Abstract
Picoeukaryotes (cells of <3 μm in diameter) contribute significantly to marine plankton biomass and productivity, and recently molecular studies have brought to light their wide diversity. Among the methods that have been used so far to quantify aquatic microorganisms, fluorescence in situ hybridization of oligonucleotide probes combined with flow cytometry offers the advantages of both high resolution for taxonomic identification and automated cell counting. However, cell losses, cell clumps, and low signal-to-background ratio have often been mentioned as major problems for routine application of this combination of techniques. We developed a new protocol associating tyramide signal amplification-fluorescence in situ hybridization and flow cytometry, which allows the detection of picoeukaryotes in cultures during both the exponential and stationary phases. The use of surfactant and sonication proved to be essential for the detection and quantification of picoeukaryotes from the natural environment, with as little as a few tenths of a milliliter of 3-μm-pore-size prefiltered sea water. The routine application of the technique was tested along a coastal transect off Brittany (France), where the different groups of picoeukaryotes were investigated using already published specific probes and a newly designed probe that targets the order Mamiellales (Prasinophyceae, Chlorophyta). Among the picoeukaryotes, Mamiellales outnumbered by 1 order of magnitude both the cyanobacteria and the non-Chlorophyta, which were represented mainly by the Pelagophyceae class. Picoeukaryote abundance increased from open toward more estuarine water, probably following changes in water temperature and stability.
Picoeukaryotes (cells smaller than 3 μm in diameter) (22) are widely distributed in aquatic environments. Their important biomass and their high productivity suggest that they play a major role in oceanic, coastal, and freshwater ecosystems (27, 28, 48). Because of their small size and their simple morphology, a detailed study of these organisms is difficult. It is only recently that their diversity has been revealed, in particular by phylogenetic studies based on 18S rDNA sequence analysis (31, 34). It is necessary to quantify the dominating phylogenetic groups of picoeukaryotes in the natural environment in order to understand their contribution to the microbial food web and biogeochemical cycles.
So far, the abundance of marine picoeukaryotes has been estimated based on their natural fluorescence by flow cytometry (28, 54) or by analysis of their pigment composition by high-performance liquid chromatography (56). The taxonomic resolution of these techniques is limited (e.g., class level only for high-performance liquid chromatography) and they do not allow the quantification of heterotrophic organisms. Molecular techniques based on full or partial sequence analysis of 18S rDNA allow us to determine species composition very precisely but are labor-intensive, and only a few samples can be analyzed at once (31, 34). To acquire a more extensive data set, van Hannen et al. (53) and Diez et al. (15) used denaturing gradient gel electrophoresis to compare rapidly the picoeukaryote species composition at various sites in natural ecosystems. Still, these techniques are not fully quantitative since they rely on PCR amplification.
Oligonucleotide probes designed to target specific taxa (phylum, division, class, or genus) have proven to be useful in estimating the abundance of nano- or picoeukaryotes (21, 30, 39). Such probes can be hybridized either on PCR-amplified rDNA immobilized on membranes (i.e., dot blot analysis) or directly on rRNA from fixed whole cells (i.e., in situ hybridization). While the dot blot method gives an approximate abundance of target organisms, in situ hybridization allows us to distinguish whole target cells within a natural assemblage of organisms. Fluorescence in situ hybridization (FISH) of probes directly labeled with a fluorochrome (i.e., monolabeled probes) has been widely used during the last 10 years to enumerate marine bacteria by epifluorescence microscopy (14, 18). Early on, scientists had thought of increasing the speed and reliability of the analysis by coupling FISH with flow cytometry (2, 57). Such coupling has also been attempted on nano- and picoeukaryotes (29, 45). However, the signal-to-background ratio was often too low for the smallest cells (<3 μm) to be detected by flow cytometry, and several methods of fluorescence amplification have been proposed (e.g., the use of multiple probes or multiple fluor labels on one probe) (1, 14). The success of these amplification techniques was limited: when nonspecific binding was not an obstacle to cell detection, only rapidly growing or large cells (>3 μm) could be detected (29, 58). Recently, tyramide signal amplification of FISH (TSA-FISH), a very powerful amplification technique that boosts thefluorescent signal of hybridized cells 20 to 40 times over that of the background, has been introduced in the field of marine microbiology (24, 40). This technique involves the deposition of multiple tyramide-bound fluorochromes in the proximity of hybridized probes that have been labeled with horseradish peroxidase (HRP) (3, 8). TSA-FISH has been successfully tested on free-living heterotrophic marine bacteria (24, 37) and more significantly on samples with a highly fluorescent background (6, 40) as well as on photosynthetic microorganisms such as cyanobacteria (41, 59) and picoeukaryotes (35). Only two of these studies have attempted to couple TSA-FISH and flow cytometry, but quantification of microorganisms was prevented by uneven labeling of cells, cell aggregation, and cell loss (24, 40).
In this paper, we propose a new protocol to allow the quantification of picoeukaryotes in the natural environment by coupling TSA-FISH and flow cytometry. First, experiments were done on cultures to assess TSA-FISH enhancement of the fluorescence intensity of hybridized cells at different growth stages. Second, tests were done on cultures and on natural samples to solve the problem of cell loss during picoeukaryote collection and hybridization. The finalized protocol allowed us to estimate the abundance of several taxa of the picoeukaryote community sampled along a coastal transect in the Bay of Morlaix (Brittany, France). This approach should be very useful for high-speed and high-precision studies of the distribution of specific taxa of microorganisms in the aquatic environment.
MATERIALS AND METHODS
Strains, cultivation, and sampling.
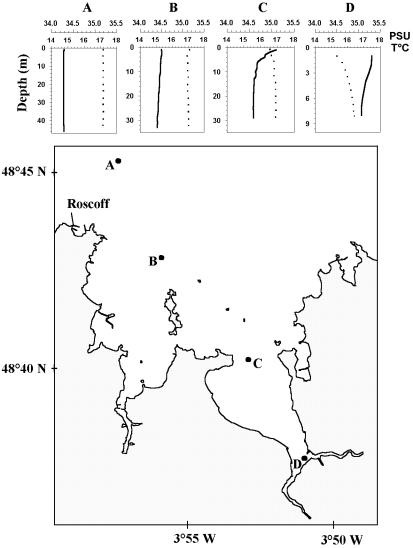
Eight marine pico- and nanoeukaryote strains were selected for in situ hybridization experiments (Table 1). Cells were grown in K medium (23) in tissue culture flasks (Sarstedt, Newton, N.C.) at 20°C, under a light-dark cycle of 14 h-10 h, with different light intensities (Table 1). For most of the experiments, cells were harvested in early stationary growth phase or at other specific growth phases when mentioned. Natural samples were harvested at a depth of 1 m with a 5 liter Niskin bottle on 17 July 2002 at four different stations along a transect in the Bay of Morlaix (Fig. 1). Temperature and salinity profiles were measured at each stations with a conductivity, temperature, and pressure profiler (CTD SBE 19; Seabird [Fig. 1]). Seawater samples were immediately prefiltered through 200-μm-mesh-size filters to remove meso- and macroplankton and then through 3-μm-pore-size Nuclepore filters (Whatman International Ltd., Maidstone, United Kingdom) to collect the picoplanktonic sea water fraction.
TABLE 1.
Strains used in this study
| Class | Order | Species | RCCa | Strain nameb | Origin | Cell diameter (μm) | Light intensity (μE m−2 s−1) |
|---|---|---|---|---|---|---|---|
| Prasinophyceae | Mamiellales | Ostreococcus tauri | 116 | OTTH 0595 | Thau Lagoon (Mediterranean sea) | 0.8 | 100 |
| Micromonas pusilla | 114 | CCMP 490 | Nantucket Sound (North Atlantic) | 1-3 | 100 | ||
| Bathycoccus prasinos | 113 | CCMP 1898 | Mediterranean Sea | 2 | 100 | ||
| Mantoniella squamata | 417 | CCMP 480 | North Sea | 3-5 | 100 | ||
| Pseudoscoufieldia marina | 261 | TAK 9801 | Takapoto Atoll (Pacific) | 4 | 100 | ||
| Prasinococcales | Prasinococcus capsulatus | 134 | CCMP 1194 | Gulf of Mexico (North Atlantic) | 3-5 | 100 | |
| Pelagophyceae | Pelagomonas calceolata | 100 | CCMP 1214 | Central Gyre (North Pacific) | 2-4 | 4 | |
| Chlorophyceae | Chlamydomonas concordia | 1 | PLY 491 | 10-15 | 300 |
RCC, Roscoff Culture Collection.
CCMP, Provasoli-Guillard National Center for Culture of Marine Phytoplankton; PLY, Plymouth Marine Laboratory.
FIG. 1.
Localization of the different stations (stations A to D) along the transect studied on 17 July 2002 in the Bay of Morlaix on the coast of Brittany. Temperature (T°C, full line) and salinity (PSU, dotted line) profiles are given for each station.
Probe labeling and design.
Oligonucleotide probes were purchased with a 5′-amino link on six atoms of carbon from Interactiva (Saint-Malo, France). Probes were labeled with HRP (Roche Diagnostic Boehringer, Maylan, France) by the methods of Urdea et al. (51) and Amann et al. (3). Oligonucleotide probes used in this study (Table 2) were available from the literature, except for PRAS 02, which was designed for this study to target the order Mamiellales (Chlorophyta, Prasinophyceae [Table 3 ]). This probe was determined with the probe design function of the ARB software (http://www.arb-home.del) using a database of rRNA gene sequence with more than 14,000 sequences. The theoretical specificity of the new probe was checked: all Mamiellales sequences available matched the probe specifically with the exception of the species Bathycoccus prasinos, which had one mismatch (Table 3). The bacterium Planctomycetales Isophaera sp. and the Charophyceae alga Oedogonium angustistomum were the first nontargeted sequences showing three central mismatches. Because of the large number of mismatches, this probe is highly unlikely to be able to hybridize both of these organisms in the marine environment. Moreover, a special treatment would be needed to perforate the Planctomycetales cell wall to allow HRP-labeled probes to penetrate, and O. angutistomum is a multicellular freshwater alga whose spores do not belong to the picoplanktonic fraction because they are larger than 7 μm in diameter (20). Specificity tests using TSA-FISH on whole cells immobilized on a filter (see below) were performed on four species belonging to the Mamiellales and four other species which did not belong to that order (Table 1). All of the Mamiellales species, apart from Bathycoccus prasinos, gave a positive signal. The fluorescence intensity yielded by B. prasinos was at least 50% lower than that of the other Mamiellales (data not shown). All the other species that were either phylogenetically close to the Mamiellales (Pseudoscourfieldia marina and Prasinococcus capsulatus) or more distant (Pelagomonas calceolata and Chlamydomonas concordia) gave a negative signal (data not shown). For all the probes, stringent conditions of hybridization were used, from 0 to 10°C above the melting temperature (Table 2). These conditions were either very close to the one used in original publications or more stringent (i.e., probe EUK1209 [Table 2]).
TABLE 2.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′-3′) of probes | Targeta site (18S rRNA position) | Tm (°C)b | Reference |
|---|---|---|---|---|---|
| EUK 1209R | Eucarya | GGG CAT CAC AGA CCT G | 1195-1211 | 27.1 | 19 |
| NCHLO 01 | Non-Chlorophyta | GCT CCA CTC CTG GTG GTG | 932-950 | 34.5 | 45 |
| CHLO 01c | Chlorophyta | GCT CCA CGC CTG GTG GTC | 932-950 | 36.8 | 45 |
| CHLO 02 | Chlorophyta | CTT CGA GCC CCC AAC TTT | 766-784 | 30 | 46 |
| PELA 01 | Pelagophyceae | ACG TCC TTG TTC GAC GCT | 730-748 | 30 | 46 |
| PRAS 02 | Mamiellales | CCC GTC CCG AGA CCA ACG | 651-669 | 37 | This study |
TABLE 3.
Theoretical specificity of probe PRAS 02
| Species | Kingdom | Class | Order | Accession no. | Position of mismatches |
|---|---|---|---|---|---|
| 5′-CCC GTC CCG AGA CCA ACG-3′a | |||||
| 3′-GGG CAG GGC UCU GGU UGC-5′b | |||||
| Micromonas pusilla | Eucarya | Prasinophyceae | Mamiellales | AJ010408 | ---c--------------- |
| Mantoniella squamata | Eucarya | Prasinophyceae | Mamiellales | X73999 | ------------------ |
| Mamiella sp. | Eucarya | Prasinophyceae | Mamiellales | AB017129 | ------------------ |
| Ostreococcus tauri | Eucarya | Prasinophyceae | Mamiellales | Y15814 | ------------------ |
| Bathycoccus prasinos | Eucarya | Prasinophyceae | Mamiellales | ZZ003565 | --- U-------------- |
| Isophaera sp. | Bacteria | Planctomycetaceae | Planctomycetales | X91958 | -----------C-CC--- |
| Oedogonium angustistomum | Eucarya | Charophyceae | Oedogoniales | U83134 | --C----UG--------- |
Probe PRAS 02.
Target sequence.
Dashes indicate bases that are identical to those of the target sequence.
Cell collection, fixation, and hybridization on cultures.
To estimate cell losses due to hybridization, cells were counted before and after hybridization (Fig. 2, also see Fig. 5). Prior to hybridization, concentrations of photosynthetic cells were determined by flow cytometry (see below). Cultures (1.5 ml) were previously fixed with a mixture of paraformaldehyde and glutaraldehyde (Sigma-Aldrich, Saint Quentin Fallavier, France) at 1 and 0.05% final concentration, respectively (modified from reference 33). Subsequently, counts were done either after a 15-min fixation at room temperature or after a quick freeze in liquid nitrogen and storage at −80°C.
FIG. 2.
Cytograms of picoeukaryotes cultures (O. tauri, M. pusilla, and P. calceolata) hybridized with specific probes (circled populations), nonspecific probes, and no probe (control). The cell hybridized with the probes were labeled with fluorescein using the TSA-FISH technique (green fluorescence). All the DNA-containing cells were labeled with PI (red fluorescence). Hybridization was performed either on nonsonicated cells or on sonicated cells. Sonication prevents cells from clumping together (arrows). b, 0.95-μm fluorescent beads; a.u., arbitrary units.
FIG. 5.
Percent cell recovery of different cultures of picoeukaryotes hybridized with probes targeting different taxonomic levels (TSA-FISH technique). Hybridized cells were counted by flow cytometry after PI counterstaining and sonication. Error bars correspond to two replicates.
For in situ hybridization, cultures of pico- and nanoplankton cells were harvested into 1.5-ml centrifuge tubes previously treated with the plastic surfactant dimethyldichlorosilane (Fluka, Sigma-Aldrich). The cell surfactant Pluronic F-68 (10% stock solution; Sigma-Aldrich) was added to the cells at a final concentration of 0.1%. The cells were concentrated to 500 μl by centrifugation (10,500 × g for 3 min at room temperature) with 5417R bench centrifuge (Eppendorf, Hamburg, Germany) and fixed with 1% paraformaldehyde at room temperature, for 15 min to 1 h. The remaining paraformaldehyde and salt from the cell medium were washed out by centrifuging the cells (10,500 × g for 3 min at room temperature) and resuspending the cell pellet into 1 ml of phosphate-buffered saline (PBS [Sigma-Aldrich] solution at 1X and pH 7.4, containing 0.1% Pluronic, stored at 4°C). For cell dehydration, cells were concentrated to 50 μl by two centrifugation steps (10,500 × g for 3 min at room temperature) to avoid cell loss and dehydrated by adding increasing amounts of 100% ethanol to reach a final concentration of 80%. Chlorophyll-less cells were either directly processed for in situ hybridization or stored at −80°C for months.
In situ hybridization was performed following a rehydration step, where PBS solution was added to 500 μl to cells kept in ethanol. At this step, cells may clump together, which can prevent efficient hybridization. When cell clumps could not be removed by vortexing, sonication was used to detach the cells from one another. For sonication, the tubes were maintained in ice and eight pulses of 10 s each (6 W with an amplitude of vibration of 20% [Bioblock, Illkirch, France]) were produced by a 2-mm probe. To complete the rehydration, the tubes were filled with PBS solution and the supernatant was subsequently discarded by centrifugation. Then 20 μl of hybridization buffer (40% deionized formamide [Fluka, Sigma-Aldrich], 900 mM NaCl, 20 mM Tris-HCl [pH 7.7] 0.01% sodium dodecyl sulfate [Bio-Rad, Ivry sur Seine, France], 20% stock blocking buffer solution, 10% [wt/vol] blocking reagent [Roche Diagnostic Boehringer] in sterile filtered maleic acid buffer; 100 mM maleic acid [Sigma-Aldrich]-150 mM NaCl [pH 7.5]) was added to the cell pellet, and the pellet was homogenized. At this step, cells were either processed further or stored at −80°C for several months. A 2-μl volume of oligonucleotide probes labeled with HRP (50 ng μl−1), and 2 μl of competitor (50 ng μl−1, not labeled with HRP) if necessary, was added, and the mixture was left to hybridize at 37°C for 2.5 h. Nonhybridized probes were then washed out by two subsequent rinses of 1 and 30 min in 1 ml of freshly made buffer (56 mM NaCl, 5 mM EDTA, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl [pH 7.5], 0.1% Pluronic) prewarmed at 37°C. The 30-min rinse was done with agitation in an oven at 37°C.
Prior to the TSA reaction, cells were equilibrated in 1 ml of TNT buffer (approximately 7% Tween 20 [Sigma-Aldrich] in 150 mM NaCl-100 mM Tris-HCl [pH 7.7]-0.1% Pluronic) at room temperature for 15 min. Most of the TNT buffer was then washed out by two steps of centrifugation. A sonication step was added when the cells were too numerous, since they have a serious tendency to clump, which can prevent even fluorescent staining. For the TSA reaction, 20 μl of TSA mix (40% [wt/vol] dextran sulfate stock solution in sterile H2O aliquoted and stored at −20°C, mixed 1:1 with the amplification diluent of the TSA-Direct kit [NEN Life Science Products Inc., Boston, Mass.] [52], and added [50:1] to fluorescein-tyramide [TSA-Direct kit]) was added, and the mixture was left to incubate for 30 min at room temperature in the dark. The unlabeled fluorochrome was then washed out by two subsequent rinses, of 1 and 20 min, in 1 ml of TNT buffer prewarmed at 55°C. The 20-min incubation was done under agitation in an oven at 55 to 60°C. Nucleic acid counterstaining was done, after the TNT buffer was discarded by centrifugation, by adding to the cell pellet 20 μl of antifade AF1 (Citifluor Ltd., University of Kent, Canterbury, United Kingdom) and 5 μl of propidium iodide (Sigma-Aldrich; 1-mg/ml stock solution, stored at 4°C). The tubes were then filled to 500 μl with PBS solution. Hybridized cells were either immediately processed for flow cytometry counts or kept at 4°C for several months. Prior to flow cytometry counts, a last sonication step was essential to prevent cell aggregation and allow quantification.
Cell collection and hybridization on natural samples.
For flow cytometry counts (see Fig. 6 and 7), cells were collected and hybridized as follows. A 35-ml volume of prefiltered seawater was collected in three replicates and added to sterile 50-ml polypropylene conical tubes (Falcon, Blue Max [Beckton Dickinson, Le Pont de Claix, France]) containing 4 ml of 10% paraformaldehyde and 400 μl of 10% Pluronic F-68. The tubes were transferred at −80°C or in liquid nitrogen. Fixed cells were then left to thaw at room temperature and concentrated from 39 ml to approximately 3 ml by three centrifugations (10,100 × g, for 10 min at 15°C) with an MR 1812 bench centrifuge (Jouan, Saint-Herblain, France) to avoid cell loss. The cells were then equally distributed into two 1.5-ml centrifuge tubes previously treated with the plastic surfactant dimethyldichlorosilane. The volume in the tubes was then reduced to 50 μl per tube by three centrifugation steps (10,500 × g for 3 min at room temperature), and the cells were washed by adding 500 μl of PBS solution. At this step, the cells were forming a dense, translucent pellet, which was first vigorously vortexed for partial disruption and subsequently sonicated. The tubes were then filled up to 1 ml with PBS solution, and 50 μl was subsampled and diluted 20- to 40-fold for cytometry counts based on natural fluorescence. These counts were compared to those for nonconcentrated picoplankton, using a similar method to the one described for cultures, in order to estimate cell loss due to centrifugation prior to hybridization. For in situ hybridization, concentrated cells were dehydrated with ethanol, stored, and hybridized as described above for the cultures.
FIG. 6.
Cytograms and confocal micrographs for the natural picoeukaryote community from station D (Fig. 1) hybridized with division-, class-, or order-specific probes (Table 3) and without a probe (control). The cell hybridized by probes were colored (TSA-FISH technique) with FITC (green fluorescence), and cell DNA was stained with PI (red fluorescence). Positively hybridized populations are circled. b, 0.95-μm fluorescent beads. Solid-head arrows, probe-labeledcells; open-head arrows, unidentified particles; bars, 5 μm; a.u., arbitrary units.
FIG. 7.
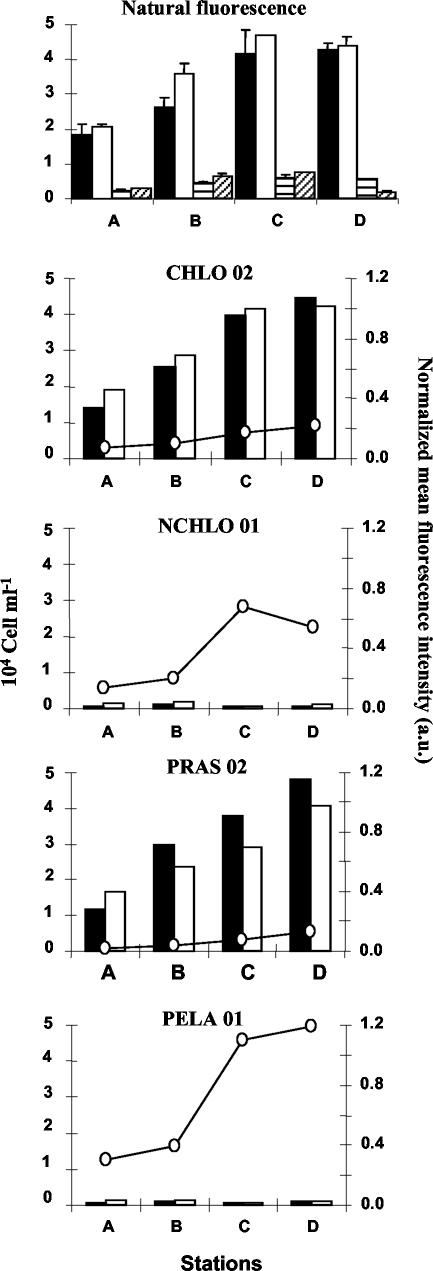
Cell counts obtained on natural picoeukaryote communities sampled at stations A, B, C, and D along a transect in the Bay of Morlaix (Fig. 1). The top panel shows a comparison of cell counts, based on the natural fluorescence of picoeukaryotes (black and whitebars) and cyanobacteria (striped bars), between two methods: concentrations of fixed cells by centrifugation prior to hybridization (black and horizontally striped bars) and cell fixation without centrifugation (white and diagonally striped bars). The bottom four panels show a comparison of cell counts and mean fluorescence intensity of hybridized cells with different probes (Table 2). Cells were counted either by flow cytometry (black bars) or by fluorescence microscopy (white bars). a.u., arbitrary units. Error bars correspond to two replicates.
For fluorescent microscopy counts (see Fig. 6 and 7), cells were collected and hybridized on filters as follows. A 100-ml volume of prefiltered seawater was fixed with paraformaldehyde (1% final concentration) for 1 h at 4°C and then filtered through a 0.2-μm-pore-size filter (Anodisc; Whatman International Ltd.) The cells were then dehydrated in an ethanol series (50, 80, and 100% for 3 min each), dried, and stored at −80°C until further analysis. In situ hybridization was done by the method of Biegala et al. (6), modified from that of Schönuber et al. (40, 41), with the difference that lysozyme step was omitted and DNA counterstaining was done with PI instead of DAPI (4′,6-diamidino-2-phenylindole [Sigma-Aldrich]). PI fluorescence is much brighter than that of DAPI and mimics the staining conditions used for flow cytometry counts (see above). PI staining was done by adding 10 μl of 10-μg/ml PI, in antifade AF1, to cells immobilized on the filter.
Flow cytometry.
Discrimination of photosynthetic cells (prior to hybridization) was achieved on the basis of their natural orange and red pigment fluorescence; and that of hybridized cells was achieved on the basis of green (fluorescein isothiocyanate [FITC]) and red (PI) fluorescence (Fig. 2, also see Fig. 6). Flow cytometry analysis was performed with a standard FACSsort flow cytometer (Becton Dickinson, San Jose, Calif.). A 488-nm laser was used for excitation. Green fluorescence was collected through a (530 ± 30)-nm band-pass filter, orange fluorescence was collected through a (585 ± 40)-nm filter, and red fluorescence was collected through a 650-nm long-pass filter. Prior to analysis, samples were diluted in PBS solution 100- to 1,000-fold for hybridized cultures and 20- to 40-fold for natural samples. Data acquisition was done at a high flow rate (approximately 100 μl min−1) for 3 to 12 min depending on the concentration of the target population. Double-distilled water was used as the sheath fluid. Cytograms were analyzed using Cytowin public software (D. Vaulot, www.sbroscoff.fr/Phyto/cyto.html) for cell counts; WinMDI software (J. Trotter, http://flowcyt.salk.edu/software.html) was used for figures. Populations of hybridized cells were counted using the bitmap function of Cytowin, which allows us to encircle precisely the target population (Fig. 2, also see Fig. 6).
Microscopy.
Cell counts were obtained with BX51 epifluorescence microscope (Olympus Optical, Rungis, France) equipped with a mercury light source and 100× oil immersion objective (Plan APO; numerical aperture 1.40). Excitation and emission filters used were 480 ± 10 nm and 515 nm long pass, respectively, for both FITC and PI. The cell cytoplasm appeared bright green (rRNA probe and FITC labeling), while the nucleus fluoresced red (PI). Counts were done on 10 randomly chosen microscopic fields, and, on average, 1,200 picoeukaryotes were observed, of which only hybridized cells were counted. Dual-color images were taken by confocal laser scanning microscopy with an argon-krypton laser (643R-OLYM-A03 Omnichrome [Melles Griot, Carlsbad, Calif.]) set at 488 nm and 568 nm to excite fluorescein and PI, respectively. The green and red emission fluorescence produced by the different fluorochromes were collected between 510 and 550 nm and above 585 nm, respectively. Confocal images were acquired with a 60× (Uplan Fi, numerical aperture 1.25 [Olympus]) oil immersion objective at a scanning speed of 23.1 s. Three slices with a thickness of 0.7 μm were added to produce the final image. Artificial colors were chosen to match the emission wavelengths.
RESULTS AND DISCUSSION
Detection of TSA-FISH-labeled picoeukaryotes by flow cytometry.
Our first step was to test the ability of the TSA amplification system to enhance the FISH, in order to improve the detection of photosynthetic picoeukaryotes by flow cytometry compared to previous work (45). For this purpose, we chose three photosynthetic picoeukaryote strains, Ostreococcus tauri, Micromonas pusilla and Pelagomonas calceolata (Table 1). O. tauri is the smallest picoeukaryote described so far (13) and for this reason allows us to assess the detection limit of the method. M. pusilla is a ubiquitous flagellate which has been frequently observed in coastal waters, including the English Channel (22, 32). Both of these species belong to the order Mamiellales (class Prasinophyceae, division Chlorophyta). The third species, P. calceolata (class Pelagophyceae, division Heterokontophyta), was chosen as a control, since it should not be labeled by the Chlorophyta probes but should be labeled by the non-Chlorophyta probes.
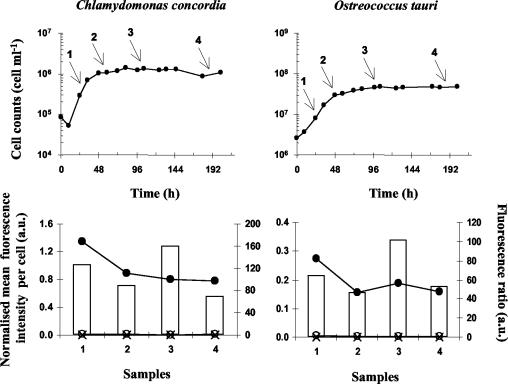
Cells hybridized with probes specific for their division, class, or order (CHLO 02 and PRAS 02 for O. tauri and M. pusilla and NCHLO 01 and PELA 01 for P. calceolata) gave a strong positive signal that could easily be distinguished from the noise or from negative controls (i.e., populations hybridized with nonspecific probes [Fig. 2]). Counterstaining with PI, which labels nucleic acids with red fluorescence, was essential for a clear discrimination of the hybridized target cells, since in some cases, particles without a nucleus, i.e., lacking red fluorescence, were strongly labeled with fluorescein for unknown reasons (these particles are not shown in Fig. 2 because they are below the threshold set on red fluorescence). Target cells, hybridized with specific probes, displayed a fluorescent signal 17 to 140 times more intense than the negative control (Fig. 2). This signal was higher for the Mamiellales O. tauri and M. pusilla than for P. calceolata. P. calceolata exhibited the lowest fluorescence ratio (i.e., 17) between positive and negative hybridization because it was slightly hybridized with the nonspecific probes CHLO 02 and PRAS 02 (Fig. 2). The reason for such hybridization is not known, but it is quite likely that it did not occur at the target site since CHLO 02 and PRAS 02 have 2 and 10 mismatches with the P. calceolata 18S rDNA sequence, respectively. However, a specific-to-nonspecific fluorescence ratio of 17 is sufficient to clearly distinguish positive from negative signals. We also observed for O. tauri that the fluorescence ratio was still very high (above 40) when hybridization was performed on cells previously stored for as long as 8 months at −80°C in ethanol or in hybridization buffer (Fig. 3A). In addition, once the cells were hybridized, it was possible to keep them at 4°C for at least 2 months, since their fluorescence was still 40 times higher than that of the negative control (Fig. 3B). The stability of the fluorescence signal was further checked during the growth of cultures of O. tauri and Chlamydomonas concordia (Fig. 4). C. concordia was initially used to guarantee a positive signal in stationary phase, because it is much larger than O. tauri and thus contains more rRNA per cell (Table 1). Both O. tauri and C. concordia were easily detected in exponential and stationary phases, showing at least 40 times more fluorescence when hybridized with the positive probe CHLO 02 than with the negative probe NCHLO 01.
FIG. 3.
Histograms representing the fluorescence ratio between populations of O. tauri hybridized with either specific (CHLO 02) or nonspecific (NCHLO 01) probes revealed by the TSA-FISH technique. Fluorescence ratios were calculated from fluorescence intensities measured by flow cytometry, either on hybridized cells previously stored in ethanol (black) or in hybridization buffer (white) for up to 8 months (A) or on hybridized cells kept for up to 2 months at 4°C in PBS (pH 7.4) (B).
FIG. 4.
Detection of nanoplanktonic (C. concordia) and picoplanktonic (O. tauri) species at different growth stages by TSA-FISH and flow cytometry. Cells were hybridized with the specific probe CHLO 02 (•) and the nonspecific probe NCHLO 01 (○). A no-probe control was also used (×). The mean fluorescence intensity is normalized to 0.95-μm fluorescent beads, and histograms show the fluorescence ratio between a specific (CHLO 02) and a nonspecific (NCHLO 01) signal. Arrows, stages at which the cells were sampled for hybridization; a.u., arbitrary units.
These data demonstrate that the combination of FISH with flow cytometry allowed the specific identification of photosynthetic picoeukaryotes in both exponential and stationary phases. In a previous study, Simon et al. (45) used monolabeled probes instead of the TSA amplification system to detect pico- and nanophytoplankton. While these authors could detect Chlamydomonas and other nanoplanktonic species in both exponential and stationary phases, they could distinguish photosynthetic picoeukaryotes from negative controls only for cultures in exponential phase (Fig. 2 and 3 in reference 45). In addition, fluorescence ratios between positive and negative hybridization were much lower (e.g., ranging only from 1 to 10 for picoeukaryotes) than were the ones obtained in this study (e.g., 41 to 101 for picoeukaryotes). Similar enhancement of fluorescence, due to TSA amplification system, has been previously mentioned by several authors for both prokaryotes (24, 40) and picoeukaryotes (35). Of these authors, only Schönuber et al. (40) successfully used flow cytometry to quantify the fluorescence intensity obtained by TSA-FISH for hybridized heterotrophic bacteria (Escherichia coli). In contrast, Lebaron et al. (24) did not succeed with flow cytometry analysis of hybridized bacteria because, according to these authors, the tyramide deposit escaped permeabilized cells following centrifugation. The results presented in this study prove, however, that for picoeukaryotes tyramide deposition inside hybridized cells is not affected by several rounds of centrifugation at speed as high as 10,500 × g.
Quantification of picoeukaryotes.
Our second step was to try to detect picoeukaryotes from natural communities and to determine their concentrations by flow cytometry. Initially, when we tried to apply TSA-FISH to natural communities of picoeukaryotes, we could not detect any hybridized cells by flow cytometry. We assumed that we were probably losing most of the cells during collection and hybridization. In the natural environment, the concentration of photosynthetic picoeukaryotes can be 3 orders of magnitude lower than in cultures (in general from 103 to 104 cells ml−1 and occasionally 107 cells ml−1) (48), which makes their collection difficult. To our knowledge, only Wallner et al. (58) and Thomas et al. (49) have managed to quantify microorganisms in the natural environment by combining FISH and flow cytometry. However, the conditions of these studies were optimal, since the targeted heterotrophic bacteria had no fluorescent background and were very abundant (in activated sludge or in sediments, at over 106 cells ml−1 or g−1). Major obstacles to quantification using both FISH and flow cytometry have been identified as cell losses and/or cell aggregation (40, 57). One possible explanation for cell losses is cell disintegration due to centrifuge forces. However, we did not observe any increase in the amount of cell debris in relation to an increase in the centrifugation speed (data not shown). In addition, live M. pusilla, a wall-less flagellated picoeukaryote, could survive several centrifugation at 10,500 × g and still show recognizable swimming behavior under the fluorescence microscope (data not shown). This led us to hypothesize that cell loss may be essentially due to the absence of sedimentation, either because picoeukaryotes were too small to migrate to the bottom of the tubes during centrifugation or because they were sticking to the tube surface and/or to invisible cracks. For this reason, we used surfactants such as Pluronic F-68 to cover live or fixed cells and silicone to coat the surfaces of centrifuge tubes. Pluronic presents the additional advantage of increasing the cell density, which makes centrifugation more efficient. If the use of both surfactants allows effective cell recovery, they do not prevent cells from clump together, which hampers proper cell quantification (arrows in Fig. 2). To solve this problem, we introduced a sonication step prior to flow cytometry counts, in order to detach hybridized cells from one another (Fig. 2).
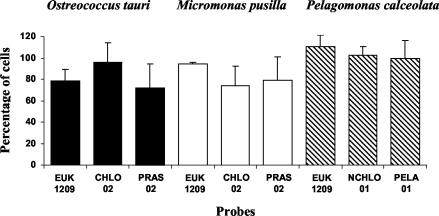
The combined use of cell surfactant and sonication allowed an excellent quantification of the three cultures (O. tauri, M. pusilla, and P. calceolata) hybridized with different probes (Fig. 5). The average cell recovery for the three species was 90% ± 18% (range, 60 to 115%), with the largest species, P. calceolata, having a better recovery than O. tauri or M. pusilla, probably because it could be more efficiently harvested by centrifugation. The use of surfactants and sonication also permitted us to detect natural populations of picoeukaryotes hybridized with most probes (i.e., CHLO 02, NCHLO 01, and PELA 01 [Fig. 6]). However, part of the population of cells labeled with probe PRAS 02 displayed very low fluorescence (images of cells hybridized on filters [Fig. 6, right]). This result was not expected because the population hybridized by PRAS 02 is a subpopulation of that hybridized by CHLO 02. However, since a fraction of the Mamiellales could be constituted by Bathycoccus prasinos (F. Not, unpublished data) which exhibits one mismatch with probe PRAS 02, this may explain the lower fluorescence of a part of the hybridized community, as observed on cultures (see Materials and Methods).
The validity of counts obtained by the new protocol on natural samples was verified in different ways. First, cell losses due to centrifugation prior to hybridization were checked by counts based on natural fluorescence (Fig. 7, top panel). Centrifugation did not induce significant cell loss for either eukaryotes or cyanobacteria, provided that surfactants and sonication were used. Second, flow cytometry counts of hybridized populations collected by centrifugation were compared with fluorescence microscopy counts of hybridized populations collected on filters (Fig. 7, lower four panels). The results were rather similar except for probe PRAS 02, for which counts with the flow cytometer were between 15 and 24% higher than with the fluorescence microscope. Since counts based on chlorophyll fluorescence (nonhybridized cells) or on hybridization with probe CHLO 02 did not indicate any accidental cell loss due to centrifugation, the discrepancies between PRAS 02 counts were probably linked to the fluorescence intensity yielded by this probe. A part of the PRAS 02 population is probably overestimated by flow cytometry, since it can be confounded by nonspecific labeling, which produces a similar low level of fluorescence, as shown on cultures (Fig. 2 and 6). Conversely, it is difficult to distinguish slightly labeled and nonlabeled cells by fluorescence microscopy, and manual counts in this case are subjective and somewhat unreliable (Fig. 6, right). It should be noticed also that probes NCHLO 01 and PELA 01 hybridized to particles which do not have any DNA (i.e., green dots not associated with red dots [Fig. 6]). Such particles were not taken into account when counts were done on filters and could correspond to cytoplasm debris (still containing 18S rRNA) resulting from sloppy feeding of larger zooplankton or from viral lysis.
Changes in picoeukaryote communities along a coastal transect.
In a final step, we used our method to describe the changes in the taxonomic composition of the picoeukaryote community along a transect in the Bay of Morlaix. Picophytoplankton was largely dominated by eukaryotes (91% ± 6%), which outnumbered the prokaryotes (Synechococcus) by 1 order of magnitude, as estimated by flow cytometry based on natural chlorophyll fluorescence (Fig. 7, top panel). At all stations, probe-based measurements indicated that Mamiellales and Pelagophyceae were dominating the Chlorophyta (92% ± 10%) and the non-Chlorophyta (100% ± 13%), respectively. Previous studies, in particular based on electron microscopy, led us to hypothesize that the members of the Prasinophyceae are the major component of photosynthetic picoeukaryote community, in particular in coastal waters (22, 42). In these early studies, two genera were dominating, the flagellate M. pusilla and a scaled nonmotile prasinophyte (22, 50), later identified as Bathycoccus prasinos (17), both belonging to the Mamiellales (Prasinophyceae). M. pusilla is very abundant around the British Isles, including the English Channel (32). This has been confirmed by recent molecular studies along the Brittany coast (55). In contrast to the Prasinophyceae, much less information is available on Pelagophyceae, since this class was first described more recently (4). Species such as the nonflagellated Pelagococcus subviridis and the flagellated Pelagomonas calceolata seem to be ubiquitous in the open ocean (4, 20, 36). Pigment data suggest that this group could be responsible for most of the new production in the tropical North Atlantic (10). In addition, some species, such as the nonflagellated Aureoumbra lagunensis and Aureococcus anophagefferens, have been implicated in brown tides, which cause serious economic damage to coastal fisheries (43, 44). Along the transect studied, it was surprising to see that this class dominates the non-Chlorophyta, since no sequences belonging to Pelagophyceae have been retrieved in any clone libraries compiled in summer in this area (K. Romari, personal communication). It will therefore be interesting to investigate the identity of the Pelagophyceae detected by FISH in future studies.
Along the transect, Chlorophyta and non-Chlorophyta populations behaved quite differently, as revealed by changes in cell concentration and probe fluorescence intensity. Chlorophyta were 14 to 61 times more abundant than non-Chlorophyta, and their numbers doubled between the coastal station (Fig. 1, station A), toward the most estuarine stations (stations C and D), while the Pelagophyceae numbers were rather stable. In contrast, the probe fluorescence intensity was 2 to 12 times lower for Chlorophyta than for non-Chlorophyta. Moreover, probe fluorescence increased regularly from stations A to D for Chlorophyta but in a stepwise fashion between stations B and C for non-Chlorophyta (Fig. 7). The change in probe fluorescence is directly related to changes in rRNA content (7, 25), which is itself related to changes in cell size, species identity, and growth rate. While differences in cell size and taxonomic composition explain the differences in fluorescence intensity between Chlorophyta and non-Chlorophyta (Fig. 6 and 7), variations within the Mamiellales (dominated by M. pusilla) probably correspond to changes in growth rates, as shown for various planktonic species (e.g., bacteria [7, 25], phytoplankton [16], zooplankton [5], and fish larvae [9]). Even if more data need to be acquired on cultures, it is interesting that changes in the fluorescence intensity of hybridized O. tauri and C. concordia followed variations in growth rates (Fig. 4) and that changes in fluorescence from the natural environment (from 0.07 to 1.19) were in the same range as those observed on cultures (from 0.16 to 1.34). Therefore, it is quite likely that Mamiellales were growing more slowly at stations A and B than at stations C and D. These variations could be explained by differences in seawater temperature and stability along the transect. At stations A and B, the water column is completely mixed; it becomes stratified only at stations C and D (Fig. 1). In contrast, changes in growth rates were probably not related to nutrient availability, since neither nitrogen nor phosphorus is limiting phytoplankton uptake in summer in this area (12, 26). For non-Chlorophyta, the stepwise increase in probe fluorescence between stations B and C contrasts with the stability in cell concentration. The reason for such a discrepancy could be either changes in species composition or changes in grazing pressure, possibly associated with differences in water column stability. Further studies of species-specific picoplankton growth rate are therefore needed to determine the origin of the differences in the population dynamics of the two groups.
Conclusion.
In this study, we developed a new method of cell collection and hybridization associated with flow cytometry, which allows the detection of picoeukaryotes from cultures in both exponential and stationary phases and from natural samples. In addition, the large flexibility in cell storage before or after hybridization allowed us to perform routine sample collection at sea and FISH manipulation in the laboratory, even if a flow cytometer was not immediately available. We also demonstrated that quantification of these organisms was possible in the natural environment with different taxon-specific probes, starting from as little as 17 ml of prefiltered seawater.
When comparing FISH quantification by using fluorescence microscopy or flow cytometry, the following points can be made. The fluorescence microscopy protocol is easier to set up and has the advantage of allowing us to see the details of hybridized cells, which is useful when working with unknown populations. In addition, when it is combined with digital image analysis, the fluorescence intensity of hybridized cells can be measured and the cell status or quality of hybridization can be estimated. However, these microscopic analyses are labor-intensive and only a limited number of cells can be measured, resulting in suboptimal statistical data. In contrast, flow cytometry is obviously much faster and provides much better statistics, especially for rare populations. In addition, flow cytometry provides objective information on the whole target population and permits us to acquire data on cell size, cell cycle status, and rRNA content from which the growth rate can be estimated (11). Finally, the sorting capacity of flow cytometers allows the acquisition of further genetic or physiological information on the targeted population (see, e.g., reference 60). Ultimately, the use of genus-specific probes detected by flow cytometry, coupled with other nucleic acid-specific dyes, will allow single-cell taxonomic and physiological information to be gathered for a variety of microorganisms, which will be extremely useful and will improve our understanding of the contribution of the different species to the food web and biogeochemical cycles.
Acknowledgments
This work was supported by the European program PICODIV EVK3-CT-1999-00021, the program PicoManche funded by Région Bretagne, and the CNRS program PROOF-BIOSOPE.
We thank the crew of the Mysis for helping with the sampling, and we thank D. Marie and E. Droucheau for assistance with flow cytometry and surfactant, respectively. Special thanks are extended to P. Morin and S. L'Helguen for useful comments and discussions.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analysing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. Zarda, D. A. Stahl, and K.-H. Schleifer. 1992. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 58:3007-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, R. A., G. W. Saunders, M. P. Paskind, and J. P. Sexton. 1993. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J. Phycol. 29:701-715. [Google Scholar]
- 5.Båmstedt, U., and H. R. Skjoldal. 1976. Studies on the deep water pelagic community of Korfjorden, western Norway. Adenosine phosphate and nucleic acids in Euchaeta norvegica (Copepoda) in relation to its life cycle. Sarsia 60:63-80. [Google Scholar]
- 6.Biegala, I. C., G. Kennaway, E. Alverca, J.-F. Lennon, D. Vaulot, and N. Simon. 2002. Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp. using tyramide signal amplification-fluorescence in situ hybridization and confocal microscopy. J. Phycol. 38:404-411. [Google Scholar]
- 7.Binder, B. J., and Y. C. Liu. 1998. Growth rate regulation of rRNA content of a marine Synechococcus (Cyanobacteria) strain. Appl. Environ. Microbiol. 64:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobrow, M. N., T. D. Harris, K. J. Shaughnessy, and G. J. Litt. 1989. Catalyzed reporter deposition, a novel method of signal amplification. J. Immuno. Methods 125:279-285. [DOI] [PubMed] [Google Scholar]
- 9.Buckley, L. J. 1984. RNA-DNA ratio: an index of larval fish growth in the sea. Mar. Biol. 80:291-298. [Google Scholar]
- 10.Claustre, H., and J.-C. Marty. 1995. Specific phytoplankton biomasses and their relation to primary production in the tropical North Atlantic. Deep-Sea Res. 142:1475-1493. [Google Scholar]
- 11.Collier, J. L. 2000. Flow cytometry and single cell in phycology. J. Phycol. 36:628-644. [DOI] [PubMed] [Google Scholar]
- 12.Colobert-Le Floch, I. 2001. Absorption et régénération de l'azote dans les systèmes côtiers; réponse à des apports massifs de nitrate. Ph.D. thesis. Université de Bretagne Occidentale, Brest, France.
- 13.Courties, C., A. Vaquer, M. Troussellier, J. Lautier, M.-J. Chrétiennot-Dinet, J. Neveux, C. Machado, and H. Claustre. 1994. Smallest eukaryotic organism. Nature 370:255. [Google Scholar]
- 14.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 15.Díez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dortch, Q., T. L. Roberts, J. R. Clayton, and S. I. Ahmed. 1983. RNA/DNA ratios and DNA concentrations as indicators of growth rate and biomass in planktonic marine organisms. Mar. Ecol. Prog. Ser. 13:61-71. [Google Scholar]
- 17.Eikrem, W., and J. Throndsen. 1990. The ultrastructure of Bathycoccus gen. nov. and B. prasinos sp. nov., a non-motile picoplanktonic alga (Chlorophyta, Prasinophyceae) from the Mediterranean and Atlantic. Phycologia 29:344-350. [Google Scholar]
- 18.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giovanonni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group specific oligonucleotide probes for identification of single microbial cell. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, L. E., and L. W. Wilcox. 2000. Algae. Prentice-Hall Press, London, United Kingdom.
- 21.Guillou, L., S.-Y. Moon-Van der Staay, H. Claustre, F. Partensky, and D. Vaulot. 1999. Diversity and abundance of Bolidophyceae (Heterokonta) in two oceanic regions. Appl. Environ. Microbiol. 65:4528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, P. W., and J. McN. Sieburth. 1982. In-situ morphology and occurrence of eukaryotic phototrophs of bacterial size in the picoplankton of estuarine and oceanic waters. J. Phycol. 18:318-327. [Google Scholar]
- 23.Keller, M. D., R. C. Selvin, W. Claus, and R. R. L. Guillard. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23:633-638. [Google Scholar]
- 24.Lebaron, P., P. Catala, C. Fajon, F. Joux, J. Baudart, and L. Bernard. 1997. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl. Environ. Microbiol. 63:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leser, T. D., M. Boye, and N. B. Hendriksen. 1995. Survival and activity of Pseudomonas sp. strain B13(FR1) in a marine microcosm determined by quantitative PCR and an rRNA-targeting probe and its effect on the indigenous bacterioplankton. Appl. Environ. Microbiol. 61:1201-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.L'Helguen, S., C. Madec, and P. Le Corre. 1996. Nitrogen uptake in permanently well-mixed temperate coastal waters. Estuarine Coast. Shelf Sci. 42:803-818. [Google Scholar]
- 27.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 28.Li, W. K. W. 1995. Composition of ultraphytoplankton in the central North Atlantic. Mar. Ecol. Prog. Ser. 122:1-8. [Google Scholar]
- 29.Lim, E. L., L. A. Amaral, D. A. Caron, and E. F. DeLong. 1993. Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl. Environ. Microbiol. 59:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim, E. L., D. A. Caron, and E. F. DeLong. 1996. Development and field application of a quantitative method for examining natural assemblages of protists with oligonucleotide probes. Appl. Environ. Microbiol. 62:1416-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-García, P., F. Rodríguez-Valera, C. Pedrós-Alió, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 32.Manton, I., and M. Parke. 1960. Further observations on small green flagellates with special reference to possible relatives of Chromulina pusilla Butcher. J. Mar. biol. Assoc. U.K. 39:275-298. [Google Scholar]
- 33.Marie, D., N. Simon, L. Guillou, F. Partensky, and D. Vaulot. 2000. DNA/RNA analysis of phytoplankton by flow cytometry. Curr. Protocols Cytometry 12(Suppl. 11):1-14. [DOI] [PubMed] [Google Scholar]
- 34.Moon-van der Staay, S. Y., R. De Watchter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 35.Not, F., N. Simon, I. C. Biegala, and D. Vaulot. 2002. Application of fluorescence in situ hybridization with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat. Microbiol. Ecol. 28:157-166. [Google Scholar]
- 36.Paul, J. H., A. Alfreider, and B. Wawrik. 2000. Micro- and macrodiversity in rbcL sequences in ambient phytoplankton populations from the southeastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 198:9-18. [Google Scholar]
- 37.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsing, N. B., M. Kühl, and B. B. Jørgensen. 1993. Distribution of sulfate-reducing bacteria, O2 anf H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, J., M. A. Sleigh, P. H. Burkill, G. A. Tarran, C. D. O'Connor, and M. V. Zubkov. 1997. Flow cytometric analysis of characteristics of hybridization of species-specific fluorescent oligonucleotide probes to rRNA of marine nanoflagellates. Appl. Environ. Microbiol. 63:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schönuber, W., B. Fuchs, S. Juretschko, and R. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schönuber, W., B. Zarda, S. Eix., R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro, L. P., and R. R. L. Guillard. 1986. Physiology and ecology of the marine eukaryotic ultraplankton. Can. Bull. Fish. Aquat. Sci. 214:371-389. [Google Scholar]
- 43.Sharma, V. K., K. B. Rhudy, and F. J. Millero. 2000. Diurnal variation of Texas “brown tide” (Aureoumbra lagunensis) in relation to metals. J. Environ. Sci. Health 35:1077-1088. [Google Scholar]
- 44.Sieburth, J. M., P. W. Johnson, and P. E. Hargraves. 1988. Ultrastructure and ecology of Aureococcus anaphageferens gen. et sp. nov. (Chrysophyceae): the dominant picoplankter during a bloom in Narragansett Bay, Rhode Island, summer 1985. J. Phycol. 24:416-425. [Google Scholar]
- 45.Simon, N., N. LeBot, D. Marie, F. Partensky, and D. Vaulot. 1995. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes to identify small phytoplankton by flow cytometry. Appl. Environ. Microbiol. 61:2506-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, N., L. Campbell, E. Örnolfsdottir, R. Groben, L. Guillou, M. Lange, and L. K. Medlin. 2000. Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J. Eukaryol. Microbiol. 47:76-84. [DOI] [PubMed] [Google Scholar]
- 47.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Inc., New York, N.Y.
- 48.Stockner, J. G., and N. J. Antia. 1986. Algal picoplankton from marine and freshwater ecosystems: a multidisciplinary perspective. Can. J. Fish. Aquat. Sci. 43:2472-2503. [Google Scholar]
- 49.Thomas, J.-C., M. Desrosiers, Y. St-Pierre, P. Lirette, J.-G. Bisaillon, R. Beaudet, and R. Villemur. 1997. Quantitative flow cytometric detection of specific microorganisms in soil samples using rRNA targeted fluorescent probes and ethidium bromide. Cytometry 27:224-232. [DOI] [PubMed] [Google Scholar]
- 50.Throndsen, J. 1976. Occurrence and productivity of small marine flagellates. Norw. J. Bot. 23:269-293. [Google Scholar]
- 51.Urdea, M. S., B. D. Warner, J. A. Running, M. Stempien, J. Clyne, and T. Horn. 1988. A comparison of non-radioisotopic hybridization assay methods using fluorescent, chemiluminescent and enzyme labeled synthetic oligodeoxyribonucleotide probes. Nucleic Acids Res. 16:4937-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Gijlswijk, R. P. M., J. Wiegant, A. K. Raap, and H. J. Tanke. 1996. Improved localization of fluorescent tyramides for fluorescence in situ hybridization using dextran sulfate and polyvinyl alcohol. J. Histochem. Cytochem. 44:389-392. [DOI] [PubMed] [Google Scholar]
- 53.van Hannen, E. J., M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1998. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J. Phycol. 34:206-213. [Google Scholar]
- 54.Vaulot, D., and D. Marie. 1999. Diel variability of photosynthetic picoplankton in the equatorial Pacific. J. Geophys. Res. 104:3297-3310. [Google Scholar]
- 55.Vaulot, D., K. Romari, and F. Not. 2002. Are autotrophs less diverse than heterotrophs in marine picoplankton? Trends Microbiol. 10:266-267. [DOI] [PubMed] [Google Scholar]
- 56.Veldhuis, M. J. W., and G. W. Kraay. 1990. Vertical distribution and pigment composition of a picoplanktonic prochlorophyte in the subtropical North Atlantic: a combined study of HPLC-analysis of pigments and flow cytometry. Mar. Ecol. Prog. Ser. 68:121-127. [Google Scholar]
- 57.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometry identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 58.Wallner, G., R. Erhart, and R. Amann. 1995. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl. Environ. Microbiol. 61:1859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West, N. J., W. A. Schönuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147:1731-1744. [DOI] [PubMed] [Google Scholar]
- 60.Zubkov, M. V., B. M. Fuchs, P. H. Burkill, and R. Amann. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]