Abstract
We screened a rice (Oryza sativa L. ‘Nipponbare’) full-length cDNA expression library through functional complementation in yeast (Saccharomyces cerevisiae) to find novel cation transporters involved in salt tolerance. We found that expression of a cDNA clone, encoding the rice homolog of Shaker family K+ channel KAT1 (OsKAT1), suppressed the salt-sensitive phenotype of yeast strain G19 (Δena1–4), which lacks a major component of Na+ efflux. It also suppressed a K+-transport-defective phenotype of yeast strain CY162 (Δtrk1Δtrk2), suggesting the enhancement of K+ uptake by OsKAT1. By the expression of OsKAT1, the K+ contents of salt-stressed G19 cells increased during the exponential growth phase. At the linear phase, however, OsKAT1-expressing G19 cells accumulated less Na+ than nonexpressing cells, but almost the same K+. The cellular Na+ to K+ ratio of OsKAT1-expressing G19 cells remained lower than nonexpressing cells under saline conditions. Rice cells overexpressing OsKAT1 also showed enhanced salt tolerance and increased cellular K+ content. These functions of OsKAT1 are likely to be common among Shaker K+ channels because OsAKT1 and Arabidopsis (Arabidopsis thaliana) KAT1 were able to complement the salt-sensitive phenotype of G19 as well as OsKAT1. The expression of OsKAT1 was restricted to internodes and rachides of wild-type rice, whereas other Shaker family genes were expressed in various organs. These results suggest that OsKAT1 is involved in salt tolerance of rice in cooperation with other K+ channels by participating in maintenance of cytosolic cation homeostasis during salt stress and thus protects cells from Na+.
High levels of salinity in the soil hinder the growth and development of crops and cause serious problems for world food production (Munns, 2005). Rice (Oryza sativa), the most important cereal crop in many parts of the world, is considered to be salt sensitive (Akbar and Ponnamperuma, 1980). Therefore, it is of agronomic importance to analyze and improve its salt tolerance.
High levels of Na+ or high Na+ to K+ ratios can disrupt various enzymatic processes in the cytoplasm owing to the ability of Na+ to compete with K+ for binding sites (Serrano, 1996; Tester and Davenport, 2003). The sensitivity of cytosolic enzymes to Na+ is similar in both glycophytes and halophytes, indicating that the maintenance of a low cytosolic Na+ to K+ ratio is a key requirement of plant growth in saline soil (Glenn et al., 1999; Apse and Blumwald, 2002). Some cation transporters mediate ion homeostasis in the cytoplasm and contribute to the salt tolerance of plants. In Arabidopsis (Arabidopsis thaliana), the plasma membrane Na+/H+ antiporter SOS1 catalyzes Na+ extrusion from cells (Shi et al., 2003). Vacuolar sequestration of Na+ is also effective at lowering Na+ concentration in the cytoplasm and is mediated by the NHX family of vacuolar Na+/H+ antiporters, such as AtNHX1 in Arabidopsis (Apse et al., 1999) and OsNHX1 in rice (Fukuda et al., 2004). HKT-type transporters are involved in regulating cation homeostasis under salt stress in Arabidopsis (Sunarpi et al., 2005) and rice (Ren et al., 2005). Genome-wide analyses indicate that additional classes of cation transporters are likely to be involved in cation homeostasis, and their further characterization is important to understanding the mechanisms responsible for salt tolerance in plants. In particular, the roles of K+ transporters in salt tolerance, which may be involved in the regulation of the cytosolic Na+ to K+ ratio, remain to be clarified.
The responses to salt stress at the whole-plant level are thought to depend to a great extent on cellular mechanisms of tolerance (Serrano, 1996). Therefore, the conservation of the basic transport mechanism makes the yeast Saccharomyces cerevisiae a model system of considerable value for the understanding of ion homeostasis in plants (Serrano and Rodriguez-Navarro, 2001). To identify novel ion transporter genes participating in salt tolerance in rice cells (‘Nipponbare’), we conducted functional complementation screening of a rice full-length cDNA library for multicopy suppressors of a salt-sensitive phenotype of yeast mutant strain G19. The strain displays salt sensitivity as a result of disruptions in the Ena1 to Ena4 genes, which encode Na+ export pumps (Quintero et al., 1996; Gobert et al., 2006). The completely sequenced full-length cDNA library, containing approximately 32,000 clones, was made by the Rice Full-Length cDNA Project from about 20 kinds of stressed tissues of sp. japonica rice (Kikuchi et al., 2003). We isolated the rice gene OsKAT1, which encodes a K+ channel protein as a multicopy suppressor of Na+ sensitivity in G19. OsKAT1-expressing yeast was the most tolerant to NaCl among the salt-tolerant transformants obtained. Inward-rectifying K+ channels, such as AKT1 and KAT1 from Arabidopsis, KST1 from potato (Solanum tuberosum), ZMK1/ZMK2 from maize (Zea mays), and TaAKT1 from wheat (Triticum aestivum), have been functionally characterized (Anderson et al., 1992; Gambale and Uozumi, 2006). These channels show high selectivity for K+ over other monovalent cations, and are reputed to specifically mediate K+ uptake and transport in plant cells (Gambale and Uozumi, 2006). K+ channels and transport systems play multiple roles in higher-plant processes, including opening and closing of stomatal pores, leaf movements, and ion uptake in roots (Hedrich and Becker, 1994; Schroeder et al., 1994; Coté, 1995). AKT1 is an inward-rectifying channel for K+ uptake in Arabidopsis roots. Elevated cytoplasmic Na+ impaired the K+ permeability mediated by AKT1 (Qi and Spalding, 2004). OsAKT1 is also reported to represent the dominant salt-sensitive K+ uptake channel in rice roots (Fuchs et al., 2005), and the expression of OsAKT1 is regulated differently in salt-sensitive and salt-tolerant cultivars of rice (Golldack et al., 2003). These results suggest the inhibition of K+ uptake mediated by these channels as a possible cause of toxicity of Na+. Meanwhile, these results make the K+ channels potential candidates for regulating cellular ion homeostasis. Here, we analyzed the functions of OsKAT1 in the maintenance of cellular levels of Na+ and K+ in yeast and rice cultured cell lines. We show that OsKAT1 functions in ion homeostasis and salt tolerance at the cellular level, and suggest OsKAT1 as a candidate halotolerance gene.
RESULTS
OsKAT1 Confers Salt Tolerance in the Salt-Sensitive Yeast Mutant
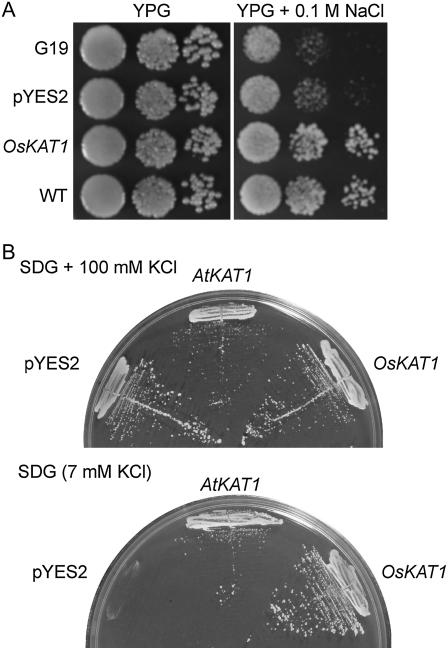
Rice full-length cDNA libraries were screened by functional complementation of yeast strain G19 to isolate rice genes conferring increased tolerance to NaCl. The screening resulted in the isolation of a cDNA clone (accession no. AK100739) encoding a putative protein of 502 amino acids. This protein shows significant homology to KAT1, a K+ channel protein of Arabidopsis; therefore, we named the protein OsKAT1. OsKAT1 expression conferred tolerance to NaCl compared with the control strain transformed with the empty pYES2 vector, but did not affect yeast growth in the absence of salt (Fig. 1A). Only the OsKAT1-expressing strain was able to grow well on the medium with 0.3 m NaCl. Some other transformants showed better growth than control strain on the medium containing up to 0.1 m NaCl.
Figure 1.
Complementation of yeast mutants by OsKAT1. A, Growth of the salt-sensitive yeast strain (G19), wild-type W303 strain (WT), and G19 transformed with either the empty plasmid (pYES2) or OsKAT1 expression plasmid (OsKAT1). Ten-fold serial dilutions of cell suspensions were spotted onto YPG plates with or without 0.1 m NaCl. Growth was followed for 5 d. B, Growth of the K+-uptake-deficient yeast strain (CY162) transformed with the empty plasmid (pYES2), Arabidopsis KAT1 (AtKAT1) expression plasmid, or OsKAT1 expression plasmid. The yeast cells were streaked onto SDG medium containing approximately 7 mm K+ (bottom) or 100 mm K+ (top). Growth was followed for 5 d.
OsKAT1 Is a Shaker Family K+ Channel Protein and Mediates K+ Uptake
Structures conserved among Shaker family members were found in OsKAT1: six transmembrane domains, a putative cyclic nucleotide-binding site, and a K+-selective pore-forming loop region with a consensus TXXTXGYG motif located between the fifth and sixth transmembrane segments (Fig. 2; Véry and Sentenac, 2002). The absence of an ankyrin repeat consensus site, a characteristic of AKT-type channels (Mäser et al., 2001), is a reason for deciding that OsKAT1 is a KAT-type channel. The homology among OsKAT1 and Shaker family proteins strongly suggests that OsKAT1 is an inward-rectifying K+ channel. To test whether OsKAT1 functions in K+ uptake, we expressed OsKAT1 and Arabidopsis KAT1 (AtKAT1) in a yeast mutant strain, CY162, lacking the K+ transporters TRK1 and TRK2 (Anderson et al., 1992). Although CY162 is lethal when the medium contains low levels of K+, expression of plant genes encoding K+ transporters such as AtKAT1 rescues the mutant strain (Anderson et al., 1992; Fu and Luan, 1998). CY162 transformed with empty pYES2 could not grow on the low-K+ medium, but the OsKAT1-expressing CY162 grew at least as well as the AtKAT1-expressing strain after incubation for 5 d (Fig. 1B). The result shows that OsKAT1 confers significant K+ uptake and growth on the mutant yeast strain at low K+ availability.
Figure 2.
Amino acid sequence alignment of the OsKAT1 and KAT-type K+ channel proteins. The amino acid sequences of OsKAT1 (gene code Os01g0756700), Arabidopsis KAT1 (AtKAT1, At5g46240), and maize Zmk2.1 (AY461584) were aligned using the program CLUSTALX 1.83. Identical amino acids are shaded. The consensus TXXTXGYG motif in the K+-selective pore-forming loop region is reversed out. The regions in the boxes drawn with solid lines are putative transmembrane domains (s1–s6). The putative cyclic nucleotide-binding domain is indicated in the boxes drawn with a dotted line.
OsKAT1 Alters the Alkali-Cation Homeostasis in Yeast Cells
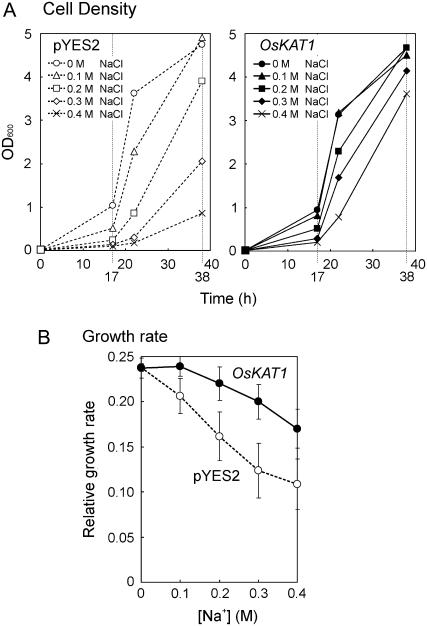
To understand why OsKAT1 increased salt tolerance, we compared the Na+ and K+ contents of yeast OsKAT1 cells (strain G19 transformed with OsKAT1 expression plasmid) and control cells (G19 transformed with pYES2) when both cell lines were cultured in SDG medium containing 0 to 0.4 m NaCl. Growth of control cells was severely affected by external NaCl. OsKAT1 cells were less affected than control cells (Fig. 3A). The increased salt tolerance by OsKAT1 was clearly shown as the higher growth rates of OsKAT1 cells under salinity stress. The growth rates were similar in both kinds of cells under nonstressed conditions (Fig. 3B).
Figure 3.
Effects of NaCl on the growth of yeast G19 strain transformed with empty plasmid (pYES2, white symbols, dotted lines) or OsKAT1 expression plasmid (OsKAT1, black symbols, solid lines). The cells were cultured in liquid SDG medium containing 0, 0.1, 0.2, 0.3, or 0.4 m NaCl. A, Time course of cell density in the culture suspension. Data represent means of three cultures grown separately in space and time. B, Dose-dependent effect of ambient NaCl on the growth rate in the exponential growth phase. Relative growth rate was determined as the slope of growth curve representing the time course of log OD600. Data represent means ± sd of three experiments.
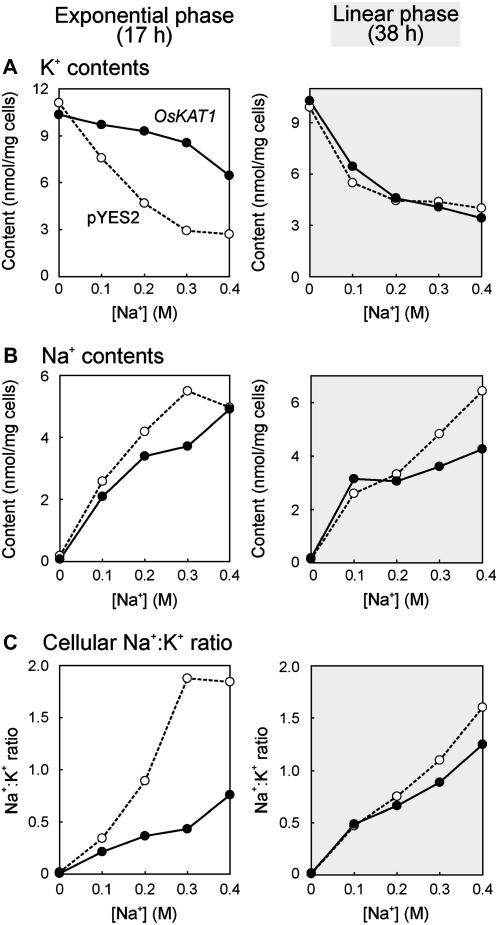
Cells were collected in the exponential growth phase (17 h) and in the linear growth phase (38 h) to analyze cellular cation contents. In the exponential growth phase (Fig. 4, left), the K+ contents of control cells were decreased by the effect of NaCl, but those of OsKAT1 cells were less affected by NaCl and remained higher under saline conditions (Fig. 4A). The cellular contents of Na+ in OsKAT1 cells were slightly lower than in control cells (Fig. 4B). In the linear phase (Fig. 4, right), OsKAT1 cells accumulated less Na+ than control cells in the medium with 0.3 or 0.4 m NaCl (Fig. 4B), whereas the K+ contents of both kinds of cells were almost the same (Fig. 4A). These results reveal that OsKAT1 not only increases cellular K+ but also decreases Na+. Consequently, OsKAT1 altered the cellular Na+ to K+ ratios. In the exponential phase, the net loss of K+ in control cells upon salt treatment resulted in higher cellular Na+ to K+ ratios. The ratios in OsKAT1 cells were then much lower due to high K+ contents (Fig. 4, A and C). In the linear phase, the ratios of both kinds of cells became similar (Fig. 4C), but those of OsKAT1 cells remained lower owing to their low Na+ contents (Fig. 4, B and C). The cellular Na+ to K+ ratios of OsKAT1 cells did not rise drastically under saline conditions, neither during the exponential nor linear growth phase (Fig. 4C), showing the function of OsKAT1 in the maintenance of alkali-cation homeostasis in cells.
Figure 4.
Cellular cation contents of yeast G19 strain transformed with empty plasmid (pYES2, white circles, dotted lines) or OsKAT1 expression plasmid (black circles, solid lines) in media containing NaCl. The cells were harvested in the exponential (17 h after inoculation, left) and linear (38 h after inoculation, right) growth phases in cultures described in Figure 3, to determine the cellular contents of cations. A, K+ contents in the cells. B, Na+ contents in the cells. C, Ratios of Na+ to K+ contents in the cells. Data points shown in this figure represent means of three cultures.
Overexpression of OsKAT1 in Rice Cultured Cell Lines Increases Growth and Cellular K+ Content during Salinity Stress
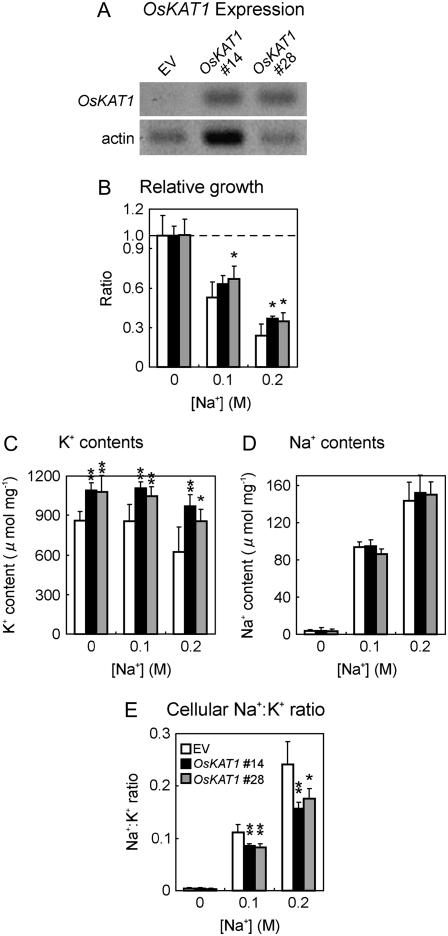
We cultured rice cells overexpressing OsKAT1 in the presence of NaCl to test whether OsKAT1 enhances salt tolerance. Expression of OsKAT1 was detected in the cell lines transformed with OsKAT1 expression plasmid (OsKAT1 #14 and #28), although it was not detected in the cell line transformed with the empty vector (EV; Fig. 5A). The EV-transformed cells were sensitive to salt and grew poorly in the presence of 0.2 m NaCl. The growth of OsKAT1-overexpressing cells was less affected (Fig. 5B). Both kinds of OsKAT1-overexpressing cells accumulated more K+ than the EV-transformed cells (Fig. 5C), yet the cellular Na+ contents of these cells were almost the same (Fig. 5D). Consequently, OsKAT1-overexpressing cells showed significantly lower Na+ to K+ ratios (Fig. 5E).
Figure 5.
Growth and cellular cation contents of the transgenic rice cells during salinity stress. Rice cell lines transformed with OsKAT1 expression plasmid (OsKAT1 #14, black bars; #28, gray bars) and EV (white bars) were cultured for 10 d in Murashige and Skoog medium containing 0, 0.1, or 0.2 m NaCl. A, RT-PCR analysis of OsKAT1 expression. B, Effects of salt stress treatment on the growth of rice cells. Relative growth is shown as ratio of cellular fresh weight in 40-mL culture suspension to that of cells grown in medium without NaCl. C, K+ contents of cells. D, Na+ contents of cells. E, Ratio of Na+ and K+ contents in cells. Each value is the mean ± sd of six samples in two independent experiments. Asterisks represent significant differences between the EV and OsKAT1 (*, P <0.05; **, P < 0.01; t test was performed in Microsoft Excel).
OsAKT1 and AtKAT1 Also Confer Salt Tolerance in Yeast
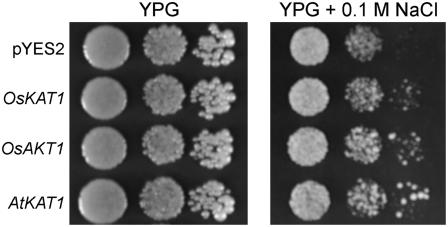
To assess whether the improvement of cellular salt tolerance is a specific function of OsKAT1 or is common among Shaker-type channels, we expressed OsAKT1 and AtKAT1, which are well-characterized inward-rectifying K+ channels (Anderson et al., 1992; Fuchs et al., 2005), in yeast strain G19. OsAKT1- and AtKAT1-expressing yeast showed better growth than control cells on plates with 0.1 m NaCl, as did OsKAT1-expressing yeast (Fig. 6). The results suggest that these channels function similarly in cellular ion homeostasis.
Figure 6.
Complementation of salt-sensitive phenotype of yeast G19 strain by Shaker family channel genes. G19 was transformed with OsKAT1, OsAKT1, Arabidopsis KAT1 (AtKAT1), or empty plasmid (pYES2). Ten-fold serial dilutions of cell suspensions were spotted onto YPG plates with or without 0.1 m NaCl. Growth was followed for 5 d.
Rice Shaker Family Genes Are Expressed in Various Organs with Gene-Specific Patterns
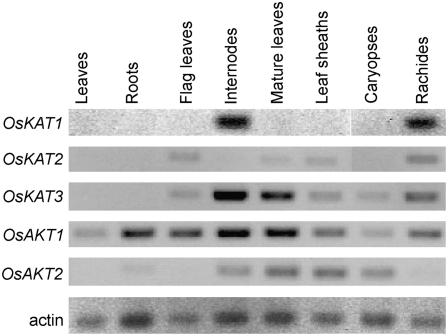
The organ-specific expression of OsKAT1 and other Shaker family genes was studied by reverse transcription (RT)-PCR using cDNAs from leaves and roots of rice seedlings and flag leaves, internodes, mature leaves, leaf sheaths, caryopses, and rachides of mature plants. The expression of OsKAT1 was restricted to internodes and rachides and was hardly detected in the other organs. OsKAT2 was not expressed in seedlings, internodes, or caryopses, but was expressed in leaves and rachides. OsKAT3 transcripts were highly accumulated in internodes and mature leaves, but were barely detected in seedlings. OsAKT2 was expressed in internodes, mature leaves, leaf sheaths, caryopses, and a little in roots. The expression of OsAKT1 was detected in all organs tested (Fig. 7).
Figure 7.
Expression of the Shaker family channel genes of rice in various tissues. Total RNA was isolated from leaves and roots of rice seedlings and flag leaves, internodes, mature leaves, leaf sheaths, caryopses, and rachides of mature plants. Transcripts were detected by RT-PCR. Rice actin was amplified as a loading control. The PCR products were separated in 0.8% (w/v) agarose gels and stained with ethidium bromide.
DISCUSSION
We have shown here that OsKAT1 induces salt tolerance in yeast and rice cultured cell lines. OsKAT1 is highly homologous to Shaker-type K+ channel proteins. Plant Shaker family members characterized to date are strongly reminiscent of most K+-selective currents in the plasma membrane described in vivo (Véry and Sentenac, 2002). The similarity between OsKAT1 and inward-rectifying K+ channels suggests that OsKAT1 mediates cellular K+ uptake as KAT1 and AKT1 (Cao et al., 1995). This was confirmed by complementation of the yeast Δtrk1Δtrk2 strain by OsKAT1 (Fig. 1B). Furthermore, presence of the TXXTXGYG motif, a hallmark for the majority of K+-selective channels (Gambale and Uozumi, 2006), in the OsKAT1 sequence suggests K+ selectivity of OsKAT1. The observation that expression of OsKAT1 increases salt tolerance in yeast (Figs. 1A and 3), while expression of nonselective ion transporters decreases salt tolerance in yeast (Amtmann et al., 2001; Horie et al., 2001; Gobert et al., 2006), is indeed evidence for high K/Na selectivity of OsKAT1.
The manner in which enhancement of K+ uptake mediated by OsKAT1 contributes to the salt tolerance of cells is an open question. OsKAT1-expressing cells accumulate more K+ and less Na+ and are more salt tolerant than control cells (Figs. 3 and 4, A and B). These are opposite phenotypes to K+-transport-defective yeast mutants (Gómez et al., 1996). In addition, the cellular Na+ to K+ ratio of OsKAT1-expressing yeast cells changed only gradually during culture, in contrast to the dramatic change in control cells (Fig. 4C). It has been suggested that the cellular Na+ to K+ ratio rather than the absolute intracellular concentration of Na+ determines salt tolerance in yeast and plant cells (Amtmann et al., 2001; Horie et al., 2001). These results suggest that OsKAT1 is likely to function in the maintenance of cellular Na+:K+ homeostasis and contribute to the salt tolerance of cells.
It should be noted that the low Na+ to K+ ratio was not only due to the increase of K+ but also the decrease of Na+ in OsKAT1-expressing yeast. The exact mechanism for the reduction of cellular Na+ level in OsKAT1-expressing cells remains to be elucidated. In the linear growth phase, the decrease of Na+ did not involve an increase of K+ (Fig. 4, A and B, right), suggesting that the changes in cation contents are not simply due to the competitive accumulation of Na+ and K+. One possibility is that enhanced membrane conductance for K+ depolarizes the membrane, thereby reducing the driving force for Na+ influx (Amtmann and Sanders, 1999). Another possibility is that high K+ concentration in the cytosol triggers Na+ export through up-regulation of nonselective cation efflux systems or down-regulation of nonselective uptake systems such as NSC1 (Bihler et al., 1998). In any case, the K+ selectivity of OsKAT1 should be essential to decrease the cellular Na+ to K+ ratio.
Overexpression of OsKAT1 also conferred salt tolerance on rice cultured cell lines (Fig. 5B). To our knowledge, this is the first report showing the enhancement of plant salt tolerance by overexpression of a K+ transporter gene. Only an increase of K+ content was observed; unlike in yeast, a decrease of Na+ content did not occur in OsKAT1-overexpressing rice cells (Fig. 5, C and D). This may be due to various differences between these kinds of cells, including growth rate, cation transport systems, and membrane potential. The observation that an increase in K+ content without an accompanying decrease in Na+ content enhances salt tolerance in rice cells further suggests cellular Na+ to K+ ratio as an important factor in salt tolerance (Fig. 5E), while Kronzucker et al. (2006) have recently shown a shoot growth unproportional to this ratio in barley (Hordeum vulgare).
It is unlikely that in planta OsKAT1 should be sufficient to confer salt tolerance because its expression is restricted to internodes and rachides. We found five putative genes coding AKT/KAT-type channels in the rice genome with diverse expression patterns. Transcripts of more than one Shaker gene were detected in all tested organs (Fig. 7). Since the improvement of cellular salt tolerance should be a common function among Shaker-type channels (Fig. 6), individual channel proteins are likely to cooperate in salt tolerance by regulating ion homeostasis in the cells in which they are localized. Formation of a heteromeric K+ channel may give rise to further variations in cation transport activities (Baizabal-Aguirre et al., 1999; Pilot et al., 2001) and confer cell-specific characteristics of Na+:K+ homeostasis.
We clearly demonstrated that OsKAT1 confers salt tolerance in rice at the cellular level. The degree to which OsKAT1 and other K+ channels contribute to salt tolerance at the whole-plant level should be revealed by studying the effect of knock-out and overexpression of Shaker K+ channels on salt tolerance in rice plants. Further analyses of the relationships among plant Shaker channels, other K+ transporters, and Na+ transporters in saline environments should reveal the mechanisms responsible for plant salt tolerance and contribute to the development of food crops that are tolerant to increased salinity.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
We used yeast (Saccharomyces cerevisiae) strains W303 (MATa ade2 ura3 leu2 his3 trp1), G19 (MATa ade2 ura3 leu2 his3 trp1 ena1Δ∷HIS3∷ena4Δ; Quintero et al., 1996), and CY162 (MATa ura3 his3 his4 trk1Δ trk2Δ1∷pCK64; Anderson et al., 1992). YPG medium consisted of 2% Difco yeast extract, 1% bacto-triptone, 2% Gal, and 1% raffinose. SDG medium contained 0.67% yeast nitrogen base, 2% Gal, 1% raffinose, and all standard amino acids except uracil. Yeast transformations were performed using LiCl as described by Chen et al. (1992). We used the AtKAT1 expression vector described by Uozumi et al. (1995). The growth temperature was 28°C.
Screening for Rice cDNA Clones That Complement the Salt-Sensitive Phenotype of Yeast Strain G19
Yeast expression libraries were constructed from the rice (Oryza sativa) full-length cDNA library of the National Institute of Agrobiological Sciences, Japan (Kikuchi et al., 2003). Inserts in the libraries were cloned into expression vector pYES2 (Invitrogen), which is a 2-μm-based multicopy yeast plasmid and contains the URA3 gene and the GAL1 promoter for selection and expression in yeast. Then the expression vector libraries were transformed into yeast strain G19. The transformants were screened under the stress condition on SDG plates containing 0.5 m NaCl. Salt-resistant colonies appearing on the plates during incubation for 7 to 12 d were suspended in water and spotted onto SDG plates with 0, 0.3, 0.5, 0.7, or 1.0 m NaCl. Plasmids were recovered from the clones that exhibited salt tolerance by transformation of Escherichia coli cells with DNA isolated from the yeast (Wen et al., 2005). cDNAs were identified from 5′- and 3′-flanking sequences determined using an ABI3100 sequencer (Applied Biosystems). Plasmids were reintroduced into fresh preparations of G19. Transformants were retested for growth on SDG plates containing 0, 0.3, 0.5, 0.7, or 1.0 m NaCl.
Construction of OsKAT1-Overexpressing Rice Cell Lines
The full-length OsKAT1 cDNA clone (AK100739) was provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan. The binary vector pBIGS-PUbi-SfiL/35SHyg (abbreviated as pRiceFOX) was constructed from a pBI-based plasmid, pBIG2113Not/dSfi (M. Fujita and K. Shinozaki, unpublished data). The pRiceFOX vector carries a hygromycin-resistance gene downstream of the cauliflower mosaic virus 35S promoter as a selectable marker, and the maize (Zea mays) Ubiquitin-1 gene promoter cassette followed by two SfiI restriction sites for the directional cloning of full-length cDNAs (H. Nakamura and H. Ichikawa, unpublished data). The OsKAT1 cDNA clone was subcloned in the forward orientation into the SfiI sites of pRiceFOX. The pRiceFOX-OsKAT1 plasmid and EV were then transferred into Agrobacterium tumefaciens strain EHA105 (Hood et al., 1986) and transformed into rice ‘Nipponbare’ as described by Toki (1997). Calluses resistant to 30 μg mL−1 hygromycin were suspended in Murashige and Skoog medium containing 3% Suc, 2 μg mL−1 2,4-dichlorophenoxyacetic acid, vitamins, and 30 μg mL−1 hygromycin. The cell lines were cultured in the dark at 26°C with rotation at 110 rpm and transferred to fresh medium every 2 weeks. Three lines (EV, OsKAT1 #14, and OsKAT1 #28) that arose from independent calluses were used in subsequent experiments. These lines grew at similar rates under the normal culture conditions.
Determination of Cation Contents in Cells
The yeast cells were collected by centrifugation of 2 mL of culture broth. After they were washed twice with ice-cold 20 mm MgCl2, cells were resuspended in 0.2 mL of distilled water and boiled for 20 min. The concentrations of the cations in the supernatant were measured by ion chromatography using an ICS90 ion chromatography system with an IonPac CS12A column (Dionex).
Rice cells were collected by filtration on Whatman Grade 114 filter paper and rinsed with distilled water at least five times. Collected cells were weighed to obtain their fresh weight and then dried at 80°C overnight. Cations were extracted from the cells in boiling water for 20 min and their concentrations were determined by ion chromatography.
Construction of OsAKT1 Expression Plasmid for Yeast
The full-length OsAKT1 cDNA clone (AK120308) was provided by the Rice Genome Resource Center. Two SfiI restriction sites were inserted into the BamHI site of pYES2 for the directional cloning of full-length cDNAs. The OsAKT1 and OsKAT1 cDNA clones were subcloned in the forward orientation into the SfiI sites. These plasmids were used for the experiment described in Figure 6.
RT-PCR
Total RNA was extracted from leaves and roots of 7-d-old seedlings of ‘Nipponbare’ grown by the method of Nakamura et al. (2006). Flag leaves, internodes, mature leaves, leaf sheaths, caryopses, and rachides were obtained from mature plants grown in a paddy field. Rice cell lines were cultured for 10 d in liquid Murashige and Skoog medium. The putative genes of Shaker channel proteins were searched by the TBLASTN algorism in the Rice Annotation Project Database (http://rapdb.lab.nig.ac.jp/) using the amino acid sequences of OsKAT1, OsAKT1 (Os01g0648000), AtKAT1 (At5g46240), and AtAKT1 (At2g26650) as queries. First-strand cDNA was synthesized using poly-T primer, and PCR was performed with primers specific for OsKAT1 (5′-gtgctcaacgcaagacagat-3′ and 5′-tttctttgacaggcaggtga-3′), OsKAT2 (Os01g0210700; 5′-tcagcaacgatgcaaagaac-3′ and 5′-gcatgtctgtttcatcatttatttc-3′), OsKAT3 (Os02g0245800; 5′-ccgctcaagcatagcataca-3′ and 5′-cggtctggttgattcatgtc-3′), OsAKT1 (5′-agagacggcgatcatcttgt-3′ and 5′-cagcagatcaacttgccaac-3′), and OsAKT2 (Os05g0428700; 5′-ccatccattcgtcagaaacc-3′ and 5′-tgcaaaaacattcccactca-3′). Rice actin cDNA was amplified as a loading control with the primers 5′-tccatcttggcatctctcag-3′ and 5′-gtacccgcatcaggcatctg-3′. The PCR products were separated in 0.8% (w/v) agarose gels, stained with ethidium bromide, and detected on a FluorImager 575 (Molecular Dynamics).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK100739 and AK120308.
Acknowledgments
We thank Dr. O. Kinclová-Zimmermannová (Academy of Science of Czech Republic, Prague) for kindly providing yeast strains W303 and G19; Dr. N. Uozumi (Tohoku University, Sendai, Japan) for kindly providing yeast strain CY162 and the AtKAT1 expression vector; and Dr. R. Gaber (Northwestern University, Chicago) for permitting us to use CY162. We also thank Drs. H. Ichikawa, H. Nakamura, and M. Hakata (National Institute of Agrobiological Sciences, Tsukuba, Japan) for providing the pBIGS-PUbi-SfiL/35SHyg vector. We thank H. Onodera (National Institute of Agrobiological Sciences, Tsukuba, Japan) for technical assistance with production of transgenic rice cells.
This work was supported by a project grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (EF1006).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yoshiyuki Tanaka (tanakayo@affrc.go.jp).
Open Access articles can be viewed online without a subscription.
References
- Akbar M, Ponnamperuma FN (1980) Saline soil of south and southeast Asia as potential rice lands. In IRRI, ed, Rice Research Strategies for the Future. IRRI, Los Baños, Laguna, The Philippines, pp 256–281
- Amtmann A, Fischer M, Marsh EL, Stefanovic A, Sanders D, Schachtman DP (2001) The wheat cDNA LCT1 generates hypersensitivity to sodium in a salt-sensitive yeast strain. Plant Physiol 126 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Sanders D (1999) Mechanisms of Na+ uptake by plant cells. Adv Bot Res 29 75–112 [Google Scholar]
- Anderson JA, Huprikar SS, Kochian LV, Lucas WJ, Gaber RF (1992) Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 89 3736–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 [DOI] [PubMed] [Google Scholar]
- Apse MP, Blumwald E (2002) Engineering salt tolerance in plants. Curr Opin Biotechnol 13 146–150 [DOI] [PubMed] [Google Scholar]
- Baizabal-Aguirre VM, Clemens S, Uozumi N, Schroeder JI (1999) Suppression of inward-rectifying K+ channels KAT1 and AKT2 by dominant negative point mutations in the KAT1 α-subunit. J Membr Biol 167 119–125 [DOI] [PubMed] [Google Scholar]
- Bihler H, Slayman CL, Bertl A (1998) NSC1: a novel high-current inward rectifier for cations in the plasma membrane of Saccharomyces cerevisiae. FEBS Lett 432 59–64 [DOI] [PubMed] [Google Scholar]
- Cao Y, Ward JM, Kelly WB, Ichida AM, Gaber RF, Anderson JA, Uozumi N, Schroeder JI, Crawford NM (1995) Multiple genes, tissue specificity, and expression-dependent modulation contribute to the functional diversity of potassium channels in Arabidopsis thaliana. Plant Physiol 109 1093–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Yang B, Kuo T (1992) One-step transformation of yeast in stationary phase. Curr Genet 21 83–84 [DOI] [PubMed] [Google Scholar]
- Coté GG (1995) Signal transduction in leaf movement. Plant Physiol 109 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HH, Luan S (1998) AtKUP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs I, Stölzle S, Ivashikina N, Hedrich R (2005) Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 221 212–221 [DOI] [PubMed] [Google Scholar]
- Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45 146–159 [DOI] [PubMed] [Google Scholar]
- Gambale F, Uozumi N (2006) Properties of shaker-type potassium channels in higher plants. J Membr Biol 210 1–19 [DOI] [PubMed] [Google Scholar]
- Glenn E, Brown JJ, Blumwald E (1999) Salt-tolerant mechanisms and crop potential of halophytes. CRC Crit Rev Plant Sci 18 227–255 [Google Scholar]
- Gobert A, Park G, Amtmann A, Sanders D, Maathuis FJM (2006) Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J Exp Bot 57 791–800 [DOI] [PubMed] [Google Scholar]
- Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ (2003) Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcript differently. Plant Mol Biol 51 71–81 [DOI] [PubMed] [Google Scholar]
- Gómez MJ, Luyten K, Ramos J (1996) The capacity to transport potassium influences sodium tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett 135 157–160 [DOI] [PubMed] [Google Scholar]
- Hedrich R, Becker D (1994) Green circuits: the potential of plant specific ion channels. Plant Mol Biol 26 1637–1650 [DOI] [PubMed] [Google Scholar]
- Hood EE, Helmer GL, Fraley RT, Chiltom MD (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region pTiBo542 outside of T-DNA. J Bacteriol 168 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27 129–138 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28 000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Kronzucker HJ, Szczerba MW, Moazami-Goudarzi M, Britto DV (2006) The cytosolic Na+:K+ ratio does not explain salinity-induced growth impairment in barley: a dual-tracer study using 42K+ and 24Na+. Plant Cell Environ 29 2228–2237 [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167 645–663 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Fukuda A, Sakai S, Tanaka Y (2006) Molecular cloning, functional expression and subcellular localization of two putative vacuolar voltage-gated chloride channels in rice (Oryza sativa L.). Plant Cell Physiol 47 32–42 [DOI] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Chérel I, Boucherez J (2001) Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem 276 3215–3221 [DOI] [PubMed] [Google Scholar]
- Qi Z, Spalding EP (2004) Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol 136 2548–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Garciadeblas B, Rodríguez-Navarro A (1996) The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′), 5′-bisphosphate nucleotidase and inositol 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encode a sodium transporter. Nat Genet 37 1141–1146 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Ward JM, Gassmann W (1994) Perspective on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct 23 441–471 [DOI] [PubMed] [Google Scholar]
- Serrano R (1996) Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int Rev Cytol 165 1–52 [DOI] [PubMed] [Google Scholar]
- Serrano R, Rodriguez-Navarro A (2001) Ion homeostasis during salt stress in plants. Curr Opin Cell Biol 13 399–404 [DOI] [PubMed] [Google Scholar]
- Shi H, Lee B, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21 81–85 [DOI] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 44 928–938 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15 16–21 [Google Scholar]
- Uozumi N, Gassmann W, Cao Y, Schroeder JI (1995) Identification of strong modification in cation selectivity in an Arabidopsis inward rectifying potassium channel by mutant selection in yeast. J Biol Chem 270 24276–24281 [DOI] [PubMed] [Google Scholar]
- Véry AA, Sentenac H (2002) Cation channels in the Arabidopsis plasma membrane. Trends Plant Sci 7 168–175 [DOI] [PubMed] [Google Scholar]
- Wen Y, Hatabayashi H, Arai H, Kitamoto HK, Yabe K (2005) Function of the cypX and moxY genes in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol 71 3192–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]