Abstract
Treatment of wild type vaccinia virus infected cells with the anti-poxviral drug isatin-β-thiosemicarbazone (IBT) induces the viral postreplicative transcription apparatus to synthesize longer-than-normal mRNAs through an unknown mechanism. Previous studies have shown that virus mutants resistant to or dependent on IBT affect genes involved in control of viral postreplicative transcription elongation. This study was initiated in order to identify additional viral genes involved in control of vaccinia postreplicative transcription elongation. Eight independent, spontaneous IBT resistant mutants of vaccinia virus were isolated. Marker rescue experiments mapped two mutants to gene G2R, which encodes a previously characterized postreplicative gene positive transcription elongation factor. Three mutants mapped to the largest subunit of the viral RNA polymerase, rpo147, the product of gene J6R. One mutant contained missense mutations in both G2R and A24R (rpo132, the second largest subunit of the RNA polymerase). Two mutants could not be mapped, however sequence analysis demonstrated that neither of these mutants contained mutations in previously identified IBT resistance or dependence genes. Phenotypic and biochemical analysis of the mutants suggests that they possess defects in transcription elongation that compensate for the elongation enhancing effects of IBT. The results implicate the largest subunit of the RNA polymerase (rpo147) in the control of elongation, and suggest that there exist additional gene products which mediate intermediate and late transcription elongation in vaccinia virus.
Keywords: vaccinia virus, genetics, drug resistance, isatin-β-thiosemicarbazone, transcription, RNA polymerase, marker rescue
Introduction
Because poxviruses are double-stranded DNA-containing viruses that replicate exclusively in the cytoplasm of infected cells, they must encode enzymes required for viral transcription and genome replication. This task is facilitated by the large coding capacity of poxviral genomes. The archetypal poxvirus, vaccinia virus, possesses a 195 kbp genome and has long been used as a model system for studying DNA replication and transcription (Moss, 2001).
Transcription in vaccinia virus is carried out by a virus-coded multi-subunit DNA-dependent RNA polymerase that shares homology with both prokaryotic and eukaryotic enzymes (Broyles and Moss, 1986). There are two forms of the vaccinia RNA polymerase: a nine-subunit form that transcribes viral early genes, and an eight subunit form that lacks an early polymerase-specific subunit (RAP94; gene H4L) but is otherwise identical to the early form (Ahn and Moss, 1992; Deng and Shuman, 1994; reviewed in Condit and Niles, 2002; Broyles, 2003). The eight-subunit polymerase functions solely after DNA replication to specifically transcribe genes of the intermediate and late (collectively termed postreplicative) classes (Wright and Coroneos, 1995).
Control of postreplicative gene transcription elongation and termination differs fundamentally from that of early genes (reviewed in Condit and Niles, 2002). Early gene termination employs a simple termination signal. The terminator sequence, UUUUUNU, is recognized with high efficiency, and early transcripts are therefore homogeneous in length (Yuen and Moss, 1987). By contrast, transcripts produced from any given postreplicative gene are heterogeneous in length (Cooper et al., 1981; Mahr and Roberts, 1984). Termination of postreplicative genes is not responsive to the early gene cis-acting element (Condit et al., 1996a); the postreplicative transcription termination cis acting sequence, if it exists, must therefore be either simple or degenerate, and must function inefficiently. Viral factors required for early termination are relatively well characterized and include RAP94 (gene H4L), NPH I (gene D11L) and VTF (genes D1R and D12L) (Shuman et al., 1987; Christen et al., 1998; Deng and Shuman, 1998; Mohamed and Niles, 2000). Elongation and termination of postreplicative gene transcription is influenced by a set of factors distinct from those which effect early termination. These factors include the negative transcription elongation factor A18, and the positive transcription elongation factors G2 and J3, described below.
Factors affecting intermediate and late transcription elongation or termination were originally identified by a phenotypic analysis of a collection of temperature-sensitive vaccinia virus mutants (Condit and Motyczka, 1981; Condit et al., 1983). Mutants Cts4, Cts22, and Cts23 exhibited a phenotype that featured sudden degradation of cellular and viral RNA late during infection (Pacha and Condit, 1985). These mutants mapped to the A18R gene (Pacha et al., 1990), which was later shown to produce an essential transcript release factor with DNA helicase and DNA-dependent ATPase activities (Bayliss and Condit, 1995; Simpson and Condit, 1995; Lackner and Condit, 2000). Due to the absence of the transcript release factor activity, A18R mutants produce postreplicative mRNAs of increased length. Some of these long mRNA molecules have large stretches of complementarity to one another since they are produced from converging transcriptional promoters. This complementarity allows the formation of long dsRNA molecules. The increase in cellular dsRNA concentration in turn activates the cellular 2-5 A antiviral pathway and ultimately leads to activation of the latent ribonuclease L which degrades both viral and cellular RNA (Pacha and Condit, 1985; Cohrs et al., 1989; Bayliss and Condit, 1993; Xiang et al., 1998).
The phenotype of mutants mapping to genes G2R or J3R is the converse of the A18R mutant phenotype. G2R and J3R mutants produce late mRNAs that are truncated from their 3′ ends and are therefore reduced in size relative to wild type mRNAs (Black and Condit, 1996; Latner et al., 2000). Thus, the truncated mRNAs produced by G2R and J3R mutants are long enough to encode the small late proteins but are too short to encode large late proteins, which are synthesized in correspondingly reduced amounts. The phenotype of these mutants suggested that G2 and J3 each function as postreplicative gene positive transcription elongation factors. Consistent with the opposing phenotypes of A18R mutants compared with G2R or J3R mutants, extragenic suppression of a temperature sensitive allele of A18R can be achieved by null mutation of G2R or J3R (Condit et al., 1996b; Latner et al., 2000). While these genetic studies suggest that G2 and J3 function as transcription elongation factors, to date they have no biochemically defined roles in elongation and no detectable sequence homology to non-poxviral elongation factors that would provide clues about their functions. J3 does perform two duties besides its role in transcription elongation: it is the (nucleoside-2′-O-)- methyltransferase for the mRNA 5′ cap maturation and the stimulatory factor for the vaccinia-encoded poly(A) polymerase, E1 (Gershon et al., 1991; Schnierle et al., 1992). The transcription elongation factor activity of J3 is independent of the methyltransferase and poly(A) polymerase stimulatory activities (Latner et al., 2002). G2 has no other known activities but has been shown to bind the H5 protein, a stimulatory factor for late transcription (Kovacs and Moss, 1996; Black et al., 1998; McCraith et al., 2000).
The treatment of wild type infected cells with the anti-poxviral drug isatin-β-thiosemicarbazone (IBT) induces the postreplicative transcription apparatus to synthesize longer-than-normal mRNAs through an unknown mechanism (Xiang, 1998). This mimics the phenotype observed during infection with an A18R mutant. Just as mutation of A18R leads to the activation of the cellular 2-5 A pathway, infection in the presence of IBT also induces the 2-5 A pathway and triggers the degradation of both viral and cellular RNA (Pacha and Condit, 1985; Cohrs et al., 1989; Bayliss and Condit, 1993). For this reason, we hypothesize that IBT functions to promote transcription elongation or inhibit transcription termination. It follows then that mutants capable of growth in the presence of IBT might leverage one of at least two potential mechanisms. A structural change in the IBT binding site could prevent the drug from binding its target. Alternatively, an impairment in transcription elongation might compensate for the elongation-promoting effects of the drug.
Several mutants have been isolated that are resistant to or dependent upon IBT for growth (Katz et al., 1973a; Katz et al., 1973b; Meis and Condit, 1991). IBT-dependent mutants mapping to G2R and J3R have been shown to produce intermediate and late transcripts in vivo that are shorter than the wild type length (Black and Condit, 1996; Latner et al., 2000; Xiang et al., 2000; Latner et al., 2002). In addition, an IBT-resistant mutant mapping to A24R, the gene that encodes the second-largest subunit of the RNA polymerase (rpo132), was identified and characterized as elongation defective in vitro (Condit et al., 1991; Prins et al., 2004). Thus, the compensatory model of defective transcription elongation appears to accurately describe several IBT-resistant and IBT-dependent mutants. While the precise mechanism of action and target of IBT are unknown, IBT has clearly been established as a valuable tool that can be used to select for mutants in factors affecting transcription elongation.
In order to further characterize the proteins which mediate intermediate and late vaccinia gene transcription elongation, we have isolated and characterized a collection of eight independently derived, spontaneous IBT-resistant mutants. These mutants display varying transcription elongation phenotypes, implicate the largest subunit of the RNA polymerase (rpo147) in the control of elongation, and suggest that there exist additional gene products which mediate intermediate and late transcription elongation in vaccinia virus.
Results
Plaque Phenotypes
Eight independently derived spontaneous IBT-resistant mutants of vaccinia virus were isolated as described in Materials and Methods. In order to visualize the ability of the mutants to grow relative to one another and to wild type virus in both the absence and presence of IBT, a plaque assay was performed. Fig. 1 shows appropriate dilutions from a 37°C plaque assay for each virus. The wild type virus is IBT-sensitive and forms large plaques only in the absence of IBT. Relative to wild type, many of the IBT-resistant mutants display a small plaque phenotype in the absence of drug. For DL1-3 and DL10-7, plaque size is not influenced by the presence or absence of drug. Each of the other viruses forms larger plaques in the absence of IBT than in the presence of IBT, though the size of those plaques varies by virus. In the case of DL5-7 and DL8-1, plaques formed in the absence of drug are equivalent in size to wild type plaques. Thus, the IBT-resistant viruses represent a spectrum of plaque phenotypes and display drug resistance to varying degrees.
Fig. 1. Plaque assay of IBT-resistant mutants in the presence and absence of IBT.
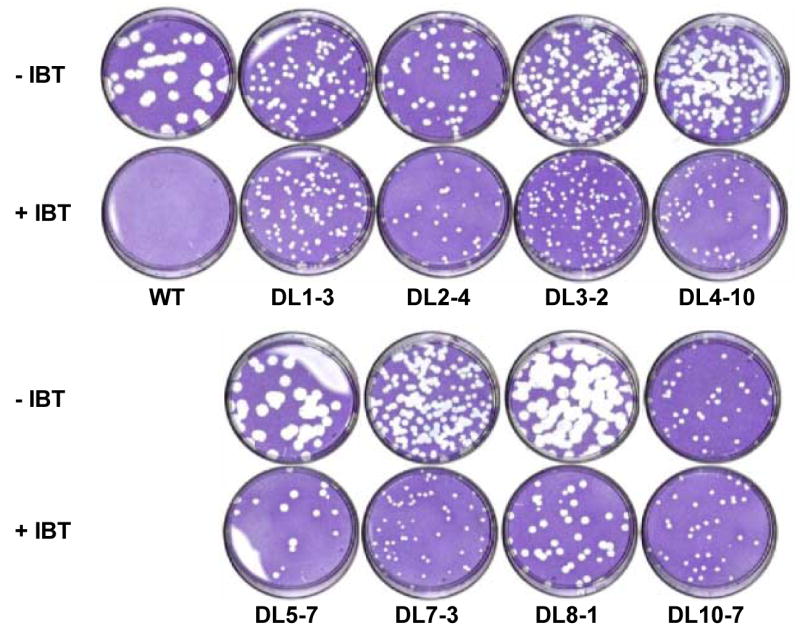
The dishes were incubated at 37°C for 6 days in the presence or absence of IBT prior to staining with crystal violet.
Genetic Mapping
To determine the specific mutation responsible for the IBT-resistance phenotype of each mutant, we used a combination of marker rescue mapping and DNA sequencing. Because of the high likelihood that a subset of these mutants would map to genes previously implicated in IBT-resistance or control of elongation, we sequenced the A18R, G2R, J3R, and A24R allele in each mutant (Table 1). None of the mutants contained mutations in genes A18R or J3R. Of the eight mutants, three contained mutations in G2R (DL2-4, DL5-7, and DL8-1), one of which (DL2-4) also contained a mutation in A24R. The five remaining mutants (DL1-3, DL3-2, DL4-10, DL7-3 and DL10-7) were of particular interest since they carry wild type alleles of A18R, G2R, J3R, and A24R, implying that the drug-resistance phenotype of these mutants maps to one or more novel IBT-resistance genes.
Table 1. Mutant Genotypes.
| Virus | Gene | DNAa | Proteinb |
|---|---|---|---|
| DL1-3 | J6R | C1604T | A535V |
| DL2-4 | G2R, A24R | G2R: G475A; A24R: A3227G | G2:A159T; A24R: D1076G |
| DL3-2 | J6R | C863A | S288Y |
| DL4-10 | unknown | unknown | unknown |
| DL5-7 | G2R | G290T | G97V |
| DL7-3 | unknown | unknown | unknown |
| DL8-1 | G2R | Δ549 | fs 184, term 187 |
| DL10-7 | J6R | C1604T | A535V |
Δ = single base deletion
fs = frameshift; term = translation termination codon
Marker rescue experiments were performed to test whether the IBT-resistance phenotype of the G2R mutants and G2R/A24R double mutant does in fact map to these genes. BSC40 cells were first infected with a temperature sensitive helper virus (Dts38), then co-transfected with wild type genomic DNA and a series of PCR products amplified from each mutant. The infected, co-transfected dishes were incubated at 37°C (non-permissive temperature for Dts38) in the presence of IBT in order to select for IBT-resistant recombinants resulting from the rescue (see Materials and Methods). Negative controls in this and all other marker rescue experiments presented include uninfected cells, cells infected with the Dts38 helper virus but not transfected, and cells infected with Dts38 and transfected with wild type genomic DNA but no PCR product. A signal over background was observed for each G2R mutant except DL2-4, the G2R/A24R double mutant (data not shown). Attempts to map the IBT-resistance phenotype of this virus to the G2R or A24R genes individually or collectively have been unsuccessful.
Because initial sequence analysis of DL1-3, DL3-2, DL4-10, DL7-3 and DL10-7 revealed no mutations in A18R, G2R, J3R, and A24R, mapping of these mutants was attempted using overlapping 5 kbp PCR products generated from the genomes of each mutant (Fig. 2A). This set of 40 primer pairs can be used to amplify virtually the entire vaccinia virus genome (Luttge and Moyer, 2005).
Fig. 2. Initial marker rescue mapping of DL1-3.
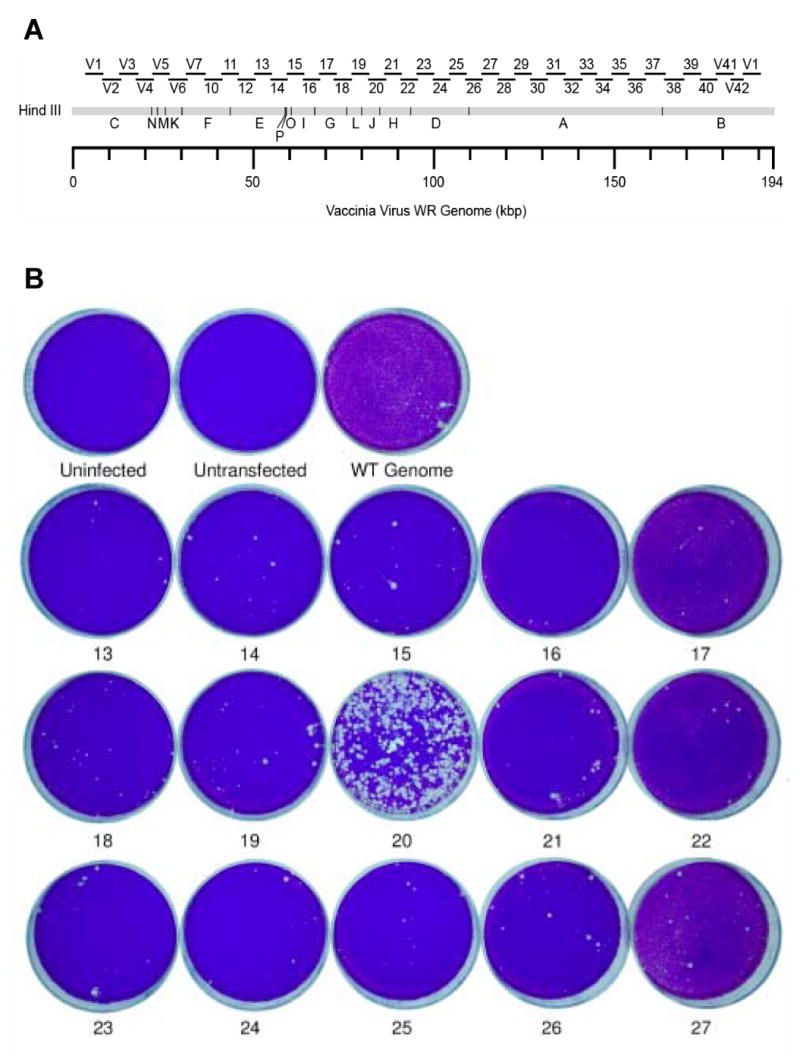
A) Primer pairs used to construct PCR products for marker rescue. Products amplified using a library of primers are shown as bars above the region they amplify on a HindIII restriction map of the vaccinia virus genome. Each product is approximately 5 kb in length. The PCR product numbers shown here are used throughout our study. B) Confluent dishes of BSC40 cells were infected with the Dts38 helper virus and co-transfected with wild-type genomic DNA and the indicated PCR product. After a 4 day incubation at 37°C in the presence of IBT, dishes were stained with crystal violet.
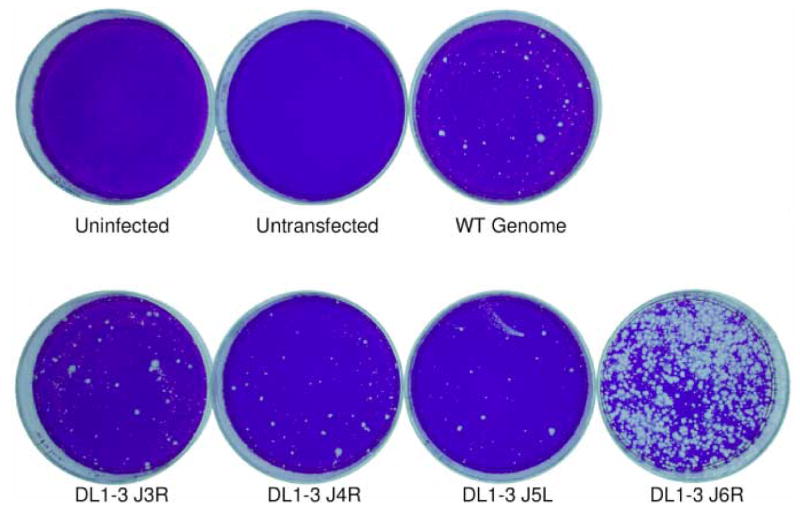
The central core region of the vaccinia genome is highly conserved throughout the poxvirus family and contains the bulk of the essential genes. Thus, initial rescues were attempted with 15 PCR products (Fig. 2A, numbers 13 to 27) that comprise this region. In the first such experiment, PCR products 13 to 27 were amplified from the DL1-3 genome. When these products were transfected in a marker rescue experiment, the result showed that DL1-3 maps to a 5 kbp region containing part of J3R, all of J4R and J5L, and part of J6R (Fig. 2B, dish “20”). Although it was known that this mutant did not map to J3R due to prior DNA sequencing, each of the J3R, J4R, J5L, and J6R open reading frames was PCR amplified and transfected in a second marker rescue experiment (Fig. 3). The result showed that DL1-3 maps to J6R, the gene encoding the largest subunit of the RNA polymerase.
Fig. 3. Marker rescue mapping of DL1-3 to the J6R gene.
Confluent dishes of BSC40 cells were infected with the Dts38 helper virus and co-transfected with wild-type genomic DNA (“WT Genome”) and the indicated PCR product. After a 4 day incubation at 37°C in the presence of IBT, dishes were stained with crystal violet. In this and some other experiments, plaques appearing in the control dish (“WT genome”) probably represent IBT resistant viruses, most likely arising from spontaneous IBT resistance alleles present in the wild type virus genomic DNA used in co-transfection experiments.
Mapping of DL3-2 was performed essentially as described for DL1-3, except that 40, 5 kbp PCR products spanning the genome were used in the initial round of mapping. Although DL3-2 did not produce a signal as robust as that seen with DL1-3, the genome-wide marker rescue indicated that DL3-2 also mapped to the 5 kbp PCR product containing all or part of genes J3R, J4R, J5L, and J6R. Fig. 4A shows the positive rescue done with PCR fragment 20 along with the WT genome control; all other dishes in this experiment, including the remaining 39 PCR fragment transfected dishes, were indistinguishable from the WT genome control. As had been done with DL1-3, genes J3R, J4R, J5L, and J6R were PCR amplified from the DL3-2 genome and used in a subsequent marker rescue experiment (Fig. 4B). In this and some other marker rescue experiments, we noticed a relatively high background of plaques in dishes that had been infected with helper virus and cotransfected with wild type genomic DNA and mutant PCR fragments, especially compared to controls. This background most likely results from residual mutant genomic DNA that was used as a template for the PCR reaction and is therefore present in transfections and can be reactivated by the helper virus. Often this background can be reduced by repeating the PCR amplification using the first round of PCR products as templates for a second round of PCR. Despite the background, the marker rescue experiment shown in Fig. 4B gave a significantly larger number of plaques after transfection with the J6R gene DNA fragment compared with other fragments. This experiment showed that DL3-2, like DL1-3, mapped to the J6R gene.
Fig. 4. Marker rescue mapping of DL3-2 to the J6R gene.
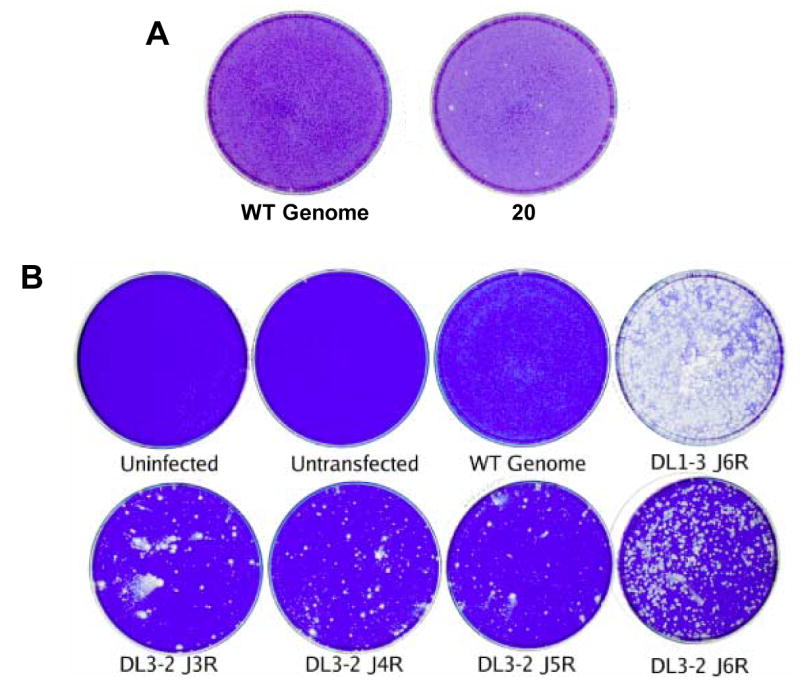
Confluent dishes of BSC40 cells were infected with the Dts38 helper virus and co-transfected with wild-type genomic DNA (“WT Genome”) and the indicated PCR product. After a 4 day incubation at 37° C in the presence of IBT, dishes were stained with crystal violet. A) Initial marker rescue mapping of DL3-2. Shown are the control dish transfected with WT genomic DNA alone (“WT Genome”), and the positive dish co-transfected with WT genomic DNA and PCR product 20 amplified from the DL3-2 genome (“20”). B) Marker rescue using gene specific PCR products spanning PCR fragment 20.
Because both DL1-3 and DL3-2 mapped to the J6R gene, the remaining mutants, DL4-10, DL7-3, and DL10-7, were assayed to determine if they mapped to J6R as well. First the J6R gene from each of these three mutants was sequenced. From this it was determined that the J6R genes of DL4-10 and DL7-3 are wild type in DNA sequence (Table 1). Combined with the initial sequencing effort, these data indicate that DL4-10 and DL7-3 must map to a gene other than A18R, G2R, J3R, A24R, and J6R. Repeated attempts to map each of these mutants have been unsuccessful (data not shown). On the other hand, DL10-7 was shown by sequence analysis to contain a mutation in J6R (Table 1). Surprisingly, the genotype of this mutant was identical to that of DL1-3. Marker rescue analysis confirmed that IBT-resistance in this mutant did in fact map to its J6R mutation and not any other gene implicated in IBT-resistance (data not shown).
Genes that gave positive results in marker rescue experiments were PCR amplified with primers flanking the coding sequence, and these products were sequenced as described in Materials and Methods. Results from the sequencing are summarized in Table 1. The sequencing revealed coding mutations in the G2R gene of DL5-7 and DL8-1 and the J6R gene of DL1-3, DL3-2, and DL10-7. These results corroborate the marker rescue mapping.
As a final confirmation that the mutations identified in the marker rescue and sequencing are responsible for the IBT resistance phenotype of each mutant, the marker rescue experiments were repeated using an agar overlay so that rescued viruses could be isolated from individual plaques. Three plaques were picked from rescues done with each of the viruses DL1-3, DL3-2, DL5-7, or DL8-1, while two plaques were picked from a DL10-7 rescue. The viruses in each of these plaques were grown in the presence of IBT, plaque purified, and grown again in the presence of IBT to produce a lysate. DNA was extracted from the lysate and used as a template for PCR. The PCR amplified G2R or J6R gene was then sequenced, and in each case the nucleotide change found in the original IBT resistant mutants was also present in the viruses derived from those mutants. Interestingly however, one of the DL1-3 derived viruses possessed a second nucleotide substitution (G1603T) in addition to the original mutation (C1604T) in J6R, resulting in a different missense mutation (A535F) relative to what is present in DL1-3 (A535V).
In Vivo Analysis of Transcription Elongation
Because IBT appears to influence transcription elongation, the in vivo transcription phenotype of the IBT-resistant mutants was analyzed. Northern blots were performed using probes specific for the 5′ ends of three vaccinia virus genes in order to measure the length of transcripts produced from these genes during infections with the eight IBT-resistant mutants. The probes used were specific for the intermediate K2L gene (Fig. 5), the highly expressed late A10L gene (not shown), and the early/late A18R gene (not shown). In each case, 9 hour post infection (h.p.i.) RNA from cells infected in the presence and absence of IBT was used. Control RNA samples included those isolated from mock-infected cells or cells infected with IBT-sensitive wild type virus, with the IBT-resistant IBTr90 virus (Condit et al., 1991; Prins et al., 2004), or with the IBT-dependent G2A virus (Meis and Condit, 1991).
Fig. 5. Northern analysis of IBT-resistant mutant RNA.
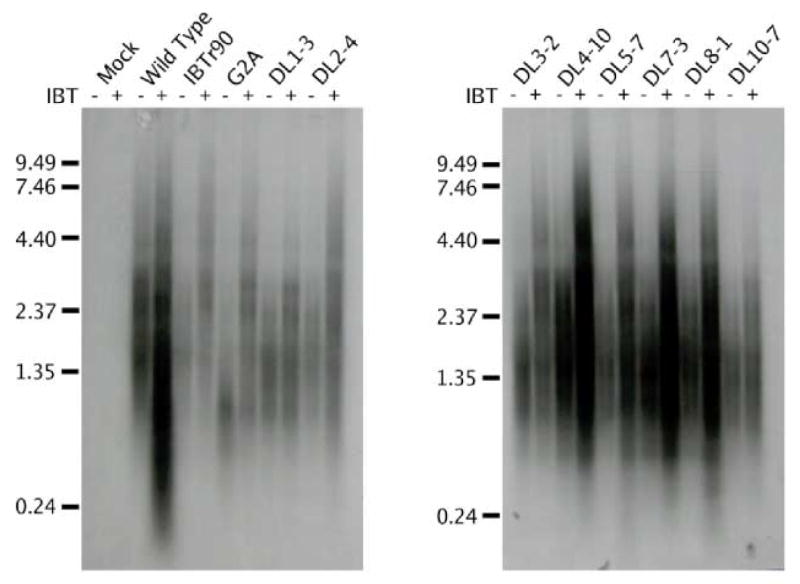
Confluent BSC40 cells were infected with the indicated virus at m.o.i. = 15 in the presence or absence of IBT and incubated at 37°C for 9 hours. Total cellular RNA was purified, fractionated by gel electrophoresis and transferred to a nylon membrane. The membrane was probed with a riboprobe specific for the intermediate K2L gene and exposed to film. The virus used for infection is indicated at the top of the autoradiogram. The presence of absence of IBT in the infection is indicated with a “+” or a “−” at the top of the autoradiogram. Size markers are indicated at the left of the autoradiagram, in kb.
Results from the northern blotting experiments confirmed the effect of IBT on transcripts produced from IBT-sensitive (wild type), IBT-resistant (IBTr90), and IBT-dependent (G2A) viruses (Fig. 5). Regardless of the virus tested, transcription of the K2L gene produces a population of transcripts that are highly heterogeneous in length and longer in the presence of IBT than in its absence. Northern blots of RNA from wild type infections done in the absence of IBT show the normal distribution of K2L transcripts. Wild type transcripts produced in the presence of IBT reach a length sufficient to trigger their degradation via the 2-5 A pathway (Pacha and Condit, 1985; Cohrs et al., 1989; Bayliss and Condit, 1993; Xiang, 1998). Products of this degradation are seen as a low molecular weight smear that is absent from each of the other lanes. In the absence of IBT, the IBT-resistant mutant IBTr90 synthesizes transcripts that are apparently similar in length to wild type. In the presence of IBT however, the mRNA degradation seen with the wild type virus is not observed in the IBTr90 infection. The IBT dependent mutant G2A produces only very short transcripts in the absence of IBT, but G2A transcripts synthesized in the presence of IBT have a normal size distribution. The phenotype of each of the remaining IBT-resistant viruses resembles that of IBTr90. Similar experiments conducted with probes specific for the A10L and A18R genes (data not shown) produced results indistinguishable from that of K2L. Consistent with the northern analysis, rRNA visualized by ethidium bromide staining of these same gels before transfer reveals rRNA breakdown in wild type infections done in the presence of IBT, while in all other infections rRNA remained intact. In summary, during infection with each of the viruses tested, whether IBT sensitive, dependent or resistant, addition of IBT to the infections resulted in synthesis of longer transcripts compared to infections done in the absence of drug. No reproducible differences were observed in size or quantity of RNAs synthesized in the absence of IBT, however subtle differences in the average distribution of sizes may exist which are beyond the sensitivity of these experiments. The size distribution of mutant RNAs made in the presence of IBT cannot be legitimately compared to the RNAs made during a wild type infection in the presence of IBT because of the RNA breakdown induced during the wild type infection.
In Vitro Analysis of Transcription Elongation
An in vitro transcription elongation system was utilized to complement and extend the analysis of transcripts synthesized in vivo. Transcription extracts were made from each of the IBT-resistant viruses (with the exception of DL10-7 because it has the same genotype as DL1-3) and assayed for their ability to promote transcription elongation compared to a wild type extract. The DNA template used in this assay, pG8GU, which was designed to analyze elongation of the transcription complexes through an artificial pause site, contains the vaccinia virus G8R gene intermediate promoter followed by a 37 bp G-less cassette, then a 40 bp T-less cassette, then a 9 bp poly-T stretch, and lastly 42 bp of plasmid DNA and is bound to streptavidin-coated paramagnetic beads (Prins et al., 2004). During a pulse reaction, virus extracts were incubated with the DNA template in transcription buffer that included ATP, UTP, 32P-CTP, and 3′-O-MethylGTP. The incorporation of the 3′-O-MethylGTP at the end of the 37 bp G-less cassette causes transcriptional arrest and allows the transcription complex to be isolated on a magnet and washed with buffer to remove unincorporated nucleotides and proteins. The pulse reaction produces labeled transcripts of varying lengths due to the addition of a 30-50 bp poly-A head on vaccinia post-replicative transcripts (Schwer et al., 1987). Elongation resumes during a chase phase of the reaction when buffer containing ATP, CTP, GTP, and varying amounts of UTP is added to the washed complexes and they are incubated at 30°C for 5 or 30 minutes. The resumption of elongation presumably follows removal of 3′-O-MethylGTP via an exonuclease mechanism (Hagler and Shuman, 1993).
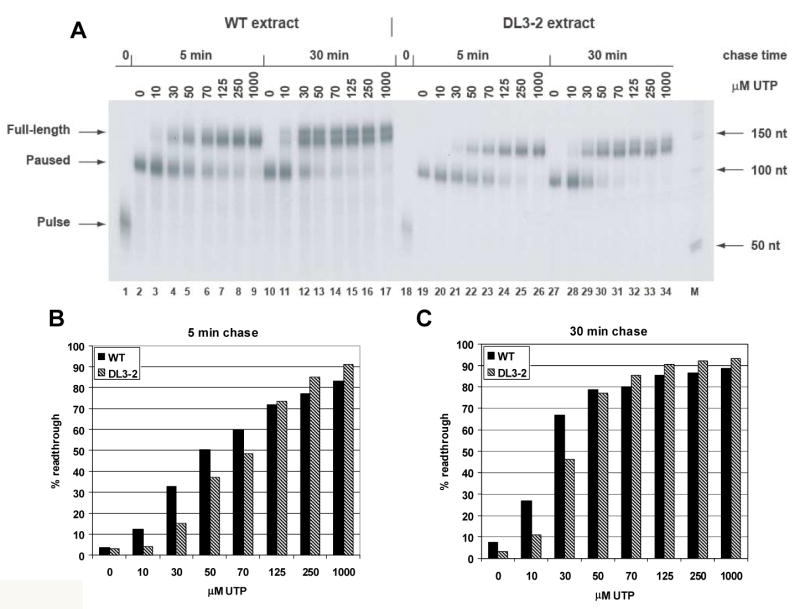
Results of the in vitro transcription elongation assay of DL1-3, a mutant whose IBT resistance maps to the largest subunit of the vaccinia RNA polymerase, rpo147, are shown in Fig. 6. For both the wild type and DL1-3 viruses, transcription elongation in the chase reaction does not proceed beyond the 9 nt poly-T pause site in the absence of UTP (Fig. 6A, lanes 2, 10, 19, 27). As UTP levels increase in the chase reaction, the amount of elongation increases for each virus, but the wild type virus produces a higher percentage of full length transcripts than the DL1-3 virus at UTP levels of 10 μM and above (Fig. 6B,C). Thus, DL1-3 exhibits a transcription elongation defect. Compared to a previous analysis of IBTr90 (Prins et al., 2004), an IBT-resistant mutant mapping to rpo132, this transcription elongation defect appears to be quite severe.
Fig. 6. In vitro transcription elongation assay of DL1-3.
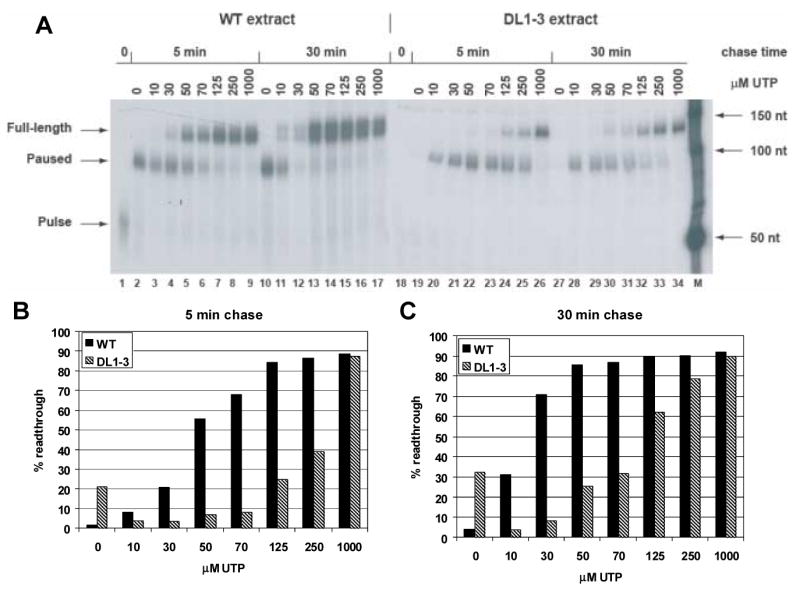
Extracts from wild type and DL1-3 infected cells were prepared and assayed for transcription elongation in vitro as described in materials and methods. A) An autoradiogram showing the amount of readthrough transcription as a function of time and UTP concentration. Source of extract, time of chase and UTP concentration are indicated at the top of the autoradiogram. Transcription products are indicated to the left of the autoradiogram. “Pulse” indicates the transcript arrested at the end of the 37 nt G-less cassette. “Pause” indicates the transcript paused at the poly-T sequence. “Full-length” indicates transcript resulting from transcription to the end of the DNA template. Size markers are indicated to the right of the autoradiogram. The autoradiogram of transcription with wild type extracts was exposed for 17 hr. The autoradiogram of transcription with DL1-3 extracts was exposed for 9 days. B) Comparative quantification of results from a 5 min chase. Gels were exposed on a phosphorimager, and percent readthrough was calculated as the amount of full length transcript divided by the total transcription (equal to the sum of the full length transcript plus the paused transcript) and multiplied by 100. C) Comparative quantification of results from a 30 min chase.
Transcription extracts from DL1-3 infected cells were also significantly decreased in overall transcription activity compared to wild type extracts; note in Fig. 6 that the autoradiogram for the DL1-3 transcription assay was exposed 12 times longer than the wild type assay. The total activity recovered from all of the mutants varied from 7 fold greater than wild type extracts (DL2-4) to 30 fold less than the wild type extract (DL1-3); however these variations showed no clear correlation with any other aspect of the mutant phenotype.
The results of transcription elongation analysis of DL3-2 infected cell extracts are presented in Fig. 7. Again, both the wild type and DL3-2 viruses do not transcribe beyond the poly-T site if UTP is not included in the chase reaction (Fig. 7A, lanes 2, 10, 19, 27) , however the wild type virus produces a higher percentage of full-length transcripts than the DL3-2 virus as UTP levels are increased in the chase reaction (Fig. 7B, C). The elongation defect of the DL3-2 mutant does not appear to be as severe as that of the DL1-3 or of IBTr90 mutants.
Fig. 7. In vitro transcription elongation assay of DL3-2.
Extracts from wild type and DL3-2 infected cells were prepared and assayed for transcription elongation in vitro as described in materials and methods. A) An autoradiogram showing the amount of readthrough transcription as a function of time and UTP concentration. Both the wild type and the DL3-2 autoradiograms were exposed for 17 hr. B and C) Comparative quantification of results from a 5 min chase and a 30 min chase. See legend to Fig. 6 for details.
Transcription elongation assays done with extracts from the remaining mutants assayed, specifically DL2-4, DL4-10, DL5-7, DL7-3, and DL8-1, showed no differences in transcription elongation compared to wild type virus infected cell extracts. In the case of the G2R mutants, DL5-7 and DL8-1, this result was not surprising since in this assay even the G2R null mutant, G2A, is indistinguishable from wild type (C. Prins, unpublished data). In fact, in our experience this transcription elongation assay is specific for mutation of the RNA polymerase and thus fails to measure defects in elongation caused by mutation of elongation factors. Therefore, the lack of an elongation defect in DL2-4, DL4-10, and DL7-3 extracts implies that the IBT-resistance loci in these mutants affect proteins other than the RNA polymerase.
Discussion
The goal of the work presented here was to explore the network of factors that collectively control the length of vaccinia virus intermediate and late mRNA transcripts. This study and those that preceded it took advantage of spontaneous IBT-resistant or IBT-dependent vaccinia virus mutants. Mutations is these viruses have generally affected one of two categories of genes: RNA polymerase subunits and elongation factors. The former category was comprised of a single mutant, IBTr90, that mapped to the second-largest subunit of the RNA polymerase, rpo132 (Condit et al., 1991; Prins et al., 2004). The latter category consisted of a number of mutants mapping to the elongation factors J3 and G2 (Meis and Condit, 1991; Black and Condit, 1996; Latner et al., 2000). In this study, we isolated and mapped additional spontaneous IBT-resistant mutants in an attempt to identify novel factors that may be involved in controlling transcription elongation. New elongation-defective alleles of G2R were identified and characterized, and for the first time IBT-resistance was mapped to the gene encoding the largest RNA polymerase subunit, rpo147. In addition, two IBT-resistant mutants (DL4-10 and DL7-3) were isolated which, although they remain unmapped, do not affect any previously identified elongation factors, implying that additional vaccinia genes exist which mediate postreplicative gene transcription elongation.
Several details of the genetic analysis presented here are noteworthy. First, the A535V IBT-resistance allele of the J6R gene was recovered in two independently isolated IBT resistant mutants. The methods used for isolation of the mutants excludes the possibility that these mutants are sibs. This finding suggests that the A535 residue is particularly sensitive to selective pressure for altered RNA polymerase elongation properties. Second, one of three viruses isolated after rescue of wild type virus to IBT resistance using the A535V J6R allele possessed a second nucleotide substitution (G1603T) in addition to the original mutation (C1604T) in J6R, resulting in a different missense mutation (A535F) relative to that which is present in DL1-3 (A535V). The most likely explanation for this observation is that the rescue resulted initially in the incorporation of the A535V J6R allele, and that the secondary mutation to the A535F allele arose spontaneously during amplification of the rescued virus under selection in the presence of IBT. In any event, this observation further emphasizes the sensitivity of the A535 residue to selection for elongation phenotypes. Third, the G2R allele identified in DL8-1 is a frameshift mutation which truncates the 220 amino acid G2 protein to 186 amino acids, where the C-terminal three amino acids are mutant owing to the frameshift at codon 184. Null mutation of the G2R gene results in IBT dependence (Meis and Condit, 1991). Mutation of G2R to IBT resistance probably represents G2 function that is crippled but not absent; the IBT resistance phenotype thus represents an intermediate condition midway between the wild type sensitive phenotype and the null mutant dependent phenotype. The DL8-1 phenotype thus implies that the C-terminal amino acids of the G2 protein contribute to but are not absolutely essential for full G2 function.
An analysis of transcript lengths in vivo implies a transcription elongation defect for each of the IBT-resistant mutants described here. As discussed previously, two models that can account for IBT-resistance include one in which mutation of the IBT binding site on its target prevents the IBT-target interaction and another in which a deleterious mutation of the transcription machinery compensates for the ability of IBT to promote transcription elongation. The existence of a compensatory deleterious mutation of the transcription machinery would theoretically be revealed in northern blots as transcripts that are shorter than the corresponding wild type transcripts synthesized in the presence or absence of IBT (excluding degradation induced by IBT during a wild type infection). However we have found that testing the compensatory model directly in vivo is made difficult by the extreme heterogeneity of transcript sizes produced from any tested gene. This compromises the ability to resolve any differences in transcript length when comparing the IBT-sensitive wild type virus and the IBT-resistant mutants. In fact, close inspection of the data in Fig. 5 suggests that the K2L transcripts synthesized during each of the mutant infections are on average shorter than the wild type transcripts; however the differences are small and not reproducibly convincing on repeated experiments with three different gene specific probes. Nevertheless, the absence of degradation products in the IBT-resistant mutant infections done in the presence of IBT implies that these transcripts are shorter than those produced during a wild type infection in the presence of IBT, where degradation products are observed. Also, since IBT exerts an effect on the length of transcripts produced by each of the IBT-resistant mutants, the drug binding site of its target likely remains intact, arguing against a drug binding deficiency model. Lastly, the observation of an elongation defect in vitro for mutants DL1-3 and DL3-2 supports the compensatory mutation model for the J6R RNA polymerase mutants.
Characterization of the J6R mutants revealed that they are defective in an in vitro transcription elongation assay at intermediate concentrations of UTP relative to wild type virus. As with IBTr90, extracts made from either DL1-3 and DL3-2 infections exhibit in an increased ratio of paused to full-length transcripts in vitro. Furthermore, the extent of the defect is greatest in DL1-3 and mildest in DL3-2, with IBTr90 falling in between (these studies and Prins et al., 2004). In an attempt to gain mechanistic understanding into the effects of these mutations, we have used homology modeling to identify residues in the yeast RNA polymerase that are equivalent to the residues mutated in each of the vaccinia mutants described here (Fig. 8). Specifically, the A535 residue that is affected in DL1-3 and DL10-7 corresponds to amino acid I608 in the rpb1 subunit of the yeast RNA polymerase, and the S228 residue that is affected in DL3-2 corresponds to amino acid T351 in the rpb1 subunit of the yeast RNA polymerase. Thus according to the model, the S288Y mutation in DL3-2 is closely positioned to the active site of the RNA polymerase, with its side chain very near one of the three universally conserved aspartate residues (D401 in rpo147, D485 in rpb1) that coordinates the active site Mg2+ ion (Zaychikov et al., 1996; Cramer et al., 2001). The potential for a mutation at this position to interfere with catalysis is readily evident. The substitution of a tyrosine for a serine presents a large R group that might, for example, interfere with the coordination of the active site Mg2+, impose steric constraints on the free passage of NTPs to the active site, or diminish the capacity of the polymerase to recover from pausing via its intrinsic endoribonuclease activity. Modeling of the A535V mutation found in both DL1-3 and DL10-7 suggests that the substituted residue is surface-exposed and within a domain of the polymerase known as pore 1 (Batada et al., 2004). This pore is critical to the functioning of the polymerase, as it is thought to serve as an entry port through which NTPs gain access to the active site and an exit port through which the 3′ end of the nascent RNA is extruded during retrograde movement of the polymerase at pause or arrest sites (Fu et al., 1999; Cramer et al., 2000). Also, in both prokaryotic and eukaryotic systems, the pore is a docking site for enzymes that modulate the transcription elongation properties of the polymerase itself (Kettenberger et al., 2003; Opalka et al., 2003). The GreA and GreB factors and their eukaryotic functional homolog TFIIS are two examples of proteins that interact via pore 1 to affect transcription elongation. These enzymes stimulate the endoribonucleolytic cleavage activity of RNA polymerase in response to RNA polymerase backtracking at arrest sites (Reines et al., 1993; Toulme et al., 2000). This activity cleaves the extruded 3′ end of RNA that is no longer aligned with the active center of the enzyme, resulting in the formation of a new 3′ end that is appropriately positioned as a substrate for further transcription elongation. Although no cleavage stimulatory factors are known to interact with the vaccinia polymerase, the polymerase itself has been demonstrated to possess this endoribonuclease activity (Hagler and Shuman, 1993). Furthermore, the RNA polymerase subunit rpo30 bears sequence similarity to TFIIS, and extra-molar rpo30 may well serve the role of stimulatory factor (Ahn et al., 1990).
Fig. 8. Structural modeling of DL1-3 and DL3-2.
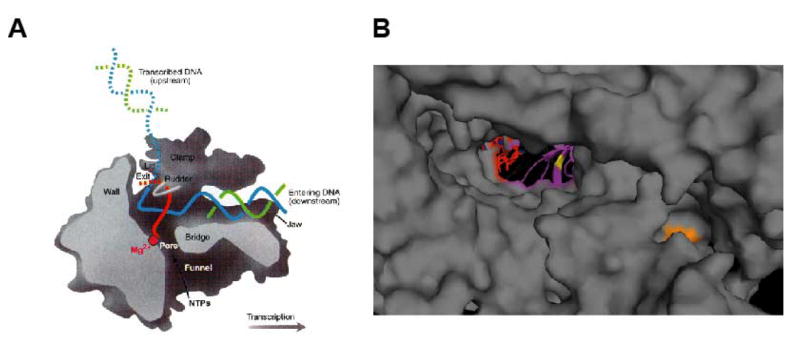
A model of the vaccinia rpo147 (gene J6R) subunit was constructed based on homology to the known structure of S. cerevisiae RNA polymerase II subunit rpb1. Yeast RNA polymerase residues homologous to the mutant vaccinia alleles were then highlighted on the structure of the elongating yeast RNA polymerase. A) Side (cutaway) view of the RNA polymerase II transcribing complex. The template DNA strand is in blue, the nontemplate DNA strand is in green, and the RNA is in red. Additional structural features of the enzyme are indicated. B) A close up view of the yeast RNA polymerase II looking down the pore to the active site of the enzyme, with residues homologous to the DL1-3 and DL3-2 alleles indicated. Colors indicate the following: RNA, red; DNA, blue; active site, purple; DL1-3 (yeast I608), orange; DL3-2 (yeast T351), yellow. A) is from Klug (2001).
Despite nearly 50 years of research concerning the antipoxviral effects of IBT, the target of the drug remains unknown. The specificity of the drug combined with the existence of both dependent and resistant viral mutants argues strongly for a viral target. The striking similarity of phenotype between an A18R viral mutant and the effects of IBT on a wild type virus infection suggests that the most likely target of the drug would be the A18 protein (Pacha and Condit, 1985; Bayliss and Condit, 1993), however none of the nearly 20 IBT sensitive and IBT resistant mutants isolated to date map to the A18R gene. IBT has no detectable effect on either A18 or the vaccinia RNA polymerase in vitro (C. Prins and R.C. Condit, unpublished observations), though it is possible that the drug is activated through some unknown modification in vivo, and thus the “prodrug” would be inactive in vitro. The entire catalogue of IBT sensitive and resistant mutants to date comprise alleles in the elongation factors G2 and J3, the RNA polymerase subunits rpo147 (J6) and rpo132 (A24), and at least one other unidentified gene. All of these mutants behave like compensatory mutants; no candidate drug binding mutants have been identified. Based on the number and character of the mutants isolated we feel that the most likely target for the drug is the RNA polymerase, however direct proof in support of this conclusion is currently lacking.
In summary, this work extends the collection of vaccinia virus mutants that can be used to study intermediate and late transcription. Genetic mapping of these mutants indicates that the collection of virus genes that can be mutated to produce IBT resistance continues to grow and now contains J6R, the gene encoding rpo147. Furthermore, a comparison of these mutants to the IBT-sensitive wild type virus, the IBT-resistant IBTr90 mutant, and the IBT-dependent G2A mutant indicates that they are defective in intermediate/late transcription elongation. Also, homology modeling of the rpo147 mutants suggests possibilities for the mechanisms underlying those defects and provides the basis for further experimentation. Lastly, the existence of the unmapped DL4-10 and DL7-3 mutants implies that additional genes exist which affect transcription elongation of postreplicative vaccinia genes.
Materials and Methods
Cells and viruses
Methods used for the propagation of the African green monkey kidney cell line BSC40, growth of vaccinia virus stocks, and plaque assays were performed as described previously (Condit and Motyczka, 1981; Condit et al., 1983). The human lung carcinoma cell line, A549, was propagated as previously described (Prins et al., 2004). Vaccinia virus strain WR was used except where noted. Dts38 is a temperature sensitive mutant of the IHDW strain of vaccinia affecting the D5R gene (Lackner et al., 2003). Dts38 does not form plaques at 37°C (D'Costa, Lackner and Condit, unpublished observations). The IBT-resistant mutant IBTr90 contains a Y462H missense mutation and has a partial defect in intermediate transcription elongation in vitro (Condit et al., 1991; Prins et al., 2004). The IBT-dependent mutant G2A has an engineered 10 bp deletion at codon 90 of the G2R gene that results in a null phenotype in vivo (Meis and Condit, 1991). Independently derived, spontaneous IBT-resistant mutants were isolated exactly as described for IBT-dependent mutants (Latner et al., 2000). Briefly, ten well-isolated plaques were picked from a plaque assay of wild type virus done in the absence of IBT and these plaques were grown to create stocks. Each of these ten wild type stocks was then plaque assayed in the presence of IBT. In each case, a number of plaques were observed due to spontaneous mutation. Ten of these plaques were picked from the plaque assay of each of the ten wild type stocks, for a total of 100 viruses capable of growth in the presence of IBT. These 100 viruses were then plaque assayed in the presence and absence of IBT to determine whether each was IBT-resistant or IBT-dependent. Eight IBT-resistant mutants that each originated from an independent stock of wild type virus were selected for further study. These viruses were given the names DL1-3, DL2-4, DL3-2, DL4-10, DL5-7, DL7-3, DL8-1, and DL10-7. Preparations of IBT were made fresh and used at a final concentration of 45 μM, as described previously (Pacha and Condit, 1985).
Polymerase chain reaction
Isolation of DNA for use as a polymerase chain reaction (PCR) template was done by either of two methods. Total cellular DNA was purified from monolayers of virus infected BSC40 cells in 6 cm dishes using the Qiagen DNeasy DNA isolation kit according to the manufacturer's protocol (Qiagen, Santa Clarita, CA). Alternatively, virus cores were prepared from 4 × 15 cm dishes of infected BSC40 cells (Esposito et al., 1981), resuspended in 200 ul PBS, and purified using the Qiagen DNeasy DNA isolation kit.
PCR reactions were optimized for primer and template, but generally consisted of 400 ng template DNA, 0.32 uM primers (each), 2 mM MgCl2, 400 μM dNTPs (each), and 2 U Deep Vent DNA polymerase (New England Biolabs, Beverly, MA). Products of PCR reactions were purified using Amicon Centricon spin filters (Millipore, Billerica, MA).
DNA Sequencing
Sequencing of viral DNA was performed by PCR amplifying the region of interest with an appropriate upstream and downstream primer and submission of the PCR product to the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR), along with appropriate sequencing primers. Individual sequencing reads were assembled into contigs using the Wisconsin package, version 10.3 (Accelrys Inc., San Diego, CA) or Vector NTI version 8.0.
Marker rescue
A one-step marker rescue mapping of IBT-resistant vaccinia virus mutants was performed as described by Condit et al (1991), with minor modifications. Briefly, confluent 60 mm dishes of BSC40 cells were infected with Dts38, which serves as a helper virus. Dishes were then co-transfected with genomic wild type vaccinia virus DNA and a DNA fragment from the IBT-resistant mutant and incubated at 37°C in the presence of IBT until 4 days post-infection. This helper virus-mediated protocol employed two selections, temperature and IBT, and thus resulted in a lower background than using wild type virus for the infections. Rather than using cloned viral DNA for the transfected IBT-resistant mutant DNA fragment, PCR products were used. For use in preliminary mapping experiments, PCR amplification was performed as described using a library of primers designed and generously donated by Benjamin Luttge and Richard Moyer to create 40 overlapping 5 kbp products that collectively span the vaccinia genome (Luttge and Moyer, 2005). Once a 5 kbp PCR product was identified as containing the IBT resistance locus, individual open reading frames in that 5 kbp region were amplified from genomic viral DNA and used in a subsequent marker rescue experiment to map each mutant to an individual gene.
RNA Isolation
Confluent 60 mm dishes of BSC40 cells were infected with wild type or mutant virus at a m.o.i. of 15 and incubated for 9 hours at 37°C. The cells were washed with PBS containing 0.01% BSA and 10 mM MgCl2, and total cellular RNA was then isolated using the RNeasy RNA isolation kit (Qiagen) according to the manufacturer's protocol. Two column elutions were each performed with 50 μl RNase-free water. Measurement of absorbance at 260 nm was used to determine RNA concentration.
Riboprobes
Riboprobes specific for the 500 bp at the 5′ end of K2L, A10L, and A18R mRNAs were transcribed from PCR products. To do this, transcription templates were PCR amplified using a forward primer complementary with the 5′ end of the open reading frame (ORF) and a reverse primer complementary to a region 500 bp downstream of the 5′ end of the ORF and tagged with the sequence “CGATTTAGGTGACACTATAGAAGCG” containing the bacteriophage SP6 promoter. (The essential promoter region is in italics, while two 5′ nucleotides and four 3′ nucleotides have been added to promote efficient transcription.) Transcription from PCR products was performed with the Ambion MAXIscript in vitro transcription system (Ambion, Inc., Austin, TX).
Northern Blotting
RNA samples were combined with RNA sample loading buffer containing ethidium bromide to give a final concentration of 20.6% formamide, 376 mM formaldehyde, and 0.4 X MOPS (1 X MOPS = 20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA, pH 7.0) and denatured by heating to 70°C for 10 minutes. Samples were then loaded onto a 1% agarose gel containing 2.2 M formaldehyde and 1 X MOPS buffer. Gels were electrophoresed at 20 V for 16 hours, during which time the 1 X MOPS running buffer was continuously recirculated using a peristaltic pump. The RNA was partially hydrolyzed by soaking the gels first in water and then in 0.05 N NaOH, each for 20 min. The gels were then soaked in 20 X SSC (transfer buffer; 1 X SSC = 15 mM sodium citrate, 150 mM sodium chloride) for 40 min prior to transfer to a GeneScreen neutral charge membrane (PerkinElmer, Boston, MA). Membranes were pre-hybridized at 55°C in a hybridization oven (Labnet International, Inc., Woodbridge, NJ) for at least 2 hours in buffer containing 50mM Tris-HCl pH 7.5, 1 M NaCl, 50% formamide, 1% SDS, 0.1% sodium pyrophosphate, 10 X Denhardt's reagent (0.2% BSA, 0.2% polyvinylpyrolidone, 0.2% Ficoll), 10% dextran sulfate, and 0.1 mg/ml salmon sperm DNA denatured by heating to 95°C for 10 minutes. Following pre-hybridization, 1 × 107 cpm fresh riboprobe was added to each blot and incubated overnight at 55°C. Blots were then quickly washed once with 1 X SSC containing 0.1% SDS at room temperature, four times with 1 X SSC containing 1% SDS at 65°C, and exposed to film.
Infected Cell Extracts for In Vitro Transcription
A549 cells were grown to confluency in 100 mm dishes and infected with either wild type vaccinia virus strain WR (Condit and Motyczka, 1981), DL1-3, DL2-4, DL3-2, DL4-10, DL5-7, DL7-3, DL8-1, or DL10-7 at a multiplicity of infection of 10 at 37°C. The infected cells were incubated in media containing 10 mM hydroxyurea, which inhibits vaccinia virus DNA replication and promotes accumulation of intermediate gene transcription factors. At 16 to 18 hours post-infection, cell extracts were prepared as described previously (Condit et al., 1996a).
Templates for In Vitro Transcription
The pG8GU template used in the in vitro transcription reaction has been previously described (Prins et al., 2004). Briefly, the bead-bound pG8GU template contains 240 bp of plasmid DNA upstream of the vaccinia virus intermediate G8 promoter, followed by a 37 bp G-less cassette, a 40 bp T-less cassette, a stretch of 9 T's, and another 42 bp of plasmid DNA for a total of 395 bp. The template is bound to streptavidin-conjugated Dynabeads M280 (Dynal), as described previously (Lackner and Condit, 2000).
In Vitro Transcription Elongation Assay
The in vitro transcription elongation assay was performed as described previously (Prins et al., 2004). Vaccinia virus infected cells extracts (15 μg) are combined with template DNA and transcription buffer for a 25 μl reaction containing 5 mM MgCl2, 25 mM HEPES pH 7.4, 1.6 mM DTT, 80 mM KOAc, 1 mM ATP, 1 mM UTP, 1 μM CTP, 200 μM 3′-O-MethylGTP, and 6 μCi [alpha-32P]-CTP (3000 Ci/mmol stock, Perkin Elmer). Reactions were incubated for 20 minutes at 30°C to form “pulse complexes”. This allows transcription elongation to proceed to the end of the 37 bp G-less cassette where the 3′-O-MethylGTP is incorporated, causing transcriptional arrest. The paramagnetic bead-bound ternary transcription elongation complexes were isolated on a magnet, and were then washed 3 times in 1-1.5 reaction volumes of low-salt wash buffer containing 5 mM MgCl2, 25 mM HEPES pH 7.4, 1.6 mM DTT, 80 mM KOAc, 200 μg/μl BSA, and 7.5% glycerol. Washed transcription complexes were resuspended in transcription buffer containing 5 mM MgCl2, 25 mM HEPES pH 7.4, 1.6 mM DTT, 80 mM KOAc, and 20 units RNAsin (Promega). The complexes were then chased in the presence of 1 mM ATP, 1 mM CTP, 1 mM GTP and differing levels of UTP at 30°C for 5 or 30 minutes. Reactions were incubated at 37°C for 30 minutes with 175 μl of PK mix (114 mM Tris-HCl pH 7.5, 14 mM EDTA, 171 mM NaCl, and 1.1% SDS, 230 μg/ml Proteins K, 800 μg glycogen) and nucleic acids were extracted with 175 μl of phenol/chloroform and then precipitated by adding 290 μl 10 M NH4OAc and 400 μl isopropyl alcohol and incubating at room temperature for 30 minutes. The precipitated DNA was pelleted by centrifuging samples for 20 minutes and removing the supernatant. Pellets were washed with cold 70% ethanol and resuspended in 10 μl of formamide loading buffer. Samples were heated at 95°C for 2 minutes and run on a 10% 8 M urea-PAGE. Gels were fixed, dried and exposed to a phosphor screen (Molecular Dynamics) that was analyzed using a Storm phosphorimager (Molecular Dynamics) and the ImageQuant (Molecular Dynamics) program.
Homology modeling
Homology models of the rpo147 and rpo132 proteins were constructed using ClustalW, Swiss-PdbViewer, version 3.7, and the Swiss-Model homology modeling server (http://swissmodel.expasy.org). An amino acid sequence alignment of rpo147 and S. cerevisiae rpb1 (Protein Data Bank codes 1i6hA and 1nikA) or rpo132 and S. cerevisiae rpb2 (Protein Data Bank codes 1i6hB and 1nikB) was constructed using ClustalW and used by Swiss-PdbViewer to generate a structural alignment. The structural alignment was edited by hand and submitted to the SwissModel as an “optimize” request for construction of a homology model. The Swiss-PdbViewer was used to export views of the model as Mega-Pov scenes that were rendered using POV-Ray (http://www.povray.org) version 3.5 for Linux.
Acknowledgments
This work was supported by NIH grant R01 AI18094 to RCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn BY, Gershon PD, Jones EV, Moss B. Identification of rpo30, a vaccinia virus RNA polymerase gene with structural similarity to a eucaryotic transcription elongation factor. Mol Cell Biol. 1990;10:5433–5441. doi: 10.1128/mcb.10.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn BY, Moss B. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci USA. 1992;89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Natl Acad Sci USA. 2004;101:17361–17364. doi: 10.1073/pnas.0408168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss CD, Condit RC. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology. 1993;194:254–262. doi: 10.1006/viro.1993.1256. [DOI] [PubMed] [Google Scholar]
- Bayliss CD, Condit RC. The vaccinia virus A18R gene product is a DNA-dependent ATPase. J Biol Chem. 1995;270:1550–1556. doi: 10.1074/jbc.270.4.1550. [DOI] [PubMed] [Google Scholar]
- Black EP, Condit RC. Phenotypic characterization of mutants in vaccinia virus gene G2R, a putative transcription elongation factor. J Virol. 1996;70:47–54. doi: 10.1128/jvi.70.1.47-54.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black EP, Moussatche N, Condit RC. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- Broyles SS. Vaccinia virus transcription. J Gen Virol. 2003 doi: 10.1099/vir.0.18942-0. on line. Ref Type: Generic. [DOI] [PubMed] [Google Scholar]
- Broyles SS, Moss B. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci USA. 1986;83:3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen LM, Sanders M, Wiler C, Niles EG. Vaccinia virus nucleoside triphosphate phosphohydrolase I is an essential viral early gene transcription termination factor. Virology. 1998;245:360–371. doi: 10.1006/viro.1998.9177. [DOI] [PubMed] [Google Scholar]
- Cohrs RJ, Condit RC, Pacha RF, Thompson CL, Sharma OK. Modulation of ppp(A2′p)nA-dependent RNase by a temperature-sensitive mutant of vaccinia virus. J Virol. 1989;63:948–951. doi: 10.1128/jvi.63.2.948-951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Easterly R, Pacha RF, Fathi Z, Meis RJ. A vaccinia virus isatin-beta-thiosemicarbazone resistance mutation maps in the viral gene encoding the 132-kDa subunit of RNA polymerase. Virology. 1991;185:857–861. doi: 10.1016/0042-6822(91)90559-t. [DOI] [PubMed] [Google Scholar]
- Condit RC, Lewis JI, Quinn M, Christen LM, Niles EG. Use of lysolecithin-permeabilized infected-cell extracts to investigate the in vitro biochemical phenotypes of poxvirus ts mutations altered in viral transcription activity. Virology. 1996a;218:169–180. doi: 10.1006/viro.1996.0177. [DOI] [PubMed] [Google Scholar]
- Condit RC, Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit RC, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Condit RC, Niles EG. Regulation of viral transcription elongation and termination during vaccinia virus infection. Biochim Biophys Acta. 2002;1577:325–336. doi: 10.1016/s0167-4781(02)00461-x. [DOI] [PubMed] [Google Scholar]
- Condit RC, Xiang Y, Lewis JI. Mutation of vaccinia virus gene G2R causes suppression of gene A18R ts mutants: implications for control of transcription. Virology. 1996b;220:10–19. doi: 10.1006/viro.1996.0280. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Wittek R, Moss B. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J Virol. 1981;39:733–745. doi: 10.1128/jvi.39.3.733-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. [Google Scholar]
- Deng L, Shuman S. A role for the H4 subunit of vaccinia RNA polymerase in transcription initiation at a viral early promoter. J Biol Chem. 1994;269:14323–14328. [PubMed] [Google Scholar]
- Deng L, Shuman S. Vaccinia NPH-I, a DExH-box ATPase, is the energy coupling factor for mRNA transcription termination. Genes Dev. 1998;12:538–546. doi: 10.1101/gad.12.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito J, Condit R, Obijeski J. The preparation of orthopoxvirus DNA. J Virol Methods. 1981;2:175–179. doi: 10.1016/0166-0934(81)90036-7. [DOI] [PubMed] [Google Scholar]
- Fu J, Gnatt AL, Bushnell DA, Jensen GJ, Thompson NE, Burgess RR, David PR, Kornberg RD. Yeast RNA polymerase II at 5 A resolution. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- Gershon PD, Ahn BY, Garfield M, Moss B. Poly(A) polymerase and a dissociable polyadenylation stimulatory factor encoded by vaccinia virus. Cell. 1991;66:1269–1278. doi: 10.1016/0092-8674(91)90048-4. [DOI] [PubMed] [Google Scholar]
- Hagler J, Shuman S. Nascent RNA cleavage by purified ternary complexes of vaccinia RNA polymerase. J Biol Chem. 1993;268:2166–2173. [PubMed] [Google Scholar]
- Katz E, Margalith E, Winer B. An isatin beta-thiosemicarbazone (IBT)-dependent mutant of vaccinia virus: the nature of the IBT-dependent step. J Gen Virol. 1973a;21:477–484. doi: 10.1099/0022-1317-21-3-477. [DOI] [PubMed] [Google Scholar]
- Katz E, Winer B, Margalith E, Goldblum N. Isolation and characterization of an IBT-dependent mutant of vaccinia virus. J Gen Virol. 1973b;19:161–164. doi: 10.1099/0022-1317-19-1-161. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- Klug A. Structural biology. A marvellous machine for making messages. Science. 2001;292:1844–1846. doi: 10.1126/science.1062384. [DOI] [PubMed] [Google Scholar]
- Kovacs GR, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner CA, Condit RC. Vaccinia virus gene A18R DNA helicase is a transcript release factor. J Biol Chem. 2000;275:1485–1494. doi: 10.1074/jbc.275.2.1485. [DOI] [PubMed] [Google Scholar]
- Lackner CA, D'Costa SM, Buck C, Condit RC. Complementation analysis of the Dales collection of vaccinia virus temperature-sensitive mutants. Virology. 2003;305:240–259. doi: 10.1006/viro.2002.1745. [DOI] [PubMed] [Google Scholar]
- Latner DR, Thompson JM, Gershon PD, Storrs C, Condit RC. The positive transcription elongation factor activity of the vaccinia virus J3 protein is independent from its (nucleoside-2′-O-) methyltransferase and poly(A) polymerase stimulatory functions. Virology. 2002;301:64–80. doi: 10.1006/viro.2002.1538. [DOI] [PubMed] [Google Scholar]
- Latner DR, Xiang Y, Lewis JI, Condit J, Condit RC. The vaccinia virus bifunctional gene J3 (nucleoside-2′-O-)- methyltransferase and poly(A) polymerase stimulatory factor is implicated as a positive transcription elongation factor by two genetic approaches. Virology. 2000;269:345–355. doi: 10.1006/viro.2000.0243. [DOI] [PubMed] [Google Scholar]
- Luttge BG, Moyer RW. Suppressors of a host range mutation in the rabbitpox virus serpin SPI-1 map to proteins essential for viral DNA replication. J Virol. 2005;79:9168–9179. doi: 10.1128/JVI.79.14.9168-9179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr A, Roberts BE. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J Virol. 1984;49:510–520. doi: 10.1128/jvi.49.2.510-520.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meis RJ, Condit RC. Genetic and molecular biological characterization of a vaccinia virus gene which renders the virus dependent on isatin-beta-thiosemicarbazone (IBT) Virology. 1991;182:442–454. doi: 10.1016/0042-6822(91)90585-y. [DOI] [PubMed] [Google Scholar]
- Mohamed MR, Niles EG. Interaction between nucleoside triphosphate phosphohydrolase I and the H4L subunit of the viral RNA polymerase is required for vaccinia virus early gene transcript release. J Biol Chem. 2000;275:25798–25804. doi: 10.1074/jbc.M002250200. [DOI] [PubMed] [Google Scholar]
- Moss B. Poxviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 2849–2884. [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Pacha RF, Condit RC. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985;56:395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacha RF, Meis RJ, Condit RC. Structure and expression of the vaccinia virus gene which prevents virus-induced breakdown of RNA. J Virol. 1990;64:3853–3863. doi: 10.1128/jvi.64.8.3853-3863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins C, Cresawn SG, Condit RC. An isatin-beta-thiosemicarbazone-resistant vaccinia virus containing a mutation in the second largest subunit of the viral RNA polymerase is defective in transcription elongation. J Biol Chem. 2004;279:44858–44871. doi: 10.1074/jbc.M408167200. [DOI] [PubMed] [Google Scholar]
- Reines D, Ghanouni P, Gu W, Mote J, Jr, Powell W. Transcription elongation by RNA polymerase II: mechanism of SII activation. Cell Mol Biol Res. 1993;39:331–338. [PubMed] [Google Scholar]
- Schnierle BS, Gershon PD, Moss B. Cap-specific mRNA (nucleoside-O2′-)- methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc Natl Acad Sci USA. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Visca P, Vos JC, Stunnenberg HG. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell. 1987;50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Broyles SS, Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J Biol Chem. 1987;262:12372–12380. [PubMed] [Google Scholar]
- Simpson DA, Condit RC. Vaccinia virus gene A18R encodes an essential DNA helicase. J Virol. 1995;69:6131–6139. doi: 10.1128/jvi.69.10.6131-6139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulme F, Mosrin-Huaman C, Sparkowski J, Das A, Leng M, Rahmouni AR. GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J. 2000;19:6853–6859. doi: 10.1093/emboj/19.24.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Coroneos AM. The H4 subunit of vaccinia virus RNA polymerase is not required for transcription initiation at a viral late promoter. J Virol. 1995;69:2602–2604. doi: 10.1128/jvi.69.4.2602-2604.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y. Ph D Thesis. 1998. [Google Scholar]
- Xiang Y, Latner DR, Niles EG, Condit RC. Transcription elongation activity of the vaccinia virus J3 protein in vivo is independent of poly(A) polymerase stimulation. Virology. 2000;269:356–369. doi: 10.1006/viro.2000.0242. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Simpson DA, Spiegel J, Zhou A, Silverman RH, Condit RC. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J Virol. 1998;72:7012–7023. doi: 10.1128/jvi.72.9.7012-7023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]