of 18:1n9 are approximately 5 times higher in modern oilseed B. napus (var. oleifera) lines than in other lines, with the corresponding absence of 22:1n9 reflecting the recent selective breeding pressure for low 22:1n9. The variety Bronowski also has relatively elevated levels of 18:1n9 and low levels of 22:1n9. Sections of the Bronowski genome have been introgressed into most modern varieties following its original selection as one of the major sources of low glucosinolates and 22:1n9 (Khachatourians et al., 2001). Other crop types within the B. napus genepool, including fodder rape, had a similar distribution of 18:1n9 and 22:1n9 content to that found in the diploid B. oleracea and B. rapa lines. A further striking feature of the modern canola oilseed lines was a 2-fold increase in levels of 18:1n7 when compared with Bronowski, or any of the non-oil varieties. This suggests the presence of a broad-specificity stearoyl acyl-carrier protein [ACP] desaturase that acts on 16:0 to produce 16:1n7 that is subsequently elongated to 18:1n7, or a highly efficient elongase acting on 16:1n7, and/or corresponding inhibition of the elongase that results in 20:1n7. This is even more striking when micromole per seed is measured. Modern oilseed canola lines had similar proportions of 18:2n6 and 18:3n3 compared to the other groups studied, although the micromole per seed values showed approximately 3-fold higher levels of 18:2n6 and 2-fold higher levels of 18:3n3. 18:2n6 esterified to phosphatidylcholine is the substrate for the synthesis of 18:3n3 (Voelker and Kinney, 2001). The line to line variation for these two fatty acids indicates variation in the activities of the FAD enzymes involved. It is striking that a similar range of variation is detected for many of the fatty acids between lines of the B. oleracea mapping populations (i.e. combining alleles from just two lines) as is found across most of the genepool, including the modern oilseed varieties. The levels of both 20:2n6 and 22:2n6 are consistently low in all species, compared to the levels of 18:2n6. However, the levels of 20:2n6 are much lower within the modern oilseed lines compared to the other groups, which suggests that there has been recent selection affecting the efficiency of metabolic steps leading to 18:2n6 elongation. The possible contribution of QTL regulating seed weight in accounting for variation among individual fatty acids was addressed in addition to enzymes in pathway E.
Figure 2.
Box plots representing the variability in the percentage contribution of each fatty acid to the total content within species (a–e) and the B. oleracea mapping populations (f and g). Groupings are as indicated in Table I. The boxes represent the interquartile range, with the median indicated by a line within the box. The maximum and minimum values are represented by the whiskers attached to the boxes. The width of the box represents the number of lines within that group. Outliers within groups are indicated as follows: (a) X = B. napus var. oliefera Bronowski; (c) X = C genome wild species; (d) X = A genome wild species; (e) X = B. carinata and O = B genome wild species; (f)♀ A12DHd, ♂ GDDH33; (g) ♀CA25, ♂AC498.
Comparative Pathway Analysis across Taxa
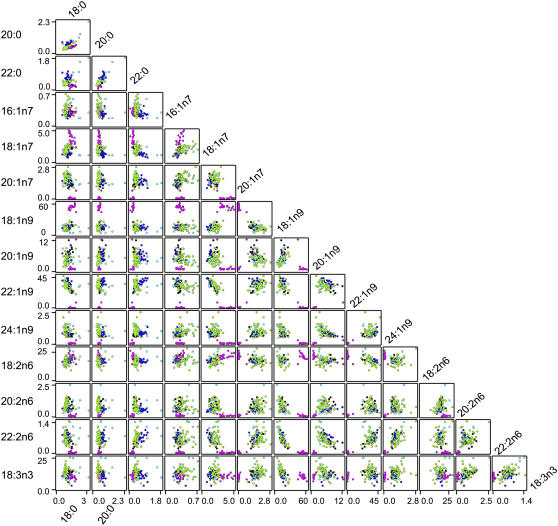
The pattern of relationships between levels of individual fatty acids across taxa can provide information on the genetic contribution to the synthesis and modification pathways. Correlation analyses between fatty acids for different sections of the genepool are shown as a scatter plot matrix (Fig. 3). These plots allow interpretation of trends within and between different taxa, and demonstrate how genetic variation within a single biochemical step can affect subsequent products. Each individual plot represents a pairwise comparison in the percentage amounts of two fatty acid products. For example, in the comparison of 18:0 and 20:0 (top left plot) the crop species are all tightly grouped with similar percentage composition of 18:0 and 20:0, however, in the wild species some lines have approximately twice as much 18:0 compared with other taxa, and the variation in 20:0 is as marked. In contrast, when 18:0 is compared to 22:0, the B. rapa varieties are characterized by having approximately twice the levels found in either B. oleracea or B. napus lines. The highest levels of 22:0 are still observed in some wild species.
Figure 3.
Matrix of scatter plots indicating correlations between the percentage contribution of each fatty acid to the total content of the different triacylglycerols. Colored dots represent individual line taxa, coded as follows: Non-oleifera B. napus, black; B. napus var. olifera, magenta; B. oleracea, green; B. rapa, blue; Brassica Wild sp., cyan. The scale for each fatty acid is indicated on the x and y axis, from zero to the maximum value indicated. Each plot shows the levels of one fatty acid compared to another with the respective scales shown on each axis. Those fatty acids with amounts below 0.5% are not shown.
Modern oilseed canola lines are the most distinct taxonomic grouping as a result of the selection they have undergone for low 22:1n9 and high 18:1n9. Concomitant with the high levels of 18:1n9, there is an increase in levels of fatty acids with chain lengths less than 20. A similar pattern is observed in the fatty acids within pathway F, where the levels of 18:2n6 are raised in modern oilseed canola. However the amounts of 20:2n6 and 22:2n6 are marginal compared to the other groups. While a 2-fold increase in 18:1n7 can be observed in modern oilseed canola lines compared to the other groups, levels of 20:1n7 are either very low or absent within these lines.
The pattern of variation found within the taxonomic groupings across the pathways suggests that concurrent with the selection for low activity of FAE (e1) there has also been a major reduction in the activities of FAE (d2) and FAE (f1), as well as a smaller reduction in the activity of FAE (c1) and FAE (c2). The activities of FAE (d1) or FAE (c3) appear unaffected. One oilseed B. napus line (var. Bronowski) is notable in having a substantial level of 20:1n9 compared with any other accession, although this is not reflected in the level of 22:1n9. This supports the presence of distinct elongase specificities, in this case for FAE (e1) and FAE (e2).
Contributions to Total Fatty Acid Synthesis
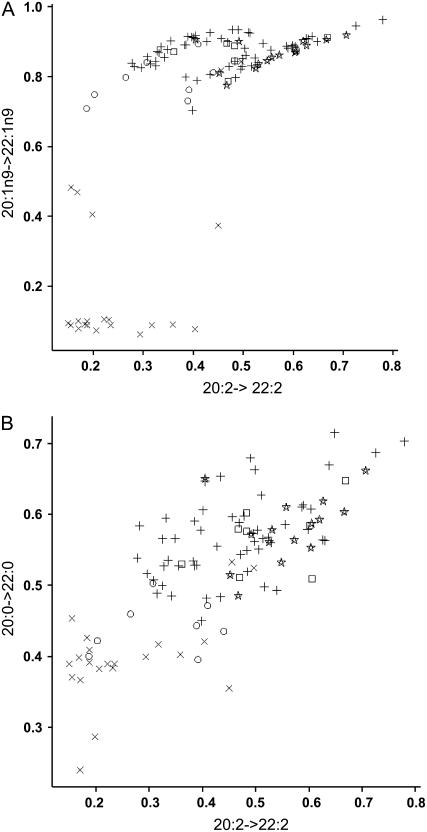
Combined step yields were calculated for all of the steps within the plastid that result in fatty acid substrates for pathway components within the endoplasmic reticulum. This confirmed the differences between the modern, low 22:1n9 oilseed canola and the other lines. The differences in pathway D indicated separate functions for FAE (d1) and FAE (d2), as well as between the step yields of FAE (f1) and FAE (f2). In pathway Ex, FAE (e1) and FAE (e2) had similar step yields, although that for FAE (e3) appeared much higher in the modern oilseed canola lines. The analysis was particularly effective at demonstrating a lack of relationship between the step yields for the two steps FAE (e2) and FAE (f2) within the endoplasmic reticulum (Fig. 4). For FAE (e2) versus FAE (f2; Fig. 4A) the step yields form four groups. The first group includes most lines that have very low step yields (<0.2) for FAE (e2). The second smaller group includes lines with step yields between 0.4 and 0.5. The remaining lines have high step yields and form two similar sized groups. Within each of these there is a strong and fairly linear relationship (r = 0.89 and 0.90) between the step yields. The group with higher yield for FAE (f2) contains most of the B. rapa lines. There does not appear to be any other consistent taxonomical relationship or environmental interaction to account for the presence of the two groupings with lines having high FAE (e2) step yields.
Figure 4.
Comparison of step yields calculated for different elongase enzymes. Inferred relative yields of two elongations are plotted for each line sampled. Yields were calculated according to the equation in “Materials and Methods.” A, y axis: 20:1n9->22:1n9, inferring activity of FAE (e2), x axis: inferred activity of 20:2-> 22:2 (FAE f2). B, y axis: 20:0->22:0 inferred activity of FAE (c2), x axis: 20:2->22:2 inferred activity of FAE (f2). The symbols represent the different taxa groupings. x, Modern oilseed B. napus; ○, nonoilseed B. napus; +, B. oleracea; ⋆, B. napus; □, wild species.
Figure 4B is a contrasting scatter plot that shows the step yields for the two steps FAE (c2) and FAE (f2). This shows a linear relationship across the taxa sampled, with a lower correlation (r = 0.75). The modern, low 22:1n9 oilseed canola lines have low step yields for both steps.
Variation within Reference Segregating B. oleracea Mapping Populations
The range of variation in the level of individual fatty acids observed within both of the B. oleracea mapping populations was of a similar order of magnitude as observed among the species diversity collections (Fig. 2). There was evidence of extensive transgressive segregation compared to parental values. The absolute levels and range of diversity for amounts of all the fatty acids measured were higher in the AG population than the NG population (data not shown). It is striking that the variation observed within the AG and NG population was comparable to, and in many cases exceeded that found within the B. oleracea and B. rapa diversity sets. Of the groups analyzed, the greatest range levels of 22:1n9 was found within segregants of the AG mapping population, where the maximum and minimum values far exceeded those found in the parental lines. Although the total amounts of the fatty acids were lower within the NG population, the variation in the percentage composition observed within the population exceeded that seen within most other taxa.
Identification of QTL in the Brassica C Genome
QTL associated with fatty acid products were detected in both B. oleracea mapping populations. Results obtained with composite interval mapping (QTL Cartographer) confirmed those obtained by multiple regression models (QTL Café), with no additional QTL detected. We were able to assign putative enzyme function to individual QTL by comparing the distribution of QTL for fatty acid products with the established synthesis and modification pathways (Fig. 1). Our interpretations are based on the premise that plastidic synthesis and modification should result in consistent QTL effects on products in any of the individual downstream sets A, B, and C. Conversely, modification in the endoplasmic reticulum should result in contrasting QTL effects on products in one or more of the sets A to F. In general, where a QTL effect has been associated with a particular fatty acid product, it is important to take into account all the preceding synthetic and modification steps. Where we are able to detect contrasting behavior in more than one set we are able to infer specificity of QTL for particular combinations of substrate and product. Out of a total of 22 QTL detected, the locations of 20 were found to be in common for fatty acid levels expressed as either micromole per seed (Table III) or percentage contribution (Table III). However, the mapping intervals for these QTL were found to differ slightly. Two additional QTL (QTL O2-b and O3-a) were detected when the levels of fatty acids were expressed as percentage contribution.
Table III.
QTL detected for each fatty acid in the reference B. oleracea segregating mapping populations, sorted by LG
Map positions are expressed relative to an integrated linkage map with common LG (e.g. O1) and QTL position (e.g. O1-a) nomenclature. The QTL shown relate to the molar concentration per seed, respectively, within the AG and the NG population, and the percentage contribution of each fatty acid within the AG population and the NG population. The midpoint and confidence intervals (min and max) are shown for each QTL in centi-Morgans where applicable, and additive effects indicated for each fatty acid, with positive effect associated with the female and negative effect associated with the male parent. QTL in bold indicate QTL identified but missing within the trait and population indicated. Significance at P < 0.05, P < 0.01, and P < 0.001 are indicated by *, **, and ***, respectively.
| LG | QTL | Pathway
|
A
|
B
|
C
|
C
|
C
|
C
|
D
|
D
|
D
|
E
|
EX
|
EX
|
EX
|
F
|
F
|
F
|
G
|
H
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Min | Max | C14:0 | C16:0 | C18:2C20 | C20:0 | C22:0 | C24:0 | C16:0C1 | C18:1n7 | C18:1n7 | C18:1n9 | C22:2C1 | C22:2C1 | C22:2C1 | C18:2 | C22 | C22:2 | C18:3n3 | C18:3n6 | ||
| cM | ||||||||||||||||||||||
| μmol FA per seed AG | ||||||||||||||||||||||
| O1 | O1-a | 37 | 17 | 55 | 31.61* | 10.49* | 62.93* | 1.55* | ||||||||||||||
| O1 | O1-b | 72 | 53 | 89 | 0.14* | 0.70** | ||||||||||||||||
| O2 | O2-a | 54 | 39 | 69 | −1.92** | |||||||||||||||||
| O2 | O2-b | (% mol only) | ||||||||||||||||||||
| O3 | O3-a | (% mol only) | ||||||||||||||||||||
| O3 | O3-b | 53 | 14 | 80 | −0.67** | |||||||||||||||||
| O3 | O3-c | |||||||||||||||||||||
| O4 | O4-a | 55 | 41 | 79 | −11.40** | −13.59* | −1.59*** | −77.43*** | ||||||||||||||
| O4 | O4-b | 83 | 64 | 104 | −1.94* | −3.89* | ||||||||||||||||
| O5 | O5-a | |||||||||||||||||||||
| O5 | O5-b | 43 | 20 | 53 | −1.74* | 1.85* | ||||||||||||||||
| O6 | O6-a | 15 | 0 | 41 | −3.48* | −26.48* | ||||||||||||||||
| O6 | O6-b | |||||||||||||||||||||
| O7 | O7-a | 16 | 0 | 30 | 11.29*** | 2.68*** | 1.65*** | 0.47* | 4.10** | 69.05** | 30.06*** | 1.51*** | 1.61* | 31.09** | ||||||||
| O7 | O7-b | 37 | 10 | 69 | 37.35** | 17.22*** | −1.59* | |||||||||||||||
| O7 | O7-c | 70 | −1.23* | |||||||||||||||||||
| O8 | O8-a | 5 | 0 | 22 | 55.94*** | 21.87** | ||||||||||||||||
| O8 | O8-b | 26 | 11 | 52 | 2.93** | 2.11*** | 0.80* | 6.43*** | 24.59** | |||||||||||||
| O8 | O8-c | 75 | 49 | 84 | −1.15*** | 0.51* | ||||||||||||||||
| O9 | O9-a | 16 | 0 | 41 | 0.78* | 0.52*** | 4.98*** | 37.35* | 11.81* | |||||||||||||
| O9 | O9-b | 50 | 34 | 62 | −0.10* | 7.35*** | ||||||||||||||||
| O9 | O9-c | 84 | 0.57** | |||||||||||||||||||
| μmol FA per seed NG | ||||||||||||||||||||||
| O1 | O1-a | |||||||||||||||||||||
| O1 | O1-b | 81 | 84 | 95 | −31.83*** | |||||||||||||||||
| O2 | O2-a | 49 | 23 | 70 | 32.55** | 9.08** | 36.12** | |||||||||||||||
| O2 | O2-b | (% mol only) | ||||||||||||||||||||
| O3 | O3-a | (% mol only) | ||||||||||||||||||||
| O3 | O3-b | 55 | 34 | 96 | −5.35* | −28.23* | ||||||||||||||||
| O3 | O3-c | 85 | 58 | 115 | −34.45* | −0.74* | ||||||||||||||||
| O4 | O4-a | 53 | 33 | 72 | −1.48*** | −0.69*** | −0.45*** | −0.33* | −0.34* | −2.03* | ||||||||||||
| O4 | O4-b | |||||||||||||||||||||
| O5 | O5-a | 9 | 0 | 33 | −40.91*** | −11.03** | ||||||||||||||||
| O5 | O5-b | |||||||||||||||||||||
| O6 | O6-a | 18 | −61.40* | |||||||||||||||||||
| O6 | O6-b | 40 | 49.80* | |||||||||||||||||||
| O7 | O7-a | |||||||||||||||||||||
| O7 | O7-b | 38 | 15 | 57 | −2.80** | |||||||||||||||||
| O7 | O7-c | |||||||||||||||||||||
| O8 | O8-a | 12 | 3 | 23 | 7.46** | |||||||||||||||||
| O8 | O8-b | 23 | 0 | 47 | 0.07* | 9.16** | 1.04* | 0.66*** | 0.68** | 0.42*** | 0.57*** | 4.60** | 3.19*** | 51.37*** | 1.82*** | 37.14*** | 1.25** | 1.52** | 25.30*** | |||
| O8 | O8-c | |||||||||||||||||||||
| O9 | O9-a | |||||||||||||||||||||
| O9 | O9-b | 45 | 32 | 54 | 0.09*** | 0.61*** | 2.21*** | |||||||||||||||
| O9 | O9-c | |||||||||||||||||||||
| QTL identified associated with the percentage contribution of each fatty acid to the total content within the AG population | ||||||||||||||||||||||
| O1 | O1-a | 40 | 24 | 56 | −0.80*** | −0.83*** | −0.06* | |||||||||||||||
| O1 | O1-b | 70 | 57 | 83 | 0.744*** | |||||||||||||||||
| O2 | O2-a | 56 | 41 | 71 | −0.09* | |||||||||||||||||
| O2 | O2-b | 86 | 74 | 98 | 0.71* | −1.03* | ||||||||||||||||
| O3 | O3-a | |||||||||||||||||||||
| O3 | O3-b | 52 | 34 | 70 | −0.20* | −0.17*** | ||||||||||||||||
| O3 | O3-c | 90 | 79 | 111 | 0.03* | −0.04** | −1.34* | |||||||||||||||
| O4 | O4-a | 60 | 44 | 76 | 0.04** | 1.58* | 0.95** | −3.07*** | ||||||||||||||
| O4 | O4-b | |||||||||||||||||||||
| O5 | O5-a | |||||||||||||||||||||
| O5 | O5-b | 36 | 19 | 53 | −0.04* | −0.03* | ||||||||||||||||
| O6 | O6-a | 12 | 0 | 30 | −1.14* | |||||||||||||||||
| O6 | O6-b | 52 | 36 | 68 | −0.21** | |||||||||||||||||
| O7 | O7-a | 16 | 2 | 30 | 0.05*** | −0.04** | −0.18*** | |||||||||||||||
| O7 | O7-b | 44 | 32 | 56 | 0.05*** | 0.50*** | ||||||||||||||||
| O7 | O7-c | |||||||||||||||||||||
| O8 | O8-a | 8 | 2 | 14 | 1.92*** | 0.85*** | −0.10*** | |||||||||||||||
| O8 | O8-b | 20 | 10 | 30 | 0.11** | 0.08*** | −0.17*** | −1.60** | ||||||||||||||
| O8 | O8-c | 68 | 50 | 86 | −0.044*** | 0.19** | 0.085*** | |||||||||||||||
| O9 | O9-a | 14 | 0 | 34 | 0.09*** | −0.83* | ||||||||||||||||
| O9 | O9-b | 30 | 20 | 40 | −0.54** | |||||||||||||||||
| O9 | O9-c | 62 | 50 | 74 | 0.31*** | 0.14*** | ||||||||||||||||
| QTL identified associated with the percentage contribution of each fatty acid to the total content within the NG population | ||||||||||||||||||||||
| O1 | O1-a | 40 | 24 | 56 | −1.97*** | |||||||||||||||||
| O1 | O1-b | 70 | 57 | 83 | −1.08* | |||||||||||||||||
| O2 | O2-a | 56 | 41 | 71 | −0.75*** | −0.06* | −0.05* | −0.33** | 0.49* | 2.03** | −1.56* | |||||||||||
| 02 | O2-b | 86 | 74 | 98 | −0.02* | −0.08* | ||||||||||||||||
| O3 | O3-a | 6 | −0.10** | 1.24** | ||||||||||||||||||
| O3 | O3-b | 52 | 34 | 70 | ||||||||||||||||||
| 03 | O3-c | 90 | 79 | 111 | 0.231** | 0.07* | 0.20* | −1.97** | ||||||||||||||
| O4 | O4-a | |||||||||||||||||||||
| O4 | O4-b | |||||||||||||||||||||
| O5 | O5-a | 2 | 0 | 22 | 0.68** | −0.76* | ||||||||||||||||
| O5 | O5-b | 36 | 19 | 53 | 0.10** | 1.04* | ||||||||||||||||
| O6 | O6-a | |||||||||||||||||||||
| O6 | O6-b | |||||||||||||||||||||
| O7 | O7-a | |||||||||||||||||||||
| O7 | O7-b | |||||||||||||||||||||
| O7 | O7-c | |||||||||||||||||||||
| O8 | O8-a | |||||||||||||||||||||
| O8 | O8-b | |||||||||||||||||||||
| 08 | 08-c | 68 | 70 | 86 | 0.16* | −3.42** | 1.91* | 0.11** | ||||||||||||||
| O9 | O9-a | |||||||||||||||||||||
| O9 | O9-b | 30 | 20 | 40 | 0.01* | 0.26** | 0.91* | |||||||||||||||
| O9 | 09-c | 62 | 50 | 74 | −0.22** | |||||||||||||||||
DISCUSSION
Based on analysis of 18 fatty acid products, we have demonstrated that considerable genetic diversity exists within the genepools of different Brassica genomes, contributing to variation in relative and absolute levels. In particular, wide variation is available among the component diploid genomes of the widely grown but relatively modern canola crop. Previous more limited surveys have indicated variation in the genepools of B. napus (among 14 accessions for four fatty acids [Kaushik and Agnihotri, 2000] and B. carinata [26 fatty acids; Genet et al., 2005]). A survey of nine fatty acids among 360 Arabidopsis accessions (O'Neill et al., 2003) also revealed significant variation, and provided information that enabled more detailed analysis of a core set of 13 accessions. Somewhat surprisingly, we also observed a similar range of variation for specific fatty acids among homozygous lines of diploid B. oleracea segregating populations that were derived from a cross between homozygous parents. Given that the range of variation within these populations sometimes exceeded that observed in the B. oleraeca or other genepools, this indicates that allelic combinations found in crop-adapted germplasm tend to mask the effect of null or active alleles that can contribute to pathway remodelling. Taken together, these results suggest that there exists within the genus an intrinsic capacity, in terms of the range of specific modification steps, to provide considerable additional scope for genetic metabolic engineering of fatty acid profiles.
At present there is no consistent genetic evidence relating Brassica seed size and oil content (Leon and Becker, 1995; Zhao et al., 2006). At least six to seven QTL contributing to oil content can be detected in B. napus populations, although the locations often differ between populations (e.g. Burns et al., 2003; Delourme et al., 2006; Qiu et al., 2006). Available variation for seed size may be under simpler genetic control, since fewer QTL have been detected to date, with only a small proportion of these coinciding and accounting for observed variation in oil content. Although this could also result from differences in population size or degree of G × E interaction, we do find similar patterns of variation and QTL identified when fatty acid data are calculated either as micromole per seed or as percent fatty acid composition.
The use of a matrix of scatterplots to infer pathway relationships by using trends in variation across a genepool is a novel approach for making inferences about the presence of steps in a metabolic pathway. Having initially identified genetic variation for such steps, we were then able to investigate them in more detail through quantitative genetic analysis. This both substantiated the genetic basis of such variation, and in many cases identified distinct loci that could then be assigned to one or more steps.
Overall, the QTL detected within our study could be divided into four categories that are discussed below.
QTL Associated with FAEs in the Endoplasmic Reticulum
FAE (c1) activity may be assigned to QTL AGO5-b, as it affected 18:0 (μmol) and 20:0 (μmol) with contrasting parental effect. This is substantiated by the consistent parental effect at this QTL for the percentage contribution of 20:0 and 22:0 to the total fatty acid pool. FAE (e3) activity may be assigned to QTL AGO2-a, as it affects both the molar concentration and the percentage contribution of 24:1, while a contrasting effect was observed at this QTL in the percentage contribution of 20:1. FAE (f2) activity may be assigned to QTL AGO7-c, as this affected the amounts of 22:2 assessed by micromole and percentage contribution, while not affecting the amounts of the other fatty acids in this pathway.
FAE activity (c2, e2, and f2) may be assigned to QTL AGO7-b. FAE (e2) or (e3) activity can be inferred from its effect on the molar concentrations of 18:1, 20:1, and 24:1, where there was consistent parental effect for 18:1 and 20:1, but with contrasting effect for 24:1. However, this QTL also affects elongation step yields for chain lengths 20 to 22 in pathways C, E, and F. This is supported by the analysis of percentage contribution, where there was an effect on 20:1 (e2) and 20:0 (c2). A similar effect was observed for AG07-a where the percentage contributions of 22:0 and 24:0 were affected with contrasting parental affect. The confidence intervals for AG07a and AG07b indicate that these are distinct QTL.
FAE activity at various steps may be assigned to QTL AGO8-a. This affected the amounts of 18:1, 20:1, and 20:2 with consistent parental effect, and 22:2 with contrasting parental effect. This was substantiated by the QTL detected with consistent parental effect for percentage contribution of 20:1 and 22:1. This QTL also affected the step yields of elongation from chain lengths 20 to 22 in pathways C, E, and F, and from chain length 18 to 20 in pathway D. FAE activity in pathway E may also be assigned to the corresponding region in the NG population (QTL NGO8a), as it affected amounts of 20:1.
FAE (d2) activity may be assigned to QTL NGO9-b as there is a significant effect on the yield of this elongation step.
QTL Associated with Early Stages of Fatty Acid Synthesis
The molar concentration of most fatty acids, as well as the total amounts of fatty acid, were affected by QTL AGO7-a, AGO9-a, and NGO8-b. This allows us to assign these QTL tentatively to early stages of synthesis. There appears to be a distinct range of synthetic activity associated with the different QTL. QTL AGO7-a had a consistent parental additive effect on all fatty acids, as did NGO8-b. In contrast, for QTL AGO9-a the parental effect on medium chain fatty acids (pathway A) was reversed compared to the effect found for other fatty acids.
A consistent parental additive effect on fatty acid components within pathway E was detected due to QTL AGO4-a, which also affected pathway B and the total fatty acid level. We therefore assign this QTL to an enzyme(s) active at an early stage in synthesis. QTL NGO4-a also showed a consistent parental effect, but in contrast to AG04-a it had no effect on total fatty acids or pathway E. However, NG04-a did affect pathways B, C, and D, consistent with enzyme activity at an early stage in synthesis.
Desaturation
All products in the Ex pathway were affected by QTL AGO1-a with consistent parental effect, as well as the total amounts of fatty acid. This QTL may therefore be assigned to enzyme activity within the plastid. However, this is inconsistent with the contrasting parental affect shown for percentage contribution of both 18:2 and 22:2 products in the Ey pathway. This QTL may therefore be assigned to FAD2. FAD2 function is also implicated, as this QTL has a significant effect on the activity of desaturating 18:1 to 18:2. The same pattern of activity is attributable to QTL NGO2-a, which affects the micromole amounts of products in the Ex pathway. This was substantiated by the effect on percentage contribution with contrasting parental effect on products in the Ey pathway. Since products in pathways C and D were also affected by this QTL, this suggests it may also represent Δ9desaturase activity. FAD3 activity may also be assigned to the same QTL region since we detect an effect for the activity of this step.
FAE (e1) activity may be assigned to QTL NGO3-c. This affected 20:1 and 22:1 with consistent parental effect, as well as the efficiency of the metabolic steps responsible for elongation from 18:1 to 20:1. However, this QTL also affected the amount of 20:2, and may therefore also account for activity of FAE (f1). However, although the percentage contributions of 14:0, 18:0, 20:0, 24:0, 16:1, and 18:1n7 were all affected with consistent parental effect at this QTL, the percentage contribution of 22:1 was also affected but with a contrasting parental effect, which suggests a potential Δ9desaturase activity.
A contrasting parental effect was observed at QTL AGO2-b for the percentage contributions to 18:2n6 and 18:3n3, which would suggest Δ6desaturase activity.
QTL Associated with Enzyme Activity in the Plastid
Ketoacyl ACP synthetase (KAS) activity may be assigned to QTL AGO9-b, indicated by contrasting parental effects on the products detected within pathways A and D. The effect on pathway D was observed for the amounts of 18:1n7, and on the percentage contribution of 16:1n7 and 20:1n7 within pathway D. NG09-b also had an affect on the molar amounts of 12:0 pathway A, and 16:1n7 and 20:1n7 pathway D, and was substantiated by observed affects on the percentage amounts within both pathway A 14:0, and pathway D 16:1n7 and 20:1n7.
An effect in the plastid prior to the synthesis of 18:0 ACP can be assigned to QTL AGO4-b and NGO1-b, as they affect pathways C and E with a consistent parental effect. Δ9Desaturase or FAT activity may be assigned to QTL AGO1-b, as it affected the molar concentration of the terminal fatty acid products of pathways A and C, as well as the percentage contributions of 16:0 and 18:3, with contrasting parental effects. A similar assignment may be made for QTL NGO5-a for products within pathways D and E.
The molar concentration of fatty acids within pathways C, D, and E was affected by QTL AGO8-b, although the same QTL did not have a detectable effect on total fatty acids. Since the parental effects are consistent for all these pathways we assign this QTL to an enzyme active (probably in the plastid) prior to Δ9desaturases. However, it is not possible to ascribe a specific enzyme activity. Eight QTL were detected within the AG population and 14 within the NG population (Table IV) for which we were unable to assign any function consistent with current understanding of the fatty acid synthesis pathways as shown in Figure 1.
Table IV.
Specific enzyme activities assigned to QTL in B. oleracea
Where the QTL could be associated with previously characterized genes, the orthologous Brassica or Arabidopsis gene has been assigned.
| Gene Category | Inferred Activity | Pathway Stage Influenced | QTL | Candidate Orthologous Gene |
|---|---|---|---|---|
| Early activity | Early stages of synthesis | Most | AGO7-a, NGO8-b | – |
| Early stages (but different to above) | Most | AGO9-a | – | |
| Early stage | (B, E) (B, C, D) | AGO4-a, NGO4-a | – | |
| One of the KAS | (A, D) (A, D) | AGO9-b, NGO9-b | – | |
| Before 18:ACP | (C, E) (C, E) | AGO4-b, NGO1-b | – | |
| Before Δ9DES(P2) | (C, D, E) | AGO8-b | – | |
| Plastid desaturases | Δ9DES or FAT | (A, C) | AGO1-b | – |
| Δ9DES or FAT | (D, E) | NGO5-a | ||
| FAD2, ?FAD3 | (Ex) | AGO1-a, NGO2-a | Canola FAD2 AthFAD2 | |
| Elongases | KCS (FAE) | (c1) | AGO5-b | Undetermined |
| KCS (FAE) | (e3) | AGO2-a | Undetermined | |
| KCS (FAE) | (f2) | AGO7-c | AthFAE | |
| KCS (FAE) | (c2, e2, f2) | AGO7-b | AthFAE | |
| KCS (FAE) | (Various) | AGO8-a | Undetermined | |
| KCS (FAE) | (e*) | NGO8-a | Undetermined | |
| KCS (FAE) | (e1,f1) | NGO3-c | AthFAE | |
| KCS (FAE) | (d2) | NGO9-b? | Undetermined | |
| Unassigned | Unassigned | 7 AG 6 NG | – |
Genes encoding many of the enzymes involved in the fatty acid synthesis and modification pathways have been identified in Arabidopsis. These include the fatty acid desaturase genes FAD3 (At2g29980) and FAD2 (At3g12120; Okuley et al., 1994), FAEs FAE1 (At4g34520) and FAE homolog (At3g52160), palmitoyl-ACP thioesterase FatB (At1g08510), and the ACP ACP1 (At3g11430; Bonaventure and Ohlrogge, 2002). Closely related orthologs of these genes have been isolated from Brassica species. The B. napus genes FAE1.1 and FAE1.2 have been shown to cosegregate with two loci that govern 22:1n9 production (Barret et al., 1998; Fourmann et al., 1998), with FAE1 genes also being isolated from B. oleracea and B. rapa (Das et al., 2002) as well as B. juncea (Gupta et al., 2004). Between four and six copies of FAD2 have been reported for B. napus (Scheffler et al., 1997) and representative orthologous genes encoding for both FAD2 and FAD3 have been cloned (Arondel et al., 1992; Okuley et al., 1994). The Brassica A genome copy of FAD2 located on LG N05 is thought to be responsible for the high 18:1n9 levels (Schierholt et al., 2000). However, loci accounting for low 18:3n3 (Tanhuanpaa et al., 1995; Jourdren et al., 1996), which were mapped to the homeologous LGs N04 and N14 (Laga et al., 2004), collocate with FAD3 (Jourdren et al., 1996).
Candidate genes can be associated with some of the specific QTL identified by analyzing the relative position of genes already characterized within the Brassicaceae. For example, an FAE gene encoding β-keto-acyl-CoA synthase (KCS) has been cloned from each of the A and C genomes of B. napus (Barret et al., 1998). These have subsequently been mapped to LGs DY7 (syn. N13 = O3) and DY9 (syn. N8) in the context of the ‘Darmor × Yudal’ B. napus map (Fourmann et al., 1998). By comparing the collinear relationships between B. napus and Arabidopsis (Parkin et al., 2005) we were able to confirm that these B. napus gene loci are located within regions that correspond to the Arabidopsis region containing the FAE gene (At4g34520). These conserved regions are located on LGs N1, N11, N3, N8, and N17, and correspond to regions where QTL were detected on LGs O1, O3, and O7 within the B. oleracea AG map. In addition, we identified two QTLs for FAE activity in the NG population. NGO3-c is collinear with the region of DY7 containing the cloned FAE gene encoding KCS, and AGO7-c corresponds to N17. QTL AGO7-b, where the mapping interval overlaps with that of AGO7-c, was also assigned as having a FAE function. We would expect that additional genes with FAE activity could be isolated from the QTL regions identified on 02-a, O5-b, O8-a, and O9-b. Some of these may provide insights into genes with novel specificity or combination of functions. For example, a QTL detected in the region of O1 described above also appears to affect the levels of 18:1n9, and so this region may contain additional genes that have a desaturase function (see Table IV).
The combined analysis has enabled us to propose the presence of novel elongases associated with specific elongation subpathways. Moreover, we have been able to infer the existence of enzymes encoded by distinct loci that are associated with specific elongation steps. It is, however, possible that factors other than enzyme-encoding genes may be responsible for such effects, such as transcription factors, as demonstrated recently in Arabidopsis (Bo et al., 2006). Our analysis provides information to guide subsequent experimental approaches to test hypotheses about pathway structure, as well as suggesting genetic approaches to modulate such pathways. Differences in specificity have already been detected for FAE1 genes cloned from Arabidopsis and B. napus (Blacklock and Jaworski, 2002). The N termini of FAE enzymes are responsible for substrate specificity, with the B. napus FAE1 gene (U50771) appearing to favor 20:1 acyl substrates, while Arabidopsis lacks this specificity. A low 22:1n9 phenotype can be attributed to a single amino acid substitution, and has been demonstrated by substituting FAE1-Phe-282 with Ser (Katavic et al., 2002). Supporting evidence for specificity of FAEs has also been observed in Nasturtium (Tropaeolum majus), where an FAE gene was found to have a strong preference for elongation of 20:1-CoA (Mietkiewska et al., 2004). Selecting for reduced enzyme activity that modulates sequestration into pathway D could be used to further modulate the desired composition of rapeseed oil. Interestingly, only a small increase in the levels of 18:0 was observed within the modern oilseed varieties that could correspond to a reduction in the levels of the longer chain fatty acids within pathway C. This suggests that the breeding programs that have produced the low 22:1n9 oilseed lines have had little impact on pathway C.
We identified QTL for 10 fatty acids that had previously been associated with B. napus genomic regions by using substitution lines (Burns et al., 2003). Three of the six QTL identified in that study corresponded to those we identified. We also appear to have been more successful in identifying QTL associated with genes known to affect fatty acid synthesis, resulting from a greater number of QTL being detected. This is likely to result from the use of diploid B. oleracea compared to amphidiploid B. napus. The region on O8 between 11 and 52 cM in both the AG and NG populations corresponds to the region on N18 that had been found to have a major effect on total fatty acid content and the levels of a range of fatty acids. This region may therefore either contain multiple QTL or be a major pleiotropic locus. Our analysis supports a diverse role for this locus, but also suggest that the variation observed could result from earlier stages in the pathway.
Our approach to the detection of new QTL and genetic variation in the diploid genomes for a range of fatty acids is substantiated by the confirmation of loci previously detected in the amphidiploid species. The degree of desaturation contributes to the economic, nutritional, and industrial value of polyunsaturated fatty acids. Copies of FAD2 genes have previously been mapped to LGs N1, N11, N3, N13, N1, and N15 of B. napus (Scheffler et al., 1997). These regions are all collinear with a 30 cM region of Arabidopsis Chromosome 3 that contains both FAD2 and FAD7 (Scheffler et al., 1997). The Brassica A genome copy of FAD2 on N5 is located within a region that contains a QTL for high 18:1n9 (Laga et al., 2004). We were able to detect/infer FAD2 function in both the diversity and QTL analyses. Within the B. oleracea AG population, one of the major QTL on LG O1 that was proposed to correspond to FAD2 function coincides with a locus defined by an Arabidopsis FAD2 gene probe on the C genome LG N11 of B. napus (Scheffler et al., 1997). Within the B. oleracea NG population, a QTL effect was also detected on O1. However, in this population the QTL with greatest effect was located between 29 to 48 cM on O5. This corresponds to the second locus defined by Arabidopsis FAD2, on N15. We were unable in this study to unequivocally identify a third locus corresponding to that found by Scheffler et al. (1997) on N13.
Multiple QTL were detected that appeared to have effects in the early stages of fatty acid synthesis, but either where a specific role could not be assigned, or where the pattern of substrate and product levels did not correspond to our current understanding of the relevant pathways. The ability of QTL analyses to resolve independent genetic loci (in genomes containing duplicated loci) may contribute to elucidating alternative routes in fatty acid synthesis, and subsequently contribute to identifying the associated genes and gene products involved. The accumulated evidence from genetic modification of specific steps indicates that the synthetic and modification pathways are more complex than originally expected. We propose that in specific cases, the function of particular loci can be resolved through development of defined experimental populations to test this hypothesis.
By combining knowledge that allows identification of sources of genetic variation, together with a genetic-metabolic model, we believe there is considerable scope for targeted modification and reallocation of fatty acid substrates to manipulate crop fatty acid profiles using existing natural variation. In particular, the observation that QTL accounting for specific fatty acids in distinct populations are located in different regions of the Brassica C genome, indicates that there is likely to be additional capacity to combine the additive effects of sets of positive or negative acting alleles to achieve even greater reallocation of substrates. This capacity is a consequence of the segmental duplicated organization of the Brassica diploid genomes, which are effectively triplicated in relation to the contemporary Arabidopsis genome (Lysak et al., 2005; Parkin et al., 2005), with both genera appearing to share a common hexaploid ancestor. Carrying out diversity and quantitative genetic analyses in diploid Brassica species appears to increase the ability to identify and resolve contributing loci. However, there is greater potential for maximizing additive effects of selective recombination in the tetraploid species B. napus and B. juncea, albeit with a contemporary narrow base of genetic diversity. These already represent the major Brassica oil crops worldwide. Metabolic pathway manipulation using marker-assisted selection to facilitate introgression has also been used to achieve significant increases in desirable isothiocyanates levels in B. oleracea var. italica broccoli (Mithen et al., 2003). Manipulation of pathways via the utilization of natural allelic diversity means that many of the problems associated with transgenic approaches, such as introducing novel nonendogenous enzymes and the associated difference in their specificities, can be avoided. This may have a range of benefits in terms of marketing, licensing, and consumer perception, as well as in management of gene flow into natural or feral populations. In addition, transgenic approaches in which multiple genes are used present severe problems of locus management for breeders when combining with elite material segregating for other performance traits.
In summary, our results demonstrate that the allelic diversity present with the Brassica gene pool can be utilized to manipulate the oil composition through pathway engineering, and that there is considerable scope for metabolic engineering for a range of industrial and nutritional end uses.
MATERIALS AND METHODS
Plant Material
A set of 107 lines was assembled to represent diversity within the Brassica genepool, primarily within the A and C cytodemes (Table I). The primary operational taxonomic unit was defined as a line. A line consists of genetic material maintained as a distinct entity within a genetic resource or research collection, and may either be uniformly homozygous (inbred or DH), uniformly heterozygous (F1 varieties), or heterozygous and heterogeneous (landraces, open pollinated varieties). An accession is defined as seed or plants of a line arising from a single generation, where seed have been harvested from one or more plants on a single occasion and location. Most seed were sourced from the Warwick HRI Genetic Resources Unit, with additional lines obtained from research collections at Warwick HRI. Four homozygous lines representing parents of the DH mapping populations were included in the diversity set.
DH lines from two Brassica oleracea reference segregating mapping populations have been described previously (Sebastian et al., 2000). The AG population was represented by 99 DH lines that had been derived by anther culture from an F1 produced from a cross between A12DHd (var. alboglabra) and GDDH33 (var. italica). The NG population was represented by 69 DH lines derived from an F1 resulting from a cross between CA25 (var. botrytis) and AC298 (var. gemmifera).
In most cases seed had been collected by hand from plants pollinated either individually or as individual lines in insect-proof cages. For material from the genetic resource unit, seed were maintained at 15°C and 15% relative humidity for 2 weeks prior to sealing in foil-laminate pouches and storing at −20°C until required. Seed from mapping populations were sampled from glasshouse grown plants.
Sampling and Measurement of Seed Fatty Acids
Seed fatty acids were analyzed on two separate occasions (February and November, 2003). To increase the sensitivity of the analysis extractions were performed on samples of five seeds. On the first occasion (February) three samples, and on the second (November), two samples of five seeds per accession were analyzed. All seed were equilibrated at 15°C and 15% relative humidity prior to sampling. Individual samples of five seeds were weighed in 2-mL glass analysis vials with sealable lids. At the sampling stage seeds were handled with nitryl gloves to reduce contamination.
For all lines but the parents of the mapping populations, a single accession was analyzed per line, on one of the two occasions. We sampled several accessions of the four parent lines of the mapping populations. These accessions were selected from harvests carried out in different years. For two lines (CA25 and AC498), two accessions were analyzed, one on each occasion. For GDDH33 four accessions were analyzed, one on the first occasion and three on the second. For A12DHd four accessions were analyzed, one on the first occasion and all four on the second, thus providing a sampling occasion replicate for the first accession.
Lipid fatty acids were converted to their methyl esters (FAMEs) using the direct transmethylation method and gas chromatography equipment as described by Larson and Graham (2001), with some modifications. To minimize seed to seed variation, five seeds were pooled and processed through the transmethylation procedure. Heptadecanoic acid (100 μg) was added as an internal standard before processing. Cooled transmethylated samples were transferred to microfuge tubes and extracted three times with 200 μL hexane, with 1 min centrifugation at 14,000g between each extraction step to clarify the partitioned layers. The hexane fractions were pooled, dried in vacuo for 10 min, and reconstituted in 1 mL fresh hexane. This procedure demonstrated comparable quantitative extraction for one to five seeds, with no losses for FAMEs with acyl chains >C10 (data not shown). A 2 μL aliquot was injected for FAME analysis. Fatty acids were identified and quantified by comparison to a 37 FAME mix (Supelco), using Chromquest 2.53 software (Thermo). The absolute and relative amounts of each fatty acid were calculated.
Calculation of Step Yields
Figure 1 outlines the current understanding of triacylglycerol synthesis within the Brassicaceae as discussed in the introduction. By comparing the total amounts of fatty acid being synthesized by a particular combination of metabolic steps, it is possible to gain a better understanding of the pathways and yields for any particular step.
To calculate the efficiencies of metabolic steps within the cytosol we calculated the ratio a/b of the total amount of the product of a step, a, to the total amount of the substrate of the step, b. a was calculated as the sum of the yields of the product of the step, together with all fatty acid products for which it is a precursor in the assumed pathway. b was calculated in a similar manner, as the sum of the yields of the substrate of the step together with all fatty acid products for which it is a precursor. b is therefore greater than or equal to a, since all the fatty acids whose products composed a are also contained in b.
For example, the step yield for FAE(e1) is calculated as:
yield = ([20:1n9] + [22:1n9] + [24:1n9]/([18:1n9] + [20:1n9] + [22:1n9] + [24:1n9] + [18:2n6] + [20:2n6] + [22:2n6] + [18:3n3] + [18:3n6]).
Analysis of Sources of Variation in the Diversity Data
To examine the sources of variability for the yields (micromole per seed) and the percentage contribution of each fatty acid, and for the efficiencies of metabolic steps, restricted maximum likelihood was carried out on the data from the seed samples within the diversity set. The progeny of the mapping populations were not included in the analysis. The data were analyzed both with and without the modern oilseed lines. The data excluding these lines are shown. Restricted maximum likelihood is a generalization of ANOVA suitable for unbalanced data, and which is particularly suitable for estimating components of variation. We considered explanatory factors of occasion, species, subspecies within species, line within subspecies, accession within line, and batch of seeds within accession. All factors were taken as random. Where negative variance components were estimated for a factor then they were forced to be zero. The analyses of fatty acid yields and efficiencies of metabolic steps for which many lines gave zeroes tended not to converge and the relevant traits were consequently not analyzed. The analyses were repeated with line taken as fixed factor, and the line means from this analysis were used as input to the QTL analyses.
To display variation of fatty acid amounts within and between species, we generated a series of box and whisker plots (Fig. 2).
Identification of Genetic Loci through QTL Analysis
Individual and integrated genetic linkage maps based on segregation of DNA markers within the B. oleracea AG and NG DH populations have been published (Sebastian et al., 2000). QTL analysis was carried out using a subset of loci evenly spaced at approximately 10 cM intervals through the linkage maps, for which the most complete genotype information was available. QTL detection and location was carried out using the multiple marker regression approach (Kearsey and Hyne, 1994) based on line means implemented in QTL Café software (http://www.biosciences.bham.ac.uk/labs/kearsey/). QTL Café software has been extensively used in published Brassica and other QTL analyses and can readily be used to compare different crosses for the presence of common QTL (Kearsey and Hyne, 1994), which is relevant to our analysis where we are comparing two populations of B. oleracea. The presence of one or more QTL was tested using ANOVAs. Usually a single QTL model on a given LG was accepted when the residual mean square in the ANOVA was not significant (P > 0.05) and the regression mean square was significant. The parental contribution to each QTL was assigned by the direction of the QTL effect, such that positive values indicated that the contribution derived from the female parents (A12DHd in AG population, CA25 in NG population), and negative values indicate a contribution from male parents (GDDH33 and AC498, respectively). At any QTL locus the contribution of an increasing allele from a female parent, indicated by a positive additive effect, is equivalent to the contribution of a decreasing allele from a male parent, as the direction of the additive effects are expressed as one parental allele relative to the other. Map positions are expressed relative to an integrated linkage map with common LG (e.g. O1) and QTL position (e.g. O1-a) nomenclature and a population-specific prefix (AG or NG). We also used interval mapping and composite interval mapping methods within QTL Cartographer (Wang et al., 2006) on the same datasets to verify the results.
Nomenclature
Throughout this study, fatty acids are written as x:ynz, where x refers to the number of carbons in the acyl chain, y to the number of double bonds, and z to the position of the first double bond from the methyl end of the fatty acid. Where more than one double bond is present, their respective positions are spaced at methylene-interrupted intervals away from the z double bond toward the carboxyl end of the fatty acid.
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council at Warwick HRI and Rothamsted Research, and by Defra at the Centre for Novel Agricultural Products and Rothamstead Research.
Open Access articles can be viewed online without a subscription.
References
- Arondel V, Lemieux B, Hwang I, Gibson S, Goodman HM, Somerville CR (1992) Map-based cloning of a gene controlling omega-3-fatty-acid desaturation in Arabidopsis. Science 258 1353–1355 [DOI] [PubMed] [Google Scholar]
- Barret P, Delourme R, Renard M, Domergue F, Lessire R, Delseny M, Roscoe TJ (1998) A rapeseed FAE1 gene is linked to the E1 locus associated with variation in the content of erucic acid. Theor Appl Genet 96 177–186 [Google Scholar]
- Blacklock BJ, Jaworski JG (2002) Studies into factors contributing to substrate specificity of membrane-bound 3-ketoacyl-CoA synthases. Eur J Biochem 269 4789–4798 [DOI] [PubMed] [Google Scholar]
- Bo S, Kerstin WS, David AS, Mitchell CT (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60 377–387 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Ohlrogge JB (2002) Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol 128 223–235 [PMC free article] [PubMed] [Google Scholar]
- Broun P, Gettner S, Somerville C (1999) Genetic engineering of plant lipids. Annu Rev Nutr 19 197–216 [DOI] [PubMed] [Google Scholar]
- Burns MJ, Barnes SR, Bowman JG, Clarke MH, Werner CP, Kearsey MJ (2003) QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) seed oil content and fatty acid composition. Heredity 90 39–48 [DOI] [PubMed] [Google Scholar]
- Cheung WY, Landry BS, Raney P, Rakow GFW (1998) Molecular mapping of seed quality traits in Brassica juncea L. Czern. & Coss. Acta Hortic 459 139–147 [Google Scholar]
- Das S, Roscoe TJ, Delseny M, Srivastava PS, Lakshmikumaran M (2002) Cloning and molecular characterization of the fatty acid elongase 1 (FAE 1) gene from high and low erucic acid lines of Brassica campestris and Brassica oleracea. Plant Sci 162 245–250 [Google Scholar]
- Delourme R, Falentin C, Huteau V, Clouet V, Horvais R, Gandon B, Specel S, Hanneton L, Dheu JE, Deschamps M, et al (2006) Genetic control of oil content in oilseed rape (Brassica napus L.). Theor Appl Genet 113 1331–1345 [DOI] [PubMed] [Google Scholar]
- Ecke W, Uzunova M, Weissleder K (1995) Mapping the genome of rapeseed (Brassica-napus L). 2. Localization of genes-controlling erucic-acid synthesis and seed oil content. Theor Appl Genet 91 972–977 [DOI] [PubMed] [Google Scholar]
- Fatland BL, Nikolau BJ, Wurtele ES (2005) Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell 17 182–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann M, Barret P, Renard M, Pelletier G, Delourme R, Brunel D (1998) The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor Appl Genet 96 852–858 [Google Scholar]
- Genet T, Viljoen CD, Labuschagne MT (2005) Genetic analysis of Ethiopian mustard genotypes using amplified fragment length polymorphism (AFLP) markers. Afr J Biotechnol 4 891–897 [Google Scholar]
- Gunstone FD (2001) Soybeans pace boost in oilseed production. Inform 11 1287–1289 [Google Scholar]
- Gupta V, Mukhopadhyay A, Arumugam N, Sodhi YS, Pental D, Pradhan AK (2004) Molecular tagging of erucic acid trait in oilseed mustard (Brassica juncea) by QTL mapping and single nucleotide polymorphisms in FAE1 gene. Theor Appl Genet 108 743–749 [DOI] [PubMed] [Google Scholar]
- Hobbs DH, Flintham JE, Hills MJ (2004) Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol 136 3341–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugly S, Kunst L, Browse J, Somerville C (1989) Enhanced thermal tolerance of photosynthesis and altered chloroplast ultrastructure in a mutant of Arabidopsis deficient in lipid desaturation. Plant Physiol 90 1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Nap JP (2001) Genetical genomics: the added value from segregation. Trends Genet 17 388–391 [DOI] [PubMed] [Google Scholar]
- Jourdren C, Barret P, Brunel D, Delourme R, Renard M (1996) Specific molecular marker of the genes controlling linolenic acid content in rapeseed. Theor Appl Genet 93 512–518 [DOI] [PubMed] [Google Scholar]
- Katavic V, Mietkiewska E, Barton DL, Giblin EM, Reed DW, Taylor DC (2002) Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. Eur J Biochem 269 5625–5631 [DOI] [PubMed] [Google Scholar]
- Kaushik N, Agnihotri A (2000) GLC analysis of Indian rapeseed-mustard to study the variability of fatty acid composition. Biochem Soc Trans 28 581–583 [PubMed] [Google Scholar]
- Kearsey MJ, Hyne V (1994) Qtl analysis—a simple marker-regression approach. Theor Appl Genet 89 698–702 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38 842–849 [DOI] [PubMed] [Google Scholar]
- Khachatourians GG, Summer AK, Phillips PWB (2001) An introduction to the history of canola and the scientific basis for innovation. In KPWB Phillips, GG Khachatourians, eds, The Biotechnology Revolution in Global Agriculture: Innovation, Invention and Investment in the Canola Industry. CABI Publishing, Wallingford, UK, pp 33–49
- King GJ, Teakle GR, Astley D, Pink DAC (2004) Development of diversity sets for long-term community-wide molecular and trait exploration of Brassica gene-pools. Rothamstead Research. http://www.brassica.info/diversity/diversity_sets.htm (November 25, 2004)
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T (2001) Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA (Suppl) 100 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laga B, Seurinck J, Verhoye T, Lambert B (2004) Molecular breeding for high oleic and low linolenic fatty acid composition in Brassica napus. Pflanzenschutz-Nachrichten Bayer 57 87–92 [Google Scholar]
- Larson TR, Graham IA (2001) Technical advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25 115–125 [DOI] [PubMed] [Google Scholar]
- Leon J, Becker HC (1995) Rapeseed Genetics. Blackwell Wiss Verl, Berlin
- Li G, Riaz A, Goyal S, Abel S, Quiros CF (2001) Inheritance of three major genes involved in the synthesis of aliphatic glucosinolates in Brassica oleracea. J Am Soc Hortic Sci 126 427–431 [Google Scholar]
- Lionneton E, Ravera S, Sanchez L, Aubert G, Delourme R, Ochatt S (2002) Development of an AFLP-based linkage map and localization of QTLs for seed fatty acid content in condiment mustard (Brassica juncea). Genome 45 1203–1215 [DOI] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Yadav S, Singh R, Begum G, Suneja P, Singh M (2002) Correlation studies on oil content and fatty acid profile of some Cruciferous species. Genet Resour Crop Evol 49 551–556 [Google Scholar]
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Byrne PF, Snook ME, Wiseman BR, Lee EA, Widstrom NW, Coe EH (1998) Quantitative trait loci and metabolic pathways. Proc Natl Acad Sci USA 95 1996–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Snook M, Lee EA, Byrne PF, Kross H, Musket TA, Houchins K, Coe EH Jr (2001) The biological basis of epistasis between quantitative trait loci for flavone and 3-deoxyanthocyanin synthesis in maize (Zea mays L.). Genome 44 667–676 [PubMed] [Google Scholar]
- Mietkiewska E, Giblin EM, Wang S, Barton DL, Dirpaul J, Brost JM, Katavic V, Taylor DC (2004) Seed-specific heterologous expression of a nasturtium FAE gene in Arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol 136 2665–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Smith MA, Kunst L (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5 95–101 [DOI] [PubMed] [Google Scholar]
- Mithen R, Faulkner K, Magrath R, Rose P, Williamson G, Marquez J (2003) Development of isothiocyanate-enriched broccoli, and its enhanced ability to induce phase 2 detoxification enzymes in mammalian cells. Theor Appl Genet 106 727–734 [DOI] [PubMed] [Google Scholar]
- Morice J (1974) Selection of a variety of Rapeseed without erucic acid and without glucosinolates. In Proceedings of the 4th International Rapeseed Congress, Geissen, Germany, pp 31–47
- Morreel K, Goeminne G, Storme V, Sterck L, Ralph J, Coppieters W, Breyne P, Steenackers M, Georges M, Messens E, et al (2006) Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J 47 224–237 [DOI] [PubMed] [Google Scholar]
- O'Neill CM, Gill S, Hobbs D, Morgan C, Bancroft I (2003) Natural variation for seed oil composition in Arabidopsis thaliana. Phytochemistry 64 1077–1090 [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Browse J, Somerville CR (1991) The genetics of plant lipids. Biochim Biophys Acta 1082 1–26 [DOI] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifico D, Francesca M, Mirta M, Andrea C, Paolo R, Giuseppe M (2006) Genetics and marker-assisted selection of the chemotype in Cannabis sativa L. Mol Breed 17 257–268 [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Ohlrogge J (1999) Testing models of fatty acid transfer and lipid synthesis in spinach leaf using in vivo oxygen-18 labeling. Plant Physiol 121 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S (1980) Cruciferous oilseeds in India. In KHS Tsunoda, C Gomez-Campo, eds, Brassica Crops and Wild Allies. Biology and Breeding. Japan Scient. Soc. Press, Tokyo, pp 151–163
- Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E, et al (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114 67–80 [DOI] [PubMed] [Google Scholar]
- Sauer FD, Kramer JKG (1983) The problems associated with the feeding of high erucic acid rapeseed oils and some fish oils to experimental animals. In JKG Kramer, FD Sauer, WJ Pigden, eds, High and Low Erucic Acid Rapeseed Oils. Production, Usage, Chemistry, and Toxicological Examination. Academic Press, Toronto, pp 253–292
- Scheffler JA, Sharpe AG, Schmidt H, Sperling P, Parkin IAP, Luhs W, Lydiate DJ, Heinz E (1997) Desaturase multigene families of Brassica napus arose through genome duplication. Theor Appl Genet 94 583–591 [Google Scholar]
- Schierholt A, Becker HC, Ecke W (2000) Mapping a high oleic acid mutation in winter oilseed rape (Brassica napus L.). Theor Appl Genet 101 897–901 [Google Scholar]
- Schwender J, Ohlrogge J, Shachar-Hill Y (2004) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7 309–317 [DOI] [PubMed] [Google Scholar]
- Sebastian RL, Howell EC, King GJ, Marshall DF, Kearsey MJ (2000) An integrated AFLP and RFLP Brassica oleracea linkage map from two morphologically distinct doubled-haploid mapping populations. Theor Appl Genet 100 75–81 [Google Scholar]
- Sharma R, Aggarwal RA, Kumar R, Mohapatra T, Sharma RP (2002) Construction of an RAPD linkage map and localization of QTLs for oleic acid level using recombinant inbreds in mustard (Brassica juncea). Genome 45 467–472 [DOI] [PubMed] [Google Scholar]
- Slabas AR, Simon JW, Brown AP (2001) Biosynthesis and regulation of fatty acids and triglycerides in oil seed rape: current status and future trends. Eur J Lipid Sci Technol 103 455–466 [Google Scholar]
- Sovero M (1993) Rapeseed, a New Oilseed Crop for the United States. Wiley, New York
- Tanhuanpaa P, Vilkki J, Vihinen M (1998) Mapping and cloning of FAD2 gene to develop allele-specific PCR for oleic acid in spring turnip rape (Brassica rapa ssp. oleifera). Mol Breed 4 543–550 [Google Scholar]
- Tanhuanpaa PK, Vilkki JP, Vilkki HJ (1995) Association of a Rapd marker with linolenic acid concentration in the seed oil of rapeseed (Brassica-napus L). Genome 38 414–416 [DOI] [PubMed] [Google Scholar]
- Thormann CE, Romero J, Mantet J, Osborn TC (1996) Mapping loci controlling the concentrations of erucic and linolenic acids in seed oil of Brassica napus L. Theor Appl Genet 93 282–286 [DOI] [PubMed] [Google Scholar]
- Van de Loo F, Fox BG, Somerville C (1993) Unusual fatty acids. In T Moore, ed, Plant Lipids. CRC, Boca Raton, FL, pp 91–126
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52 335–361 [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B (2006) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
- Yan Z (1990) Overview of rapeseed production and research in China. In Proceedings of the International Canola Conference, Potash & Phosphate Institute, Atlanta, pp 29–35
- Zhao J, Becker HC, Zhang D, Zhang Y, Ecke W (2006) Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113 33–38 [DOI] [PubMed] [Google Scholar]