Abstract
Molecular imprinting is a technique used to create specific recognition sites on the surface of materials. Although widely developed for chromatographic separation of small molecules, this approach has not been adequately investigated for biomaterial applications. Thus, the objective of these experiments was to explore the potential of molecular imprinting for creating biomaterials that preferentially bind specific proteins. Macroporous polysiloxane (silica) scaffolds were imprinted with either lysozyme or RNase A using sol–gel processing. The quantity of surface-accessible protein, which was related to the number of potential binding sites, was varied by changing the amount of protein loaded into the sol. Up to 62% of loaded protein was accessible. The amount of protein per unit surface area ranged from 0.3 μg m−2 for low loading of RNase to 152 μg m−2 for high loading of lysozyme. Protein-imprinted scaffolds were then evaluated for their ability to preferentially recognize the template biomolecule when incubated in mixtures containing both the imprinted protein and a competitor protein of comparable size (approximately 14 kD). In solutions containing a single protein, up to 3.6 times more template bound compared with the competitor. Furthermore, in solutions containing equal amounts of both molecules, the porous scaffolds bound up to three times more template than the competitor protein, which is a level of preferential binding similar to values reported in the molecular imprinting literature for both organic and inorganic materials.
Keywords: Molecular imprinting, Silica, Scaffold, Sol–gel techniques, Protein adsorption
1. Introduction
A variety of metals, ceramics, polymers and composites are used as biomaterials. It has been estimated more than 5% of the U.S. population have implants [1]. Although these devices have benefited millions of people, the fact remains that most of the “biomaterials” were not originally intended to be placed in the human body. Consequently, biological reactions to these substrates tend to be characterized by nonspecificity, sluggish kinetics, a broad spectrum of active processes simultaneously occurring and an opportunistic, rather than designed, outcome [2]. Hundreds of biomolecules can adsorb from tissues and body fluids onto the biomaterial surfaces, and the molecules can exist in several conformational and orientational states [3]. Subsequent cellular and tissue responses will depend on which particular molecules adsorb and their states.
The design of materials that promote biomolecular recognition may be able to stimulate adhesion, growth, differentiation, and activity of particular cell types. A common approach, as reviewed by Shin et al. [4], is surface or bulk modification of biomaterials to attach short peptides, fragments or whole chains of extracellular matrix molecules. Use of large molecules, however, poses problems with long-term shelf-stability of the immobilized polypeptides and can be expensive. On the other hand, use of small peptides leads to difficulties with selectively binding or stimulating only one type of cell.
Molecular imprinting is a technique for preparing substrates to selectively bind particular biomolecules. In the more common noncovalent approach, functional monomers assemble with the template biomolecule in solution. Subsequent crosslinking of monomers “locks in” the shape and chemical functionality of the template. Removal of the template leaves nanocavities of specific shape and with a defined arrangement of functional groups. Analogous to the way an antibody recognizes an antigen, the combination of geometry and chemistry of the cavities can make them selective for the template molecule. Most molecular imprinting studies have used methacrylic polymers imprinted with low molecular weight compounds, such as sugars, steroids and pesticides (e.g. Refs. [5–10]). Comparatively few reports describe imprinting inorganic materials [11–16].
The aims of this study were to begin development and characterization of protein-imprinted biomaterials to enable preferential binding of particular biomolecules.
2. Materials and methods
2.1. Fabrication of the polysiloxane scaffolds
Polysiloxane scaffolds were fabricated by a two-step sol–gel procedure. Tetraethoxysilane (TEOS; Fluka, Milwaukee, WI) was used as a crosslinker, γ-aminopropyltriethoxysilane (APS; Sigma, St. Louis, MO) as a functional monomer, HCl as a catalyst, ethanol as a co-solvent and sodium docecylsulfate (SDS; Sigma) as a foaming agent to generate macroporosity for cell ingrowth. In the first step, a solution was mixed to include 1.32 ml of TEOS, 0.235 ml of deionized water, 0.33 ml of 0.1 M HCl and 0.4 ml of absolute ethanol in a cylindrical plastic vial. After prehydrolysis of TEOS at room temperature for 24 h, this solution was mixed with a solution containing 0.33 ml of APS and 1 ml of 0.1 M SDS. Vials were vortexed to mix the silane components and to foam the sol.
After gelation, the vials were capped, and the gels were aged at room temperature for 24 h and then dried at 40 °C for 48 h. To remove nonuniform glassy layers, the top and bottom surfaces of the scaffolds were ground using 600 grit silicon carbide paper on an ECOMET 3 grinder-polisher (Buehler). After grinding, all samples were thoroughly washed with phosphate-buffered saline (PBS), pH 7.4, to remove debris, and the scaffolds were stored in the dark until used.
2.2. Protein loading
The model template/competitor molecules lysozyme (chicken egg white; Sigma) and RNase A (Sigma) were fluorescently labeled with Alexa Fluor 350 (Molecular Probes; Eugene, OR) according to the manufacturer’s protocol before imprinting. Specific fluorescence of the labeled proteins was determined through construction of standard curves relating fluorescence (Spectra MAX Gemini XS, Sunnyvale, CA; λex = 346 nm and λem = 442 nm) to protein concentration as quantified using the BCA protein assay (Pierce, Rockford, IL). The labeled template molecules were incorporated in the scaffolds by addition to the APS-containing solution prior to mixing with the TEOS solution. To determine the amount of protein available on the surface of scaffolds after loading, samples were gently shaken in a solution of 0.4 mg protease (Pronase E; Sigma) per ml of 0.1 M carbonate–bicarbonate buffer, pH 8.5, for up to 24 h. The amount of protein released from the scaffold surfaces was quantified by measuring fluorescence at 3 and 24 h.
2.3. Characterization of scaffolds
Blank scaffolds and scaffolds loaded with 1 or 10 mg of lysozyme were examined using scanning electron microscopy (SEM). Samples were sputter-coated with gold in argon gas using an Emscope sc 400 before visualization with a Hitachi S-3200 (Tokyo, Japan) at an accelerating voltage of 20 kV.
The surface area of scaffolds was measured by nitrogen gas adsorption. Scaffolds (blanks and imprinted with 0.1, 1 and 6 mg of lysozyme or RNase A) were placed in an oven at 40 °C for 3 days and then kept under vacuum for 24 h. The weight of each scaffold was measured after desiccation in a sample tube at 120 °C for 24 h. The surface area was then measured with a TriStar 3000 (Micromeritics, Norcross, GA) instrument and calculated using the BET (Brunauer–Emmett–Teller) method.
Porosity of scaffolds was measured with an Autopore IV 9500 mercury intrusion porosimeter (Micromeritics). Blank scaffolds and scaffolds imprinted with 0.05 mg of lysozyme or RNase A were completely dried at 40 °C for 3 days before testing. Three samples were placed in a 5cc penetrometer. After testing, total intrusion volume, average pore diameter, density and porosity were calculated.
2.4. Preferential protein binding
In order to examine the ability of imprinted scaffolds to preferentially bind their template proteins, scaffolds were immersed in single or double protein solutions following digestion of the attached biomolecules. The proteins in the solutions were labeled with fluorophores that have different spectra compared with the labels used for protein loading. Lysozyme was labeled with Alexa Fluor 488 (Molecular Probes; λex = 495 nm and λem = 519 nm), and RNase A was labeled with Alexa Fluor 594 (Molecular Probes; λex = 590 nm and λem = 617 nm). For the lysozyme-imprinted scaffolds, lysozyme was used as template protein and RNase A was used as competitor protein in the rebinding solution. The total amount of protein in the rebinding solutions was kept constant at the maximum amount of protein released from each type of sample. For example, if 100 μg of template protein was digested from the scaffold, the total amount of template plus competitor in the rebinding solutions was 100 μg. The ratios of template to competitor protein were 1:0, 1:1 and 0:1. In order to check the selectivity of scaffolds for another protein, RNase A-imprinted scaffolds were exposed to rebinding solutions containing template protein (RNase A) and competitor protein (lysozyme). The template to competitor protein ratios tested were again 1:0, 1:1 and 0:1. After immersion in the rebinding solutions for 24 h on an orbital shaker, the solutions were aspirated and scaffolds were rinsed three times with PBS, pH 7.4. As in the protein loading experiments, surface-bound protein was digested with 0.4 mg protease per ml of 0.1 M carbonate–bicarbonate buffer, pH 8.5. The amount of protein released from scaffolds into the protease solution was determined after 24 h by measuring fluorescence of the two labels.
2.5. Statistical analysis
One-way analysis of variance was conducted using the computer application InStat (Graphpad Software, San Diego, CA). Post-hoc comparisons were made using the Tukey–Kramer test when the p-value was significant (p<0.05). A minimum of three replicates was used for each experiment, and experiments were repeated at least once.
3. Results
3.1. Fabrication and characterization of scaffolds
The finished polysiloxane scaffolds were white in color and had a cylindrical geometry of 4.3 mm in height and 9 mm in diameter. The scaffolds were highly textured and included micro-, meso- and macropores (Fig. 1). Fig. 2(a) is a higher magnification image of an imprinted scaffold, with noticeable protein embedded on the surface, and Fig. 2(b) shows that submicron-sized pores were revealed on the surface after the scaffold was exposed to protease solution to digest the surface-accessible protein. Protease did not affect the blank silica materials (data not shown).
Fig. 1.
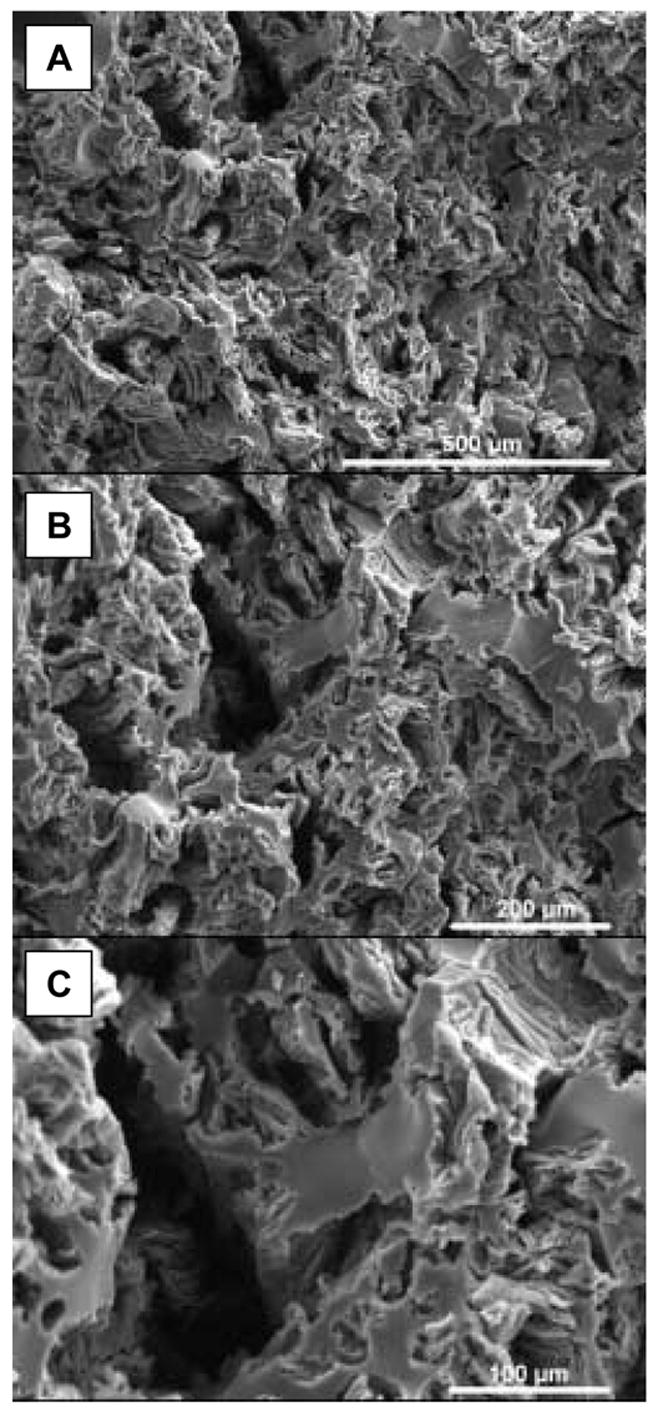
SEM images showing morphology of imprinted polysiloxane scaffolds.
Fig. 2.
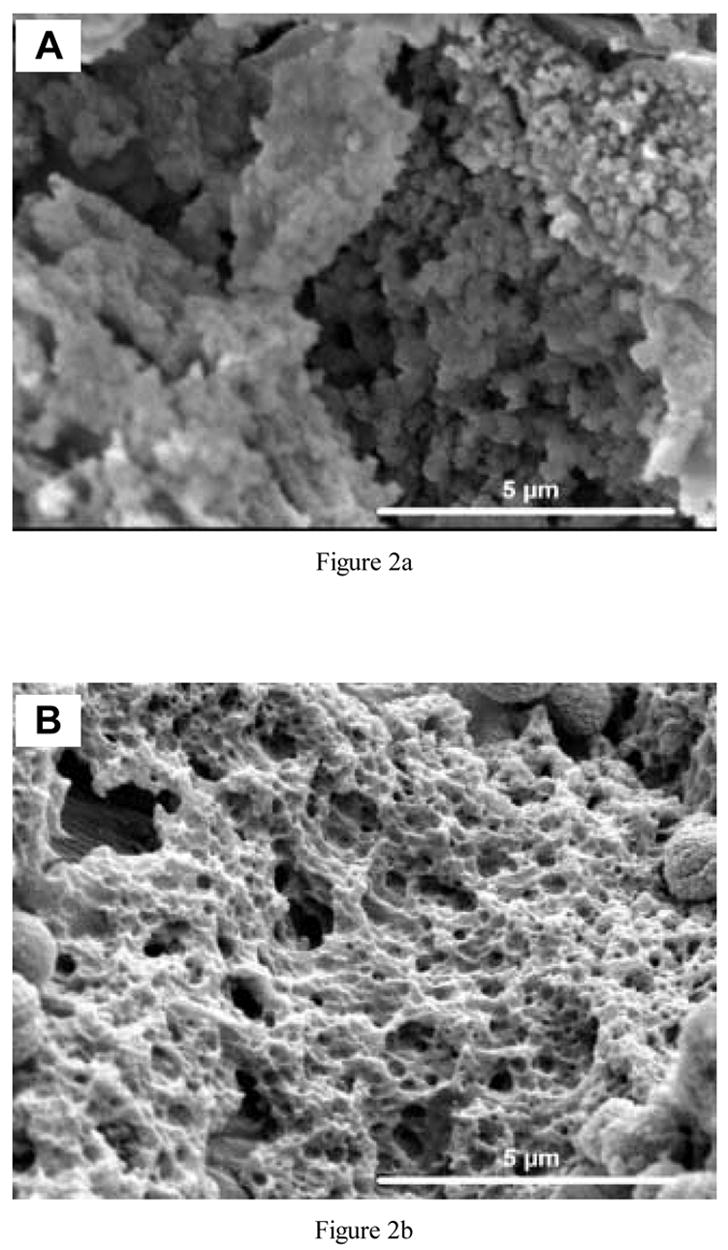
Higher magnification SEM images of protein (lysozyme)-imprinted scaffolds (a) before and (b) after exposure to protease to digest surface-accessible protein.
As shown in Table 1, the average total pore volume for both the blank and protein-imprinted scaffolds was approximately 40%. The average pore size was 6.19 μm for blank scaffolds, 5.56 μm for 0.05 mg RNase A-imprinted scaffolds and 5.46 μm for 0.05 mg lysozyme-imprinted scaffolds. There were no statistically significant differences of porosity or pore size for the different types of samples fabricated.
Table 1.
Total porosity and pore size of scaffolds
| Scaffolds | Blank (non-imprinted) | 0.05 mg RNase A-imprinted | 0.05 mg lysozyme-imprinted |
|---|---|---|---|
| Porosity (%) | 43±1.18 | 42±3.06 | 41±2.73 |
| Average pore size (μm) | 6.19±0.58 | 5.56±1.70 | 5.46±0.70 |
Table 2 shows the average surface area for the blank and imprinted scaffolds. There were no clear trends, and all samples had statistically similar surface areas. In contrast to some other porous scaffolds, the polysiloxane samples sustained the high (150 °C) temperature needed for degassing without damage.
Table 2.
Surface area of scaffolds
| Protein added (mg) | Imprinted scaffolds | Non-imprinted scaffolds | |
|---|---|---|---|
| Lysozyme (m2 g−1) | RNase A (m2 g−1) | Blank (m2 g−1) | |
| 0.1 | 182±18 | 191±20 | 202±19 |
| 1 | 129±48 | 174±31 | |
| 6 | 186±12 | 212±2 | |
3.2. Protein loading
The amount of protein released from the scaffolds’ surface by digestion with protease depended on both the amount of protein added to the sol before gelation and on the time of digestion. For both template proteins, the amount released increased with the quantity loaded into the scaffolds. For lysozyme-loaded scaffolds (Fig. 3), there was no statistically significant difference between the amounts at 3 and 24 h when less than 1 mg of protein was added to the scaffolds. However, when 1 mg or more was loaded, the amount of lysozyme released after 24 h was approximately five times greater than that at 3 h (p<0.001). At the highest loading, approximately 62% of lysozyme added to the sol was released from the materials, and even greater percentages were released at lower loadings. For RNase A-imprinted scaffolds (Fig. 4), a much smaller amount of protein was released from the surface, with up to only 11% of the loaded protein released after 24 h of digestion. Comparing the results for lysozyme- and RNase A-imprinted scaffolds, the quantity of lysozyme released was approximately 10 times more than that of RNase A after 24 h. Table 3 shows the average amount of protein released from each type of scaffold normalized by surface area. The amount of protein per unit surface area for 1 mg lysozyme-imprinted scaffolds (55 μg m−2) was almost six times greater than that for 0.1 mg lysozyme-imprinted scaffolds (8.8 μg m−2). Similarly, 1 mg RNase-imprinted scaffolds had about 10 times greater surface density of protein compared with those loaded with 0.1 mg RNase (3.5 vs. 0.32 μg m−2, respectively).
Fig. 3.
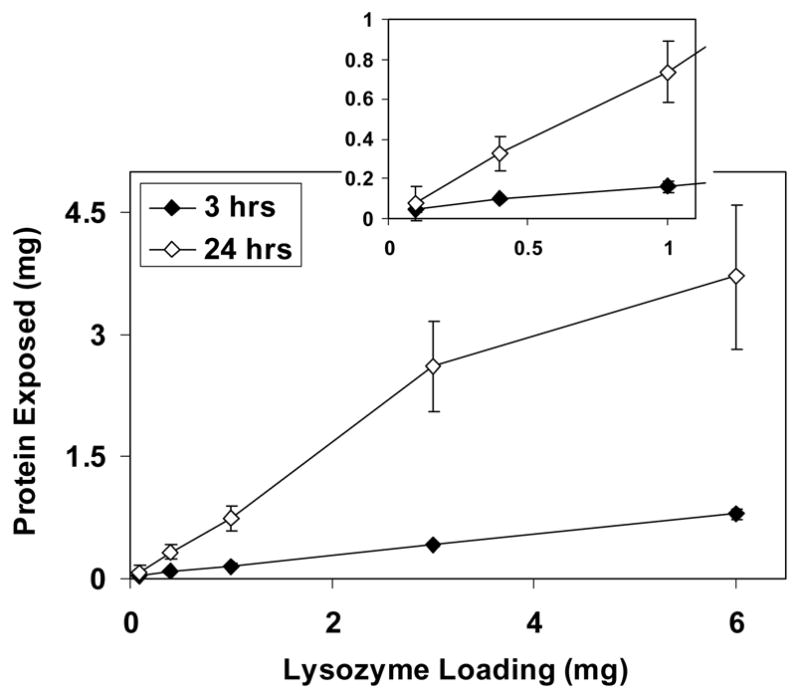
Amount of lysozyme released from the surface of scaffolds after 3 and 24 h of digestion. The inset is an expanded view of the low loading region.
Fig. 4.
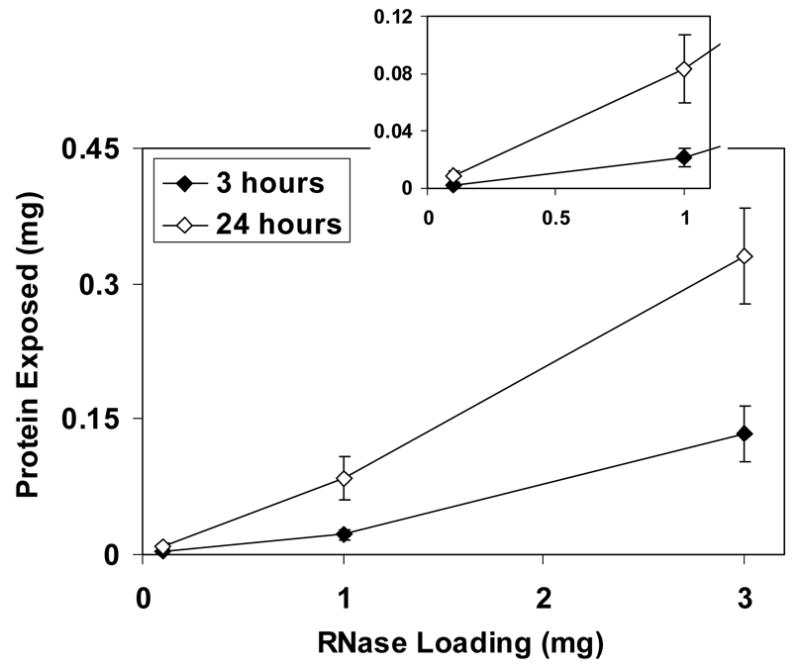
Amount of RNase A released from the surface of scaffolds after 3 and 24 h of digestion. The inset is an expanded view of low loading region.
Table 3.
Average amount of surface-accessible protein per unit surface area
| Protein added (mg) | Imprinted scaffolds | |
|---|---|---|
| Lysozyme (μg m−2) | RNase A (μg m−2) | |
| 0.1 | 8.85±3.5 | 0.319±0.15 |
| 1 | 55.1±8.7 | 3.53±1.0 |
| 6 | 152±35 | – |
3.3. Preferential protein binding
Blank (nonimprinted) scaffolds did not discriminate between lysozyme and RNase, and bound similar amounts of the different proteins (data not shown). Differences were observed, however, for imprinted materials. In a solution containing only lysozyme (1:0), approximately 45% of the available protein re-bound to the surface of scaffolds imprinted with 0.1 mg lysozyme (Fig. 5(a)). In a solution of just RNase A (0:1), the amount of protein re-bound was approximately 27%. Thus, almost twice as much template protein than competitor was re-bound to the surface (p<0.05). When equal amounts of the proteins (1:1) were present in the rebinding solution, about 2.5 times more lysozyme than RNase A was present on the surface (p<0.03).
Fig. 5.
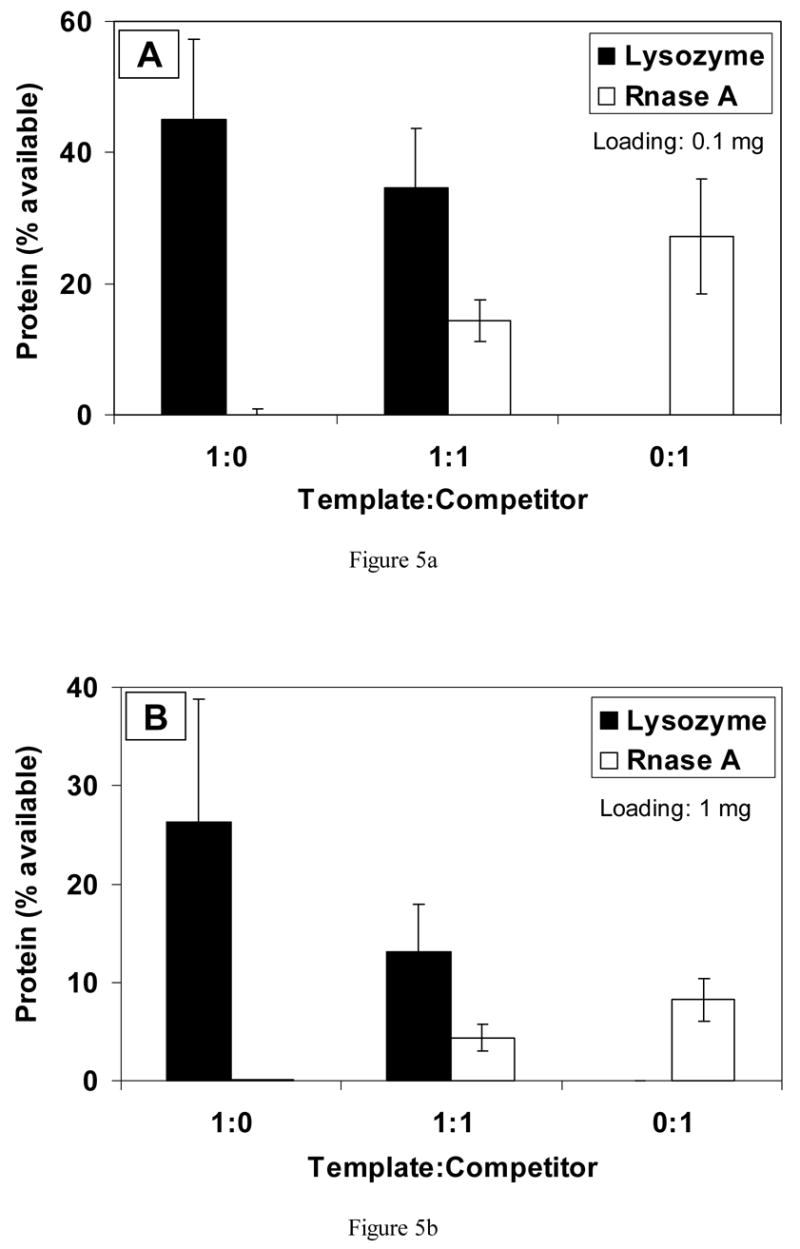
Competitive binding of protein to scaffolds imprinted with (a) 0.1 mg, (b) 1 mg and (c) 3 mg of lysozyme.
Fig. 5(b) shows results for scaffolds imprinted with 1 mg of lysozyme. In single protein solutions of just the template or competitor protein, nearly three times more template (lysozyme) protein bound to the surface compared with competitor protein (p<0.001). In 1:1 solutions of lysozyme and RNase A, approximately 2.6 times more template protein bound (p<0.03).
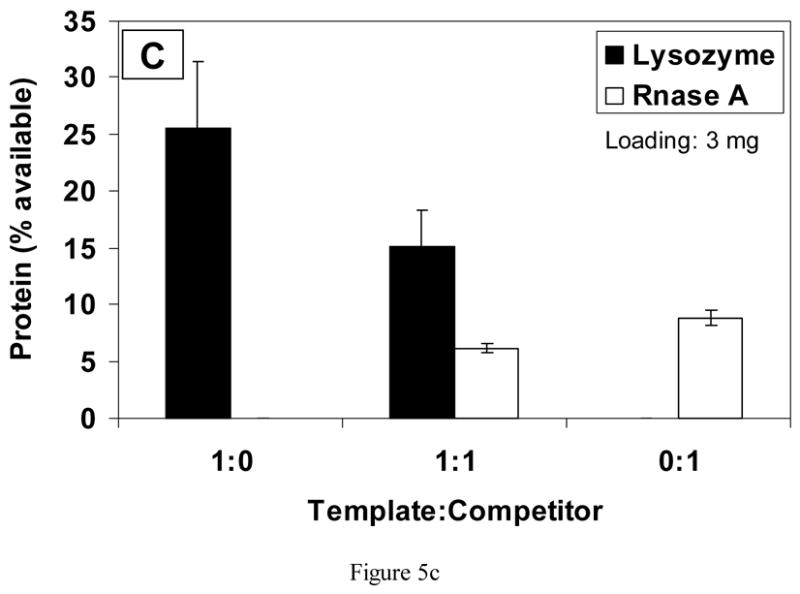
On scaffolds loaded with a higher amount of lysozyme (3 mg), approximately 26% of available lysozyme bound from the 1:0 solution, but only 12% of RNase bound from the 0:1 solution (Fig. 5(c)). In the 1:1 rebinding solution, almost three times more lysozyme (template) than RNase A (competitor) bound to the scaffolds (p<0.01).
RNase A was also used as a template protein, with lysozyme serving as a competitor. With a loading of 0.05 mg per scaffold, approximately 40% of available RNase was re-bound from the 1:0 solution and 11% of lysozyme was bound from the 0:1 solution (Fig. 6(a)). Almost four times more template adsorbed compared with the competitor (p<0.001). When equal amounts of the two proteins were present (1:1), approximately two times more template protein re-bound, but the difference was not statistically significant.
Fig. 6.
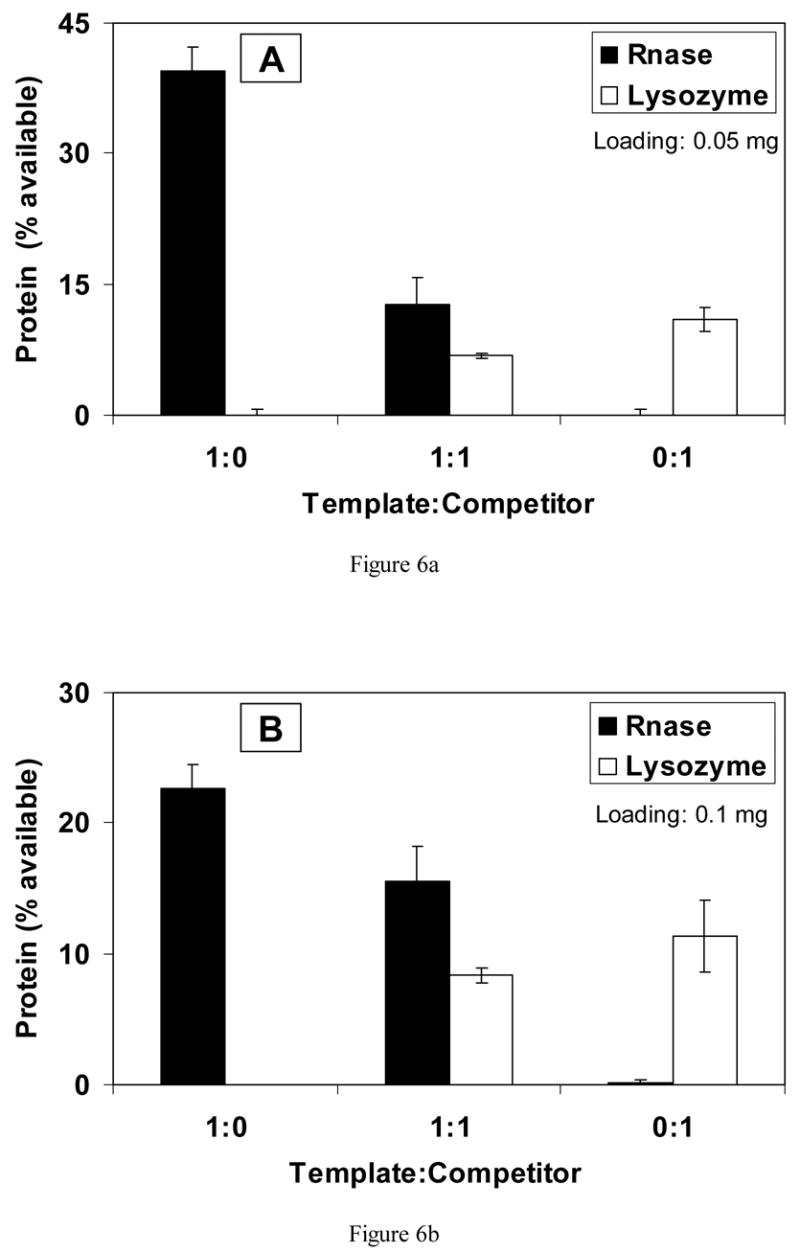
Competitive binding of protein to scaffolds imprinted with (a) 0.05 mg and (b) 0.1 mg of RNase A.
In Fig. 6(b), results are shown for scaffolds imprinted with 0.1 mg RNase A. In solutions containing only template, approximately 23% of available protein bound, and in solutions of only competitor, about 11% of protein re-bound (p<0.001). When rebinding solutions contained both proteins at the ratio of 1:1, the percentage of RNase A that re-bound was almost 16%, with only approximately 8% of lysozyme binding (p<0.02). Therefore, the scaffolds bound two times more template (RNase A) than competitor (lysozyme).
4. Discussion
Molecular imprinting techniques have been widely investigated for chromatography, which seeks to separate or analyze complex mixtures. Methacrylic polymers are commonly imprinted in bulk form, after which they are pulverized and loaded into columns. A goal of such studies is to create “artificial antibodies”, polymers that have affinity and specificity approaching those of biomolecules used in immunoseparations (reviewed in Ref. [17]). If successful, these materials would offer significant advantages in durability and stability compared with natural antibody methods. Molecular imprinting has yet to be used for influencing cell behavior. As a long-term goal, scaffolds imprinted with specific biomolecules may lead to desired cellular responses.
Polysiloxane (silica; SiO2) materials have been used to encapsulate biomolecules, such as cytochrome c, myoglobin and hemoglobin, in porous, glassy matrices [18–20]. Even when entrapped, the biomolecules can detect and react with substrate molecules by diffusion [19,21]. Compared with organic materials, silica is more robust, in terms of chemical, mechanical and thermal stability. In the present work focusing on molecular imprinting, a similar initial approach was used for depositing template protein in the polysiloxane. This resulted in protein that was embedded within the material as well as exposed on surfaces of the pores. Removal of surface-accessible template protein leaves binding sites that may provide molecular recognition.
The scaffolds were formed by sol–gel processing. This process was used because of its ease of use and versatility. TEOS is readily hydrolyzed and condensed into silica gel, and therefore was employed in this study. In these first steps toward developing protein-imprinted scaffolds, APS, with its terminal amino group, was used as a functional monomer to associate with the template biomolecules. Experiments to determine the mechanism(s) of interaction between protein and substrate are ongoing. Silanes with other functional groups are commercially available and may be used in future research to refine selectivity of the scaffolds.
Effects of polycondensation on conformation of the proteins used as template molecules remain unknown. Acid catalyst, ethanol solvent and other sol–gel parameters can disturb protein structure. Similarly, solvents, initiators, etc. in methacrylate-based polymer systems can affect protein conformation. In future studies, analytical techniques such as circular dichroism may be useful for understanding protein structure during the templating process. Nonetheless, as shown in the results and discussed later, undenatured enzyme molecules in solution were able to bind to imprinted scaffolds in a preferential manner.
Lysozyme was used as a model template biomolecule before imprinting with more relevant proteins because of the large number of formulations tested. In addition, the structure (PDB code 1AZF) and molecular weight (14.3 kD) of lysozyme are well documented [22]. RNase A (PDB code 1AFU) was chosen as a model competitor protein because its molecular weight (13.7 kD) is comparable to that of lysozyme. Thus, the two proteins have similar overall mass but differ in chemistry and geometry. These differences are useful for determining selectivity of the imprinted scaffolds for their template molecules.
When protein was added to the sol for fabricating molecularly imprinted polysiloxane scaffolds, loading was varied from 0 to 10 mg of protein per sample. However, protein can alter properties of scaffolds because of interaction between biomolecules and the silane mixture, especially APS, affecting the condensation reactions [23]. During preliminary experiments to identify useful compositions of the sol, gelation time of the mixture was found to be influenced by the amount of protein added (data not shown). Additionally, scaffold properties were also affected by drying conditions. The pores collapsed and the connected network of polysiloxane was disrupted because of excessive stresses incurred at high drying temperatures [18]. Another consideration for drying is the effect on protein structure. Pooart et al. investigated thermal stability of lytic activity of rhea and ostrich lysozyme. The enzymes were stable up to 40 °C, but only 20% activity remained after treatment at high temperature (80 °C) [24]. Therefore, the drying temperature was fixed at 40 °C in the present work.
In order to expose the maximum number of imprinted binding sites, scaffolds were treated with protease to digest imprinted biomolecules rather than simply washed with water. Because they are stabilized by siloxane bonds, the scaffolds are not affected by protease treatment. If the template could be removed by washing with water, the binding strength between protein and the material is weak, and protein retention on the surface would be low [25]. Other strong washing solutions reported for removing template molecules include acetone, acetic acid [16], acetonitrile [26] and ammonia [27]. The submicron-sized features observed on the scaffolds after treatment with protease may indicate the imprinted binding sites on the surface in which the template protein can rebind. Thus, the quantity of protein available on the surface of scaffolds is important.
Protein loading experiments showed that as the quantity added to the sol increased, an increasing amount was exposed at the surface. However, at the highest loading, only 60% of the lysozyme loaded was released from scaffolds and only 11% of the added RNase was released. The difference in digestion of the two proteins may be related to their different chemical properties and consequent effects on embedding vs. imprinting of the biomolecules. The pI of lysozyme is about 11.4 [28], whereas it is 9.6 for RNase A [29]. This may have contributed to differential electrostatic interactions between protein and polysiloxane, which contained amino groups. Overall, scaffolds contained embedded as well as surface-imprinted protein, and such effects will need to be accounted for in future studies with more relevant proteins.
Porosity of scaffolds is also important. Generally, large pores (tens to hundreds of micrometers) are needed for cell and tissue ingrowth, micron-sized pores may contribute to cell adhesion, and very small pores (nm) may play a role in protein binding [30]. The polysiloxane scaffolds formed in the present studies were about 40% porous and contained a wide distribution of pore sizes, ranging from the nanometer scale up to about 200 μm. The blank and protein-imprinted materials had statistically similar porosity and average pore size. In other work, composition of the sol prior to gelation (e.g. catalyst, TEOS:APS and surfactant) was found to have a more dramatic effect on scaffold properties (manuscript in preparation).
Lysozyme or RNase A-imprinted scaffolds were tested in rebinding solutions that contained different ratios of template to competitor proteins. Blank (nonimprinted) scaffolds adsorbed both lysozyme and RNase in comparable quantities. The objective of imprinting was to enhance binding of a particular protein. Table 4 summarizes the preferential binding ratios of template to competitor for the scaffolds developed thus far. As would be expected for most inorganic substrates, all scaffolds demonstrated some level of nonspecific protein adsorption, i.e. RNase A bound to lysozyme-imprinted materials and vice versa. Grafting of nonadhesive moieties is possible in future work; however, there were no attempts at this stage to make the surfaces nonfouling. Still up to nearly four times as much of the template adsorbed from single protein solutions. In samples loaded with larger amounts of protein, it is possible for imprinted protein to be distributed heterogeneously through the material, with aggregates of protein exposed at the surface. Rebinding results, though, showed binding of more template than competitor on highly loaded scaffolds. Most importantly, when equal amounts of template and competitor were present, scaffolds preferentially bound two to three times more template compared with competitor. This was found for both the lysozyme- and RNase-imprinted materials, with high loading scaffolds exhibiting a little more apparent preference for their template than did the low loading scaffolds. Incorporation of additional functional groups into polysiloxane scaffolds may enhance their preference for the template biomolecules.
Table 4.
Ratios of rebound templates to competitor for imprinted scaffolds
| Protein added (mg) | Lysozyme-imprinted | Protein added (mg) | RNase A-imprinted | ||
|---|---|---|---|---|---|
| Template/competitor (lysozyme)/(RNase A) | Template/competitor (RNase A)/(lysozyme) | ||||
| 1:0/0:1 | 1:1 | 1:0/0:1 | 1:1 | ||
| 0.1 | 1.66 | 2.4 | 0.05 | 3.6 | 1.87 |
| 1 | 2.9 | 2.44 | 0.1 | 2 | 1.86 |
| 3 | 3.18 | 3.01 | |||
Methacrylate-based polymers imprinted with small organic molecules have most commonly been explored for molecular imprinting. For example, Gore et al. [31] showed 1.4–3.1 times selectivity for cholesterol compared with other steroids. Oral and Peppas [32] described substantial nonspecific binding of glucose to hydrophilic polymers; however, up to 3.5 times more glucose than galactose bound to glucose-imprinted materials. Recently, imprinted organic surface coatings were shown to discriminate between the proteins trypsin and lysozyme in a concentration-sensitive manner [33]. Cummins et al. [16] compared the selectivity of acrylic and sol–gel polymers, both of which were imprinted with 2-aminopyridine. Three different solvents, chloroform, acetonitrile and methanol, were used for the rebinding experiments. Although there was a high degree of nonspecific binding, the sol–gel materials re-bound more template than did acrylic polymers, especially in a more polar environment. Furthermore, silica showed higher selectivity for the template, with twice as much 2-aminopyridine retained compared with an isomer of the molecule. The present results of two to three times enhancement in preferential binding on porous polysiloxane scaffolds under competitive conditions compare well with these aforementioned findings reported in the molecular imprinting literature.
Using a completely different method involving plasma polymerization, Shi et al. investigated selectivity of surfaces imprinted with lysozyme and RNase A, the same proteins as in the current study [25,34]. Selectivity was assessed by calculating an R50 value, defined as the competitor to template ratio at which imprinted protein adsorption decreased to 50% of its non-competing value. Lysozyme-imprinted surfaces exhibited a 26-fold enhanced selectivity, while selectivity on RNase-imprinted surfaces was increased 20-fold. Although preferential binding was not as high in the present work, the methodology for preparing the three-dimensionally porous scaffolds is simple and flexible, and does not require specialized equipment.
5. Conclusion
Porous protein-imprinted polysiloxane (silica) scaffolds were developed. The density of binding sites can be readily altered by changing protein loading. The imprinted scaffolds exhibited up to three times preference for binding their template molecules, even in the presence of a similar-sized competitor. The level of preferential binding was similar to values reported in the molecular imprinting literature for interaction of simple biomolecules with both organic an inorganic substrates. Ongoing research is directed at enhancing selectivity and at imprinting molecules relevant to cell interactions with biomaterials.
Acknowledgments
This work was supported by NIH/NIBIB (EB02958).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moss AJ, Hamburger S, Moore RM, Jr, Jeng LL, Howie LJ. Use of selected medical device implants in the United States, 1988. Hyattsville, MD: National Center for Health Statistics; 1991. [PubMed] [Google Scholar]
- 2.Ratner BD. New ideas in biomaterials science--a path to engineered biomaterials. J Biomed Mater Res. 1993;27:837–850. doi: 10.1002/jbm.820270702. [DOI] [PubMed] [Google Scholar]
- 3.Horbett TA, Brash JL. Proteins at interfaces: current issues and future prospects. In: Brash JL, Horbett TA, editors. Proteins at Interfaces: Physiochemical and Biochemical Studies. Washington, D.C: American Chemical Society; 1987. pp. 1–33. [Google Scholar]
- 4.Shin H, Jo S, Mikos AG. Biomimetic materials for tissue engineering. Biomaterials. 2003;24:4353–64. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 5.Sellergren B. Noncovalent molecular imprinting: antibody-like molecular recognition in polymeric network materials. Trends Anal Chem. 1997;16:310–320. [Google Scholar]
- 6.Buchmeiser MR. New synthetic ways for the preparation of high-performance liquid chromatography supports. J Chromatogr A. 2001;918:233–66. doi: 10.1016/s0021-9673(00)00129-1. [DOI] [PubMed] [Google Scholar]
- 7.Kugimiya A, Kuwada Y, Takeuchi T. Preparation of sterol-imprinted polymers with the use of 2-(methacryloyloxy)ethyl phosphate. J Chromatogr A. 2001;938:131–5. doi: 10.1016/s0021-9673(01)01384-x. [DOI] [PubMed] [Google Scholar]
- 8.Batra D, Shea KJ. Combinatorial methods in molecular imprinting. Curr Opin Chem Biol. 2003;7:434–42. doi: 10.1016/s1367-5931(03)00060-7. [DOI] [PubMed] [Google Scholar]
- 9.Ikegami T, Lee WS, Nariai H, Takeuchi T. Synthetic polymers adsorbing bisphenol A and its analogues prepared by covalent molecular imprinting using bisphenol A dimethacrylate as a template molecule. Anal Bioanal Chem. 2004;378:1898–902. doi: 10.1007/s00216-004-2490-8. [DOI] [PubMed] [Google Scholar]
- 10.Ye L, Haupt K. Molecularly imprinted polymers as antibody and receptor mimics for assays, sensors and drug discovery. Anal Bioanal Chem. 2004;378:1887–97. doi: 10.1007/s00216-003-2450-8. [DOI] [PubMed] [Google Scholar]
- 11.Glad M, Norrlow O, Sellergren B, Siegbahn N, Mosbach K. Use of silane monomers for molecular imprinting and enzyme entrapment in polysiloxane-coated porous silica. J Chromatogr. 1985;347:11–23. [Google Scholar]
- 12.Wulff G. Molecular imprinting in cross-linked materials with the aid of molecular templates -- a way towards artificial antibodies. Angew Chem Int Ed Engl. 1995;34:1812–1832. [Google Scholar]
- 13.Hunnius M, Rufiska A, Maier WF. Selective surface adsorption versus imprinting in amorphous microporous silicas. Micropor Mesopor Mater. 1999;29:389–403. [Google Scholar]
- 14.Katz A, Davis ME. Molecular imprinting of bulk, microporous silica. Nature. 2000;403:286–9. doi: 10.1038/35002032. [DOI] [PubMed] [Google Scholar]
- 15.Dai S. Hierarchically imprinted sorbents. Chemistry. 2001;7:763–8. doi: 10.1002/1521-3765(20010216)7:4<763::aid-chem763>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 16.Cummins W, Duggan P, McLoughlin P. A comparative study of the potential of acrylic and sol–gel polymers for molecular imprinting. Anal Chim Acta. 2005;542:52–60. [Google Scholar]
- 17.Bruggemann O. Molecularly imprinted materials--receptors more durable than nature can provide. Adv Biochem Eng Biotechnol. 2002;76:127–63. doi: 10.1007/3-540-45345-8_4. [DOI] [PubMed] [Google Scholar]
- 18.Dave BC, Miller JM, Dunn B, Valentine JS, Zink JI. Encapsulation of proteins in bulk and thin film sol–gel matrices. J Sol-Gel Sci Technol. 1997;8:629–634. [Google Scholar]
- 19.Liu DM, Chen I. Encapsulation of protein molecules in transparent porous silica matrices via an aqueous colloidal sol–gel process. Acta Mater. 1999;47:4535–4544. [Google Scholar]
- 20.Gill I, Ballesteros A. Bioencapsulation within synthetic polymers (Part 1): sol–gel encapsulated biologicals. TIBTECH. 2000;18:282–296. doi: 10.1016/s0167-7799(00)01457-8. [DOI] [PubMed] [Google Scholar]
- 21.Jin W, Brennan JD. Properties and applications of proteins encapsulated within sol–gel derived materials. Anal Chim Acta. 2002;461:1–36. [Google Scholar]
- 22.Pembroke JT. Bio-molecular modeling utilizing RasMol and PDB resources: a tutorial with HEW lysozyme. Biochem Molec Biol Educ. 2000;28:297–300. [Google Scholar]
- 23.Venton DL, Gudipati E. Influence of protein on polysiloxane polymer formation: evidence for induction of complementary protein–polymer interactions. Biochim Biophys Acta. 1995;1250:126–136. doi: 10.1016/0167-4838(95)00080-e. [DOI] [PubMed] [Google Scholar]
- 24.Pooart J, Torikata T, Araki T. Enzymatic properties of rhea lysozyme. Biosci Biotechnol Biochem. 2005;69:103–12. doi: 10.1271/bbb.69.103. [DOI] [PubMed] [Google Scholar]
- 25.Shi H, Ratner BD. Template recognition of protein-imprinted polymer surfaces. J Biomed Mater Res. 2000;49:1–11. doi: 10.1002/(sici)1097-4636(200001)49:1<1::aid-jbm1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Rachkov A, Minoura N. Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim Biophys Acta. 2001;1544:255–266. doi: 10.1016/s0167-4838(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 27.Kunitake T, Lee S. Molecular imprinting in ultrathin titania gel films via surface sol–gel process. Anal Chim Acta. 2004;504:1–6. [Google Scholar]
- 28.Wetter LR, Deutsch HF. Immunological studies on egg white proteins. IV. Immunochemical and physical studies of lysozyme. J Biol Chem. 1951;192:237–42. [PubMed] [Google Scholar]
- 29.Tanford C, Hauenstein JD. Identification of the chemical difference between chromatographic components of ribonuclease. Biochim Biophys Acta. 1956;19:535–9. doi: 10.1016/0006-3002(56)90477-2. [DOI] [PubMed] [Google Scholar]
- 30.Kim BS, Mooney DJ. Development of biocompatible synthetic extracellular matrices for tissue engineering. Trends Biotechnol. 1998;16:224–30. doi: 10.1016/s0167-7799(98)01191-3. [DOI] [PubMed] [Google Scholar]
- 31.Gore MA, Karmalkar RN, Kulkarni MG. Enhanced capacities and selectivities for cholesterol in aqueous media by molecular imprinting: role of novel cross-linkers. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;804:211–21. doi: 10.1016/j.jchromb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 32.Oral E, Peppas NA. Hydrophilic molecularly imprinted poly(hydroxyethyl-methacrylate) polymers. J Biomed Mater Res A. 2006;78:205–10. doi: 10.1002/jbm.a.30725. [DOI] [PubMed] [Google Scholar]
- 33.Hayden O, Haderspock C, Krassnig S, Chen X, Dickert FL. Surface imprinting strategies for the detection of trypsin. Analyst. 2006;131:1044–50. doi: 10.1039/b608354b. [DOI] [PubMed] [Google Scholar]
- 34.Shi H, Tsai WB, Garrison MD, Ferrari S, Ratner BD. Template-imprinted nanostructured surfaces for protein recognition. Nature. 1999;398:593–597. doi: 10.1038/19267. [DOI] [PubMed] [Google Scholar]


