Abstract
The marine Roseobacter clade comprises several genera of marine bacteria related to the uncultured SAR83 cluster, the second most abundant marine picoplankton lineage. Cultivated representatives of this clade are physiologically heterogeneous, and only some have the capability for aerobic anoxygenic photosynthesis, a process of potentially great ecological importance in the world's oceans. In an attempt to correlate phylogeny with ecology, we investigated the diversity of Roseobacter clade strains from various marine habitats (water samples, biofilms, laminariae, diatoms, and dinoflagellate cultures) by using the 16S rRNA gene as a phylogenetic marker gene. The potential for aerobic anoxygenic photosynthesis was determined on the genetic level by PCR amplification and sequencing of the pufLM genes of the bacterial photosynthesis reaction center and on the physiological level by detection of bacteriochlorophyll (Bchl) a. A collection of ca. 1,000 marine isolates was screened for members of the marine Roseobacter clade by 16S rRNA gene-directed multiplex PCR and sequencing. The 42 Roseobacter clade isolates found tended to form habitat-specific subclusters. The pufLM genes were detected in two groups of strains from dinoflagellate cultures but in none of the other Roseobacter clade isolates. Strains within the first group (the DFL-12 cluster) also synthesized Bchl a. Strains within the second group (the DFL-35 cluster) formed a new species of Roseovarius and did not produce Bchl a under the conditions investigated here, thus demonstrating the importance of genetic methods for screening of cultivation-dependent metabolic traits. The pufL genes of the dinoflagellate isolates were phylogenetically closely related to pufL genes from Betaproteobacteria, confirming similar previous observations which have been interpreted as indications of gene transfer events.
Phototrophy through bacteriochlorophyll (Bchl) a-mediated aerobic anoxygenic photosynthesis (40) has been estimated (based on in situ measurements of Bchl a) to be responsible for as much as 5 to 10% of the energy generation in the upper layers of the tropical oceans (12, 18, 19). The bacteria responsible for the process are thought to be related to the uncultivated marine SAR83 cluster within the alpha subclass of the Proteobacteria, which represents the second most abundant lineage of marine picoplankton bacteria after SAR11 (2, 11, 29). However, aerobic phototrophs have also been found in other genera of Alphaproteobacteria (Erythrobacter, Roseivivax, and Sphingomonas) and Betaproteobacteria (Roseateles) as well as among as yet uncultivated marine bacteria (2). In recent years, a large number of bacteria which are distantly related to the SAR83 cluster, and which form the Roseobacter clade, have been cultivated. Presently, 12 genera are recognized within this clade: Ketogulonicigenium, Marinosulfonomonas, Jannaschia, Octadecabacter, Ruegeria, Staleya, Roseobacter, Sulfitobacter, Antarctobacter, Sagittula, Roseovarius, and Silicibacter. The genus Roseivivax (36) forms a separate lineage outside the Roseobacter clade (see Fig. 1) (E. Stackebrandt, personal communication). These bacteria were isolated from diverse marine or saline habitats, e.g., from marine sand, sediments, and macroalgae (31), toxin-producing dinoflagellates (22, 27), the Black Sea (33), the North Sea (39), a meromictic lake (42), black smokers (40), and hot springs (26). Accordingly, their described physiological properties are highly diverse, ranging from sulfite reduction (33) to metabolism of dimethylsulfoniopropionate (43, 44) and production of toxins (10, 15). Only four species with the capability for aerobic anoxygenic photosynthesis are known to date, namely, Roseobacter litoralis and Roseobacter denitrificans, isolated from marine algae and sediments (31), and Staleya guttiformis (21) and Roseovarius tolerans (20), both from the hypersaline antarctic Ekho Lake.
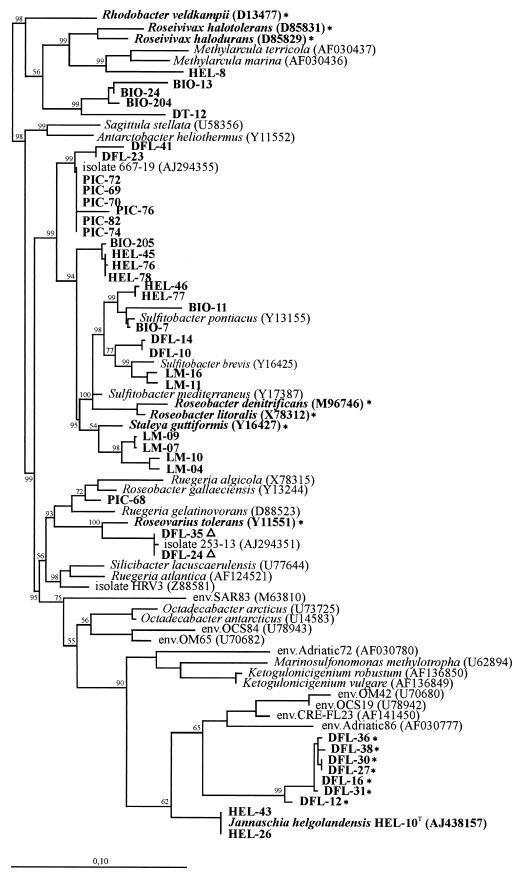
FIG.1.
Phylogenetic tree of the Roseobacter clade within the alpha subclass of the Proteobacteria. Reference organisms are italicized, and marine isolates described in this work are boldfaced. Representative environmental clones from uncultivated organisms of the Sargasso Sea (env.SAR), the Pacific Ocean (env.OCS), the Mediterranean Sea (env.Adriatic), the continental shelf of the Atlantic Ocean (env.OM), and the Columbia River estuary (env.CRE) were included (29). In addition, isolates from toxic dinoflagellates (667-19 and 253-13) and a marine sponge (HRV3) were also included. Strains in which the pufLM genes as well as Bchl a were detected are marked with asterisks. Strains which contained the pufLM genes but not bchla are marked with a Δ. Except for environmental clones and isolate HRV3, the tree is based on near-complete 16S rRNA gene sequences and was calculated using the maximum-likelihood algorithm (8). Bootstrap values above 50% are given. The bar corresponds to 10 base substitutions per 100 nucleotide positions. Rhodobacter veldkampii was used as an outgroup.
Speciation in bacteria is thought to be driven by the occupation of a new ecological niche (4). Thus, within a described species, ecotypes can be found with highly similar or even identical 16S rRNA gene sequences but clearly different ecological adaptations. Several examples have been described among pathogenic bacteria (7). In Neisseria meningitidis, 10 ecotypes, which occurred in different geographical areas and had different outbreak characteristics, could be identified by multilocus sequence typing (6). In surveys of as yet uncultured bacteria, clusters of highly related 16S rRNA gene sequences have frequently been described. In some instances, experimental evidence showed that these corresponded to different populations inhabiting distinct microenvironments. For example, clusters of uncultured Synechococcus strains from Yellowstone hot springs inhabited distinct microniches defined by temperature, photic zone, and stage of succession (28). Similar observations have been made among cultured marine organisms; small differences in 16S rRNA gene sequences of Prochlorococcus strains have been shown to correspond to high-light- and low-light-adapted populations (24), which have major genomewide genetic differences (http://web.mit.edu/chisholm/www/index.html).
The spectrum of ecological niches described in the literature for Roseobacter clade bacteria is huge, ranging from oligotrophic picoplankton (an uncultivated SAR83 cluster) to microalgal symbioses, but no systematic survey has been undertaken so far. Here we attempted to correlate ecology with phylogeny, by using the 16S rRNA gene as a phylogenetic marker gene and the genetic potential for aerobic anoxygenic photosynthesis as a cultivation-independent functional trait, for a large collection of marine isolates from various North Sea habitats. Our aims were to identify phylogenetic clusters of Roseobacter clade bacteria which were typical for certain habitats and to determine the capability for aerobic anoxygenic photosynthesis throughout these clusters.
MATERIALS AND METHODS
Sample collection and processing.
Samples were obtained from the North Sea around the island of Helgoland during sampling trips in 1998, 1999 and 2002. The research vessel Aade or Diker was used for sampling except for the rocky littoral, which could be reached by foot. Water samples were taken with a Ruttner sampler at the specified depth. A list of strains, sampling dates, sampling locations, and media used for first cultivation of isolates is given in Table 1. With the exception of strains designated PIC (for picoplankton), isolates were obtained by direct plating of aliquots (50 μl) of the highest sample dilution on the media indicated. Serial dilutions were made in artificial seawater or phosphate-buffered saline (PBS) (93.2 ml of 1 M Na2HPO4, 6.8 ml of 1 M NaH2PO4, and 5.8 g of NaCl per 1,000 ml).
TABLE 1.
List of isolated Roseobacter clade strains and sampling informationa
| Strainb | Type of sample | Sampling date (mo/day/yr) | Sampling site | Mediumc |
|---|---|---|---|---|
| Bacterioplankton | ||||
| HEL-8 | Water sample | 9/23/98 | Deep ridged | 1 |
| HEL-10 | 1 | |||
| HEL-26 | 1 | |||
| HEL-43 | 1 | |||
| HEL-45 | 1 | |||
| HEL-46 | 1 | |||
| HEL-76 | 2 | |||
| HEL-77 | 2 | |||
| HEL-78 | 2 | |||
| PIC-68 | Filtered water sample | 9/23/98 | Deep ridge | 1 |
| PIC-69 | 1 | |||
| PIC-70 | 1 | |||
| PIC-72 | 1 | |||
| PIC-74 | 1 | |||
| PIC-76 | 1 | |||
| PIC-82 | 1 | |||
| Biofilms | ||||
| BIO-7 | Suspended particles | 7/26/99 | Dunee | 3 |
| BIO-11 | In situ biofilm (1 wk)f | South harborg | 3 | |
| BIO-13 | In situ biofilm (1 wk) | Dune | 1 | |
| BIO-24 | In situ biofilm (2 wk) | Dune | 1 | |
| BIO-204 | In situ biofilm (1 wk) | Dune | 10 | |
| BIO-205 | In situ biofilm (2 wk) | Dune | 9 | |
| Diatoms, DT-12 | Thalassiosira sp. | 4/4/02 | Cable tonh | 4 |
| Macroalgal surface | ||||
| LM-4 | Laminaria sp. | 4/7/02 | Rocky littorali | 5 |
| LM-7 | 8 | |||
| LM-9 | 8 | |||
| LM-10 | 8 | |||
| LM-11 | 8 | |||
| LM-16 | 8 | |||
| Dinoflagellates | ||||
| DFL-10 | A. ostenfeldii | April 2002 | Culture KO287 | 8 |
| DFL-12 | P. lima | Culture ME130 | 6 | |
| DFL-14 | A. ostenfeldii | Culture KO287 | 6 | |
| DFL-16 | A. ostenfeldii | Culture KO287 | 6 | |
| DFL-23 | A. lusitanicum | Culture ME207 | 5 | |
| DFL-24 | A. ostenfeldii | Culture KO287 | 8 | |
| DFL-27 | A. ostenfeldii | Culture KO287 | 7 | |
| DFL-30 | A. ostenfeldii | Culture KO287 | 6 | |
| DFL-31 | A. ostenfeldii | Culture KO287 | 6 | |
| DFL-35 | A. ostenfeldii | Culture KO287 | 8 | |
| DFL-36 | A. ostenfeldii | Culture KO287 | 6 | |
| DFL-38 | A. ostenfeldii | Culture KO287 | 6 | |
| DFL-41 | A. ostenfeldii | Culture KO287 | 6 |
Samples were taken in the North Sea around the island of Helgoland during trips in 1998, 1999, and 2002. For details on isolation of strains, see Materials and Methods.
For each set of related strains (with designations beginning with the same letters), the type of sample, sampling date, or sampling site is given only in the first row if it is the same in all cases.
For descriptions of media, see Table 2.
For the deep ridge (Tiefe Rinne), samples were taken at 54°08′N, 7°52′E; depth, 10 m; temperature, 15.5°C; oxygen, 8.1 mg/liter; Secchi disk visibility, 5.5 m.
For the dune (Düne), samples were taken at 54°11.42′N, 7°53.51′E; depth, 3 m; temperature, 19.6°C; pH 8.22; conductivity, 44 mS/cm; oxygen, 6.2 mg/liter.
In situ biofilms were grown for 1 or 2 weeks.
For the south harbor (Südhafen), samples were taken at 54°10.13′N, 7°53.55′E; depth, 3 m.
For the cable ton (Kabeltonne), samples were taken at 54°11.18′N, 7°54.00′E; depth, 0.5 to 1.5 m; temperature, 6.5°C; Secchi disk visibility, 2.8 m.
For the rocky littoral (Felswatt), samples were taken at a depth of 0.5 m and a water temperature of 8.5°C.
For enrichment of PIC strains, 50 liters of the water sample was filtered through 5-μm-pore-size nylon mesh filters on the sampling day and stored in polyvinyl chloride containers in the dark at 4°C until enrichment was started (38). The isolates from the Roseobacter clade were enriched as follows. Aliquots (20 ml) of the filtered North Sea water sample were amended with tryptone and yeast extract (each at 1.0 g/liter) and incubated either at room temperature in natural daylight without shaking (PIC-68, PIC-69, and PIC-70), at room temperature in natural daylight with shaking (PIC-74), at room temperature in the dark without shaking (PIC-76), or at 4°C in the dark without shaking (PIC-82). After growth was visible, serial dilutions were plated on medium 1 (see Table 2).
TABLE 2.
List of mediaa used for the first cultivation of marine Roseobacter clade strains
| Medium no. | Medium description | Main carbon sources | Other components | Salts |
|---|---|---|---|---|
| 1 | Cytophaga (marine) medium, DSMZ medium 172b | Tryptone (1.0 g), yeast extract (1.0 g) | Sea salts | |
| 2 | Marine R2A mediumc | Peptone (0.5 g), yeast extract (0.5 g) | Casamino Acids (0.5 g), dextrose (0.5 g), soluble starch (0.5 g), sodium pyruvate (0.3 g) | 75% artificial seawater |
| 3 | M13 Verrucomicrobium medium, DSMZ medium 607 | Peptone (0.25 g), yeast extract (0.25 g) | Glucose (0.25 g), vitamin solution (10 ml), Hutner's salts (20 ml) | 25% artificial seawater |
| 4 | Marine broth 2216 (Difco), 1/10 strength | Peptone (0.5 g), yeast extract (0.1 g) | Tap water | |
| 5 | Luria-Bertani broth (Sigma) in North Sea water | Tryptone (10.0 g), yeast extract (5.0 g) | Unfiltered North Sea water | |
| 6 | Luria-Bertani broth (Sigma) in North Sea water, 1/10 strength | Tryptone (1.0 g), yeast extract (0.5 g) | Unfiltered North Sea water | |
| 7 | Peptone, yeast extract, North Sea water | Peptone (0.04 g), yeast extract (0.008 g) | Filtered North Sea water (pore size, 3 μm) | |
| 8 | Peptone, yeast extract, glucose, vitamins, 30% North Sea water | Peptone (0.25 g), yeast extract (0.25 g) | Glucose (0.25 g), vitamin solution (10 ml), Hutner's salts (20 ml) | Unfiltered North Sea water (300 ml) |
| 9 | Hg medium | Peptone (0.25 g), yeast extract (0.25 g) | HgCl2 (1.0 mg) | Filtered North Sea water (pore size, 0.2 μm) |
| 10 | North Sea water | Unfiltered North Sea water |
Amounts are given for a total volume of 1 liter. All media additionally contained 15 g of agar (Difco) and 25 mg of cycloheximide (Sigma).
Information on DSMZ media is available at http://www.dsmz.de/media/media.htm.
From the work of Suzuki et al. (35).
For isolation of strain BIO-7, a water sample of 7 liters was taken from a depth of 3 m and allowed to settle overnight. Sedimented particles were collected by using a glass pipette, serially diluted, and plated. For strains BIO-11, BIO-13, BIO-24, BIO-204, and BIO-205, biofilms were grown in situ by exposing sterilized glass plates (200 by 200 by 3 mm) mounted in a biofilm collector (3) to the North Sea on 26 July 1999 at two different locations at a depth of 3 m. After 1 or 2 weeks, biofilms were aseptically scraped off, serially diluted, and plated.
For isolation of strain DT-12, a plankton sample was taken by using a 170-μm-mesh-size net pulled behind a boat for 5 min. Diatoms (Thalassiosira sp.) were picked under a dissecting microscope and washed three times (see below). Approximately 30 cells were suspended in 100 μl of artificial seawater, and aliquots were plated without dilution.
For isolates designated LM, seaweeds (a Laminaria sp.) from a small pool (depth, 60 cm) in the rocky littoral zone of the island of Helgoland (“Felswatt”) were sampled during low tide. Cauloids were cut off. Phylloids were washed several times in artificial seawater. The surface biofilm was scraped off with a sterile blade, suspended in 5 ml of artificial seawater, mixed vigorously, serially diluted, and plated.
Strains designated DFL were obtained from the following dinoflagellate cultures of the Biological Research Institute of Helgoland: Prorocentrum lima ME130, Alexandrium lusitanicum ME207, and Alexandrium ostenfeldii KO287. Cultures were grown in F-2 medium (14) at 18°C with a cycle of 12 h of light and 12 h of darkness. The algal cells were picked by using a 1.5-μl Gilson pipette and transferred to a petri dish with artificial seawater (ca. 5 ml). They were washed by vigorous mixing, and the individual cells were subsequently transferred to a new petri dish. This procedure was repeated twice. Approximately 10 washed cells were finally pooled in 100 μl of artificial seawater and plated without dilution.
Isolation and cultivation of strains.
Strains were isolated on solid marine media with relatively low concentrations of carbon sources (Table 2). Samples for isolation of HEL strains were plated on medium 1, 2, 3, or 9; those for PIC and BIO strains were plated on medium 1, 3, 9, or 10 or the medium given by Uphoff et al. (38); and those for DT, DFL, and LM strains were plated on media 4 to 8. For the composition of artificial seawater, Hutner's salt solution, and vitamin solution, see the information on DSMZ media 607, 590, and 603, respectively, on the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) website (http://www.dsmz.de/media/media.htm). All media contained cycloheximide (25 mg liter−1) to prevent the growth of eukaryotes and were solidified with agar (15 g liter−1). Media were sterilized by autoclaving at 121°C for 20 min. Unfiltered seawater was autoclaved before use. Heat-sensitive components, such as antibiotics and vitamins, were added to autoclaved media from filter-sterilized (pore size, 0.2 μm) stock solutions. Picked colonies were restreaked on the same medium two to three times until they were pure. Subsequently, they were cultivated on Marine Broth 2216 (Difco). Roseovarius tolerans (DSMZ 11457), S. guttiformis (DSMZ 11458), Roseobacter litoralis (DSMZ 6996), and Roseobacter denitrificans (DSMZ 7001) were used as reference strains. The Roseobacter strains were grown on DSMZ medium 695 (http://www.dsmz.de/media/media.htm), while Roseovarius tolerans and S. guttiformis were grown on medium 8 (Table 2) with 25 and 10% artificial seawater, respectively. In addition to the type strain of Roseovarius tolerans (EL-172T), seven other isolates of Roseovarius tolerans were investigated (EL-52, EL-78, EL-83, EL-90, EL-164, EL-171, and EL-222) (personal gifts from Matthias Labrenz).
DNA extraction.
Genomic DNA was extracted from a loopful of bacterial cells from a fresh plate by using the NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany) with an additional step for RNase digestion after cell lysis. To this end, 4 μl of RNase (Roche, Basel, Switzerland) was added and incubated at 37°C for 30 min, followed by incubation at 95°C for 10 min to denature the enzyme. Genomic DNA was eluted by using 100 μl of elution buffer and stored at −20°C. Strains which did not yield enough genomic DNA by this method were extracted by using the FastDNA SPIN Kit for Soil from Bio 101 (now called Qbiogene, Heidelberg, Germany). For cell lysis, a bead beater (Retsch, Haan, Germany) was used instead of the FastPrep instrument. Genomic DNA was eluted with 50 μl of DNase-pyrogen-free water and stored at −20°C.
Amplification of 16S rRNA and pufLM genes.
The primer pair 16F 27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16R 1492 (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) was used for PCR amplification of the nearly complete 16S rRNA gene (23). The reaction mixture contained 1.5 mM MgCl2, 0.25 μM each deoxynucleoside triphosphate, 0.5 μM each primer, 1 μl of template DNA, and 5 U of Taq DNA polymerase (Qiagen, Hilden, Germany) in a total volume of 50 μl. PCR was performed in a Landgraf (Langenhagen, Germany) thermocycler or a Mastercycler gradient (Eppendorf, Hamburg, Germany) by using an initial denaturation step at 94°C (2 min), followed by 30 cycles of denaturation at 94°C (1 min), annealing at 54°C (1 min), and extension at 72°C (2 min). A final extension at 72°C (10 min) and subsequent cooling at 4°C completed the reaction. The amplified DNA was purified using the QIAquick PCR Purification kit (Qiagen), eluted with 50 μl of H2O, and stored at −20°C.
The primer pair pufLF (5′-CTK TTC GAC TTC TGG GTS GG-3′) and pufMR (5′-CCA TSG TCC AGC GCC AGA A-3′) (K is G or T and S is G or C) was used for amplification of the pufLM genes from the bacterial photosynthesis reaction center (2). The reaction mixture was the same as that described above, except that 2 U of Taq DNA polymerase was used. The reaction volume was 20 μl (for screening of strains) or 50 μl (for excision of bands). The PCR program was the same as that described above, except that initial denaturation at 94°C was performed for 3 min, and the annealing temperature was 60°C. The amplified pufLM fragment (ca. 1.5 kb) was excised from the agarose gel under UV light and extracted by using the QIAquick Gel Extraction kit (Qiagen) according to the manufacturer's instructions. Extracted DNA was eluted with 30 μl of H2O and stored at −20°C.
Sequencing.
Sequencing was performed with an automated capillary sequencer, the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany), according to the manufacturer's instructions. The sequencing reaction contained 5 μl of DNA, 1 μl of primer (10 μM), 5 μl of the reagent from the sequencing kit (ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems), 1 μl of dimethyl sulfoxide, and 8 μl of H2O. The sequencing PCR was performed with an Eppendorf (Hamburg, Germany) Mastercyler gradient by using 25 cycles consisting of denaturation (at 96°C for 15 s), annealing (at 60°C for 15 s), and extension (at 60°C for 240 s). For sequencing of the 16S rRNA gene, primers 16F 27 and 16R 1492 were used; for sequencing of the pufLM genes, primers pufLF and pufMR were used. After amplification, the DNA was cleaned by ethanol precipitation as follows. The reaction mixture was transferred to a 0.5-ml Eppendorf reaction tube containing 10 μl of sodium acetate (3 M) and 250 μl of ethanol (98%, vol/vol). After vigorous mixing, the DNA was precipitated by centrifugation (at 14,000 × g for 30 min). The supernatant was discarded, and the pellet was washed in 300 μl of ethanol (70%, vol/vol). The DNA was precipitated by centrifugation (at 14,000 × g for 10 min), the supernatant was discarded, and the pellet was dried for 5 min in a SpeedVac.
Phylogenetic inferences.
Sequences were analyzed by using the ARB software package (http://www.arb-home.de). Partial 16S rRNA gene sequences were imported into the ARB database of ca. 12,600 aligned reference sequences. Partial sequences were automatically aligned and assembled into full sequences, and alignments were corrected manually. Full sequences were integrated into the ARB phylogenetic tree by using the maximum parsimony algorithm tool (9). This tool does not correct for evolutionary distances and does not change the tree topology. It served as a basis for identification of the reference organisms. These were subsequently selected for tree calculation by means of the maximum-likelihood algorithm (8) by using the fastDNAml program (25). Partial pufLM sequences were aligned using ClustalW (37). The alignment was imported into ARB. A phylogenetic tree was calculated in ARB using the maximum-likelihood method. Almost full length 16S rRNA gene sequences and partial pufLM sequences were checked for homologies to published sequences by BLAST searches (www.ncbi.nlm.nih.gov). Similar sequences were downloaded, imported into ARB, and used for calculation of phylogenetic trees.
Determination of Bchl a.
Bchl a was detected spectrophotometrically in vivo or in vitro. For in vivo measurements, 2 ml of liquid culture (Marine Broth 2216; Difco) incubated in the dark or in a natural daylight rhythm was washed (at 7,000 rpm for 5 min, with resuspension of the pellet in 1 ml of PBS). To reduce light scattering (32), 1 ml of bovine serum albumin (30%, wt/vol; Sigma) was added. For in vitro measurements, Bchl a was extracted by the procedure of Cohen-Bazire et al. (5). Two microliters of a liquid culture was centrifuged (at 7,000 rpm for 5 min), and the pellet was resuspended in a drop of remaining medium. A 1.5-ml volume of an ice-cold (−20°C) acetone-methanol solution (7:2, vol/vol) was added, mixed well, and incubated at room temperature in the dark for 12 h. Cell fragments were then removed by centrifugation (at 2,500 rpm for 5 min). Spectrophotometric measurements were performed with an Ultrospec K 4053 instrument from LKB Biochrom (Cambridge, United Kingdom). Relative absorption was determined after calibration at 900 nm against air in the range from 400 to 900 nm (in vivo) or 600 to 900 nm (in vitro).
Nucleotide sequence accession numbers.
Sequences of Roseobacter clade bacterium 16S rRNA genes sequenced in this study can be found in the EMBL data bank under accession numbers AJ534205 to AJ534244 and AJ532580. pufL sequences can be found under accession numbers AJ532572 to AJ532579.
RESULTS AND DISCUSSION
Isolation of strains.
A total of 1,019 isolates were obtained from different marine habitats, mainly from the North Sea around the island of Helgoland. Of these, 71 were isolated from an untreated seawater sample (strains designated HEL), 412 were enriched from the same water sample after filtering (PIC strains), 463 were obtained from biofilms grown in situ in the North Sea on glass plates (BIO strains), 12 were isolated from the surfaces of Thalassiosira sp. diatoms (DT strains), 44 were isolated from cultures of A. ostenfeldii, A. lusitanicum, or P. lima (Dinophyta) (DFL strains), and 17 were isolated from Laminaria sp. surfaces (LM strains). In the latter three cases, pigmented colonies were picked selectively, while HEL, BIO, and PIC strains were picked independently of their pigmentation. Bacteria were isolated by direct plating of the fresh, untreated sample or the highest sample dilution on solid marine media, with the exception of PIC strains, which were enriched under a wide spectrum of incubation and amendment conditions (38). The media used for isolation of strains had carbon contents that were relatively low, though high in comparison to those used for the first cultivation of SAR11 (30), and differed in their salt contents (Table 2). Unamended, autoclaved North Sea water was also used as a culture medium.
Screening for Roseobacter clade strains.
All strains were screened for Roseobacter clade phylogenetic affiliation by multiplex PCR targeting the 16S rRNA gene. This PCR results in amplicons of a certain size for selected phylogenetic groups. The primer mix used produced a 650-bp amplicon in Alphaproteobacteria and a 250-bp fragment in Roseobacter clade bacteria (38). Positive strains were partially sequenced, and false positives were excluded from further analysis. Forty-two strains that were assigned to the Roseobacter clade based on partial 16S rRNA gene sequencing were fully sequenced.
Phylogenetic affiliation of strains.
Of the 42 strains analyzed, 9 had been isolated from a North Sea water sample, 7 from picoplankton enrichment cultures, 6 from biofilms, 6 from Laminaria surfaces, 1 from Thalassiosira surfaces, and 13 from dinoflagellate cultures (Table 1). Figure 1 shows that the isolates tended to form clusters of related organisms from the same habitat. These clusters were distributed over the whole Roseobacter clade and represented new genera (e.g., Jannaschia helgolandensis [39]) or species. Only in one case did a cluster contain a strain highly related, but not identical, to a strain which had been isolated from a different habitat: BIO-205, from biofilms in the cluster around HEL-45 from the water column. Among isolates from dinoflagellates, strains derived from A. ostenfeldii cultures (DFL-36, DFL-38, DFL-30, DFL-27, DFL-16, and DFL-31) were more closely related to each other than to the isolate from a P. lima culture (DFL-12). Similar observations have been interpreted previously as the result of coevolution (1). The data might point to a process of evolution driven by ecological adaptation which has resulted in clusters of phylogenetically related strains specifically adapted to the ecological niche in question, e.g., biofilm, dinoflagellate, or picoplankton.
Almost all cloned environmental sequences within the Roseobacter clade fell into a separate branch containing SAR83 from the Sargasso Sea as well as sequences from the Mediterranean Sea, Atlantic Ocean, and Pacific Ocean. They formed three subclusters, which were affiliated with recently described genera, i.e., Octadecabacter (13), isolated from arctic and antarctic ice; Marinosulfonomonas methylotropha (16); and J. helgolandensis from the North Sea (39). Even more similar to the third and largest cluster of SAR83-related sequences than J. helgolandensis were isolates obtained from dinoflagellate cultures (the DFL-12 cluster). The level of similarity to J. helgolandensis was 92 to 93%, indicating at least genus level divergence. However, the phylogenetic affiliation of the environmental sequences, as well as the grouping of genera within the Rosebacter clade, was not robust, as indicated by the low bootstrap values, and thus should not be overestimated.
Genetic potential for aerobic anoxygenic photosynthesis in Roseobacter clade marine isolates.
All marine isolates shown in Fig. 1 as well as several type strains were screened for the presence of the pufLM genes of the photosynthesis reaction center by PCR amplification and sequencing. Their presence was confirmed in Roseobacter denitrificans, Roseobacter litoralis, and S. guttiformis. Interestingly, the pufLM genes were found in all eight strains of Roseovarius tolerans tested. When first describing Roseovarius tolerans, Labrenz et al. (20) could detect Bchl a in only four strains (EL-78, EL-83, EL-171, and EL-172T) and therefore named the genus Roseovarius. In addition, pufLM genes were found in two dinoflagellate isolates (DFL-24 and DFL-35) within the genus Roseovarius (probably representing a new species) and in the DFL-12 cluster of strains, which were also isolated from dinoflagellate cultures. Interestingly, none of the other Roseobacter clade marine isolates had pufLM genes. However, these genes were also detected in other Alphaproteobacteria (Fig. 2A). Some strains that had the pufLM genes and were also isolated from dinoflagellates formed a cluster related to Ahrensia kielensis (DFL-13, DFL-44, DFL-42, and DFL-43), and another isolate was related to Roseibium hamelinense (DFL-11). Moreover, a water sample isolate related to Sphingomonas alaskensis (HEL-42) also had pufLM genes.
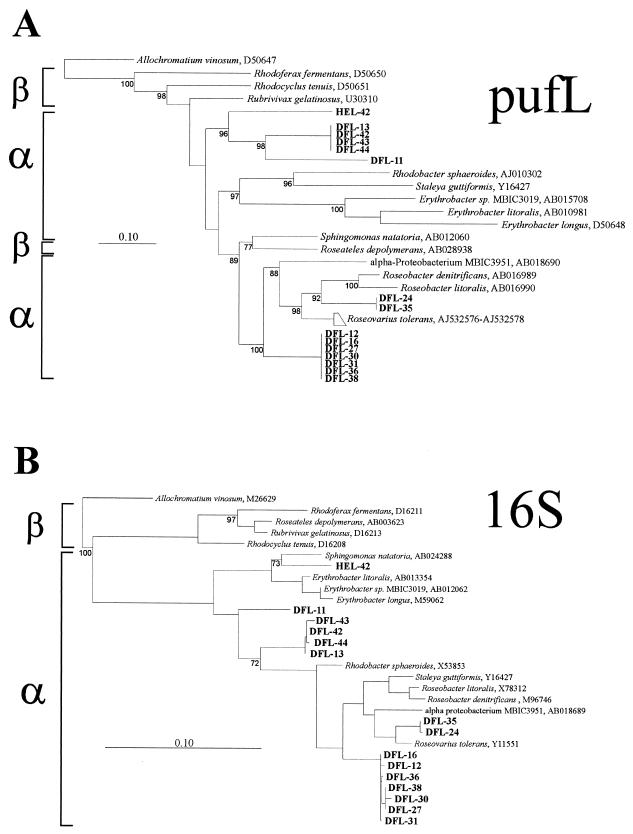
FIG. 2.
Sequence similarities among pufL partial sequences (A) and 16S rRNA gene sequences (B) from reference strains of phototrophic alpha- and beta-subclass proteobacteria, marine bacteria described in this study (boldfaced), and phototrophic bacteria described by Beja et al. (2). The tree was calculated by using the ARB software package (http://www.arb-home.de) and is based on the maximum-likelihood algorithm (8). For the 16S rRNA gene-based tree, near-complete sequences were used and a protobacterial filter was applied. Bootstrap values above 70% are given in the pufL gene-based tree, and those above 50% are given in the 16S rRNA gene-based tree. The bar corresponds to 10 base substitutions per 100 nucleotide positions. The gamma proteobacterium Allochromatium vinosum was used as an outgroup in both trees.
Production of Bchl a.
Bchl a was detected in freshly grown cells and methanol extracts of all DFL-12 cluster strains. It was not found in the two other Roseobacter clade strains that had the pufLM genes (DFL-24 and DFL-35) or in the isolates outside the Roseobacter clade that had the pufLM genes (DFL-13, DFL-44, DFL-42, DFL-43, DFL-11, and HEL-42). Moreover, of the eight strains of Roseovarius tolerans, only four were reported to produce Bchl a (20). Thus, of the 16 strains analyzed in this study which had the pufLM genes, only 7 produced Bchl a under the conditions investigated, i.e., growth in liquid culture in the dark or in the natural light-dark cycle in full-strength marine broth. Expression of Bchl a synthesis in aerobic, anoxygenic phototrophs is regulated by environmental parameters (41). It is completely suppressed by light. In addition, oxygen concentration and carbon limitation have been shown to be involved in Roseateles depolymerans photosynthesis (34). It might be expected that production of Bchl a can be induced in most of the isolates that have pufLM genes if the “right” conditions are found. For example, by growing the dinoflagellate isolates in 1/10 marine broth, pigmented cultures could be obtained for some strains, indicating that nutrient concentration and/or salt concentration may play a role in induction of Bchl a synthesis.
Phylogeny of pufL.
Sequence alignment and phylogenetic analysis of the pufL fragments from the marine isolates and of closely related published pufL sequences (Fig. 2) showed several differences between the 16S rRNA gene phylogenetic tree and the pufL-based tree, except for the highest level of similarity. Thus, clusters of related or identical pufL sequences (the DFL-12 cluster, Roseovarius cluster, DFL-35 cluster, and DFL-13 cluster) corresponded to clusters of related or identical 16S rRNA gene sequences. However, the DFL-35 cluster was most closely related to Roseovarius tolerans on the basis of 16S rRNA gene sequence analysis but to Roseobacter denitrificans on the basis of pufL alignment. Similarly, S. guttiformis was monophyletic with Roseobacter litoralis and Roseobacter denitrificans on the basis of 16S rRNA gene analysis, while Rhodobacter formed a separate lineage. By contrast, S. guttiformis and Rhodobacter sphaeroides were monophyletic in the pufL gene-based tree, while Roseobacter litoralis, Roseobacter denitrificans, and Roseovarius tolerans formed a separate lineage. For phylogenetic groups at even higher taxonomic levels, the tree topology showed differences between the two marker genes. Subgroups 1 to 4 of the alpha proteobacteria were intermixed in the pufL gene-based tree, and the pufL sequence from the beta proteobacterium Roseateles depolymerans affiliated with those of Alphaproteobacteria. This phenomenon has been observed before (17) and has been interpreted as the result of gene transfer of the photosynthesis gene cluster from an ancient alpha proteobacterium into the Betaproteobacteria, especially since it turned out that the photosynthesis genes are highly clustered and arranged in a similar fashion in different phototrophic bacteria (17). Recently it was suggested that there is a large diversity of uncultured aerobic phototrophic proteobacteria, some of them within the beta-proteobacterial subgroup (2). It remains to be seen how these new sequences will affect overall tree topology.
Conclusions.
The data show that dinoflagellates represent an important ecological niche for aerobic anoxygenic phototrophic Alphaproteobacteria. Since dinoflagellates are photoautotrophic organisms themselves and can swim actively, they probably provide a suitable light regimen for the associated bacteria.
Acknowledgments
We thank Gunnar Gerdts for logistic support. Thanks to the divers of the BAH Helgoland, Udo Schilling and Carsten Wanke, and the crew of the research vessels Diker and Ade for sampling. Special thanks to Matthias Labrenz for providing strains and to Hanno Biebl for many helpful discussions.
REFERENCES
- 1.Ashen, J. B., and L. J. Goff. 2000. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl. Environ. Microbiol. 66:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beja, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 3.Brümmer, I. H. M., W. Fehr, and I. Wagner-Dobler. 2000. Biofilm community structure in polluted rivers: abundance of dominant phylogenetic groups over a complete annual cycle. Appl. Environ. Microbiol. 66:3078-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 6.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 7.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 9.Fitch, W. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20:406-416. [Google Scholar]
- 10.Gallacher, S., K. J. Flynn, J. M. Franco, E. E. Brueggemann, and H. B. Hines. 1997. Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp. (Dinophyta) in culture. Appl. Environ. Microbiol. 63:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni, S. J., and M. S. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the ocean. John Wiley & Sons, Inc., New York, N.Y.
- 12.Goericke, R. 2002. Bacteriochlorophyll a in the ocean: is anoxygenic bacterial photosynthesis important? Limnol. Oceanogr. 47:290-295. [Google Scholar]
- 13.Gosink, J. J., R. P. Herwig, and J. T. Staley. 1997. Octadecabacter arcticus gen. nov., sp. nov., and O. antarcticus, sp. nov., nonpigmented, psychrophilic gas vacuolate bacteria from polar sea ice and water. Syst. Appl. Microbiol. 20:356-365. [Google Scholar]
- 14.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 15.Hold, G. L., E. A. Smith, M. S. Rappé, E. W. Maas, E. R. B. Moore, C. Stroempl, J. R. Stephen, J. Prosser, T. H. Birkbeck, and S. Gallacher. 2001. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 37:161-173. [Google Scholar]
- 16.Holmes, A. J., D. P. Kelly, S. C. Baker, A. S. Thompson, P. DeMarco, E. M. Kenna, and J. C. Murrell. 1997. Methylosulfonomonas methylovora gen. nov., sp. nov., and Marinosulfonomonas methylotropha gen. nov., sp. nov.: novel methylotrophs able to grow on methanesulfonic acid. Arch. Microbiol. 167:46-53. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi, N., J. Harada, S. Nagashima, K. Matsuura, K. Shimada, and K. V. Nagashima. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J. Mol. Evol. 52:333-341. [DOI] [PubMed] [Google Scholar]
- 18.Kolber, Z. S., C. L. Van Dover, R. A. Niederman, and P. G. Falkowski. 2000. Bacterial photosynthesis in surface waters of the open ocean. Nature 407:177-179. [DOI] [PubMed] [Google Scholar]
- 19.Kolber, Z. S., F. G. Plumley, A. S. Lang, J. T. Beatty, R. E. Blankenship, C. L. Van Dover, C. Vetriani, M. Koblizek, C. Rathgeber, and P. G. Falkowski. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492-2495. [DOI] [PubMed] [Google Scholar]
- 20.Labrenz, M., M. D. Collins, P. A. Lawson, B. J. Tindall, P. Schumann, and P. Hirsch. 1999. Roseovarius tolerans gen. nov., sp. nov., a budding bacterium with variable bacteriochlorophyll a production from hypersaline Ekho Lake. Int. J. Syst. Evol. Microbiol. 49:137-147. [DOI] [PubMed] [Google Scholar]
- 21.Labrenz, M., B. J. Tindall, P. A. Lawson, M. D. Collins, P. Schumann, and P. Hirsch. 2000. Staleya guttiformis gen. nov., sp. nov. and Sulfitobacter brevis sp. nov., alpha-3-Proteobacteria from hypersaline, heliothermal and meromictic antarctic Ekho Lake. Int. J. Syst. Evol. Microbiol. 50:303-313. [DOI] [PubMed] [Google Scholar]
- 22.Lafay, B., R. Ruimy, C. R. Detraubenberg, V. Breitmayer, M. J. Gauthier, and R. Christen. 1995. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45:290-296. [DOI] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S-23S rRNA sequencing, p. 125-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 24.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed]
- 25.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 26.Petursdottir, S. K., and J. K. Kristjansson. 1997. Silicibacter lacuscaerulensis gen. nov., sp. nov., a mesophilic moderately halophilic bacterium characteristic of the Blue Lagoon geothermal lake in Iceland. Extremophiles 1:94-99. [DOI] [PubMed] [Google Scholar]
- 27.Prokic, I., F. Brummer, T. Brigge, H. D. Gortz, G. Gerdts, C. Schutt, M. Elbrachter, and W. E. G. Muller. 1998. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist 149:347-357. [DOI] [PubMed] [Google Scholar]
- 28.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rappé, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 30.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 31.Shiba, T. 1991. Roseobacter litoralis gen. nov. sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol. 14:140-145. [Google Scholar]
- 32.Sojka, G., H. H. Freeze, and H. Gest. 1970. Quantitative estimation of bacteriochlorophyll in situ. Arch. Biochem. Biophys. 136:578-580. [DOI] [PubMed] [Google Scholar]
- 33.Sorokin, D. Y. 1995. Sulfitobacter pontiacus gen. nov., sp. nov., a new heterotrophic bacterium from the Black Sea, specialised on sulfite oxidation. Microbiology 64:295-305. [Google Scholar]
- 34.Suyama, T., T. Shigematsu, T. Suzuki, Y. Tokiwa, T. Kanagawa, K. V. Nagashima, and S. Hanada. 2002. Photosynthetic apparatus in Roseateles depolymerans 61A is transcriptionally induced by carbon limitation. Appl. Environ. Microbiol. 68:1665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, M. T., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Strobel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, T., Y. Muroga, M. Takahama, and Y. Nishimura. 1999. Roseivivax halodurans gen. nov., sp. nov., and Roseivivax halotolerans sp. nov., aerobic bacteriochlorophyll-containing bacteria isolated from a saline lake. Int. J. Syst. Bacteriol. 49:629-634. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uphoff, H., A. Felske, and I. Wagner-Döbler. 2001. The microbial diversity in picoplankton enrichment cultures: a molecular screening of marine isolates. FEMS Microbiol. Ecol. 35:249-258. [DOI] [PubMed] [Google Scholar]
- 39.Wagner-Döbler, I., H. Rheims, A. Felske, R. Pukall, and B. Tindall. 2003. Jannaschia helgolandensis, gen. nov., sp. nov., a novel abundant member of the marine Roseobacter clade from the North Sea. Int. J. Syst. Evol. Microbiol. 53:731-738. (First published 4 October 2002; 10.1099/ijs.0.02377-0.) [DOI] [PubMed]
- 40.Yurkov, V., and J. T. Beatty. 1998. Isolation of aerobic anoxygenic photosynthetic bacteria from black smoker plume waters of the Juan de Fuca Ridge in the Pacific Ocean. Appl. Environ. Microbiol. 64:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yurkov, V. V., and J. T. Beatty. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yurkova, N., C. Rathgeber, J. Swiderski, E. Stackebrandt, J. T. Beatty, K. J. Hall, and V. Yurkov. 2002. Diversity, distribution and physiology of the aerobic phototrophic bacteria in the mixolimnion of a meromictic lake. FEMS Microbiol. Ecol. 40:191-204. [DOI] [PubMed] [Google Scholar]
- 43.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
- 44.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2002. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep Sea Res. Part I 49:3017-3038. [Google Scholar]