Abstract
A transient rise in chlorophyll fluorescence after turning off actinic light reflects nonphotochemical reduction of the plastoquinone (PQ) pool. This process is dependent on the activity of the chloroplast NAD(P)H dehydrogenase (NDH) complex, which mediates electron flow from stromal reductants to the PQ pool. In this study, we characterized an Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutant pifi (for postillumination chlorophyll fluorescence increase), which possesses an intact NDH complex, but lacks the NDH-dependent chlorophyll fluorescence increase after turning off actinic light. The nuclear gene PIFI (At3g15840) containing the T-DNA insertion encodes a chloroplast-targeted protein localized in the stroma and is annotated as a protein of unknown function. The pifi mutant exhibited a lower capacity for nonphotochemical quenching, but similar CO2 assimilation rates, photosystem II (PSII) quantum efficiencies (ΦPSII), and reduction levels of the primary electron acceptor of PSII (1 − qL) as compared with the wild type. The pifi mutant grows normally under optimal conditions, but exhibits greater sensitivity to photoinhibition and long-term mild heat stress than wild-type plants, which is consistent with lower capacity of nonphotochemical quenching. We conclude that PIFI is a novel component essential for NDH-mediated nonphotochemical reduction of the PQ pool in chlororespiratory electron transport.
In addition to photosynthetic electron transport in the chloroplast, a respiratory electron flow [from NAD(P)H to plastoquinone (PQ) and O2], named chlororespiration and involving both nonphotochemical reduction and reoxidation of the PQ pool, was indicated to exist in the chloroplast of higher plants and originate from a bacterial ancestor (Garab et al., 1989; Gruszecki et al., 1994; Scherer, 1990). This concept is supported by recent identification and characterization of several components that could function in the chlororespiratory pathway (for reviews, see Bennoun, 2002; Peltier and Cournac, 2002). A membrane-associated plastid NAD(P)H dehydrogenase (NDH) complex was shown to mediate nonphotochemical reduction of the PQ pool by stromal reductants, which can be monitored by a far-red light-quenchable postillumination chlorophyll (Chl) fluorescence increase after turning off actinic light (AL; Burrows et al., 1998; Shikanai et al., 1998). Other pathways, involving a putative ferredoxin PQ reductase (FQR), might also be involved in nonphotochemical reduction of the PQ pool (Burrows et al., 1998; Shikanai et al., 1998; Peltier and Cournac, 2002). The reoxidation of reduced PQ possibly by O2 was recently shown to involve carbon monoxide-sensitive oxidases (Feild et al., 1998), a membrane-associated plastid-terminal oxidase (PTOX; Carol et al., 1999; Wu et al., 1999; Joët et al., 2002b), and cytochrome b559 (Bondarava et al., 2003). However, very limited information is currently available on the soluble stromal electron carriers or stroma side components of the chlororespiratory pathway.
Although chlororespiration in higher plants is estimated to be very low in comparison with photosynthetic activity, it could function as an additional protective mechanism (such as avoiding overreduction of PSII acceptors through PTOX and facilitating thermal dissipation or providing extra ATP through cyclic electron transport [CET] in response to environmental stresses) and it might also be involved in reoxidation of stromal reductants for various metabolic pathways in the chloroplast (such as carbon metabolism in the dark; Peltier and Cournac, 2002). In addition to its role in chlororespiration, the NDH complex was also suggested to be involved in CET (Burrows et al., 1998; Shikanai et al., 1998; Joët et al., 2002a). Tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana) ndh mutants that lack a functional NDH complex did not exhibit any visible growth phenotypes under nonstressful conditions (Burrows et al., 1998; Shikanai et al., 1998; Hashimoto et al., 2003). However, NDH-mediated CET may help to alleviate the effects of environmental stresses, such as excess light (Endo et al., 1999), drought (Horváth et al., 2000), chilling (Li et al., 2004), heat, and oxidative stresses (Wang et al., 2006) on photosynthetic performance. Although NDH-mediated CET was suggested to protect PSII and PSI from photoinhibition (Endo et al., 1999), the protection mechanism is not clear (Burrows et al., 1998).
Chlororespiration activity is not only controlled by the redox state of its electron carriers, but also by posttranslational modification and likely by redox regulation of its components, such as the NDH complex. For example, activity of the NDH complex was closely correlated with the phosphorylation level of its NDH F subunit under photooxidative stress caused by high light or the presence of hydrogen peroxide (Lascano et al., 2003). The NDH complex can be activated by illumination and was proposed to be controlled by redox regulation (Teicher and Scheller, 1998). However, direct evidence for redox regulation of chlororespiration is still not available.
In this article, we characterized an Arabidopsis T-DNA insertion mutant, pifi (for postillumination chlorophyll fluorescence increase), that apparently lacks NDH-dependent nonphotochemical reduction of the PQ pool, but possesses an intact NDH complex. Initial interest in the PIFI protein arose from the fact that it has a similar redox-modulated carboxyl terminus to that of the 46-kD isoform of Rubisco activase (Fig. 1A; Zhang and Portis, 1999; Wang and Portis, 2006) and that it is present in plant species (e.g. tobacco and maize [Zea mays]) that lack the large isoform of activase. Although in vitro and in vivo studies have not supported our original hypothesis that PIFI could be involved in the redox regulation of Rubisco activase (data not shown), lack of a postillumination Chl fluorescence increase (CFI) in the pifi mutant similar to that of ndh mutants redirected our research interest to investigate involvement of PIFI in chlororespiration. By comparing the Chl fluorescence parameters of the pifi mutant with wild type and their photosynthetic performance under stressful conditions, we conclude that PIFI is a novel component of the chlororespiratory electron transport pathways and can help to protect plants from stressful environmental conditions (such as high light and mild heat stress).
Figure 1.
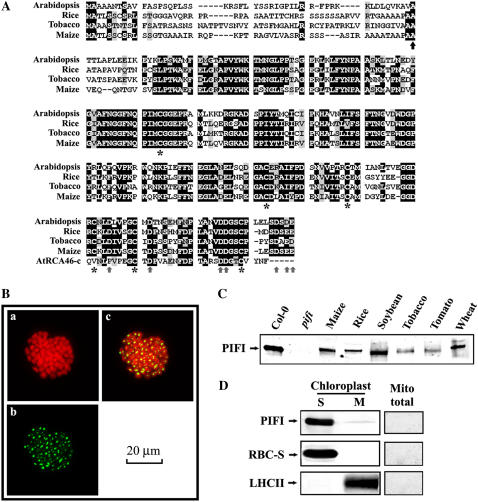
Sequence alignment of PIFI protein and its subcellular localization. A, Alignment of the amino acid sequences deduced from PIFI cDNA from Arabidopsis, rice (Oryza sativa), tobacco, and maize and carboxyl-terminal peptide sequence of a 46-kD Rubisco activase (RCA46-C). The predicted cleavage site of chloroplast target peptide (ChloroP) is indicated by a black up arrow. Six conserved Cys residues and several negative residues are marked with asterisks and gray up arrows, respectively. B, Subcellular localization of PIFI protein. The localization of PIFI protein was investigated by transient expression of GFP-tagged PIFI gene in Arabidopsis protoplasts. Images of auto (Chl) fluorescence (a), GFP fluorescence (b), and an overlap of auto and GFP fluorescence (c) are shown. C, Immunodetection of PIFI proteins from various higher plants. Total soluble proteins (15 μg) from leaves of seven different higher plants and the Arabidopsis pifi mutant were used for western-blot assays using antibodies raised against Arabidopsis PIFI protein. D, Localization of PIFI protein in the chloroplast stroma. Stromal soluble proteins (10 μg), thylakoid membranes (10 μg Chl), and total mitochondrial (Mito total) proteins (5 μg) from wild-type Arabidopsis chloroplasts were analyzed using PIFI antibodies. Rubisco small subunit (RBC-S) and light-harvesting complex II (LHCII) were analyzed using specific antibodies as controls for the stromal (S) and membrane (M) fractions, respectively, or for examining the possible chloroplast contamination in the mitochondria sample.
RESULTS
PIFI Gene Encodes a Chloroplast Stromal Protein Specific to Higher Plants
Arabidopsis PIFI cDNA encodes a 268-amino acid protein with a predicted 49-amino acid chloroplast-targeting peptide (ChloroP) and seven Cys residues (six of which are highly conserved; Fig. 1A). A BLAST search revealed that homologs of the PIFI gene are only found in higher plants and not in algae, such as Chlamydomonas reinhardtii, which does not have the chloroplast NDH complex (Maul et al., 2002) or cyanobacteria from which the chloroplast NDH complex is thought to originate. The homologous PIFI genes from maize (GenBank accession no. DQ854728) and tobacco (GenBank accession no. DQ854729) were cloned and their deduced full-length amino acid sequences exhibited 72% and 73% homology, respectively, to the Arabidopsis PIFI mature protein, respectively (Fig. 1A). An immunoblot assay using antibodies raised against Arabidopsis PIFI protein confirmed the expression of PIFI protein in several higher plants (Fig. 1C). These results indicate that PIFI is specific to higher plants.
Transient expression of a GFP-tagged PIFI in protoplasts (Fig. 1B) and subsequent immunoassay (data not shown) confirmed that PIFI is expressed in the chloroplasts. The GFP-tagged PIFI protein was detected inside chloroplasts and appeared to aggregate possibly due to the association of the GFP tag (Ma et al., 2004; S. Ma, personal communication). The native PIFI protein was also detected in the stromal soluble fraction of chloroplasts and not in mitochondria (Fig. 1D). Searches of public protein domain databases (Prosite, Pfam, and National Center for Biotechnology Information Conserved Domain Database) did not reveal any predicted transmembrane domains or other functional motifs in the PIFI sequence, but its carboxyl terminus exhibits 44% identity to the redox-modulated carboxyl terminus of the 46-kD isoform of Rubisco activase, including two conserved Cys and several negatively charged residues (Fig. 1A; Zhang and Portis, 1999; Wang and Portis, 2006). However, no redox regulatory effects on the activities of Rubisco activase were detected with purified recombinant PIFI proteins (data not shown).
Growth and Photosynthesis of pifi Mutants Are Similar to Wild Type
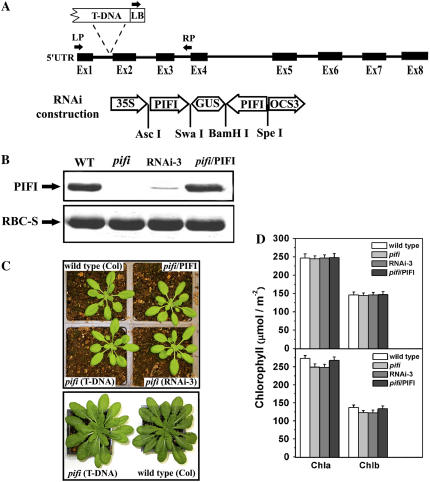
To elucidate the function of the PIFI protein, we characterized a homozygous Arabidopsis pifi mutant obtained by T-DNA insertion (Salk_085656) that does not have detectable PIFI protein (Fig. 2, A and B). To further confirm the phenotypes of the T-DNA insertion pifi mutant, RNA interference (RNAi) of the PIFI gene and complementation of the pifi mutant with a wild-type PIFI gene were performed (Fig. 2A). In one RNAi line (RNAi-3), expression of PIFI was less than 10% of that in the wild type (estimated by band intensity on immunoblots), whereas a comparable level of PIFI was observed in a complemented line (Fig. 2B). When grown with 50 μmol photon m−2 s−1 light, the pifi mutant, RNAi-3, and complemented line exhibited similar Chl content, growth, and developmental phenotypes as those of the wild type (Fig. 2, C and D). When grown at higher light intensity (150 μmol photon m−2 s−1), both the pifi mutant and RNAi-3 line exhibited slightly lower Chl content than the wild-type and complemented lines (Fig. 2, C and D). The steady-state photosynthetic rates (as determined by CO2 assimilation rate) of the pifi mutant were similar to those of wild-type plants under either 50 or 150 μmol photon m−2 s−1 growth lights (Supplemental Fig. S1).
Figure 2.
T-DNA insertion, RNAi, and complementation studies. A, Schematic presentation of the T-DNA insertion site (Salk_085656) and vector construction of PIFI-RNAi. Exons and introns of the PIFI gene (At3g15840) are shown as thick and thin lines, respectively. LB, LP, and RP are primers used for PCR analysis from the left border of the T-DNA, the first exon, and the junction of third intron and fourth exon, respectively. For RNAi construction, PIFI represents a partial coding sequence of At3g15840 cDNA, 35S represents the p35S promoter, GUS represents the GUS reporter gene, and OCS3 represents the OCS3′ terminator. B, Immunoblot assay of PIFI protein in the wild type (WT), the pifi mutant (pifi), an RNAi line (RNAi-3), and a complemented line (pifi/PIFI). Each lane was loaded with 15 μg total soluble protein and antibodies to PIFI or the Rubisco small subunit (RBC-S) were used. C, Growth of wild-type and pifi mutant plants. Image at the top shows the 4-week-old wild-type, T-DNA insertion mutant (pifi), RNAi (RNAi-3), and complemented line (pifi/PIFI) grown with a 12-h light/12-h dark photoperiod at 50 μmol photons m−2 s−1. The image at the bottom shows an 8-week-old wild type and pifi mutant grown with a 12-h light/12-h dark photoperiod at 150 μmol photons m−2 s−1. D, Chl content of Arabidopsis leaves grown under 50 (top) and 150 (bottom) μmol photons m−2 s−1 light. Leaf discs (1.5 cm2) were cut out and extracted with 80% acetone. Chl content was measured and calculated according to Porra et al. (1989).
Nonphotochemical Reduction of the PQ Pool Is Altered
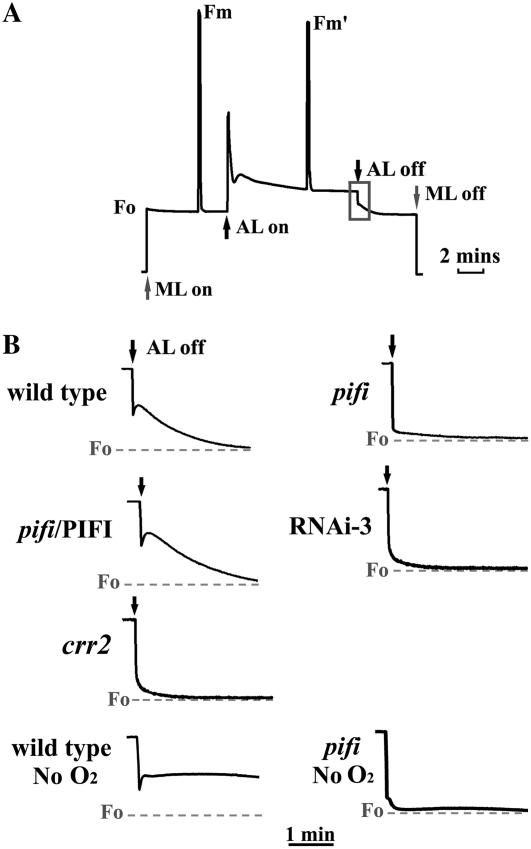
Transient CFI after turning off AL is attributed to the nonphotochemical reduction of the PQ pool and depends on the activity of a plastid NDH complex (Groom et al., 1993; Burrows et al., 1998; Feild et al., 1998; Shikanai et al., 1998; Hashimoto et al., 2003). For Arabidopsis leaves, a measuring light (ML) of low intensity is required to observe postillumination CFI (Hashimoto et al., 2003). Postillumination CFI is present in wild-type plants after turning off 100 μmol photons m−2 s−1 AL in an 8 μmol photons m−2 s−1 ML background, but not in the pifi mutant grown under 50 μmol photons m−2 s−1 (Fig. 3, A and B). This phenotype is similar to that of an ndh-defective Arabidopsis mutant (chlororespiratory reduction mutant crr2; Fig. 3B; Hashimoto et al., 2003). To further confirm that the absence of CFI is due to the T-DNA insertion in PIFI, we examined the effects on CFI of PIFI silencing via RNAi and complementation of the pifi mutant with a wild-type PIFI gene. CFI was absent in an RNAi line (RNAi-3) in which expression of PIFI protein is minimal (Fig. 2C) and was restored by complementation (Fig. 3B). To examine whether the loss of CFI in the pifi mutant is due to stimulation of the activity of an oxidase like PTOX (Joët et al., 2002b), Chl fluorescence measurements were conducted without O2 in the gas stream (380 μL L−1 CO2 with extrapure N2). Under such conditions, postillumination CFI in the wild type was enhanced with very slow quenching (Fig. 3B), which is consistent with decreased nonphotochemical reoxidation of the PQ pool by an oxidase. However, no apparent change in CFI was observed in the pifi mutant (Fig. 3B) and the NDH-defective crr2 mutant (data not shown). In addition, the redox state of QA (1 − qL) in the pifi and crr2 mutants as measured by applying saturating flash after turning off the AL was less reduced than wild type (Supplemental Fig. S2). These results together clearly indicate that the absence of PIFI in the mutant results in impairment of NDH-mediated nonphotochemical reduction of the PQ pool.
Figure 3.
Postillumination Chl fluorescence of wild-type and pifi mutant leaves. A, Typical Chl fluorescence trace of a wild-type leaf. The transient CFI that occurs after switching off AL (100 μmol photon m−2 s−1) with ML (8 μmol photon m−2 s−1) background is highlighted by the black box. The up and down arrows indicate turning on and off AL (black) and ML (gray), respectively. B, Postillumination Chl fluorescence after switching off AL with plants grown with 50 μmol photon m−2 s−1 light and preilluminated for 5 h before measurement. Chl fluorescence was recorded on attached leaves of the wild type, the pifi mutant (pifi), complemented (pifi/PIFI), RNAi line (RNAi-3), and an ndh-defective (crr2) mutant in ambient air or without O2 (wild type and pifi only) after switching off AL. The gray dashed lines indicate the initial F0 levels before switching on AL.
The Gross Nonphotochemical Quenching Is Lower at High Light Intensities
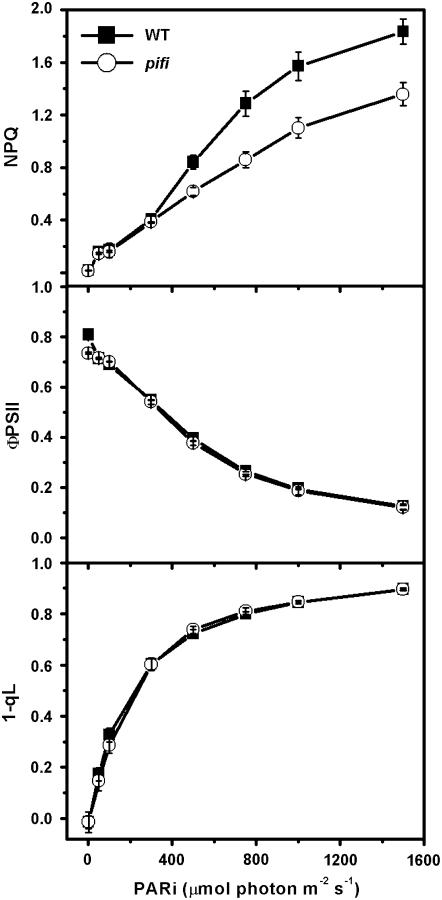
Phenotypes of the pifi mutant were further investigated by characterizing the modulated Chl a fluorescence of PSII (Fig. 4). The gross nonphotochemical quenching (NPQ) of Chl fluorescence is mainly caused by thermal dissipation that dissipates excess light energy as heat and protects plants from photooxidation damage (Niyogi, 1999). At elevated light intensities, the pifi mutant exhibited lower (26% at 1,500 μmol photons m−2 s−1 AL) steady-state value of gross NPQ than wild type grown with 50 μmol photons m−2 s−1 light (Fig. 4). However, the quantum yields of PSII (ΦPSII) and reduction level of QA (1 − qL) were similar in the pifi mutant and wild type (Fig. 4).
Figure 4.
Light dependence of steady-state Chl fluorescence parameters of wild-type and pifi mutant leaves. Plants were grown with 50 μmol photon m−2 s−1 light and preilluminated for 4 to 5 h before measurement. Gross NPQ, quantum efficiency of PSII photochemistry (ΦPSII), and reduction state of QA (1 − qL) were measured on attached leaves under various AL intensities.
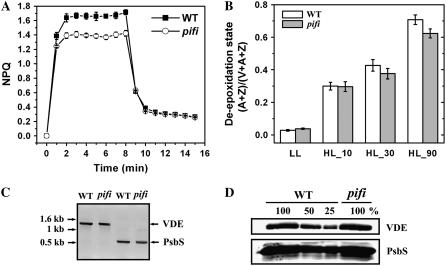
For both the wild type and the pifi mutant, the major portion of NPQ was attributed to qE because nearly complete relaxation of NPQ was observed during the first 4 min of darkness following an 8-min period of 1,000 μmol photons m−2 s−1 high light illumination (Fig. 5A). This reversible portion of NPQ was 28% lower in the pifi mutant compared with that in wild type. Because the xanthophyll cycle contributes to qE significantly (Niyogi et al., 1998), we compared the deepoxidation state, which is conversion of violaxanthin (V) to antheraxanthin (A) and zeaxanthin (Z), of the pifi mutant and wild type. A slightly lower (14%) ratio of (A + Z)/(V + A + Z) was observed in the pifi mutant than in the wild type after 90-min exposure to high light (Fig. 5B). The low qE in pifi could not be attributed to lower levels of either PsbS or violaxanthin deepoxidase (VDE), which are essential for qE in Arabidopsis (Niyogi et al., 1998; Li et al., 2000), as measured by quantitative PCR (data not shown), reverse transcription (RT)-PCR (Fig. 5C), and immunoblot assays (Fig. 5D).
Figure 5.
Characterization of altered NPQ in the pifi mutant. A, Induction and relaxation of NPQ. NPQ was measured on predarkened leaves upon switching on AL (1,000 μmol photon m−2 s−1) for 8 min and subsequently off for 7 min. B, Deepoxidation state of the xanthophyll cycle. Pigment content of the xanthophyll cycle was analyzed by HPLC. Deepoxidation of violaxanthin (V) to antheraxanthin (A) and zeaxanthin (Z) was compared between the wild type and the pifi mutant under low light (LL; 50 μmol photon m−2 s−1) and subsequently exposed to high light (HL; 800 μmol photon m−2 s−1) for the indicated times. The deepoxidation state is presented as (A + Z)/(A + Z + V). C, RT-PCR analysis of expression of PsbS and VDE genes in the wild type (WT) and the pifi mutant. D, Immunoblot analysis of PsbS and VDE proteins in the wild type (WT) and the pifi mutant. Thylakoid membrane samples equivalent to 10 μg Chl were loaded as 100%, with a series of dilutions as indicated for the wild type.
Protein Levels and in Vitro Activity of NDH in the pifi Mutant Are Similar to Wild Type
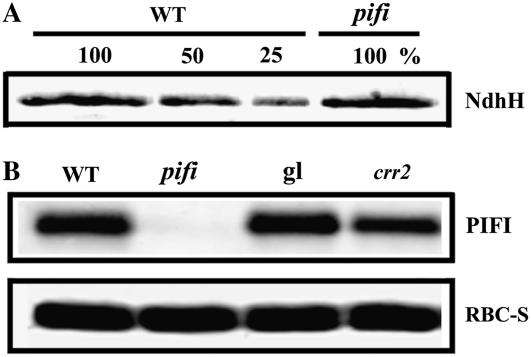
Immunoblot assays were performed to investigate whether the phenotypes observed in pifi are due to impaired accumulation of NDH. No significant differences from the wild type in the levels of the Ndh H subunit (Fig. 6A) were observed. It has been shown that the Ndh H subunit is unstable in the absence of other NDH subunits (Munekage et al., 2004; Rumeau et al., 2005) and can be used to estimate the accumulation of the NDH complex (Hashimoto et al., 2003). Therefore, the pifi mutant possesses an intact NDH complex (Fig. 6A). Conversely, a slightly lower level of PIFI was present in the NDH-defective crr2 mutant as compared with wild-type Columbia-0 (Col-0) and Col-gl plants (Fig. 6B).
Figure 6.
Immunoblot analysis of NdhH in the pifi mutant (A) and PIFI in the crr2 mutants (B). For detection of NdhH, thylakoid membrane samples of the wild-type Col (WT) and the pifi mutant equivalent to 20 or 10 μg Chl, respectively, were loaded as 100%, with a series of dilutions for wild-type Col (WT). Detection of PIFI in wild-type Col (WT) and the pifi mutant, wild-type gl1 (gl), and crr2 was performed the same as described in Figure 2B.
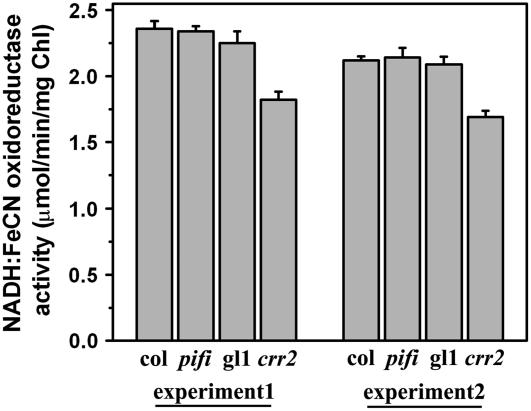
To determine whether the NDH complex in the pifi mutant is functional, the NADH dehydrogenase activities of thylakoid membranes isolated from pifi and wild type in the presence of electron donor NADH and electron acceptor ferricyanide (FeCN) were compared in vitro (Sazanov et al., 1998; Rumeau et al., 2005). It has been shown that about 30% of the NADH-FeCN activity measured in this system is associated with the plastidial NDH in pea (Pisum sativum) thylakoid membranes (Sazanov et al., 1998). NADH-FeCN activity of the thylakoid membranes from the pifi mutant was 2.3 μmol min−1 mg−1 Chl, similar to those of wild-type plants (Col and gl1; Fig. 7). As a positive control, NADH-FeCN activity of the thylakoid membranes from the crr2 (NDH-defective) mutant was found to be about 23% less than that of the wild type and pifi mutant. Residual NADH-FeCN activity bound to thylakoid membranes in the crr2 mutant can be attributed to other enzyme activities (Sazanov et al., 1998). Therefore, the results indicate that the NDH complex in the pifi mutant is functional when NADH is provided in vitro and further suggest that PIFI protein is involved in either electron donation to NDH or regulation of NDH in vivo.
Figure 7.
NADH-FeCN oxidoreductase activity. Thylakoid membranes (10 μg Chl) from either the pifi mutant (pifi) and its wild-type background (Col) or crr2 mutant (crr2) and its wild-type background (gl1) were used in the assay. NADH-FeCN oxidoreductase activity was assayed at 25°C by spectrophotometrically measuring the reduction of FeCN at 420 nm (see “Materials and Methods”). The assay was repeated with two groups of plants grown at a different date (experiment 1 and experiment 2).
The pifi Mutant Is More Sensitive to Photoinhibition
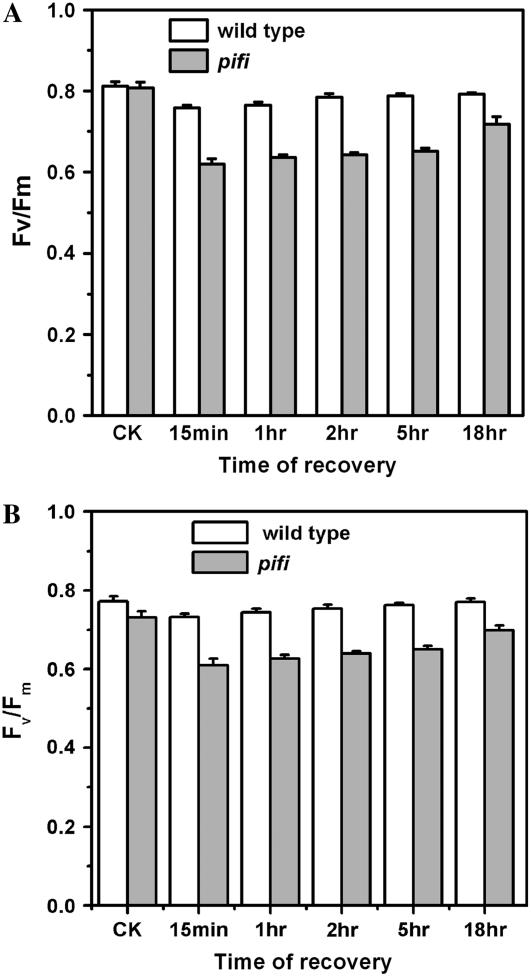
Because both gross NPQ and qE are lower in the pifi mutant than wild type at high light intensities (Figs. 4 and 5A) and CET via the NDH complex was proposed to function in photoprotection (Burrows et al., 1998; Endo et al., 1999), the sensitivity of the pifi mutant to photoinhibition was compared to the wild type by measuring the recovery of the maximal quantum yield of PSII (Fv/Fm) after high light illumination (Fig. 8). Prior to exposure to high light, Fv/Fm was similar in the pifi mutant (0.81 ± 0.01) and wild-type plants (0.82 ± 0.02) grown with 50 μmol photons m−2 s−1 light, whereas lower in the pifi mutant (0.73 ± 0.01) and wild type (0.78 ± 0.02) grown with 150 μmol photons m−2 s−1 light. During the dark recovery after exposure to high light (approximately 1,600 μmol photons m−2 s−1) for 4 h, the pifi mutant exhibited more severe depression and a slower recovery of PSII quantum efficiency than wild-type plants (Fig. 8). After 1-h darkness, wild-type plants exhibited 97% (low light) and 98% (high light) of control Fv/Fm, whereas the pifi mutant only exhibited 77% (low light) and 84% (high light) of the control. These data are consistent with the lower gross NPQ and qE observed in the pifi mutant (Figs. 4 and 5A) and further indicate that the pifi mutant is more sensitive to photoinhibition than wild type.
Figure 8.
Sensitivity of wild-type and pifi mutant plants to photoinhibition grown under 50 (A) and 150 (B) μmol photon m−2 s−1 light. The quantum efficiency of PSII (Fv/Fm) was measured after 12-h darkness to serve as control (CK). The recovery of Fv/Fm was monitored at the indicated times from 15 min to 18 h after exposure to full sunlight for 4 h (see “Materials and Methods”).
The pifi Mutant Is More Sensitive to Extended Heat Stress
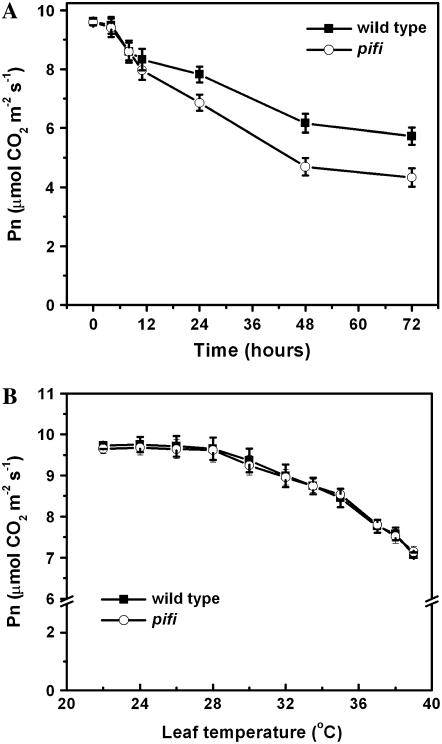
It was reported that NDH-mediated electron transport plays a role in alleviation of drought stress (Horváth et al., 2000) and oxidative damage caused by heat stress (Wang et al., 2006). To test whether the PIFI protein plays a similar role, we compared the CO2 assimilation rates of the pifi mutant and wild type under mild heat stress. Although the steady-state photosynthetic rates of the pifi mutant and wild type grown under the nonstressful condition were similar (Supplemental Fig. S1), the photosynthetic performance of the pifi mutant was more sensitive to long-term mild heat stress than wild type (Fig. 9A). After exposure to 32°C/30°C (day/night) for 48 h, wild-type plants retained 60% (measured at leaf temperature of 34°C) of the control photosynthetic rate (measured before heat stress), whereas the pifi mutant retained only 46% of the control. However, when exposed to short-term (<12 h) heat stress (Fig. 9A) or rapidly increased temperatures (Fig. 9B), the pifi mutant did not exhibit significant differences in photosynthetic performance compared with wild type.
Figure 9.
Effect of long-term (A) and short-term (B) heat stress on the photosynthetic rate (Pn) of the wild type and the pifi mutant. All measurements were performed with ambient air and 350 μmol m−2 s−1 of AL as described in “Materials and Methods.” For long-term heat stress, Pn was measured at the indicated times after the temperature in the growth chamber reached 32°C, with a leaf temperature setting of 34°C in the LI-COR 6400. Pn at time zero was measured at room temperature as control. For short-term heat stress, the leaf temperature was increased at a rate of 2°C per 8 min. Pn of three or four independent plants for each line was averaged and the error bars are shown.
DISCUSSION
It is generally accepted that the transient postillumination fluorescence increase after turning off AL is due to nonphotochemical reduction of PQ via chlororespiration. The absence of this fluorescence increase in the pifi mutant resembles the phenotype of both ndh mutants with directly disrupted NDH genes (Burrows et al., 1998; Shikanai et al., 1998; Takabayashi et al., 2002; Martín et al., 2004; Rumeau et al., 2005) and other mutants with defective assembly, expression, or stability of the NDH complex (Hashimoto et al., 2003; Kotera et al., 2005; Munshi et al., 2005; Muraoka et al., 2006). The phenotype of the pifi mutant is consistent with defective NDH-mediated nonphotochemical reduction of the PQ pool (Fig. 3). However, in marked contrast to the other mutants previously reported, the pifi mutant possesses an intact and apparently functional NDH complex (Figs. 6A and 7), which clearly indicates that PIFI plays a novel role in chlororespiration. Our results also indicate that PIFI is not involved in regulation of oxidation of the PQ pool because no apparent changes in postillumination Chl fluorescence were observed in pifi (Fig. 3B) and crr2 (data not shown) mutants exposed to air without O2 in contrast to the wild type. Interestingly, when grown under higher light (150 μmol photons m−2 s−1), the pifi mutant exhibited a novel postillumination CFI (Supplemental Fig. S3), which is not present in the wild type or the crr2 mutant. However, a similar phenotype was reported in tobacco ndh mutants after a short 1,200 μmol photon m−2 s−1 high light illumination (Takabayashi et al., 2002). The underlying mechanism is not clear and further study is required.
Sequence analysis of PIFI did not reveal any typical motifs present in electron carrier proteins, which makes it difficult to ascertain the function of PIFI in the nonphotochemical reduction of the PQ pool. Considering the stromal localization of PIFI and its putative redox-modulated C-terminal domain, we speculate that PIFI might be involved in the control of electron donation to NDH, possibly by redox modulation of unknown stromal electron carriers or the NDH complex itself. Teicher and Scheller (1998) reported light dependency in initial activation of NDH in vitro and proposed that this process might be related to oxidation of a redox mediator. However, no other evidence is currently available on the redox regulation of NDH or chlororespiration. Further work exploring complementation of the pifi mutant with mutated PIFI genes (e.g. replacing the conserved C-terminal Cys residues) can examine this possibility. In addition, absence of a PIFI gene in algae and cyanobacteria indicates that PIFI is specific to higher plants and further suggests that electron transport through NDH differs in higher plants from that in cyanobacteria. It is possible that the PIFI gene appeared during evolution to meet special needs of higher plants. This speculation is also supported by a recent report that a nuclear-encoded chloroplast protein CRR3, which is required for accumulation of the NDH complex in Arabidopsis, also is not found in algae and cyanobacteria (Muraoka et al., 2006).
Disruption of PIFI gene expression by T-DNA insertion or RNAi did not cause any serious growth retardation or decrease in photosynthetic rates under the 50 or 150 μmol photon m−2 s−1 light conditions (Fig. 2, C and D; Supplemental Fig. S1), which indicates that PIFI is dispensable or its roles can be sufficiently complemented by other processes under such conditions. However, under stressful environments, such as excess light, PIFI appears to be important in the protection of plants from photooxidative stress, which can clearly be seen from the decreased photosynthetic performance of the pifi mutant under such conditions (Fig. 8). The low gross NPQ in the pifi mutant under high light cannot be attributed to differences in PsbS and VDE. One possible explanation is that CET in the pifi mutant does not build up a sufficient proton gradient (ΔpH) across the thylakoid membrane to fine tune the photoprotection mechanisms in response to excess light (Heber and Walker, 1992). The moderately lower qE (28%) and deepoxidation state (14% after 90-min high light) in the pifi mutant than the wild type suggest a lower ΔpH in the pifi mutant.
Although the NDH complex was proposed to be involved in photoprotection (Burrows et al., 1998; Endo et al., 1999), the role of this pathway in thermal dissipation of excess energy in PSII remains unclear because an increase in gross NPQ (Rumeau et al., 2005), no change (Hashimoto et al., 2003), and a decrease in gross NPQ (Martín et al., 2004) have been observed in several NDH-defective mutants as compared with wild-type plants. These differences are possibly due to the varying extent of NDH deficiency in the mutants (Rumeau et al., 2005). An alternative way to examine the roles (such as photoprotection) of NDH-mediated electron transport is to identify and characterize mutants deficient in other components of this route. However, to our knowledge, all mutations documented so far either directly disrupt genes encoding NDH subunits or affect ndh RNA editing or stability of the NDH complex in an indirect manner. Therefore, the pifi mutant as a likely new mutant in the NDH-mediated electron transport pathway should be valuable in accessing the physiological significance of this pathway in the future.
In addition to impaired thermal dissipation, the pifi mutant exhibited higher sensitivity to mild heat stress than wild type (Fig. 9A), which is consistent with previous reports with ndh mutants under comparable conditions (Horváth et al., 2000; Wang et al., 2006). It has been proposed that, under conditions of drought and heat stress, where the requirements of various biochemical reactions for ATP increase, the NDH-mediated electron transport pathway along with others (e.g. FQR-mediated CET) is important to provide extra ATP to maintain a proper ATP-to-NADPH ratio (Bendall and Manasse, 1995; Kramer et al., 2004b). It is likely that under stressful conditions the mechanism mentioned above does not function properly in the pifi mutant. We predict that more severe phenotypes will be observed in double mutants obtained by crossing the pifi with crr2 or the pgr5 (proton gradient regulation) mutant, which is deficient in FQR-mediated CET (Munekage et al., 2002).
In conclusion, we investigated several possible physiological functions of a novel chloroplast protein PIFI specific to higher plants by characterizing a pifi mutant. Our results support the involvement of PIFI in nonphotochemical reduction of the PQ pool mediated by NDH in chlororespiration and demonstrate important roles of PIFI in adjusting photosynthetic performance in response to fluctuating environmental factors (such as light and temperature). Further characterization of the pifi mutant will benefit our understanding of the chlororespiratory electron transport pathways in higher plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotypes Col-0 and Col-gl1 wild type and mutants were grown in Sunshine number 1 potting soil under either 50 or 150 μmol photon m−2 s−1 light and a 12-h light/12-h dark photoperiod at 22°C/19°C, except when indicated otherwise. The T-DNA insertion mutant line Salk_085656 was obtained from the Arabidopsis Biological Resource Center (Ohio State University). The seeds of Arabidopsis crr2 and Col-gl1 were gifts from Dr. Shikanai. For growth on agar medium, seeds were surface sterilized and sown on agar-solidified one-half-strength Murashige and Skoog medium, including 2% Suc (Sigma-Aldrich).
T-DNA Insertion, RNAi, Complementation, and Plant Transformation
For PCR analysis of the T-DNA insertion line (Salk_085656), the following primers were used: left primer (LP, 5′-ACATATTTTTCTCCTTCTTTCATCC-3′), right primer (RP, 5′-TTTCAACTTTTCACCCTAATCCA-3′), and left border primer (LB, 5′-GCGTGGACCGCTTGCTGCAACT-3′). Primers (forward, 5′-TCCTAGTATTGTGGGTCGTCCTCG-3′; reverse, 5′-GCTCATTCTGTCGGCGATTCCAGG-3′) were used for PCR of the actin3 gene as a control. For PIFI-RNAi vector construct, the partial coding region for the Arabidopsis PIFI gene was cloned into pFGC1008 as described (Ma et al., 2004), using a pair of primers (forward, 5′-TAACTAGTGGCGCGCCACGACTCTTGCACCTCTCGAA-3′ and reverse, 5′-GCGGATCCATTTAAATTCATCAGAATCGGATAACTC-3′). For complementation of the pifi mutant, a 1.1-kb wild-type At3g15840 cDNA flanked by a pair of primers (forward, 5′-GGTCTAGAATCTTGAAGCCATATATCAATTACTTACAT-3′ and reverse, 5′-AAGAGCTCGATCAACAAGTGAACATTTATAATATTGGG-3′) was cloned into the XbaI and SstI sites in pBI121 vector, resulting in pBI121-PIFI. PIFI-RNAi and pBI121-PIFI vectors were transferred into Agrobacterium tumefaciens GV3101 by electroporation and used to transform either the wild-type Arabidopsis (Col-0) or the pifi mutant by floral dipping (Clough and Bent, 1998).
Subcellular Localization of GFP Fusion Proteins
A full-length cDNA of PIFI without a stop codon was amplified from Arabidopsis (Col-0) by RT-PCR and inserted into the pRZ238 vector to give the pRZ238-PIFI vector, encoding a carboxy-terminal fusion of PIFI onto GFP under control of a cauliflower mosaic virus 35S promoter (Ma et al., 2004). Arabidopsis mesophyll protoplast isolation and polyethylene glycol-mediated transformation were performed essentially according to Sheen (2002). After 16 to 20 h of incubation, GFP images of protoplasts were obtained using a Zeiss Axiovert 200-m microscope equipped with a 100-W Hg arc lamp. Image acquisition and analysis were performed using Axiovision software (version 4.2).
Chl Fluorescence and Gas Exchange
All experiments were conducted in ambient air at 22°C, except as indicated elsewhere. The fully expanded attached leaves were placed into a chamber with a leaf chamber fluorometer head on the LI-COR 6400 portable photosynthesis system (LI-COR). AL supplied with light-emitting diodes (90% red light, 630 nm; 10% blue light, 470 nm) were used to record the steady-state Chl fluorescence level (Fs). Chl fluorescence changes after turning off AL were monitored as described (Burrows et al., 1998; Shikanai et al., 1998; Hashimoto et al., 2003). ML (630 nm; 1 μmol photon m−2 s−1) was used to determine minimal Chl fluorescence at the open PSII center (F0). An 800-ms saturating pulse was applied to measure maximal Chl fluorescence at the closed PSII center in the dark (Fm) or during AL illumination (Fm′). The Fv/Fm of predarkened leaves and quantum efficiency of PSII (ΦPSII) of illuminated leaves were calculated as (Fm − F0)/Fm and (Fm′ − Fs)/Fm′, respectively (Genty et al., 1989). NPQ was calculated by (Fm − Fm′)/Fm′. Reduction levels of QA (1 − qL) were calculated as [Fm′ × (Fs − F0′)]/[(Fm′ − F0′) × Fs] (Kramer et al., 2004a).
The light-dependent photosynthetic rates were measured simultaneously with Chl fluorescence parameters at increased AL intensities with a LI-COR 6400 (equipped with standard 2 cm2 leaf chamber) under ambient CO2 concentrations and 21% O2. Measurement of photosynthesis rate in response to short-term heat stress was performed as previously described (Kim and Portis, 2005) with the following modifications. All measurements were conducted in ambient air and under 350 μmol m−2 s−1 of light illumination. The temperature was increased from 22°C to 39°C at a rate of 2°C/8 min and was allowed to stabilize for 3 min at each temperature point when five replicates of data were recorded. For long-term heat stress experiments, the temperature of the growth chamber was gradually increased from 22°C to 32°C and humidity was decreased gradually from 55% to 35% in about 1 h to avoid heat shock. Other settings were 12-h light, 32°C/30°C and 35% to 40% relative humidity. Photosynthesis performance was measured as described above at indicated times for 3 d in the same growth chamber, with leaf temperature setting of 34°C and relative humidity setting of 40%.
Deepoxidation State
Wild-type and pifi mutant Arabidopsis plants were illuminated at 800 μmol photons m−2 s−1 for the indicated times. Then leaf discs were cut out and quickly transferred into liquid N2. For pigment extraction, frozen leaf discs were ground in a mortar with 80% (v/v) acetone. After short centrifugation, the acetone extract was used for HPLC pigment analysis using a Spherisorb ODS-1 column (Alltech) on a Waters liquid chromatographic system (Milford) according to Gilmore and Yamamoto (1991). The deepoxidation activity was calculated as (A + Z)/(A + Z + V) (Niyogi et al., 1998; Li et al., 2000).
Photoinhibition
Photoinhibition was estimated by monitoring the recovery of Fv/Fm after illumination by high light as described (Niyogi et al., 1998) with the following modifications. Fv/Fm was first measured on attached leaves after overnight darkness and used as control. The wild type and the pifi mutant were then illuminated at either 50 μmol photon m−2 s−1 (for low light-grown plants) or 150 μmol photon m−2 s−1 (for high light-grown plants) for 1 h in the growth chamber before being moved to the greenhouse. Plants were exposed to full sunlight with incident photon flux density of 1,600 μmol photon m−2 s−1 for 4 h. Plants were then moved back to the growth chamber and kept in the dark. Fv/Fm was then measured at indicated times during the recovery.
Chloroplast and Mitochondria Preparation
Chloroplasts were prepared from leaves of 6-week-old Arabidopsis plants according to Rensink et al. (1998) with the following modifications. All procedures were conducted at 4°C. The extraction buffer contains 330 mm sorbitol, 20 mm HEPES-KOH, pH 7.6, 5 mm MgCl2, 3 mm EDTA, 0.1% (w/v) bovine serum albumin, and 330 mg L−1 ascorbate. The crude chloroplast pellet was gently suspended in a resuspension buffer (330 mm sorbitol, 20 mm HEPES-KOH, pH 7.6, 5 mm MgCl2, and 3 mm EDTA) before purification through discontinuous layers (40%/50%/70%) of Percoll (Amersham). To obtain soluble and membrane fractions, the intact chloroplasts were broken osmotically in a cold lysis buffer containing 20 mm HEPES-KOH, pH 7.6, and 1% (v/v) protease inhibitor cocktail (Sigma-Aldrich) on ice for 30 min, followed by centrifugation at 30,000g for 20 min at 4°C. Supernatants were collected as a stromal soluble fraction and pellets as a thylakoid membrane fraction. The isolation and purification of mitochondria from Arabidopsis leaves essentially followed a protocol described by Keech et al. (2005).
NADH-FeCN Oxidoreductase Assays
The thylakoid membranes were prepared as described (Sazanov et al., 1998; Rumeau et al., 2005) to minimize mitochondrial contamination. NADH-FeCN oxidoreductase activity was assayed at 25°C by spectrophotometrically measuring the reduction of FeCN at 420 nm (Sazanov et al., 1998). The assay buffer (1 mL) consisted of 50 mm Tris-HCl, pH 7.6, 0.5 mm EDTA, 0.1% dodecyl maltoside, 0.1 mm NADH, 0.5 mm FeCN, and thylakoid membrane samples (equal to 10 μg Chl).
Immunoblot Analysis
Thylakoid membrane proteins or stromal soluble proteins were separated by 12% Tris-Gly SDS-PAGE (15% Tris-tricine SDS-PAGE for immunoassay of Rubisco small subunit RBS-S), and blotted onto the polyvinylidene difluoride membrane. PIFI protein was detected using polyclonal rabbit antibodies raised against purified recombinant Arabidopsis PIFI protein (Immunological Resource Center at the University of Illinois, Urbana and Champaign). PsbS antibodies were a gift from Dr. K.K. Niyogi, VDE antibodies from Dr. A.D. Hieber, and NdhH antibodies from Dr. T. Endo.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ854728 and DQ854729.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Photosynthetic rates of the wild type and the pifi mutant.
Supplemental Figure S2. Redox state of QA (1 − qL) after turning off AL.
Supplemental Figure S3. Postillumination Chl fluorescence measured with 50 μmol photons m−2 s−1 AL and 3 or 0.1 μmol photons m−2 s−1 ML.
Supplementary Material
Acknowledgments
We thank Dr. R.E. Zielinski for access to a Zeiss Axiovert 200-m microscope, Dr. D.R. Ort, Dr. E.A. Ainsworth for reviewing the manuscript, and Dr. T.D. Sharkey for fruitful discussion. We thank Dr. T. Endo, Dr. A.D. Hieber, Dr. K.K. Niyogi, and Dr. T. Shikanai for providing antibodies.
This work was supported in part by the U.S. Department of Energy (grant no. 97ER20268).
Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that may also be suitable.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bendall DS, Manasse RS (1995) Cyclic phosphorylation and electron transport. Biochim Biophys Acta 1229 23–38 [Google Scholar]
- Bennoun P (2002) The present model for chlororespiration. Photosynth Res 73 273–277 [DOI] [PubMed] [Google Scholar]
- Bondarava N, Pascalis LD, Al-Babili S, Goussias C, Golecki JR, Beyer P, Bock R, Krieger-Liszkay A (2003) Evidence that cytochrome b559 mediates the oxidation of reduced plastoquinone in the dark. J Biol Chem 278 13554–13560 [DOI] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J 17 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntza M (1999) Mutations in the Arabidopsis gene immutans cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Endo T, Shikanai T, Takabayashi A, Asada K, Sato F (1999) The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett 457 5–8 [DOI] [PubMed] [Google Scholar]
- Feild TS, Nedbal L, Ort DR (1998) Nonphotochemical reduction of the plastoquinone pool in sunflower leaves originates from chlororespiration. Plant Physiol 116 1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garab G, Lajko F, Mustardy L, Marton L (1989) Respiratory control over photosynthetic electron transport in chloroplasts of higher plant cells—evidence for chlororespiration. Planta 179 349–358 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990 87–92 [Google Scholar]
- Gilmore AM, Yamamoto HY (1991) Resolution of lutein and zeaxanthin using a non-endcapped, lightly carbon-loaded C18 high-performance liquid chromatographic column. J Chromatogr 543 137–145 [Google Scholar]
- Groom QJ, Kramer DM, Crofts AR, Ort DR (1993) The non-photochemical reduction of plastoquinone in leaves. Photosynth Res 36 205–215 [DOI] [PubMed] [Google Scholar]
- Gruszecki WI, Bader KP, Schmid GH (1994) Light-induced oxygen uptake in tobacco chloroplasts explained in terms of chlororespiratory activity. Biochim Biophys Acta 1188 335–338 [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36 541–549 [DOI] [PubMed] [Google Scholar]
- Heber U, Walker D (1992) Concerning a dual function of coupled cyclic electron transport in leaves. Plant Physiol 100 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth EM, Peter SO, Joët T, Rumeau D, Cournac L, Horváth GV, Kavanagh TA, Schäfer C, Peltier G, Medgyesy P (2000) Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol 123 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët T, Cournac L, Peltier G, Havaux M (2002. a) Cyclic electron flow around photosystem I in C(3) plants: in vivo control by the redox state of chloroplasts and involvement of the NADH-dehydrogenase complex. Plant Physiol 128 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët T, Genty B, Josse EM, Kuntz M, Cournac L, Peltier G (2002. b) Involvement of a plastid terminal oxidase in plastoquinone oxidation as evidenced by expression of the Arabidopsis thaliana enzyme in tobacco. J Biol Chem 277 31623–31630 [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardeströma P (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124 403–409 [Google Scholar]
- Kim K, Portis AR Jr (2005) Temperature dependence of photosynthesis in Arabidopsis plants with modifications in Rubisco activase and membrane fluidity. Plant Cell Physiol 46 522–530 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE (2004. a) New fluorescence parameters for the determination of QA redox state and excitation energy. Photosynth Res 79 209–218 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Kanazawa A, Cruz JA, Ivanov B, Edwards GE (2004. b) The relationship between photosynthetic electron transfer and its regulation. In GC Papageorgiou, X Govindjee, eds, Chlorophyll Fluorescence: The Signature of Green Plant Photosynthesis. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 251–278
- Lascano HR, Casano LM, Martín M, Sabater B (2003) The activity of the chloroplastic Ndh complex is regulated by phosphorylation of the NDH-F subunit. Plant Physiol 132 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-P, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403 391–395 [DOI] [PubMed] [Google Scholar]
- Li XG, Duan W, Meng QW, Zou Q, Zhao SJ (2004) The function of chloroplastic NAD(P)H dehydrogenase in tobacco during chilling stress under low irradiance. Plant Cell Physiol 45 103–108 [DOI] [PubMed] [Google Scholar]
- Ma S, Quist TM, Ulanov A, Joly R, Bohnert HJ (2004) Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. Plant J 40 845–859 [DOI] [PubMed] [Google Scholar]
- Martín M, Casano LM, Zapata JM, Guéra A, del Campo EM, Schmitz-Linneweber C, Maier RM, Sabater B (2004) Role of thylakoid Ndh complex and peroxidase in the protection against photo-oxidative stress: fluorescence and enzyme activities in wild-type and ndhF-deficient tobacco. Physiol Plant 122 443–452 [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110 361–371 [DOI] [PubMed] [Google Scholar]
- Munshi MK, Kobayashi H, Shikanai T (2005) Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J 44 1036–1044 [DOI] [PubMed] [Google Scholar]
- Muraoka R, Okuda K, Kobayashi Y, Shikanai T (2006) A eukaryotic factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Physiol 142 1683–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50 333–359 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophylls cycle in the regulation of photosynthetic energy conversion. Plant Cell 10 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G, Cournac L (2002) Chlororespiration. Annu Rev Plant Biol 53 523–550 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P (1998) Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol 118 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumeau D, Bécuwe-Linka N, Beyly A, Louwagie M, Garin J, Peltier G (2005) New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA, Burrows PA, Nixon PJ (1998) The plastid ndh genes code for an NADH-specific dehydrogenase: isolation of a complex I analogue from pea thylakoid membranes. Proc Natl Acad Sci USA 95 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S (1990) Do photosynthetic and respiratory electron transport chains share redox proteins? Trends Biochem Sci 15 458–462 [DOI] [PubMed] [Google Scholar]
- Sheen J (2002) A transient expression assay using Arabidopsis mesophyll protoplasts. Massachusetts General Hospital Department of Molecular Biology Genetics Web Server. http://genetics.mgh.harvard.edu/sheenweb/protocols.html. (June 6, 2005)
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A (1998) Directed disruption of the tobacco ndhB gene impaired cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi A, Endo T, Shikanai T, Sato F (2002) Post-illumination reduction of the plastoquinone pool in chloroplast transformants in which chloroplastic NAD(P)H dehydrogenase was inactivated. Biosci Biotechnol Biochem 66 2107–2111 [DOI] [PubMed] [Google Scholar]
- Teicher HB, Scheller HV (1998) The NAD(P)H dehydrogenase in barley thylakoids is photoactivatable and uses NADPH as well as NADH. Plant Physiol 117 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Portis AR Jr (2006) Increased sensitivity of oxidized large isoform of Rubisco activase to ADP inhibition is due to an interaction between its carboxyl-extension and nucleotide-binding pocket. J Biochem (Tokyo) 281 25241–25249 [DOI] [PubMed] [Google Scholar]
- Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye JY, Mi H (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol 141 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Wright DA, Wetzel C, Voytas DF, Rodermel S (1999) The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Portis AR Jr (1999) Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci USA 96 9438–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.