Abstract
Oligomerization has been proposed as one of several mechanisms to regulate the activity of G protein-coupled receptors (GPCRs), but little is known about the structure of GPCR oligomers. Crystallographic analyses of two new crystal forms of rhodopsin reveal an interaction surface which may be involved in the formation of functional dimers or oligomers. New crystallization conditions lead to the formation of two crystal forms with similar rhodopsin-rhodopsin interactions, but changes in the crystal lattice are induced by the addition of different surfactant additives. However, the intermolecular interactions between rhodopsin molecules in these crystal structures may reflect the contacts necessary for the maintenance of dimers or oligomers in rod outer segment membranes.Similar contacts may assist in the formation of dimers or oligomers in other GPCRs as well. These new dimers are compared with other models proposed by crystallography or EM and AFM studies. The inter-monomer surface contacts are different for each model, but several of these models coincide in implicating helix I, II, and H-8 as contributors to the main contact surface stabilizing the dimers.
Introduction
G protein-coupled receptors (GPCRs) provide a way for cells to evaluate and respond to environmental signals. Members of this large family of transmembrane receptors (950 genes in the human genome) are crucial molecular sensors in important physiological processes, and they make up the largest class of targets for therapeutic agents (Bjenning et al., 2004; Dahl and Sylte, 2005; Doggrell, 2004). Despite the importance of this family, X-ray crystallographic studies of GPCRs have been limited to rhodopsin due to difficulties in the expression, purification and crystallization of most integral membrane proteins.
GPCRs, in the form of rhodopsin and the cone opsins, provide the initial molecular response to light in visual systems (Filipek et al., 2003; McBee et al., 2001). In rhodopsin's ground state, a buried chromophore, 11-cis-retinal, is covalently bound to the protein via a Schiff base linkage to a lysine side-chain. When a photon is absorbed, the chromophore changes its conformation to all-trans-retinylidene. The polypeptide portion of the receptor responds to the chromophore's conformational changes and undergoes further structural changes which include alterations of the intracellular surface. In this activated state (Meta II), the protein can activate its cognate heterotrimeric G protein, transducin (Gt) through the exchange of GTP for GDP on the Gαt subunit (Okada et al., 2001; Palczewski, 2006). The activated Gαt then participates in a signaling cascade that gives rise to a signal recognized by the visual cortex of the brain (Rodieck, 1998).
For nearly all GPCRs, the external signal is the binding of a small molecule ligand (agonist), and this leads to conformational changes in the protein that transmit the signal across the plasma membrane. In the case of rhodopsin, binding of a small molecule agonist has been replaced by absorption of a photon by the protein's covalently bound retinal chromophore. Structural changes in the retinal and protein result in formation of the active state of the receptor. The steps following the initial signal are believed to be very similar for all GPCRs. Sequence similarities among the GPCRs and among the heterotrimeric G-proteins argue for a common activation mechanism for these receptors (Ballesteros et al., 2001).
Rhodopsin is the best characterized GPCR (Menon et al., 2001), largely because it is relatively easily purified from bovine eyes. It has been extensively studied using biophysical and structural methods (Palczewski, 2006). Rhodopsin was the first GPCR studied using cryo-electron microscopic techniques which showed the seven transmembrane helices characteristic of this protein family (Davies et al., 2001; Schertler et al., 1993; Unger et al., 1997). The first X-ray crystallographic structure of a GPCR was that of ground-state rhodopsin (PDB entry 1F88 (Palczewski et al., 2000)). Improved models for that crystal form have resulted from further refinement (entries 1HZX (Teller et al., 2001) and 1L9H (Okada et al., 2002)), and higher resolution studies (2.2 Å) (1U19 (Okada et al., 2004)). The ground-state structure in a trigonal crystal form has also been determined (1GZM (Edwards et al., 2004; Li et al., 2004)).
The structure of Meta I rhodopsin, the photostate immediately preceding Meta II, has been studied using cryo-EM approaches (Ruprecht et al., 2004). Crystal structures of the early photo states, batho-rhodopsin (Nakamichi and Okada, 2006a) and lumi-rhodopsin (Nakamichi and Okada, 2006b), have been reported that are isomorphous with the ground-state tetragonal structure. Only small conformational differences have been found among these structures of rhodopsin.
What is not yet available from these studies is how the rhodopsin structure changes from Meta I rhodopsin to Meta II, the transducin activating conformation. A number of biophysical and biochemical approaches have allowed the identification of the α-helices and loops that change upon activation (Choi et al., 2002; Hamm, 2001; Meng and Bourne, 2001).
In pursuit of the structure of photo-activated rhodopsin, we have found two new crystal forms that diffract poorly, but remain ordered after absorption of light (Salom et al., 2006a; Salom et al., 2006b) (see Table 1). Examination of the molecular packings in these two new crystal forms, one rhombohedral and one trigonal, reveals the existence of rhodopsin dimers formed by subunit-subunit interactions that can be physiologically relevant to the quaternary structure of rhodopsin and other GPCRs.
Table 1.
Summary of data set and refinement statistics
| rhombohedral, ground state | trigonal, ground state | trigonal, photoactivated | |
|---|---|---|---|
| space group | R32 | P3112 | P3112 |
| unit cell dimensions, | a=277.5 Å
c=66.3 Å |
a=159.5 Å
c=141.4 Å |
a=161.1 Å
c=144.8 Å |
| resolution | 3.8 Å | 4.1 Å | 4.2 Å |
| Rmerge | 0.080 | 0.087 | 0.060 |
| Rwork | 0.373 | 0.382 | 0.373 |
| Rfree | 0.418 | 0.411 | 0.384 |
| Wilson B factor | 227 Å2 | 185 Å2 | 183 Å2 |
| PDB ident | 2I35 | 2I36 | 2I37 |
Resolution limit defined as when <F2>=<σ(F2)>
Rmerge on F2
Data sets were obtained at ALS, SSRL, and SLS.
Results and Discussion
Packing Analysis of the Rhombohedral Crystal Form
The rhombohedral crystal form (space group R32) contains a single rhodopsin molecule in the asymmetric unit. Figure 1a shows the rhombohedral crystal structure (in projection). All of the molecules in this crystal form are related by crystallographic symmetry operations and located in equivalent packing environments.
Figure 1.
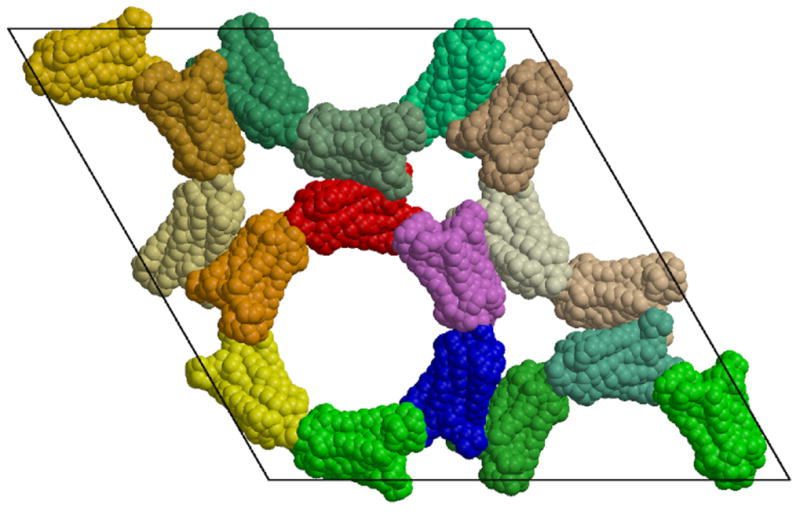
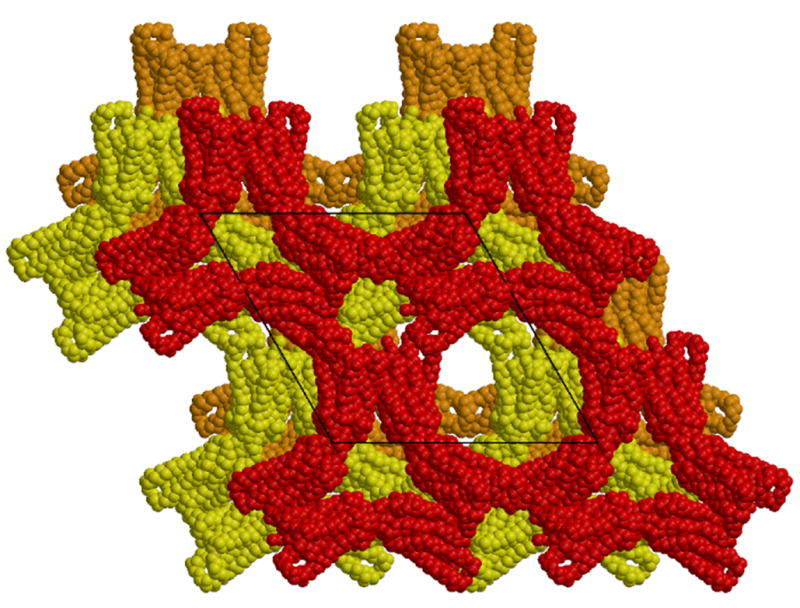
a). Arrangement of 18 rhodopsin monomers in the non-primitive, hexagonal setting of the rhombohedral unit cell, projected along the z axis. The space group is R32, with three-fold screw axes along the z axis located in the centers of the rings of subunits (in projection). One ring is colored red through violet. This crystal form contains a single rhodopsin monomer per asymmetric unit. Dimers of rhodopsin hold the rings together. The molecules colored red and dark-green form one such dimer. b). Packing diagram for the P3112 trigonal crystal form. Each of the three layers of molecules in the trigonal unit cell is related to the layer of molecules in the rhombohedral cell. This packing leads to a crystal lattice containing less solvent. The rhombohedral cell contains ∼80% solvent while the trigonal crystal form contains ∼71% solvent. Figure drawn with Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
The crystal is made up of tubes of rhodopsin molecules surrounding three-fold screw axes (parallel to the z axis of the unit cell). The tubes are formed by helical arrangements of monomers interacting through their extracellular and cytoplasmic surfaces with alternating orientations of the molecules along the helix. The helix extending across the entire crystal is formed by unit cells stacked along the z axis. Thus the pitch of this helix is the same as the c unit cell dimension, i.e., 66.3Å. The helical arrangement of the molecules making up the walls of these tubes is shown in Figure 2. Adjacent tubes forming the crystal are related by two-fold rotation axes and have the same helical sense.
Figure 2.
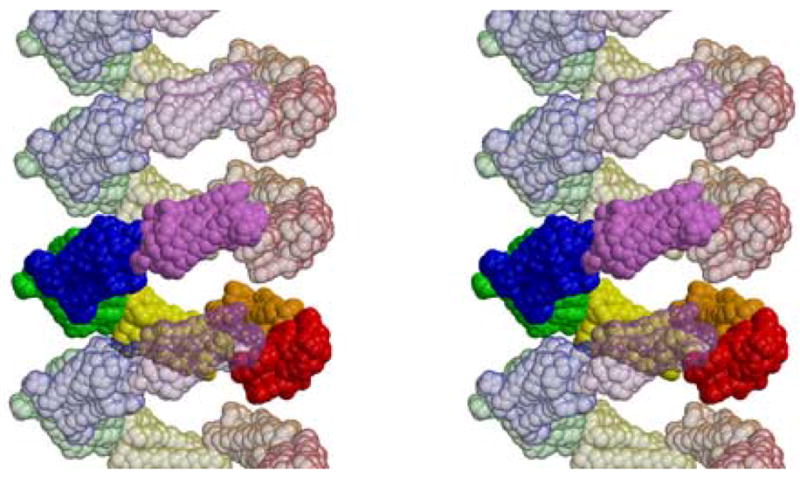
Stereoview of the rings of rhodopsin monomers in the rhombohedral crystal form showing that they are helical arrangements around the three-fold screw axes. One ring is show in this stereoscopic side view, with adjacent rings shown as transparent molecules. Subunits in one ring do not contact subunits directly above or below them. The three-dimensional structure is stablized by contacts to neighboring helices. Dimers form these contacts. Figure drawn with Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
The lateral connections between the tubes are formed by side-by-side interactions between pairs of rhodopsin molecules. These interactions are described in more detail below. They involve the hydrophobic surfaces of the transmembrane helices. The orientations of the molecules in the dimer are consistent with the parallel orientations in their native membranes. The subunits within each dimer are related by a crystallographic two-fold rotation axis that would be perpendicular to the plane of the membrane bilayer containing the rhodopsin molecules.
The protein molecules occupy about 20% of the crystal volume. At this resolution, there is no evidence for the existence of detergent micelles covering the hydrophobic surfaces of the protein. The large amount of solvent space in the crystals likely contributes to the disorder in the structure giving rise to the low resolution of the diffraction pattern.
Packing Analysis of the Trigonal Crystal Form
The molecules in the rhombohedral crystal form (Figure 1a) make up a layer in an extended structure formed by the unit cell translations along z. Similar layers are seen in the new trigonal crystal form (space group P3112) (Figure 1b). Each trigonal unit cell contains three layers, with adjacent layers along the z axis shifted by a third of the repeat distance along the x direction (and directions related by the trigonal symmetry). This shifting of the layers limits the helical tubes to only one turn of helix, forming a "lock-washer" structure in three-dimensions.
The trigonal crystal form grows consistently in the presence of a wide variety of additives (Salom et al., 2006a). To obtain both crystal forms, rhodopsin was solubilized with nonylglucoside, purifed/delipidated by immunochromatography, and concentrated by ammonium sulfate-induced phase separation. The main difference in the growing conditions is that trigonal crystals were grown in the presence of the surfactant Merpol HCS, whereas rhombohedral crystals contained Merpol DA
The staggered packing of layers is likely more stable than the stacked layers in the rhombohedral lattice. The protein occupies 29.0% of the trigonal unit cell, somewhat more than the volume occupied in the rhombohedral crystal form (20%). The contacts between the subunits in the dimers in both crystal forms involve residues in helices I and II as well as H-8 and are discussed in more detail below.
Hexameric Packing Units
While the helical tubes and lock-washers provide the major crystal packing contacts along the z axis, a more fundamental packing unit in both crystal forms is a hexamer as shown in Figure 3 for the trigonal crystals. The three crystallographically unique subunits (shown as red, blue and beige subunits in Figure 3) are related by a crystallographic two-fold axis to three others, forming a hexamer containing three parallel dimers. In the trigonal crystals, a non-crystallographic three-fold rotation axis is oriented perpendicular to the view shown in Figure 3. The hexamer has approximate 32 symmetry (D3), but only one of the two-fold axes is crystallographic. The other two are non-crystallographic axes and apply only to the particular layer containing the hexamer.
Figure 3.
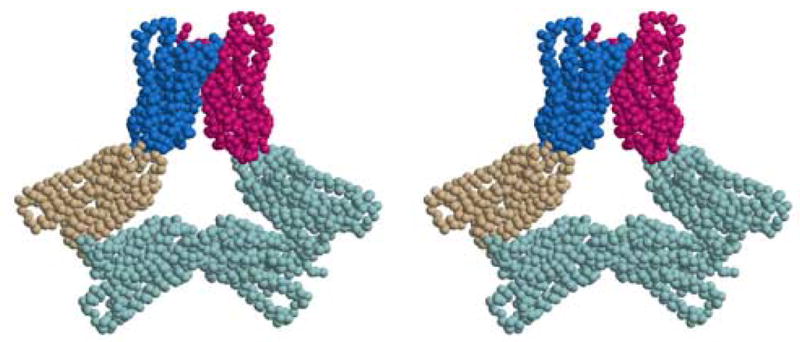
Stereoview of the hexamer building block from the trigonal crystals. The hexamer shows pseudo-D3 symmetry, but only one of its two-fold axes coincides with a crystallographic symmetry axis. The three subunits (red, blue, and beige) are crystallographically inequivalent. A similar hexamer is the fundamental building block for the R32 crystal form. Figure drawn with Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
A similar hexamer is found in the rhombohedral structure, but because the subunits are all identical in that crystal structure, that hexamer possesses exact D3 symmetry. The three-fold and two-fold axes are all crystallographic and apply to adjacent layers as well as to the layer in which they are found.
While the hexamers are very similar in the two crystal forms, they differ slightly, and this might be associated with the relative stabilities of the crystal forms. These differences are all manifestations of a slight difference in the subunit-subunit interface within each of the side-by-side dimers. The trigonal hexamers possess only one crystallographic two-fold rotation axis, so the twisting of the monomer-monomer contact within the dimer interface results in a reorientation of the arms of the hexamer. This reduces the pitch of the lock-washers in the trigonal crystals to 34.5Å. It is unclear how or whether this would limit the stacking of the lock washers to favor the tri-layered trigonal crystal packing.
While the hexamer is a structural unit common to both crystal forms, there is no evidence for its existence in the rhodopsin solutions. Rhodopsin dimers have been observed in other studies (Jastrzebska et al., 2006), but the oligomeric state of rhodopsin in the solutions used to generate these crystal forms is unknown.
Rhodopsin-Rhodopsin Interactions
Three types of inter-protein interactions suffice to build up the two structures. One type of interaction involves the extracellular surfaces of two rhodopsin molecules and contributes to the formation of the hexamers as well as the rings or lock washers. The accessible surface buried in this interaction in the rhombohedral crystals is 677 Å2 per molecule.
The cytoplasmic faces of adjacent rhodopsin molecules also interact, but with a significantly smaller accessible surface of 165 Å2. This forms the connection between hexamers, but the strength of this interaction can not be high. This weak interaction could contribute to the relatively poor crystal quality. Because this interface is minimal, the rhodopsin is largely unconstrained, and the crystal is capable of accommodating the conformational changes associated with photoactivation (unlike previously described crystals that show no diffraction pattern upon photoactivation). The small extent of the inter-monomer contacts could also account for the limited resolution.
Rhodopsin Dimers
The third type of rhodopsin-rhodopsin interaction seen in these new crystal forms is between side-by-side molecules in a parallel dimer that could serve as a model of rhodopsin dimers in their native membranes (Figure 4). GPCRs are believed to function as homo- or hetero-dimers (Angers et al., 2002; Lee et al., 2000; Milligan et al., 2003; Terrillon and Bouvier, 2004), but the quaternary arrangement of the individual GPCR molecules in these dimers is not known. Earlier X-ray crystallographic studies of rhodopsin contained anti-parallel or head-to-tail dimers with the molecules' cytoplasmic faces oriented toward what would be opposite sides of the membrane.
Figure 4.
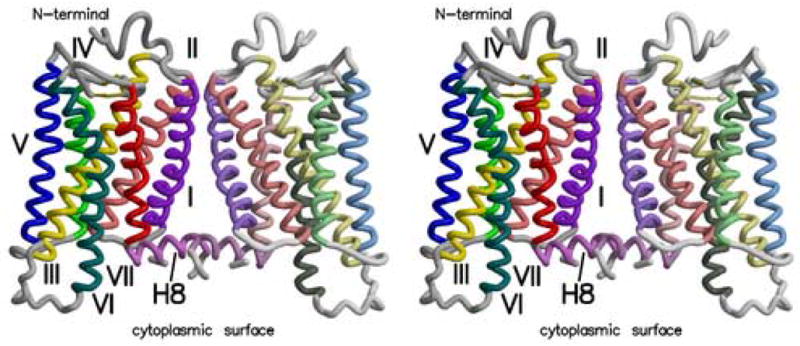
Stereoview of the rhodopsin dimer found in the rhombohedral crystal form (PDB ident 2I35). Transmembrane helices are numbered from I to VII. Helix 8 is denoted by H-8. Helices are colored as follows: Helix I - purple, II – orange, III – yellow, IV – green, V – blue, VI – violet, VII – red, H-8 – violet. Figure drawn with Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
The side-by-side dimers in the new crystal forms described here are appropriately aligned, but the low accessible surface area involved in their interaction (362 Å2, again for the rhombohedral structure) indicates that their interactions may be weak and dimerization through this surface may be reversible in membranes. It should be noted that the rhodopsin in these crystals is almost completely delipidated, and perhaps in the membrane, specific lipid interactions could also stabilize these dimers. The protein-protein interactions in these dimers involve residues from helices I, II and H-8, as well as the palmitoyl chains covalently bound to the protein (Salom et al., 2006b). The primary contact between helix I in one subunit and helix I in the other involves residues Phe45, Leu46, Met49 and Phe52. These four residues, and the same ones in the other subunit, form a cluster at the closest contact between helices I in each molecule. Tyr96 and His100 from helix II in each subunit also form another cluster of side-chains connecting the two subunits. Finally, the palmitoyl groups attached to Cys322 and Cys323, are close to the H-8 helix and the palmitoyls from the adjacent subunit. The locations of the palmitoyls are uncertain, given the low resolution of these structure analyses, but their hydrocarbon chains could occupy the space between the subunits and contribute to the stability of the dimer.
Given the size of the contact surfaces in this dimer, it is difficult to imagine that the dimer would be a stable entity. If this were a soluble protein, that would certainly be the case. However rhodopsin is a membrane protein, and membrane phospholipids must complicate considerations of dimer stability in these environments. Local concentration effects due to the location of the proteins in a two-dimensional membrane will also affect the stability of the dimer. Also, buried solvent accessible surface might not be an appropriate metric for understanding the strength of membrane protein interactions. Accessible surface areas are useful when hydrophobic surfaces are removed from interactions with water, but the surfaces involved here are hydrophobic surfaces being removed from interactions with the hydrophobic components of the membrane. Burial of hydrophobic residues plays a smaller role in the folding of membrane proteins than for the folding of soluble proteins (DeGrado et al., 2003). This likely holds as well for the aggregation of membrane proteins, be it the formation of oligomers or crystals.
Comparison of Observed and Proposed Rhodopsin Dimers
Candidate regions for the interfaces between subunits in GPCR dimers have come from a number of biochemical studies (Fotiadis et al., 2006; Javitch, 2004; Park et al., 2004), and these experiments indicate that transmembrane helices I, IV, V and VI are located at the subunit-subunit boundary.
In addition to the dimers seen in these crystal forms (Figure 5a), dimers are observed in cryo-EM studies of two dimensional crystals of various rhodopsins. The Meta I rhodopsin dimer (Ruprecht et al., 2004) (Figure 5b) provides the best opportunity for comparison. Helices I and H-8 contribute to the inter-subunit contacts in this dimer, just as they do in the dimers seen in our crystal forms. However, as can be seen in comparing Figures 5a and 5b, the orientations of the subunits relative to each other and the membrane surface in the two types of dimers are significantly different. Helix I in each monomer interacts with helix I in the other subunit, but different faces of the helices interact in the two dimers. This changes the residues in helix I that are in closest contact with the other subunit by shifting the interacting surface about 10 residues towards the N-terminus when compared with the contacts in the rhombohedral and trigonal dimers. This corresponds to a shift of several turns of α-helix and makes the Meta I dimer substantially different from that seen in the three-dimensional crystals.
Figure 5.
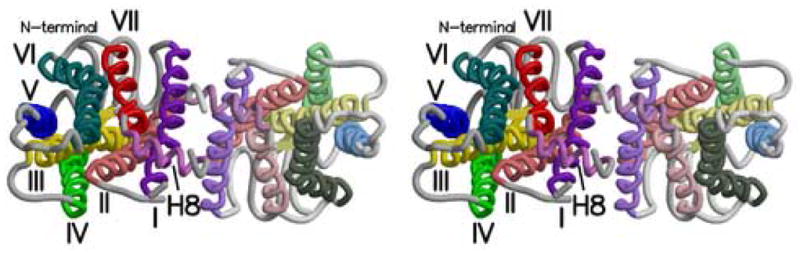
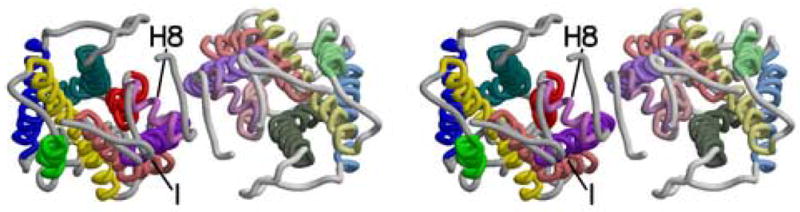
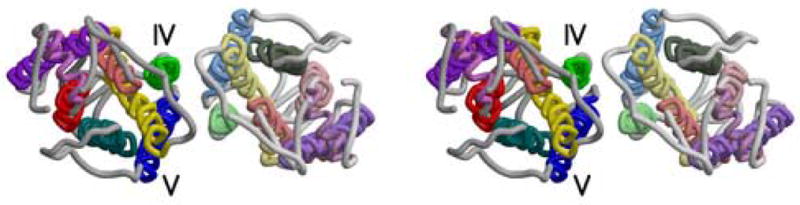
Stereoviews of three parallel rhodopsin dimers as seen from the cytoplasm. a) Rhodopsin dimer observed in the rhombohedral crystal form (PDB ident 2I35). b) Dimer from the Meta I cryo-EM structure. Coordinates were generated by arranging monomers from 1GZM in a manner identical to figures in (Ruprecht et al., 2004; Vogel et al., 2004). c) Model dimer based on AFM images of rhodopsin in native rod outer segment membranes (PDB ident 1N3M) (Liang et al., 2003). Figure drawn with Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
Another dimer model (Figure 5c) has been developed to account for AFM and EM images of rhodopsin in native rod disc membranes (Fotiadis et al., 2003; Fotiadis et al., 2004; Liang et al., 2003). Helices IV and V play major roles in the inter-subunit contacts within this dimer. Javitch and colleagues (Guo et al., 2005) have recently reported that the subunit interface in dopamine D2 receptor dimers is similar to that modeled from the AFM results. This dimer is formed using parts of the transmembrane surface that are diametrically opposed to the surfaces seen in the dimers from the rhombohedral and trigonal crystals.
However, a secondary dimer interface stabilized by weaker interactions was also proposed in the AFM study to be present in the two-dimensional arrays of rhodopsin in rod outer segment membranes. This secondary dimer forms the contacts between the double rows of rhodopsin molecules seen in the AFM images. The interactions between the double rows are not as regular as those within the rows, so presumably these secondary contacts are more easily broken and perhaps are less stable (Fotiadis et al., 2006; Fotiadis et al., 2004; Liang et al., 2003). These interactions were proposed to involve helix I and helix H-8 as do the Meta I rhodopsin dimer (Ruprecht et al., 2004) and the crystallographic dimers presented here.
Conclusions
Rhodopsin protein crystals have been grown in two new three-dimensional crystal forms, with slightly different crystal packing and intermolecular contacts. This is not altogether surprising as the contacts leading to crystalline lattices are largely hydrophobic and utilize contact surfaces that are normally masked by the rod outer segment membrane. Interestingly, there are commonalities seen in several of the crystal forms. In the 1F88 tetragonal structure, rhodopsin was arrayed in a head to tail dimer, which utilized helix I as a contact surface. In both the trigonal and rhombohedral crystal forms described here, contacts along this same helix make up the core of the observed dimers, albeit in a parallel orientation more consistent with the orientation found in native membranes.
The growth of our trigonal and rhombohedral crystal forms required small quantities of the surfactants Merpol HCS and Merpol DA respectively (Salom et al., 2006b). These studies demonstrate that the varieties of crystal forms that can be obtained for a single membrane protein can depend on the level of delipidation (Guan et al., 2006) as well as the use of specific additives and detergents, suggesting that the inclusion of lipid as well as surfactant additives may be beneficial for the crystallization of membrane proteins.
A characteristic of oligomers of globular proteins is that the subunit-subunit interactions are strong and specific. This leads to well-formed, regular oligomers. In the case of membrane proteins, restricted largely to inter-subunit interactions within the two-dimensional membrane and thus to interactions between their hydrophobic surfaces, you might expect to find considerable variability in their quaternary structures. Given our examination of rhodopsin-rhodopsin interactions to date, it is clear that the hydrophobic transmembrane surfaces of rhodopsin and other GPCRs are capable of forming more than one kind of dimer or higher order oligomer. Further structural studies will likely produce examples of dimers using other helices and surfaces in their rhodopsin-rhodopsin interactions. This leaves open the possibility that GPCRs might take on a number of different homo- and hetero-oligomeric structures, depending on the particular GPCR and its particular environment.
Acknowledgments
This research was supported in part by the U.S. Public Health Service grants EY01730 and EY08061 from the National Eye Institute, and GM63020 from NIGMS, National Institutes of Health, Bethesda, MD. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Other portions were carried out at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under Contract No. DE-AC03–76SF00098 at Lawrence Berkeley National Laboratory. Part of this work was performed at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angers S, Salahpour A, Bouvier M. Dimerization: An emerging concept for G protein-coupled receptor ontogeny and function. Annu Rev Pharmacol Toxicol. 2002;42:409–435. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Shi L, Javitch JA. Structural mimicry in G protein-coupled receptors: Implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol Pharmacol. 2001;60:1–19. [PubMed] [Google Scholar]
- Bjenning C, Al-Shamma H, Thomsen W, Leonard J, Behan D. G protein-coupled receptors as therapeutic targets for obesity and type 2 diabetes. Curr Opin Investig Drugs. 2004;5:1051–1062. [PubMed] [Google Scholar]
- Choi G, Landin J, Galan JF, Birge RR, Albert AD, Yeagle PL. Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin. Biochemistry. 2002;41:7318–7324. doi: 10.1021/bi025507w. [DOI] [PubMed] [Google Scholar]
- Dahl SG, Sylte I. Molecular modelling of drug targets: the past, the present and the future. Basic Clin Pharmacol Toxicol. 2005;96:151–155. doi: 10.1111/j.1742-7843.2005.pto960302.x. [DOI] [PubMed] [Google Scholar]
- Davies A, Gowen BE, Krebs AM, Schertler GF, Saibil HR. Three-dimensional structure of an invertebrate rhodopsin and basis for ordered alignment in the photoreceptor membrane. J Mol Biol. 2001;314:455–463. doi: 10.1006/jmbi.2001.5167. [DOI] [PubMed] [Google Scholar]
- DeGrado WF, Gratkowski H, Lear JD. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 2003;12:647–665. doi: 10.1110/ps.0236503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggrell SA. New drugs and new targets. Drug News Perspect. 2004;17:615–632. [PubMed] [Google Scholar]
- Edwards PC, Li J, Burghammer M, McDowell JH, Villa C, Hargrave PA, Schertler GF. Crystals of native and modified bovine rhodopsins and their heavy atom derivatives. J Mol Biol. 2004;343:1439–1450. doi: 10.1016/j.jmb.2004.08.089. [DOI] [PubMed] [Google Scholar]
- Filipek S, Stenkamp RE, Teller DC, Palczewski K. G protein-coupled receptor rhodopsin: A Prospectus. Annu Rev Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: A working model for G-protein-coupled receptors. Curr Opin Struct Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564:281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Smirnova IN, Verner G, Nagamori S, Kaback HR. Manipulating phospholipids for crystallization of a membrane transport protein. Proc Natl Acad Sci USA. 2006;103:1723–1726. doi: 10.1073/pnas.0510922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm HE. How activated receptors couple to G proteins. Proc Natl Acad Sci, USA. 2001;98:4819–4821. doi: 10.1073/pnas.011099798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitch JA. The ants go marching two by two: Oligomeric structure of G-protein-coupled receptors. Mol Pharmacol. 2004;66:1077–1082. doi: 10.1124/mol.104.006320. [DOI] [PubMed] [Google Scholar]
- Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and Structural Characterization of Rhodopsin Oligomers. J Biol Chemistry. 2006;281:11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript - a program to produce both detailed and schematic plots of protein structures. J Appl Crystallography. 1991:946–950. [Google Scholar]
- Lee SP, Xie Z, Varghese G, Nguyen T, O'Dowd BF, George SR. Oligomerization of dopamine and serotonin receptors. Neuropsychopharmacology. 2000;23:S32–40. doi: 10.1016/S0893-133X(00)00155-X. [DOI] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBee JK, Palczewski K, Baehr W, Pepperberg DR. Confronting complexity: The interlink of phototransduction and retinoid metabolism in the vertebrate retina. Prog Retin Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- Meng EC, Bourne HR. Receptor activation: what does the rhodopsin structure tell us? Trends in Pharmacological Sciences. 2001;22:587–593. doi: 10.1016/s0165-6147(00)01825-3. [DOI] [PubMed] [Google Scholar]
- Menon ST, Han M, Sakmar TP. Rhodopsin: Structural basis of molecular physiology. Physiol Rev. 2001;81:1659–1688. doi: 10.1152/physrev.2001.81.4.1659. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon JD. Raster3D: Photorealistic Molecular Graphics. Methods in Enzymology. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Milligan G, Ramsay D, Pascal G, Carrillo JJ. GPCR dimerisation. Life Sci. 2003;74:181–188. doi: 10.1016/j.lfs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed. 2006a;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006b doi: 10.1073/pnas.0601765103. Epub, ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: New insights from structural and biochemical studies. Trends in Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Åcrystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Palczewski K. G protein-coupled receptor rhodopsin. Annual Review of Biochemistry. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Park PS, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW. The first steps in seeing. Sinauer Associates; Sunderland, Mass: 1998. [Google Scholar]
- Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GF. Electron crystallography reveals the structure of metarhodopsin I. Embo J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D, Le Trong I, Pohl E, Ballesteros JA, Stenkamp RE, Palczewski K. Improvements in G protein-coupled receptor purification yield light stable rhodopsin crystals. J Structural Biology. 2006a;156:497–504. doi: 10.1016/j.jsb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin, Proc. Natl Acad Sci, USA. 2006b;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler GF, Villa C, Henderson R. Projection structure of rhodopsin. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM, Hargrave PA, Baldwin JM, Schertler GF. Arrangement of rhodopsin transmembrane alpha-helices. Nature. 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- Vogel R, Ruprecht J, Villa C, Mielke T, Schertler GF, Siebert F. Rhodopsin photoproducts in 2D crystals. J Mol Biol. 2004;338:597–609. doi: 10.1016/j.jmb.2004.03.006. [DOI] [PubMed] [Google Scholar]


