Abstract
The Scaffold attachment factor B1 (SAFB1) is an estrogen receptor (ESR1) repressor that has been proposed to inhibit breast tumorigenesis. To obtain insight into the functions of SAFB1 we utilized a yeast two-hybrid screen and identified the Ret finger protein (RFP) as interacting with the SAFB1 C-terminus. RFP is a member of the trimotif (TRIM) family of proteins, which we found widely expressed in a series of breast cancer cell lines. We confirmed the interaction between SAFB1 and RFP through in vitro (GST-pulldown) and in vivo (coimmunoprecipitations) assays. We hypothesized that SAFB1 functions as a scaffolding protein to recruit proteins such as RFP into proximity with ESR1. Consequently, we asked whether RFP would modulate ESR1 activity and we discovered that RFP was important for the ESR1-dependent expression of cyclin D1 (CCND1) and the progesterone receptor (PR) but not IRS1 or MYC. Although RFP did interact with ESR1 directly, it does coimmunoprecipitate with ESR1 demonstrating that RFP is found within the same protein complex. Chromatin immunoprecipitation assays (ChIP) located RFP to the TFF1 promoter, a known ESR1-regulated gene. Taken together, our study provides further evidence that coactivation and corepression are integrally linked processes and that RFP is a component of an ESR1 regulatory complex.
Keywords: RET finger protein, estrogen receptor, scaffold attachment factor-B1, gene regulation, and breast cancer
Introduction
The ovarian steroid hormone estradiol (E2) is key regulator of epithelial cell proliferation and is essential for the normal functioning and development of mammary epithelial cells [1]; however, the mechanisms by which E2 influences epithelial cells have not been fully established. The effects of E2 are predominantly mediated through estrogen receptors α (ESR1) and β (ESR2) [1, 2]. Estrogen receptors are members of the steroid receptor family of transcription factors that regulate the expression of genes involved in proliferation, apoptosis, migration, and other cellular processes in a ligand-dependent or -independent manner [3]. The activation or repression of transcription by ESR1 is mediated through the respective and opposing actions of coactivator and corepressor protein complexes which regulate activity of ancillary transcription factors, the basal transcription apparatus, RNA polymerase II, and chromatin structure [4–6]. Coactivators and corepressors may be found within the same protein complex, highlighting the close physical and functional association between proteins with opposing activities [7].
We have previously shown that the members of the scaffold attachment factor B family, termed SAFB1 and SAFB2, can function as potent ESR1 corepressors [8, 9]. SAFB1/2 are nuclear matrix proteins characterized by an N-terminal DNA-binding domain (SAF-box) that binds to scaffold/matrix attachment regions (S/MARs), and a central RNA recognition motif (RRM) [10]. SAFB1 interacts with the RNA processing machinery and RNA polymerase II, suggesting that it is part of a “transcriptosome” complex coupling chromatin structure to transcription and RNA processing [11]. We have also shown that the C-terminus of SAFB1/2 harbors a strong independent repression domain that can mediate repression when transferred to a heterologous DNA-binding protein. This repression by SAFB1 is in part mediated via interactions with NCOR1 [12] and hTAfII68 (TAF15) [8], and is sensitive to histone deacetylase inhibitors [13].
The RET finger protein (RFP), originally identified as the N-terminal fusion partner with the RET tyrosine kinase proto-oncogene [14]. RFP, (TRIM 27), is a member of the tripartite motif (TRIM) superfamily (reviewed in [15]). The TRIM family is characterized by a combination of a RING finger, one or two B-Box zinc fingers, and a coiled-coil motif, followed by one of several C-terminal domains. Half of the TRIM family members, including RFP, have a C-terminal domain similar to PRY or SPRY domains [16]. RFP is widely expressed and depending upon the cell type, may be localized to the nucleus, cytoplasm, or cell membrane [17]. Evidence suggests that RFP is important for male germ cell tumors [18] and is expressed in a wide range of other tumor types [19]. While the membrane and cytoplasmic function(s) of RFP are obscure, several nuclear functions have been identified.
RFP is a component of PML nuclear bodies through direct interaction with PML [20] and a transcription factor [21]. Subsequent studies have demonstrated an interaction between RFP and several transcription factors, including enhancer of polycomb (EPC1) [22], Mi-2β [23], the retinoblastoma protein (RB1) [19], and bHLH family members [24]. It has been suggested that RFP contains multiple repression domains [22], yet RFP-containing complexes and interacting proteins are associated with both gene expression and repression [22, 23, 25], suggesting that the function(s) of RFP may be promoter dependent. The observation that RFP inhibits RB1-dependent gene expression, but does not effect RB1-dependent gene repression [19] provides further evidence that RFP acts within a context dependent manner. Lastly, an association between RFP and the structural maintenance chromosome 3 protein (SMC3) points to an additional role for RFP in chromatin stability [26].
To gain further insight into the functions of SAFB1, we performed a yeast two-hybrid screen for proteins interacting with the SAFB1 C-terminus (amino acids 600–915) [8]. In this screen, we isolated three independent RFP clones. Here we show that the SAFB1 C-terminal Glu/Arg-rich domain (a coiled-coil structure) directly interacts with the coiled-coil region of RFP and that the interaction between these two proteins occurs in vitro and in vivo. Although we were able to confirm that RFP functions as a repressor in heterologous transfection assays, we did not detect any synergistic interaction between SAFB1 and RFP which would point towards a role of RFP in SAFB1-mediated repression or corepression of ESR1. Surprisingly, we made the novel observation that reduction of RFP is required for expression of a subset of ESR1-regulated genes in vitro. We discuss a role for RFP in ESR1-mediated gene regulation and its possible contribution to breast tumorigenesis.
Materials and methods
Cell culture
Cell lines CV-1 (African green monkey kidney cells); NIH-3T3 (mouse fibroblast); HeLA (cervical adenocarcinoma); HEK293 (primary human embryonal kidney); and the human breast cancer cell line MCF-7, were maintained in improved MEM (IMEM) (Invitrogen, Carlsbad, CA) supplemented with 200U/ml penicillin, 200ug/ml streptomycin, 6ng/ml insulin and 5–10% fetal bovine serum (Hyclone, Logan, UT). Stripped-serum medium was composed of phenol red-free IMEM with 5% charcoal stripped fetal bovine serum (Hyclone), antibiotics, and insulin.
Plasmid constructs
GST-fusion genes were constructed by cloning digested PCR fragments into pGEX-2TK and Gal4DBD-fusion proteins by cloning into pCMX-Gal4N. Yeast two-hybrid and other constructs for SAFB1 have been previously described [8]. All constructs were confirmed by restriction digestion and/or sequencing (Seqright, Houston TX). HA-tagged RFP was generated by PCR with a 3′ primer that contained the HA epitope and cloned into the expression vector pCDNA3.1 (Invitrogen).
Yeast two-hybrid screening
The yeast two-hybrid screen and interaction tests were undertaken using the Matchmaker3 system (BD Biosciences) with the appropriate controls according to the manufacturer’s instructions as previously described [8, 9]. The bait and test constructs were cotransformed into yeast strain AH109 which was then plated onto SD-Leu/Trp and SD-Leu/Trp/His/Ade medium containing X-α-gal as a substrate for α-galactosidase. Yeast plasmids were isolated using the Yeastmaker yeast plasmid isolation kit (BD Biosciences) and retransformed into DH5α (Stratagene). β-Galactosidase (β-Gal) assays were performed according to the Clontech yeast protocols handbook.
Transient and stable transfections
To generate MCF-7 cells stably expressing HA-RFP, we first cloned full-length RFP into pCDNA3.1 using a C-terminal HA-epitope tag. Stable transfected MCF-7 clones were selected in the presence of 1000μg/ml G148 and maintained in 500μg/ml. Control cells were transfected with empty pCDNA3.1 (Invitrogen) vector. To determine the effect of RFP siRNA on ESR1-mediated gene regulation, MCF-7 cells were transfected for 16–24 hrs with siRNAs (10–25nM) which were generated using the Silencer siRNA kit (Ambion, Austin TX) (siRNA#1: 5′A GAA CCA GCT CGA CCA TT-3′; siRNA#2: 5′-AAG AGG CGA TAC TCA TGC TCC-3′). Sixteen hours later, cells were stimulated with E2 or vehicle and harvested for Western blots 2–8 hrs later.
Antibodies and immunoblotting
Western blot experiments using anti-HA and anti-SAFB antibodies have recently been described [8]. Other antibodies used for immunoblotting were anti-ESR1 (Novocastra), anti-PR (Dako), anti-p85/PI3K (Upstate), anti-β-actin (AC-15) (Sigma), anti-p42/44 MAPK (Cell Signaling), anti-CCND1 (Santa Cruz Biotechnology), anti-RFP (IBL America, and anti-MYC (Santa Cruz). Polyclonal rabbit anti-RFP antibody was obtained from Dr. Elkin from M.D. Anderson Cancer Center [20]. Immunohistochemistry was performed according to Townson et al. [9] using anti-RFP antibodies (IBL America).
In vitro protein and communoprecipitation assays
GST pulldown experiments were performed as recently described [8, 9]. MCF-7 cells stabley expressing HA-RFP were used to coimmunoprecipitate RFP, ESR1, and SAFB. Cells were plated in IMEM with 5% charcoal-stripped serum. After 24hrs growth, the cells were stimulated with vehicle or 10−8M E2 and cross-linked with dithiobis (succinimidylpropionate) (DSP) (Pierce). Anti-ESR1 (H-184), -SAFB, and -HA antibodies were purchased from Santa Cruz, Upstate, and Covance, respectively.
Reverse transcriptase-PCR
RNA was isolated from cells using the RNEasy kit from Qiagen and RT-PCR was performed using a polydT(18)N primer and Superscript II reverse transcriptase (Invitrogen) followed by PCR for 25–35 cycles. RFP and GAPDH were amplified using 5′-TGC CAA CAT CTC CCA CCT CAG-3′ and 5′-CCA AGA CAC AGG GAA ACA GAT TG-3′, and 5′-GAC AAC TTT GGT ATC GTG GAA GG-3′ and 5′-CCC TGT TGC TGT AGC CAA ATT CG-3′, primer sets respectively. RT-PCR primers for discriminating between RFP isoforms α and β were 5′-CTT GCA ACA TCT CCC ACC TCA G-3′ and 5′-GGC CCA CAA AAG GTA GCA TGA G-3′.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed in MCF-7 cells stably expressing HA-RFP as described for the TFF1 gene [27]. PCR for the TFF1 ERE site was performed using the primers 5′-GGC CAT CTC TCA CTA TGA ATC ACT TCT GCA G-3′ and 5′-GGC AGG CTC TGT TTG CTT AAA GAG CGT TAG ATA-3′ at promoter positions from −353 to −30 to amplify a 323 bp genomic fragment. Antibodies used in the ChIP assays were rabbit anti-ESR1 (H-184) (Santa Cruz), mouse anti-SAFB (Upstate), and mouse anti-HA (Covance). The negative control primers for the TFF1 gene 5′-GGC TGT CAG GAA ATG C-3′ and 5′-AAT GCT GGC TGC TCT TCT ACG-3′, amplify a 385 bp fragment from position +871 to +1256 [28].
Results
SAFB1 and RFP interact in vitro and in vivo
To identify proteins that interact with the SAFB1 C-terminus (amino acids 599–915), we performed a yeast two-hybrid screen using a mammary gland Matchmaker3 library (Clontech) [8] and isolated three RFP clones. The two longest clones started at amino acid 111 and the shorter clone at 181. The two longer clones contained part of the B-box zinc finger and the complete coiled-coil domain, while the shorter more weakly interacting clone started within the coiled-coil domain and contained the complete third coiled region. All clones were of the RFPα splice form. RFP was the only TRIM family member identified and the only protein that contained any of the TRIM domains in combination.
At first, we set out to confirm the results in a directed yeast-two hybrid assay. As shown in Figure 1A, we mapped the minimal SAFB1-interaction site to the coiled-coil domain, which is composed of three individual coil regions, using a series of C-terminal truncations in combination with N- and C-terminal regions, and the complete coiled coil domain (see interaction of amino acids 113–313, and data not shown). Quantitative beta-galactosidase assays confirmed the interaction between SAFB1 and RFP (data not shown). Further refinement of the SAFB1-interaction domain within the coiled-coil region, using the coils individually or in combination, failed to detect a robust interaction with SAFB1. Additional control experiments were performed showing that an out-of-frame RFP mutant failed to interact with SAFB1 and that the SAFB1-RFP interaction was bi-directional with respect to both bait and prey.
Fig. 1.

SAFB1 interacts with RFP in vitro and in vivo. A) SAFB1 and RFP interact in yeast two-hybrid assays. Yeast expression plasmids for RFP and SAFB1 were transformed into yeast cells that were then plated onto selective media. The figure represents growth of colonies after they had been streaked successively three times on selective plates with X-α-Gal for α-galactosidase activity. The SAFB1 and RFP interaction domains were mapped using directed yeast two-hybrid assays with the indicated RFP and SAFB1 domains. These experiments show that the SAFB1 C-terminus interacts with the RFP coiled-coil domain (amino acids 114-330). B) RFP interacts with the SAFB1 C-terminal Glu/Arg-rich domain (amino acids 599–720). C) SAFB1 and RFP directly interact in GST-pulldown assays. The input lanes represents 20% of total in vitro transcribed and translated RFP. D) Expression of HA-RFP in transfected MCF-7 cells. Western blot performed on cell lysates of MCF-7 cells stably expressing HA-RFP using anti-HA and β-actin antibodies. E) SAFB and HA-RFP are found within the same protein complex in vivo. Coimmunoprecipitation experiments were performed using lysates from MCF-7 cells stably expressing HA-RFP protein using anti-SAFB, -HA, -ESR1, and -IgG antibodies, followed by immunoblotting using antibodies for ESR1, HA, and SAFB.
Next we confirmed and fine-mapped the interaction domain in SAFB1. As expected, RFP interacted with the SAFB1 C-terminus of (amino acids 599–915), but not with the N-terminus (amino acids 1–260) or central region (amino acids 260–600) (data not shown and Figure 1B). Using SAFB1 C-terminal polypeptides, we also determined that RFP interacted specifically with SAFB1 amino acids 599–720, which contains a Glu/Arg-rich coiled-coil domain [8]. Thus, the central region harboring the coiled-coil domain in RFP is required for interaction with the Glu/Arg-rich coiled domain in the SAFB1 C-terminus.
The interaction between SAFB1 and RFP was also confirmed in GST-pulldown assays using an in vitro translated full-length RFP and GST-SAFB1 fusion proteins (Figure 1C). RFP failed to interact with the GST control while it did interact with GST-SAFB1 fusion proteins encompassing SAFB1 amino acids 600–915 and 599–720.
Finally, we wanted to confirm the interaction in vivo. Coimmunoprecipitation experiments were performed using an MCF-7 cell line stably expressing an HA epitope-tagged RFP (Figure 1D, top panel). The level of RFP within these cells is approximately 2.5 times higher than in the parental control cells with a stably integrated empty pcDNA3.1 vector. Moreover, the level of RFP protein within this MCF-7 cell line as detected with anti-RFP antibodies is within the biological range of RFP expression from different epithelial tumor cell lines (data not shown) and is considerably less than when cells are transiently transfected with an RFP expression construct. Consistent with the in vitro data, we were readily able to immunoprecipitate HA-RFP with SAFB antibodies, and likewise, SAFB with HA-RFP antibodies (Figure 1D, bottom panel). Given that SAFB1 is a component of an ESR1 complex, we asked whether RFP may also associate with ESR1 in vivo. We investigated an in vitro association between RFP and ESR1, using coimuoprecipitation experiments with anti-HA and -ESR1 antibodies in our RFP cell lines. Our results demonstrate that RFP can be immunoprecipitated with anti-ESR1 antibodies and that ESR1 can be immunoprecipitated with anti-HA antibodies. We then investigated whether RFP would interact directly with ESR1 using recombinant GST-ESR1 (ESR1 domains B–E) and in vitro translated RFP; however, RFP failed to bind to ESR1 in the presence or absence of E2 (Figure 2).
Fig. 2.
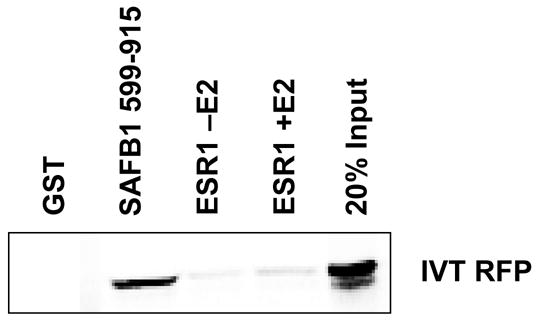
RFP does not interact with ESR1 in vitro. GST-pulldown assays were performed with in vitro translated RFP, and GST-SAFB1 (aa 600-715) and GST-ESR1. Interactions with ESR1 were performed in the presence or absence of 10−6M E2.
In summary, these data demonstrate that SAFB1 and RFP interact in vitro and in vivo, that this interaction is direct, and mediated through the coiled-coil region in RFP and the C-terminal region of the repression domain in SAFB1. Furthermore, RFP is found within an ESR1 complex but does not directly interact with ESR1.
RFP is widely expressed in human breast cancer cell lines
Given the role of SAFB1 in breast cancer tumorigenesis, we next asked whether RFP is expressed within breast cancer cell lines and therefore in breast tumors. RT-PCR analysis in 10 routinely cultured breast cancer cells showed that RFP was widely expressed (Fig 3A). Furthermore, RFP was expressed in three different lines of MCF-7 cells. Expression was not limited to cancer cells as we also detected RNA expression in MCF10A cells, which are immortalized but not transformed breast cells.
Fig. 3.

Expression of RFP in human breast cancer cell lines. A) RFP RNA is widely expressed in breast cancer cells. Agarose gel of RT-PCR products for RFP and GAPDH expression performed on total RNA isolated from breast cancer cell lines. PCR using the RFP expression plasmid served as a positive control. B) Western blot of cell extracts from ESR1-positive and –negative breast cancer cell lines probed with an anti-RFP antibody. C) RFP is localized to the nucleus in MCF-7 cells. MCF-7 cells were stained with an anti-RFP antibody followed by a fluorescent secondary antibody, and then counterstained with DAPI.
To demonstrate that RFP protein was present in some of these cell lines, we probed cell extracts from three ESR1-positive and three ESR1-negative cell lines with an anti-RFP antibody by Western blotting. As shown in Figure 3B, we were able to detect protein expression in all cell lines analyzed. Interestingly, we detected a second band, similar in size to the reported 45kDa size of the RFPβ splice form, which differs from RFPα splice form in the C-terminal SPRY domain [15]. This band was only present in the ESR1-positive breast cell lines. To determine whether this band corresponded to the RFPβ splice form, we designed RT-PCR primers to discriminate between the two RFP isoforms. Since we were unable to detect any RFPβ mRNA in these cells (data not shown), we concluded that this additional band is either a new RFP isoform or a protein cross-reacting with the RFP antibodies only expressed in ESR1-positive cells. Subsequent experiments have further demonstrated that this 45kDa band is present in several different epithelial tumor cell lines regardless of hormone receptor status (data not shown).
Since RFP may be localized to both the cytoplasm and nucleus depending on cell type we were also interested in the subcellular localization of RFP in breast cancer cells [17]. Staining of MCF-7 cells with an anti-RFP antibody demonstrated that RFP was localized predominantly to the nucleus (Figure 3C). To demonstrate that transfected HA-tagged RFP was also nuclear, we also stained the MCF-7 and T47D, Hela, and CV-1 cells transfected with HA-RFP using the anti-HA antibody, and observed a strong nuclear staining in all cell lines, demonstrating that HA-tagged RFP nuclear localization recapitulates that of the normal protein. However, when RFP was coupled to the green fluorescent protein it did not localize to the cell nucleus in any cell line we tested (data not shown). Thus, RFP is widely expressed in human breast cancer cell lines, and can be found predominantly in the nucleus as would be predicted for a transcription factor.
RFP does not enhance SAFB1-mediated repression
Next we asked the question whether the interaction between SAFB1 and RFP would alter SAFB1-mediated repression or corepression of ESR1. We first confirmed that RFP functions as a transcriptional repressor [22]; however, we were unable to detect any synergistic interaction between SAFB1 and RFP in heterologous transient transfections using expression constructs for Gal4 DNA-binding domain (DBD) fusion proteins and a UAS-TK-Luciferase reporter [8] (data not shown). Likewise, we could not shown an RFP effect on SAFB1-mediated ESR1 corepression [29].
In the absence of identifying an RFP-deficient cell line, we then asked if depletion of RFP would affect SAFB1-mediated repression or ESR1 activity and designed several siRNAs directed against RFP. We tested the efficacy of our siRNAs in MCF-7 cells stabley transfected with an HA-RFP expression construct. As shown in Figure 4A, we identified one siRNA (siRNA#1) which resulted in an efficient down regulation of RFP protein whereas the control (siRNA#2) produced little to no effect on RFP levels. Transfection of siRNA#1 resulted in an approximate 70% reduction in RFP mRNA as measured by semi-quantitative RT-PCR analysis (4B). These siRNAs had no effect on endogenous SAFB1 levels (Figure 4C), ESR1 levels, or degradation of ESR1 by the proteasome (see Figure 4D).
Fig. 4.
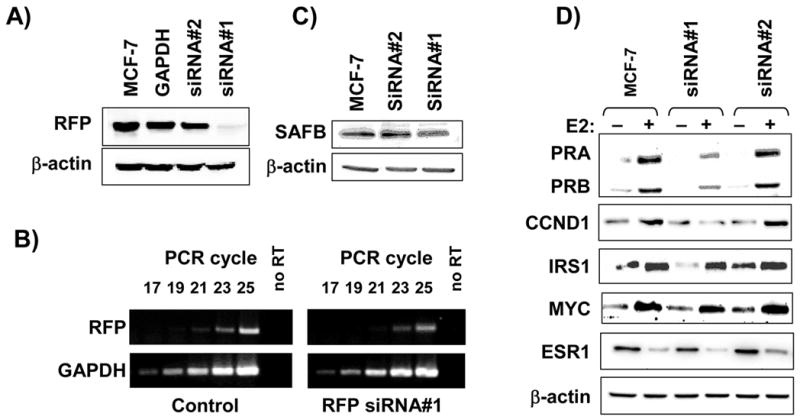
RFP functions in ESR1-mediated gene regulation. A) siRNA reduces RFP protein levels. MCF-7 cells stably expressing with RFP and GAPDH siRNAs, protein was extracted, and analyzed by immunoblotting for RFP. B) siRNA reduces RFP RNA levels. MCF-7 cells were transfected with control or RFP siRNA, and RNA was isolated 24 hrs later. RT-PCR was performed for 17–25 cycles with primers for RFP and GAPDH. C) RFP siRNAs do not alter SAFB protein levels. MCF-7 cells were transfected with GAPDH, siRNA#1 or siRNA#2 and 24hrs later proteins were extracted and analyzed by Western blot for SAFB and β-actin protein levels. D) Decrease of RFP inhibits E2-mediated expression of ESR1-dependent genes. activity in transient reporter assays. MCF-7 cells were cultured in stripped serum medium for three days, transfected with siRNA#1 or siRNA#2 and 24hrs later stimulated with 10−8M E2 for 2–8 hrs followed by protein extraction for Western blots.
We then investigated using transient transfection experiments whether RFP was required for SAFB1-mediated repression or ESR1 activity in MCF-7 cells using the Gal4DBD or ERE-TK-Luc reporter systems respectively. Our results showed that depletion of RFP lead to a near complete loss of UAS-TK-Luc and ERE-TK-Luc reporter activity. Consequently, it became clear that assessing RFP activity using siRNA knockdown and/or transient transfections were not suitable experimental systems for determining RFP function due to nonspecific effects in these assays.
RFP is important for the ESR1-dependent regulation of endogenous genes
Given that RFP failed to effect ESR1 activity using transient transfections we investigated the role of RFP on the ESR1-mediated regulation of endogenous genes. MCF-7 cells were treated with E2, and we measured protein expression of candidate genes known to be E2- and ESR1-regulated including CCND1, PR, insulin receptor substrate 1 (IRS1), and MYC (Figure 4D). As expected, E2 treatment resulted in induction of progesterone receptors A and B (PRA and PRB), CCND1, IRS1, and MYC. Decrease of RFP by our siRNA resulted in diminished induction of PRA, PRB, and CCND1. MYC and BCL2 (data not shown) expression were not effected by decreased RFP. Interestingly, IRS1 basal levels were sensitive to decreased RFP whereas the E2-mediated induction was not. We did not see any significant change in protein levels for a number of control proteins including β-actin, MAPK, and PI3K (Figure 4D, and data not shown), confirming that the effects seen are due specifically to an unidentified function of RFP in E2-mediated transcription and not on generalized transcription. In summary, we have demonstrated that siRNA can decrease RFP mRNA and protein levels resulting in reduced ESR1 activity, suggesting that RFP is important for ESR1–dependent regulation of specific genes.
RFP is localized to the TFF1 promoter
Our conclusion that RFP may function in the regulation of ESR1 activity, prompted us to determine if RFP could be localized to the promoter of an ESR1-regulated genes. Using chromatin immunoprecipitation assays (ChIP), we investigated whether RFP could be found at the +90 AP-1/ESR1 site in the PR promoter [30]. However, since we were unable to clearly establish that RFP was localized to this promoter site we selected to investigate RFP localization to the previously characterized trefoil factor 1 (TFF1) promoter [27].
MCF-7 cells stably expressing HA-RFP treated with either vehicle or E2 (10−8M) for 1 hr were employed in ChIP experiments as described by Shang et al. [27] and Burakov et al. [28] (Figure 5). As expected, the addition of E2 resulted in the recruitment of ESR1 to the TFF1 promoter and SAFB was DNA-bound in the absence of E2 with a significant fraction released upon ligand treatment. Conversely, RFP was present before and after ligand treatment. The binding of these factors was not non-specific since we did not detect RFP, ESR1, or SAFB binding to a region 900bp downstream of the TFF1 transcription start site.
Fig. 5.
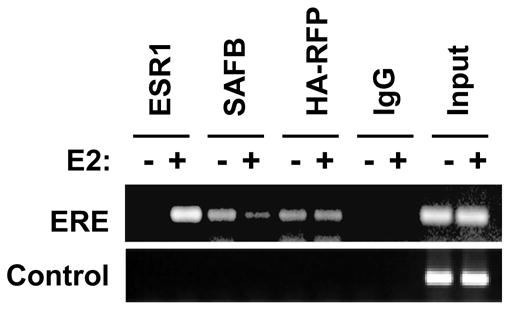
RFP colocalizes with ESR1 to the TFF1 promoter. MCF-7 cells expressing HA-RFP cells were cultured in stripped-serum medium for 3 days, followed by treatment with E2 (10−8M) or vehicle for 1 hr. ChIP assays were performed using TFF1 primers [28].
Analysis of the TFF1 promoter by ChIP has revealed that ESR1 and protein complexes are cyclically recruited to and removed from the promoter during transcription [27, 28]. Likewise, we hypothesized that a similar dynamic recruitment and loss of RFP to the TFF1 promoter may occur. ChIP assays were performed with cells treated with E2 over the course of an hour. Analysis of RFP and ESR1 localization in 20 minute increments showed that there was little difference in the level of RFP localized to the TFF1 promoter at these time points. In summary, we have shown that RFP is spatially and temporally present on the promoter of an ESR1-regulated gene in agreement with a possible role in regulating ESR1 activity.
Discussion
The mechanisms by which coactivators and corepressors coordinate the control of ESR1–regulated gene expression are still not completely defined [5]. Likewise, not all proteins that perform in this exquisite “molecular ballet” have been identified. Here, we provide evidence for a new function for RFP in gene regulation. We show that the ESR1 corepressor SAFB1 interacts with RFP in vitro and in vivo. While we were able to validate that RFP functions as a repressor in heterologous transient transfections, we failed to detect synergism between SAFB1 and RFP. We also show for the first time that RFP itself is a critical player in E2 action and is vital for selected ESR1 transcriptional activity. RT-PCR and Western blot analyses have revealed that RFP is expressed in routinely cultured breast cancer cell lines regardless of ESR1 status, and that RFP is localized to the nucleus consistent with its role in gene expression.
To characterize the interaction between SAFB1 and RFP, we first mapped the RFP binding site in SAFB1 to the C-terminal amino acids 599–720 harboring a Glu/Arg rich coiled structure [8]. The RFP binding site differs from the SAFB1-domains involved in interaction with TAF15 (amino acids 720–915), ESR1 (amino acids 426–600) [8], and SAFB1/2 (amino acids 720–808) [8]. Amino acids 111–132 of the B-box and the complete coiled-coil domain of the RFP TRIM motif were required for interaction with SAFB1. The individual coils had minimal binding to SAFB1; presumably, the integrity of the RFP coiled-coil domain is necessary for the interaction with other coiled structures somewhat similar to what was determined for the interaction between RFP and PML [20].
Using Gal4DBD-fusion proteins and a UAS-TK-Luc reporter plasmid in heterologous transfection assays, we confirmed previous data [22] that RFP is a repressor of transcription when tethered to a reporter construct and that the major region responsible for this activity is localized within the coiled-coil motif. However, we were unable to demonstrate that RFP would enhance SAFB1-mediated transcriptional repression or that SAFB1 would enhance RFP-mediated repression. A comparable lack of cooperation in transcriptional repression was documented between RFP and EPC1 [22], suggesting that over expression of cofactors and/or cofactor-interacting proteins in cells that already express the gene of interest might result in false-negative results.
There is increasing evidence that transcription factors function within the context of particular promoters and some cofactors have been described as coactivators by some groups and as corepressors by others, possibly reflecting possible cell- and promoter-specific activity. Examples are RIP140 [31, 32, 33] and FKHR [34, 35]. Recent evidence has demonstrated that the RFP/Mi-2β complex is associated with both gene repression [23] and expression [25] in a context-dependent manner. Moreover, the RFP-interacting protein EPC1 has activities in both gene expression and gene repression [36, 37]. Thus, we propose that RFP is not necessarily a repressor in the context of ESR1 as demonstrated by our findings that RFP is required for ESR1-regulated genes and our ChIP data where RFP and ESR1 colocalize to the promoter of an actively transcribed gene. Such a model would clarify our counterintuitive observation that while RFP is a repressor in some assays, that decreasing amounts of RFP lead to inhibition of ESR1 activity.
Transcriptional repression and activation of nuclear receptors are closely linked processes. For example, the coactivator AIB1 has been found in a complex with the corepressors NCOR1 [7]. We would like to hypothesize that RFP provides a molecular scaffold which allows the integration of various cofactor proteins at different stages in gene regulation. Peng et al. [38] have shown that members of the TRIM family can form homo- or hetero-dimers allowing them to function as a switch between coactivation and corepression. Similarly, RFP may also serve as a molecular switch between as shown for TBL1/TBL1R and PPARG [31, 33, 39].
Additionally, there is some evidence that RFP may also have intrinsic activity as an E3 ubiquitin ligase. Other TRIM family members with a similar structure to RFP such as RET finger protein like 4 (RFPL4) [40] are putative E3 ubiquitin ligases. Although depletion of RFP appeared not to effect affect global ESR1 or SAFB protein levels, RFP may be required for the necessary degradation of other ESR1-associated proteins.
Currently, how RFP contributes to breast tumorigenesis is unclear. Such a role, however, would not be totally surprising since our studies clearly show that RFP is necessary for the ESR1–dependent regulation of CCND1 and PR. Our data also suggest that RFP’s role depends on the promoter context since for some genes (PR and CCND1) RFP was necessary for E2 induction, while for others it was involved in ligand-independent regulation of promoter activity (IRS1). Failure to demonstrate an effect of RFP on MYC and BCL2 revealed that there is yet another subset of ESR1-regulated genes that do not require RFP. Thus, we can define subsets of ESR1-regulated genes based upon the combination of coactivator and corpressor proteins required for their control. Moreover, given the role of ESR1 coactivators and corepressors in breast cancer, analysis of such gene sets may have predictive and/or prognostic value [41].
In summary, we propose a testable model where RFP is a necessary factor for the expression of a subset of ESR1-regulated genes. In this model, a promoter bound SAFB1/RFP complex defines a chromatin state unreceptive to ESR1. Upon binding of ligand, ESR1 is recruited to the promoter concurrently with the release of SAFB1. An altered RFP structure, potentially through altered homo- or hetero-oligomerization, enables interactions with chromatin remodeling complexes to convert the repressed chromatin to an open and transcriptionally active chromatin state. Ongoing studies in our laboratory will aid our understanding of the complex machinery required to regulate ESR1 and define how RFP contributes to breast tumorigenesis.
Acknowledgments
We are grateful to Drs. M. Takahashi (Nagoya University Graduate School of Medicine) and Ron Evans (Salk Institute) for plasmids. We also thank Dr. Gary Chamness for critical reading of this manuscript, and helpful suggestions.
Footnotes
Funding: This work was supported by an R01 (CA097213) to SO, a Department of Defense breast cancer fellowship (DAMB-17-01-1-0146) to SMT.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson E, Clarke RB. Steroid receptors and cell cycle in normal mammary epithelium. J Mammary Gland Biol Neoplasia. 2004;9:3–13. doi: 10.1023/B:JOMG.0000023584.01750.16. [DOI] [PubMed] [Google Scholar]
- 2.Matthews J, Gustafsson JA. Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol Interv. 2003;3:281–292. doi: 10.1124/mi.3.5.281. [DOI] [PubMed] [Google Scholar]
- 3.Osborne CK, Schiff R, Fuqua SA, Shou J. Estrogen receptor: current understanding of its activation and modulation. Clin Cancer Res. 2001;7:4338s–4342s. discussion 4411s– 4412s. [PubMed] [Google Scholar]
- 4.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 5.Dobrzycka KM, Townson SM, Jiang S, Oesterreich S. Estrogen receptor corepressors -- a role in human breast cancer? Endocr Relat Cancer. 2003;10:517–536. doi: 10.1677/erc.0.0100517. [DOI] [PubMed] [Google Scholar]
- 6.Edwards DP. The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J Mammary Gland Biol Neoplasia. 2000;5:307–324. doi: 10.1023/a:1009503029176. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Kimbrel EA, Kenan DJ, McDonnell DP. Direct interactions between corepressors and coactivators permit the integration of nuclear receptor-mediated repression and activation. Mol Endocrinol. 2002;16:1482–1491. doi: 10.1210/mend.16.7.0860. [DOI] [PubMed] [Google Scholar]
- 8.Townson SM, Kang K, Lee AV, Oesterreich S. Structure-function analysis of the ER alpha corepressor SAFB1: Identification of a potent transcriptional repression domain. J Biol Chem. 2004;279:26074–26081. doi: 10.1074/jbc.M313726200. [DOI] [PubMed] [Google Scholar]
- 9.Townson SM, Dobrzycka KM, Lee AV, Air M, Deng W, Kang K, Jiang S, Kioka N, Michaelis K, Oesterreich S. SAFB2, a New Scaffold Attachment Factor Homolog and Estrogen Receptor Corepressor. J Biol Chem. 2003;278:20059–20068. doi: 10.1074/jbc.M212988200. [DOI] [PubMed] [Google Scholar]
- 10.Oesterreich S. Scaffold attachment factors SAFB1 and SAFB2: Innocent bystanders or critical players in breast tumorigenesis? J Cell Biochem. 2003;90:653–661. doi: 10.1002/jcb.10685. [DOI] [PubMed] [Google Scholar]
- 11.Nayler O, Stratling W, Bourquin J-P, Stagljar I, Lindemann L, Jasper H, Hartmann A, Fackelmayer F, UA, SS SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998;26 doi: 10.1093/nar/26.15.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang S, Meyer R, Kang K, Kent Osborne C, Wong J, Oesterreich S. Scaffold attachment factor SAFB1 suppresses ER{alpha}-mediated transcription in part via interaction with N-CoR. Mol Endocrinol. 2005 doi: 10.1210/me.2005-0100. [DOI] [PubMed] [Google Scholar]
- 13.Oesterreich S, Zhang QP, Lee AV. Inhibition of oestrogen receptor activity by the co-repressor HET/SAF-B is relieved by blockade of histone deacetylase activity. Eur J Cancer. 2000;36(Suppl 4):S43–44. doi: 10.1016/s0959-8049(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 15.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry J, Mather IH, McDermott MF, Pontarotti P. B30.2-like domain proteins: update and new insights into a rapidly expanding family of proteins. Mol Biol Evol. 1998;15:1696–1705. doi: 10.1093/oxfordjournals.molbev.a025896. [DOI] [PubMed] [Google Scholar]
- 17.Tezel G, Nagasaka T, Iwahashi N, Asai N, Iwashita T, Sakata K, Takahashi M. Different nuclear/cytoplasmic distributions of RET finger protein in different cell types. Pathol Int. 1999;49:881–886. doi: 10.1046/j.1440-1827.1999.00957.x. [DOI] [PubMed] [Google Scholar]
- 18.Tezel G, Nagasaka T, Shimono Y, Takahashi M. Differential expression of RET finger protein in testicular germ cell tumors. Pathol Int. 2002;52:623–627. doi: 10.1046/j.1440-1827.2002.01401.x. [DOI] [PubMed] [Google Scholar]
- 19.Krutzfeldt M, Ellis M, Weekes DB, Bull JJ, Eilers M, Vivanco MD, Sellers WR, Mittnacht S. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol Cell. 2005;18:213–224. doi: 10.1016/j.molcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Cao T, Duprez E, Borden KL, Freemont PS, Etkin LD. Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J Cell Sci. 1998;111:1319–1329. doi: 10.1242/jcs.111.10.1319. [DOI] [PubMed] [Google Scholar]
- 21.Isomura T, Tamiya-Koizumi K, Suzuki M, Yoshida S, Taniguchi M, Matsuyama M, Ishigaki T, Sakuma S, Takahashi M. RFP is a DNA binding protein associated with the nuclear matrix. Nucleic Acids Res. 1992;20:5305–5310. doi: 10.1093/nar/20.20.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimono Y, Murakami H, Hasegawa Y, Takahashi M. RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J Biol Chem. 2000;275:39411–39419. doi: 10.1074/jbc.M006585200. [DOI] [PubMed] [Google Scholar]
- 23.Shimono Y, Murakami H, Kawai K, Wade PA, Shimokata K, Takahashi M. Mi-2 Associates with BRG1 and RET Finger Protein at the Distinct Regions with Transcriptional Activating and Repressing Abilities. J Biol Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- 24.Bloor AJ, Kotsopoulou E, Hayward P, Champion BR, Green AR. RFP represses transcriptional activation by bHLH transcription factors. Oncogene. 2005;24:6729–6736. doi: 10.1038/sj.onc.1208828. [DOI] [PubMed] [Google Scholar]
- 25.Shimono K, Shimono Y, Shimokata K, Ishiguro N, Takahashi M. Microspherule protein 1, Mi-2beta, and ret finger protein associate in the nucleolus and up-regulate ribosomal gene transcription. J Biol Chem. 2005 doi: 10.1074/jbc.M507356200. [DOI] [PubMed] [Google Scholar]
- 26.Patel CA, Ghiselli G. The RET finger protein interacts with the hinge region of SMC3. Biochem Biophys Res Commun. 2005;330:333–340. doi: 10.1016/j.bbrc.2005.02.162. [DOI] [PubMed] [Google Scholar]
- 27.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 28.Burakov D, Crofts LA, Chang CP, Freedman LP. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem. 2002;277:14359–14362. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- 29.Oesterreich S, Lee AV, Sullivan TM, Samuel SK, Davie JR, Fuqua SA. Novel nuclear matrix protein HET binds to and influences activity of the HSP27 promoter in human breast cancer cells. J Cell Biochem. 1997;67:275–286. [PubMed] [Google Scholar]
- 30.Petz LN, Ziegler YS, Loven MA, Nardulli AM. Estrogen receptor alpha and activating protein-1 mediate estrogen responsiveness of the progesterone receptor gene in MCF-7 breast cancer cells. Endocrinol. 2002;143:4583–4591. doi: 10.1210/en.2002-220369. [DOI] [PubMed] [Google Scholar]
- 31.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO-J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata A, Hayashi Y, Imai T, Funahashi H, Nakao A, Seo H. Somatic gene alteration of AIB1 gene in patients with breast cancer. Endocr J. 2001;48:199–204. doi: 10.1507/endocrj.48.199. [DOI] [PubMed] [Google Scholar]
- 33.Wei LN, Hu X, Chandra D, Seto E, Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J Biol Chem. 2000;275:40782–40787. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 34.Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, Brown PH, Fuqua SA, Osborne CK. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem. 2001;276:27907–27912. doi: 10.1074/jbc.M104278200. [DOI] [PubMed] [Google Scholar]
- 35.Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ. Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J Biol Chem. 2001;276:33554–33560. doi: 10.1074/jbc.M105555200. [DOI] [PubMed] [Google Scholar]
- 36.Mahmoudi T, Verrijzer CP. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene. 2001;20:3055–3066. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 38.Peng H, Feldman I, Rauscher FJ., 3rd Hetero-oligomerization among the TIF family of RBCC/TRIM domain-containing nuclear cofactors: a potential mechanism for regulating the switch between coactivation and corepression. J Mol Biol. 2002;320:629–644. doi: 10.1016/S0022-2836(02)00477-1. [DOI] [PubMed] [Google Scholar]
- 39.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 40.Suzumori N, Burns KH, Yan W, Matzuk MM. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc Natl Acad Sci U S A. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cottone E, Orso F, Biglia N, Sismondi P, De Bortoli M. Role of coactivators and corepressors in steroid and nuclear receptor signaling: potential markers of tumor growth and drug sensitivity. Int J Biol Markers. 2001;16:151–166. doi: 10.1177/172460080101600301. [DOI] [PubMed] [Google Scholar]


