Abstract
This translational research program applies a working model of advanced functional genomics/proteomics and bioinformatics to human peripheral arterial occlusive disease (PAOD). It is a multidisciplinary collaborative effort of clinicians, scientists, and statisticians with an advisory panel comprised of experts in inflammation biology, vascular biology, molecular genetics, bioinformatics, clinical trial design, and epidemiology. The proposed human initiative is designed to study 300 symptomatic patients with PAOD undergoing medical management with or without vascular intervention by either lower extremity angioplasty/stenting or vein graft bypass. The study aims to test the hypothesis that the systemic inflammatory response following vascular intervention influences the local milieu responsible for vascular repair and adaptation. The expectation is that this response is not uniform in all patients, but rather, is modulated by either preoperative genetic predisposition or post-procedure differential regulation of the innate immune response to injury that promotes a maladaptive phenotype leading to intervention failure. Therefore, some of these differences may be present and detectable pre-intervention amenable to class prediction and prospective treatment strategies, while others may be detectable in the early post-procedure period, prior to the onset of clinical failure, permitting interventions to prevent an adverse outcome. The combination of genomic/proteomic data together with functional and quality of life outcome measures to define a critical model for class prediction and analysis should lead to new knowledge about failure mechanisms of vascular intervention and new strategies to improve existing approaches to lower extremity revascularization.
Introduction
Outcomes following lower extremity revascularization for peripheral arterial occlusive disease (PAOD) continue to be disappointing. Conventional wisdom suggests 5-year bypass patency rates of 60–80%,1–6 but more recent information suggets a concerning 1–year primary patency rate of only 61% for vein bypass.7 Outcomes are less well defined for angioplasty/stenting, but primary patency rates of 70–90% at 3 months that drop to an unacceptable 20–50% at 1 – 3 years have been described.8–10 Furthermore, these results are continually being scrutinized in the context of ~80% improvement in patients with intermittent claudication treated with conservative measures (i.e. smoking cessation, risk factor modification, and structured exercise),11––13 and reports of poor functional and quality of life outcomes despite successful revascularization.14, 15 Unfortunately, there is a poor understanding of the disease process of lower extremity PAOD, the arterial response to angioplasty, the vein graft response to arterial hemodynamics, or what metrics constitute the definition of success or failure of such interventions. Consequently, without a defined evidence-based approach to symptomatic lower extremity PAOD, management decisions are frequently made without a clear understanding of how to individualize the treatment to optimize patient outcomes.16–18
Research over that last decade has shifted away from a focus on local mediators at sites of vascular injury as the stimulus for vascular smooth muscle cell pathology leading to inward vessel remodeling and end organ ischemia. Current theory holds that the blood vessel response to injury may be intimately linked to the host's systemic inflammatory response, and that negative remodeling may be driven by these systemic factors.19–22 In patients with atherosclerosis, this association has been established globally (i.e. serum C-reactive protein),23 but a detailed understanding of the systemic pathways and mechanisms that direct local blood vessel wall adaptation to physical perturbations remains lacking. The critical role that systemic inflammation plays in directing local responses to vascular injury at the time of intervention is the topic of another component of this Supplement (see Ozaki, Cytokines and the Early Vein Graft –Strategies to Enhance Durability). However, despite the important findings reviewed in that article, decades of focus on local vascular wall events have failed to yield substantial progress toward more durable peripheral interventions.
Genomic/Proteomic Application to Human Vascular Disease
A paradigm shift has occurred recently, away from the focused study of local factors in the vessel wall towards the study of the influence of systemic inflammation on these local events that ultimately lead to revascularization failure. This new approach has been fueled in part by a broad based human initiative taking advantage of advances in the sequencing of the human genome and the development of high throughput genomic and proteomic analyses. This sort of approach opens further avenues of discovery into specific single nucleotide polymorphisms (SNPs) linked to intervention failure, gene expression profiles or molecular signatures in the interactome that predict successful or unsuccessful outcomes, pharmacogenomic markers that might help direct anti-inflammatory treatments, and complex genotype-environmental interactions that ultimately determine outcome.24 In this way, through a systems biology approach, we are empowered to translate changes in basic building blocks (i.e. gene sequence), to changes in gene function (functional genomics), and finally to changes in organ function or clinical phenotype (physiological genomics).25
Little exists in the literature at present describing application of these methods to patients with symptomatic lower extremity PAOD.26, 27 However, due to the potential impact and importance of this type of investigation, the National Heart, Lung, and Blood Institute (NHLBI) began a Genomics Initiative in 2000 designed to provide funding for programs looking to correlate the vast information, technology, and resources made available from the Human Genome Project with the physiology and pathophysiology of human cardiovascular disease.28 As a result, several Programs for Genomic Applications (PGAs) and Centers of Excellence in Genomic Studies (CEGS) have been funded to study areas ranging from animal models of cardiovascular disease, to application of high throughput genomics, to cardiovascular system development and disease. What is available in the literature are a number of observational studies that have linked a putative SNP with some aspect of cardiovascular disease – most commonly hypertension or heart failure, or the response to a particular pharmacologic intervention. Genes associated with cardiovascular disease in these studies include myocyte enhancer factor-2 (MEF2A)29, connexin 37 gene in men, PAI-1 and stromelysin genes in women,30 5-lipoxygenase activating protein,31 leukotriene A4 hydrolase,32 lymphotoxin-α gene,33 HMG-CoA reductase and ADAMTS-1 metalloproteinase in statin therapy,34, 35 β-adrenergic receptors with β-blockade response,36 and CYP2C9 and vitamin K epoxide reductase-1 in warfarin therapy.37, 38 The limitations to these studies and their findings lie in the fact that they often offer little biological or functional link from the specific gene to the disease process studied. Furthermore, they are single institution observational studies with no subsequent confirmatory studies, validation, or intervention.39, 40 This underappreciated limitation emphasizes the importance of a systems-wide approach to define genomic signatures and pattern recognition of genomic classifiers, with validation coming from the application of such classifiers to other populations (i.e. to related patient cohorts or between similar cohorts in a multicenter study design).24
The CardioGene Study is an example of an investigation using comprehensive high throughput genome-wide molecular approaches to study clinical restenosis in bare metal stents used in the treatment of coronary artery disease.41 The goal is to identify genetic determinants or predictors of inward remodeling and in-stent restenosis to explain the dichotomous outcome of failure following percutaneous coronary intervention. The study is a collaborative initiative between the NHLBI and two clinical sites in the US, and plans to enroll 350 patients. Blood is sampled pre-intervention and then again at 2 weeks and 6 months following intervention. Clinical endpoints include symptomatic restenosis at 6 months and at 12 months. Genomic studies are performed on circulating leukocytes and mononuclear cells using the Affymetrix U133A GeneChip™ platform. Plasma proteomic studies are performed using multidimensional liquid chromatography and tandem mass spectroscopy. The investigators' initial focus is on gene regulatory regions and transcriptomes associated with modulation of gene expression. They are then planning a secondary genome-wide analysis to identify genes or clusters of genes related to in-stent restenosis and unfavorable outcomes. This will include investigation of candidate SNPs linked to stent failure. They plan a complex bioinformatics approach to define genomic biomarkers that would allow riskstratification prior to intervention and may lead to development of new techniques to prevent coronary stent restenosis and failure. Results from this trial are not yet available, but are eagerly awaited due to the parallel nature of our study design for application of these methodologies to failure following lower extremity revascularization.
Study Design
Patient Selection
Eligible patients are identified amongst all patients being evaluated for symptomatic PAOD in our current vascular surgical practice. The summary of specific inclusion and exclusion criteria are listed in Table I.
Table I.
Inclusion/Exclusion Criteria
| Inclusion criteria | Exclusion criteria |
|---|---|
|
|
Study Overview
A five-year study is underway in which 300 patients undergoing evaluation for symptomatic PAOD will be enrolled. The study cohort will be comprised of 50 patients treated with medical management alone, 125 patients undergoing additional lower extremity angioplasty with or without stenting, and 125 patients undergoing additional lower extremity vein bypass (Figure 1). Data is collected prospectively with longitudinal evaluation to determine success or failure of the intervention with corresponding quality of life (QOL) measures. At the same time points as clinical assessment, blood sampling is performed for high throughput genomic and proteomic analyses. (Figure 2) Bioinformatics tools are then applied to reconcile the molecular data with clinical outcomes to arrive at molecular profiles that correspond to success or failure of intervention. All study patients sign informed consent under an Institutional Review Board approved protocol. An Access™ database (HIPAA-defined “limited data set”) is currently in use to collect and store all study data.
Figure 1.
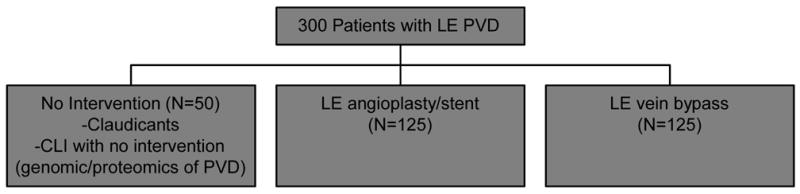
Study Overview
Figure 2.
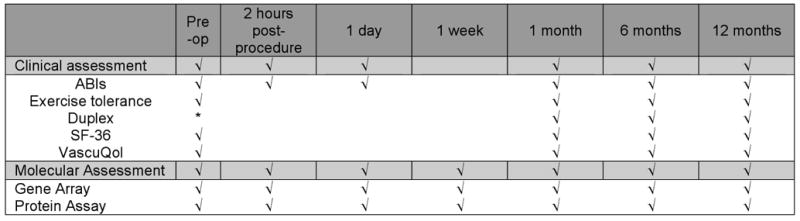
Study Timeline
Clinical Outcomes
Evaluation of patients for symptomatic PAOD follows current standards of practice.42–44 Generally, patients with severe claudication or critical limb ischemia (Rutherford Grade I Category 3 level disease or greater)42 are considered for intervention. In an effort to optimize patient outcomes45, 46 and standardize patients with respect to medications that likely influence systemic inflammatory response profiles, all patients are placed on antiplatelet therapy (at least 81 mg ASA daily) and statin (HMG-CoA reductase inhibitor) therapy (at least atorvastatin 10 mg daily). Statin therapy is adjusted according to the recent National Cholesterol Education Program (NCEP) Adult Treatment Panel (ATP) III revised national guidelines and recommendations.47 Those with a normal cholesterol profile will be maintained on 10 mg atorvastatin daily except in patients with documented intolerance to statins.
Patients requiring intervention will then go on to lower extremity arteriography with treatment decisions guided by the TransAtlantic Inter-Society Consensus (TASC) I and II recommendations.42, 44 For patients undergoing percutaneous intervention, primary angioplasty is the preferred initial approach for superficial femoral and popliteal artery stenoses with subintimal recanalization and angioplasty for chronic total occlusions.48–50 Primary angioplasty is also performed for infragenicular tibial artery stenoses or occlusions in patients with critical limb ischemia.51–53 Selective stenting is indicated for unacceptable results following angioplasty (i.e. significant lesion recoil with residual stenosis or flow limiting dissection).54 For patients undergoing vein bypass surgery, a non-reversed anatomic bypass using ipsilateral long saphenous vein is the approach of choice with other alternatives considered as the specific case warrants. No synthetic bypass patients will be included. All patients are placed on an antiplatelet regimen consisting of ASA 81 mg daily with the addition of clopidogrel 75 mg daily for 30 days in patients undergoing angioplasty/stent procedures. Warfarin is initiated for bypass grafts with compromised outflow,55 and continued in all patients with preexisting indications.
Clinical and laboratory data will be collected prospectively from all patients to determine preoperative risk factors and postoperative response to revascularization. The timing of assessment and data collection is summarized in Figure 2 and is scheduled in accordance with current standard practice for surveillance following lower extremity intervention. As indicated, evaluation includes review of symptoms, pulse exam with ABIs, and duplex ultrasound examination of the revascularized region. Repeat angiography is performed selectively in patients undergoing evaluation for re-intervention or salvage with failure of revascularization defined according to the Society for Vascular Surgery Recommended Standards for Reports Dealing with Lower Extremity Ischemia (revised version).43
Functional and Quality of Life Assessments
Functional and QOL measures are obtained at the same time intervals indicated in Figure 2. Subjective patient data regarding function is obtained through history and reconciled with objective data obtained using walking exercise tolerance. Baseline values are assessed pre-intervention using a timed, monitored walk to failure. Distance at the six-minute mark is documented for subsequent comparison. Post-intervention, the six-minute walk is then utilized for follow-up functional assessment.56, 57 Distance achieved as well as pre- and post-exercise ABIs are recorded and compared to pre-intervention measurements. Quality of life instruments used in this study include both generic health and disease-specific QOL questionnaires. The Medical Outcomes Short Form-36 (SF-36)58 serves as the generic health questionnaire and the Vascular Quality of Life (VascuQol)59 questionnaire to measure specific elements of PAOD to capture more subtle disease-specific effects of intervention. Results are compared to pre-intervention to determine the impact of intervention on QOL. These data are then compiled with clinical outcome data into the time course correlative analysis in concert with genomic and proteomic information as part of the class prediction model discussed below.
Genomic/Proteomic Analysis
Molecular analyses are performed on peripheral venous blood and includes evaluation of the transcriptome from a total leuckocyte population and an enriched monocyte population, as well as the proteome from the plasma fraction. Initial blood samples are obtained in the pre-operative holding area immediately prior to the procedure. Subsequent samples are taken 2 hours and 1 day postoperatively and then at 1 week, 1, 6, and 12-months of follow-up.(Figure 3) At each time point, 15 ml of blood is sampled to establish genomic and proteomic inflammatory response profiles. All samples are de-identified and assigned a study-specific identification number to assure confidentiality and allow sample tracking. A 7 ml collection of EDTA anticoagulated whole blood is obtained for flow cytometric analysis of the peripheral blood leukocyte phenotype, genomic analyses on the total leukocyte preparation, and proteomic analyses of the plasma fraction. Simultaneously, an 8ml whole blood collection is collected in a Becton-Dickinson CPT™ tube containing sodium citrate to be processed further for the isolation of an enriched blood monocyte fraction. Plasma and leukocyte RNA are also stored for additional future analysis if needed. The actual protocols are detailed in two recent publications including a discussion of the advantages and limitations of these analytical approaches.25, 60 They are also available through the Large Scale Collaborative Research Program (www.gluegrant.org).
Figure 3.
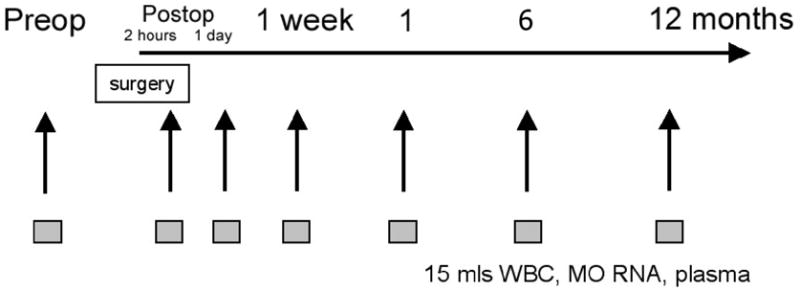
Timing of phlebotomy for genomic and proteomic analysis See Figure 2 for correlation with clinical outcome assessment
Briefly, for the isolation of whole blood leukocytes, each sample will be centrifuged and the plasma fraction recovered. Erythrocytes in the buffy coat fraction are lysed and the leukocytes captured by centrifugation. Total leukocyte RNA is then isolated using a commercial kit (RNeasy™, Qiagen, Inc) with yields of 2 – 6 μg/ml of blood. This will be ample since quantities required for microarray analyses have now been reduced to less than 50 ng of total cellular RNA. Monocytes are isolated by negative selection with a commercial preparation (RosetteSep™) added directly to the CPT™ tubes. All leukocyte populations other than monocytes are removed by rosetting and centrifugation across the density gradient. Purity of cell preparation using this technique is approximately 90%.25, 61 The advantage of this approach is that the desired population is isolated without the same degree of cell activation associated with positive selection techniques. RNA yields from monocyte enriched fractions are between 500 and 1000 ng of RNA.
The technology for hybridization and microarray analysis is constantly changing. Whereas, several months ago μg quantities of total cellular RNA were required for starting material, the amount now required is at the mere ng level. Using commercially available kits that generate a labeled cDNA product for hybridization (i.e. NuGen Ovation™ labeling scheme), the quantity of starting material has been reduced several logs to as little as 5 ng total cellular RNA. This permits much smaller sampling volumes and/or archival of larger quantities of RNA for validation analysis.
Similarly, there has been dramatic progress in the microarrays themselves. Increased density and better manufacturing have yielded commercial arrays that demonstrate significantly improved reliability and reduced variance. The most dramatic development has been that of the exon arrays that have increased the density of current chip iterations to nearly 10 million tiles, and the ability to look simultaneously at alternative splicing of mRNA products. In these current studies, an exon array (GeneChip® Exon ST 1.0 Array System) with over 1 million exon clusters will be used. Representative examples of heatmap data output from microarray analyses are displayed in Figure 4.
Figure 4.
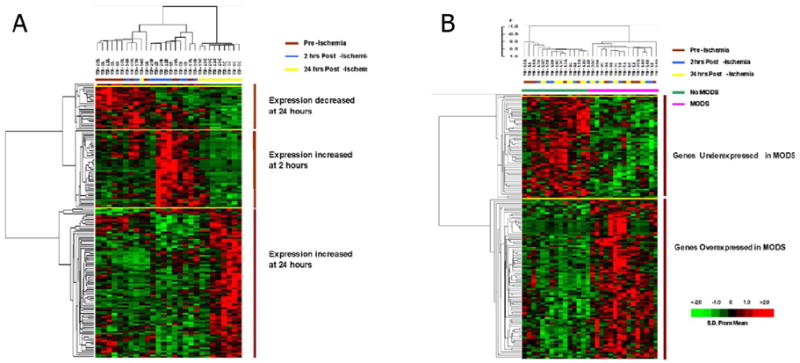
Representative heat map display of microarray data using time series (A) and supervised (B) analytic approaches. (Used by permission, J Immunology)76
Plasma samples will be analyzed using the Luminex® 100™ xMAP (Multi-Analyte Profiling)® System. This bead-based assay system is essentially a flow cytometric analysis employing novel fluorescent beads that are covalently linked (in the case of cytokine measurements) to antibodies specific for individual analytes. By coupling the specificity of antibody-based capture of specific cytokines using chromophore-labeled antibodies with flow cytometric analyses of individual reactions identified by unique fluorescent beads, the analytical system can multiplex the analysis of theoretically an unlimited number of cytokines simultaneously from a single sample. Using a two laser system, the Luminex technology simultaneously identifies the quantity of an analyte bound to a specific antibody, as well as its identity, critical for a multiplex approach. Our current working Luminex platform determines simultaneously the concentrations of the following analytes: eotaxin, G-CSF, GM-CSF, IFN-γ, IL-10, IL-12p70, IL-13, IL-15, IL-17, IL-18, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IP-10, MIP1α, MCP1 and TNFα. A representative output of the proteomic data is displayed in Figure 5.
Figure 5.
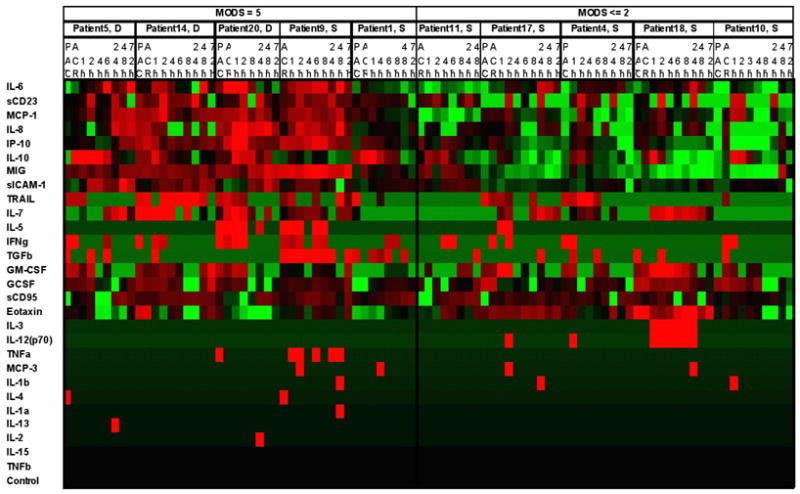
Representative proteomic heat map. (used by permission , J Immunology)76
Flow cytometric analysis of monocyte phenotype is initiated by treating 1.5 ml of blood with an equal volume of FACS Lysing Solution (Becton Dickinson). Monocyte analysis employs a combination of forward and side scatter characteristics as well as the CD14/CD33 markers in order to isolate CD16bright/dim monocytes which are then analyzed for their expression of activation markers (CD11b, CD18, and CD69), and human MHCII (HLA-DR). Events are evaluated for both their proportion of positive cells as well as for their mean fluorescence intensity (MFI) for each antibody to determine the number of cells positive for a cell surface marker and the expression quantity of that marker on the cell surface membrane, respectively. Samples will be acquired and analyzed on a six-parameter FACSCaliburTM machine with CellquestTM Software (Becton Dickinson Systems, San Jose, CA).
Bioinformatics
A laboratory information management system (LIMS) has been designed to track all aspects of this study from patient specific clinical information to specimen collection, RNA isolation and labeled target cRNA preparation through hybridization, staining and scanning to publication.
The large datasets that result from microarray experiments and proteomics determinations, and the associated noise generated through their analyses present ongoing challenges. Initially, low level analyses are performed to normalize and filter the datasets. A number of academic software packages, including dChip and RMA, are available to perform these critical analyses. Once these are complete, there are two major approaches for the analysis of microarray-based gene expression studies: unsupervised and supervised analysis methods (Figure 4). Unsupervised approaches such as multidimensional scaling, cluster analysis and self-organizing maps are used in class discovery exercises to discover relationships among genes that were not previously recognized without any a priori assumptions. Supervised approaches are used to identify gene expression differences in predefined classes (i.e. control vs. experimental) or subsequent groupings of the data (i.e. patients who develop failure of their revascularization versus those who do not). Unsupervised and supervised analytical approaches are not mutually exclusive and when used in conjunction with one another represent a very powerful method for identifying relationships among genes.
Unsupervised Analysis of Gene Expression Patterns
Unsupervised analytical methods are useful tools for class discovery and identification purposes. One goal of unsupervised analyses is to identify similarities between specimens that were previously unrecognized. Principal component analysis (PCA), a form of multidimensional scaling, is often useful in the analysis of gene expression datasets to identify similarities between specimens and to identify outliers.62 In PCA, the dimensionality of the dataset is reduced through identification of the principal components. These principal components can be used to identify similarities in expression patterns among arrays without imposing any pre-defined structure. This first step in our analysis is therefore designed to identify similarities and differences in gene expression patterns among patients with symptomatic PAOD. The next analytical step then utilizes academic software packages such as Cluster and TreeView63 that are capable of performing several widely used clustering methods including hierarchical clustering and k-means clustering. Hierarchical clustering constructs a hierarchical tree of the gene expression data in which genes and arrays whose expression are most similar to each other are placed on adjacent branches of a tree. In an attempt to arrive at the “true” relationship between genes, hierarchical clustering is combined with k-means clustering, a reiterative clustering method that partitions members into a number of a priori user-specified bins. The dataset is clustered with k-means into the appropriate number of bins and the members of each individual bin are then subsequently clustered using hierarchical algorithms. The ultimate goal is to identify clusters that are biologically and physiologically relevant. Clustering approaches are powerful tools for identifying genes in related pathways and with related function 63, 64. Software tools are now available including MAPPFinder 65, GenMAPP 66, and Ingenuity Pathway Analysis (Ingenuity® Systems, www.ingenuity.com)67 that identify and display related groups of genes identified by microarray experiments based on gene ontology classifications and biological systems networks.(Figure 6)
Figure 6.
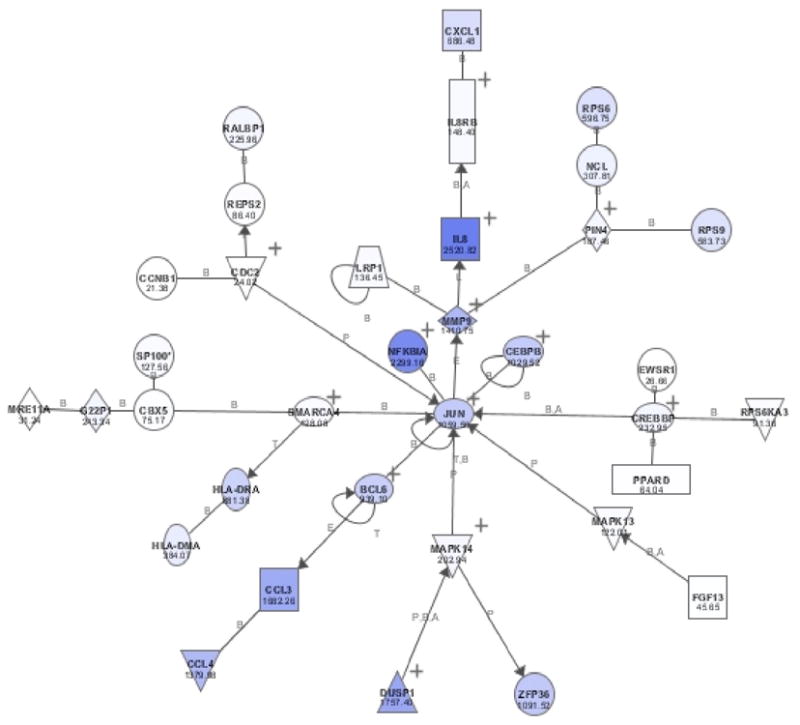
Representative display of inflammatory pathway analysis (Ingenuity Pathway Analysis, Ingenuity® Systems, www.ingenuity.com).
Supervised Analysis of Gene Expression Patterns
Supervised learning approaches are next used to overcome the limitations of unsupervised approaches in describing expression profiles consistent with a specific phenotype.68 The goal of our supervised analyses is to identify gene expression differences between patients who fail following lower extremity revascularization and those who have a successful outcome. To minimize error associated with these approaches and optimize our discovery of real differences that are potentially both statistically and biologically significant we use several methods. Significance Analysis of Microarrays (SAM) is used to minimize false discovery rates through thousands of permutations of the dataset. Analysis of variance using BRB tools is used to identify gene expression differences between the patient outcome classes with a significance level set at a p-value of < 0.001 to identify significant probe sets with a false positive rate of 1:1000 probe sets analyzed.
Next, leave-one-out cross validation is used to minimize the potential to over fit data by identifying patterns resulting from chance alone without any biological relevance or significance. Leave-one-out-cross-validation using four different prediction models (1KNN, 3KNN, linear discriminant analysis, and nearest centroid) is a reiterative process where each array is left out of the training set in turn, and the remaining arrays are used to identify probe sets significant at the p < 0.001 level between the various clinical outcomes. The significant probe sets so identified will then be used to predict the outcome label of the array omitted from the set. The number of correct predictions will be used to evaluate the ability of the predictor probe sets (genes) to accurately predict a specific clinical outcome (phenotype). Monte Carlo simulations with 2000 permutations of the dataset are then performed to determine if the cross-validation misclassification rate observed with “predictor probe sets” derived from the test dataset is lower than that expected by chance alone.
Finally, since longitudinal time-course data is being collected, time series analyses allows us the ability to discover dynamic information about time-dependent gene expression patterns, apparent co-regulation of genes or gene clusters, and potential gene regulatory pathways that coincide with success or failure of revascularization.69 It is particularly important to model the early profiles of the patients that may engender specific delayed endpoints. It is typical to analyze time-course data using procedures based on parametric time-series analysis, perhaps using Fourier type models or other parametric forms.70, 71 However, recent reports demonstrate progress in using nonparametric models, particularly those based on functional data analysis.72–74 Using such a functional data analysis approach offers us the ability to capture subtle profile changes in patient response, and, combined with functional PCA, to uncover and describe the differences in the genomic temporal profile of patients who have a successful outcome vs. those that go on to failure.
Discussion
The critical questions asked in this study are derived from the study’s original aims. There are two broad sets of variables to consider: (1) clinical - comprised of anatomic, functional and quality of life variables, and (2) genomic/proteomic profile results. With such a comprehensive study design and the use of state-of-the-art high throughput molecular and bioinformatics techniques, we expect to arrive at answers to the questions that follow.
First, can we prospectively develop a class prediction model for gene expression profiles in 300 patients to identify a pattern of host gene expression in either the total leukocyte or monocyte population that discriminates between patients who go on to develop failure of their revascularization with either angioplasty/stenting or vein bypass and those who do not? Furthermore, can we detect perhaps subtle but critical differences in the response to intervention between patients undergoing angioplasty/stenting vs. those undergoing vein bypass surgery? Class prediction is perhaps the low-hanging fruit, and ultimately, we intend to apply these technologies to better understand the underlying biology that determines outcome. For example, from these genomic and proteomic patterns, what can we learn about the underlying inflammatory processes in blood monocytes and leukocytes in patients with PAOD? Using a pathway analysis tool, such as Ingenuity Pathway Analysis, or gene ontologies, can we identify cell signaling pathways or functional modules that are differentially expressed in patients with varying clinical outcomes, and use that information to better define the inflammatory state of the patient? Importantly then, can the temporal expression of the inflammatory cell transcriptome provide insight into the biologic mechanisms of inward remodeling and occlusive adaptation at sites of angioplasty or in the implanted vein graft leading to intervention failure? If so, can we use this new knowledge to alter selection of patients for intervention, help delineate what intervention strategy is best suited for an individual patient, and/or identify potential tools for therapeutic intervention (i.e. antinflammatory regimens) to engineer the inflammatory response and improve outcomes following lower extremity revascularization?
At the same time, can we discover and interpret patterns of gene expression apparent before surgery that are predictive of outcome such that they may reflect the presence of pre-existing leukocyte activation or immunological “priming”? Are these patterns evident in the total leukocyte population, or as we might predict, in a selected cell population directly involved in the inflammatory process, such as blood monocytes? Ultimately, can we use this pre-intervention information to guide treatment decision making prospectively?
Next, are patterns of gene expression from the peripheral blood monocytes reflective of their phenotypic changes, as determined by cell surface expression? Are the patterns of gene expression observed in peripheral blood monocytes indicative of either monocyte activation or the presence of a systemic inflammatory response to vascular intervtnion? Does the pattern of gene expression in blood monocytes better discriminate outcome than the gene expression pattern from total leukocyte populations in terms of identifying patients who go on to develop revascularization failure and those who do not?
Finally, using comparable genomic chip and proteomic plasma measurements, is there an association between the changes in peripheral blood leukocyte or monocyte gene expression and their relative protein concentrations in plasma? Although it is well established that the primary source of inflammatory mediators in the plasma compartment are not blood leukocytes, but are cells of the reticuloendothelial system, particularly in the splanchnic bed,75 what can we infer about an associative relationship between plasma cytokine concentrations and leukocyte gene expression?
With so many important questions to be answered, we are committed to making our results available to interested members of the scientific community as quickly as possible. Once results from this study have been peer reviewed and are published in the scientific literature, we will post the complete reference of the published work and experimental details conforming to the proposed “minimum information about a microarray experiment” (MIAME) standard on our research web site (www.surgery.ufl.edu/Research). This site will also include the complete dataset and *.dat files along with supplemental material and other background information pertaining to the published work.
Conclusion
This translational research model allows us to (1) unify a definition of outcome following peripheral lower extremity revascularization using combined metrics of standard clinical endpoints (i.e. patency, objective anatomic and hemodynamic measurements), functional endpoints (i.e. exercise tolerance and subjective assessment of functionality), and quality of life endpoints (as measured by a combination of generic and disease-specific questionnaires); (2) define molecular evidence that a differential inflammatory state and/or response to vascular injury contributes to inward remodeling and revascularization failure secondary to restenosis or occlusion at the site of intervention through advanced high throughput genomic and proteomic analysis; and (3) establish class prediction models for patients prone to failure of lower extremity revascularization through specific transcriptome identification and analysis. This will form the basis for what is currently unavailable - an evidence-based approach to peripheral intervention and revascularization for symptomatic lower extremity PAOD.
Acknowledgments
Work supported in part by NHLBI grant 1K23HL084090–01 (PRN) and NIGMS grant 2T32GM08721–06 (KAO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dalman RL, Taylor LM., Jr Basic data related to infrainguinal revascularization procedures. Ann Vasc Surg. 1990;4(3):309–12. doi: 10.1007/BF02009464. [DOI] [PubMed] [Google Scholar]
- 2.Lam EY, Williamson WK, Nicoloff AD, Porter JM. Distal bypass: is it effective for limb salvage? Adv Surg. 2000;34:383–92. [PubMed] [Google Scholar]
- 3.Chew DK, Conte MS, Belkin M, et al. Arterial reconstruction for lower limb ischemia. Acta Chir Belg. 2001;101(3):106–15. [PubMed] [Google Scholar]
- 4.Dougherty MJ, Calligaro KD, DeLaurentis DA. The natural history of "failing" arterial bypass grafts in a duplex surveillance protocol. Ann Vasc Surg. 1998;12(3):255–9. doi: 10.1007/s100169900149. [DOI] [PubMed] [Google Scholar]
- 5.Gupta AK, Bandyk DF, Cheanvechai D, Johnson BL. Natural history of infrainguinal vein graft stenosis relative to bypass grafting technique. J Vasc Surg. 1997;25(2):211–20. doi: 10.1016/s0741-5214(97)70344-6. discussion 220–5. [DOI] [PubMed] [Google Scholar]
- 6.Mills JL, Sr, Wixon CL, James DC, et al. The natural history of intermediate and critical vein graft stenosis: recommendations for continued surveillance or repair. J Vasc Surg. 2001;33(2):273–8. doi: 10.1067/mva.2001.112701. discussion 278–80. [DOI] [PubMed] [Google Scholar]
- 7.Conte MS, Bandyk DF, Clowes AW, et al. Results of PREVENT III: a multicenter, randomized trial of edifoligide for the prevention of vein graft failure in lower extremity bypass surgery. J Vasc Surg. 2006;43(4):742–751. doi: 10.1016/j.jvs.2005.12.058. discussion 751. [DOI] [PubMed] [Google Scholar]
- 8.Borozan PG, Schuler JJ, Spigos DG, Flanigan DP. Long-term hemodynamic evaluation of lower extremity percutaneous transluminal angioplasty. J Vasc Surg. 1985;2(6):785–93. [PubMed] [Google Scholar]
- 9.Surowiec SM, Davies MG, Eberly SW, et al. Percutaneous angioplasty and stenting of the superficial femoral artery. J Vasc Surg. 2005;41(2):269–78. doi: 10.1016/j.jvs.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 10.Harris RW, Dulawa LB, Andros G, et al. Percutaneous transluminal angioplasty of the lower extremities by the vascular surgeon. Ann Vasc Surg. 1991;5(4):345–53. doi: 10.1007/BF02015295. [DOI] [PubMed] [Google Scholar]
- 11.Murphy TP. Medical outcomes studies in peripheral vascular disease. J Vasc Interv Radiol. 1998;9(6):879–89. doi: 10.1016/s1051-0443(98)70415-4. [DOI] [PubMed] [Google Scholar]
- 12.Donnelly R. Assessment and management of intermittent claudication: importance of secondary prevention. Int J Clin Pract Suppl. 2001;(119):2–9. [PubMed] [Google Scholar]
- 13.Hiatt WR. Medical treatment of peripheral arterial disease and claudication. N Engl J Med. 2001;344(21):1608–21. doi: 10.1056/NEJM200105243442108. [DOI] [PubMed] [Google Scholar]
- 14.Nicoloff AD, Taylor LM, Jr, McLafferty RB, et al. Patient recovery after infrainguinal bypass grafting for limb salvage. J Vasc Surg. 1998;27(2):256–63. doi: 10.1016/s0741-5214(98)70356-8. discussion 264–6. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SM, Kalbaugh CA, Blackhurst DW, et al. Determinants of functional outcome after revascularization for critical limb ischemia: an analysis of 1000 consecutive vascular interventions. J Vasc Surg. 2006;44(4):747–55. doi: 10.1016/j.jvs.2006.06.015. discussion 755–6. [DOI] [PubMed] [Google Scholar]
- 16.Keefer A, Davies MG, Illig KA. Can endovascular therapy of infrainguinal disease for claudication be justified? Expert Rev Cardiovasc Ther. 2004;2(2):229–37. doi: 10.1586/14779072.2.2.229. [DOI] [PubMed] [Google Scholar]
- 17.Aquarius AE, Denollet J, Hamming JF, et al. Impaired health status and invasive treatment in peripheral arterial disease: a prospective 1-year follow-up study. J Vasc Surg. 2005;41(3):436–42. doi: 10.1016/j.jvs.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 18.Brothers TE, Cox MH, Robison JG, et al. Prospective decision analysis modeling indicates that clinical decisions in vascular surgery often fail to maximize patient expected utility. J Surg Res. 2004;120(2):278–87. doi: 10.1016/j.jss.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340(2):115–26. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 20.Hoch JR, Stark VK, van Rooijen N, et al. Macrophage depletion alters vein graft intimal hyperplasia. Surgery. 1999;126(2):428–37. [PubMed] [Google Scholar]
- 21.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–95. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka T, Ozaki K. Inflammation as a risk factor for myocardial infarction. J Hum Genet. 2006;51(7):595–604. doi: 10.1007/s10038-006-0411-8. [DOI] [PubMed] [Google Scholar]
- 23.Libby P, Ridker PM. Novel inflammatory markers of coronary risk: theory versus practice. Circulation. 1999;100(11):1148–50. doi: 10.1161/01.cir.100.11.1148. [DOI] [PubMed] [Google Scholar]
- 24.Barth A, Hare J. The potential for the transcriptome to serve as a clinical biomarker for cardiovascular diseases. Circ Res. 2006;98(12):1459–1461. doi: 10.1161/01.RES.0000231257.15059.d7. [DOI] [PubMed] [Google Scholar]
- 25.Cobb JP, Mindrinos MN, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102(13):4801–6. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrell DK, Neverova I, Van Eyk JE. Cardiovascular proteomics: evolution and potential. Circ Res. 2001;88(8):763–73. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 27.Rubin EM, Tall A. Perspectives for vascular genomics. Nature. 2000;407(6801):265–9. doi: 10.1038/35025236. [DOI] [PubMed] [Google Scholar]
- 28.Lenfant C. NHLBI genomics initiatives : looking beyond the human genome project. Circulation. 2000;101(5):468–9. doi: 10.1161/01.cir.101.5.468. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Fan C, Topol SE, et al. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302(5650):1578–81. doi: 10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada Y, Izawa H, Ichihara S, et al. Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med. 2002;347(24):1916–23. doi: 10.1056/NEJMoa021445. [DOI] [PubMed] [Google Scholar]
- 31.Helgadottir A, Manolescu A, Thorleifsson G, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36(3):233–9. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 32.Helgadottir A, Manolescu A, Helgason A, et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2006;38(1):68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki K, Inoue K, Sato H, et al. Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature. 2004;429(6987):72–5. doi: 10.1038/nature02502. [DOI] [PubMed] [Google Scholar]
- 34.Chasman DI, Posada D, Subrahmanyan L, et al. Pharmacogenetic study of statin therapy and cholesterol reduction. Jama. 2004;291(23):2821–7. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 35.Gerdes LU, Gerdes C, Kervinen K, et al. The apolipoprotein epsilon4 allele determines prognosis and the effect on prognosis of simvastatin in survivors of myocardial infarction : a substudy of the Scandinavian simvastatin survival study. Circulation. 2000;101(12):1366–71. doi: 10.1161/01.cir.101.12.1366. [DOI] [PubMed] [Google Scholar]
- 36.Mialet Perez J, Rathz DA, Petrashevskaya NN, et al. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med. 2003;9(10):1300–5. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 37.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. Jama. 2002;287(13):1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 38.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 39.Sabatine MS, Seidman JG, Seidman CE. Cardiovascular genomics. Circulation. 2006;113(11):e450–5. doi: 10.1161/CIRCULATIONAHA.105.560151. [DOI] [PubMed] [Google Scholar]
- 40.Weng L, Kavaslar N, Ustaszewska A, et al. Lack of MEF2A mutations in coronary artery disease. J Clin Invest. 2005;115(4):1016–20. doi: 10.1172/JCI24186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganesh SK, Skelding KA, Mehta L, et al. Rationale and study design of the CardioGene Study: genomics of in-stent restenosis. Pharmacogenomics. 2004;5(7):952–1004. doi: 10.1517/14622416.5.7.949. [DOI] [PubMed] [Google Scholar]
- 42.Dormandy JA, Rutherford RB. Management of peripheral arterial disease (PAD). TASC Working Group. TransAtlantic Inter-Society Concensus (TASC) J Vasc Surg. 2000;31(1 Pt 2):S1–S296. [PubMed] [Google Scholar]
- 43.Rutherford RB, Baker JD, Ernst C, et al. Recommended standards for reports dealing with lower extremity ischemia: revised version. J Vasc Surg. 1997;26(3):517–38. doi: 10.1016/s0741-5214(97)70045-4. [DOI] [PubMed] [Google Scholar]
- 44.Norgren L, Hiatt WR, Dormandy JA, et al. Inter-Society Consensus for the Management of Peripheral Arterial Disease (TASC II) Eur J Vasc Endovasc Surg. 2006 doi: 10.1016/j.jvs.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 45.Hiatt WR. Pharmacologic therapy for peripheral arterial disease and claudication. J Vasc Surg. 2002;36(6):1283–91. doi: 10.1067/mva.2002.129654. [DOI] [PubMed] [Google Scholar]
- 46.Daskalopoulou SS, Daskalopoulos ME, Liapis CD, Mikhailidis DP. Peripheral arterial disease: a missed opportunity to administer statins so as to reduce cardiac morbidity and mortality. Curr Med Chem. 2005;12(4):443–52. doi: 10.2174/0929867053363009. [DOI] [PubMed] [Google Scholar]
- 47.Grundy SM, Cleeman JI, Merz CN, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110(2):227–39. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 48.Lazaris AM, Tsiamis AC, Fishwick G, et al. Clinical outcome of primary infrainguinal subintimal angioplasty in diabetic patients with critical lower limb ischemia. J Endovasc Ther. 2004;11(4):447–53. doi: 10.1583/03-1159.1. [DOI] [PubMed] [Google Scholar]
- 49.Nadal LL, Cynamon J, Lipsitz EC, Bolia A. Subintimal angioplasty for chronic arterial occlusions. Tech Vasc Interv Radiol. 2004;7(1):16–22. doi: 10.1053/j.tvir.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Bolia A, Brennan J, Bell PR. Recanalisation of femoro-popliteal occlusions: improving success rate by subintimal recanalisation. Clin Radiol. 1989;40(3):325. doi: 10.1016/s0009-9260(89)80231-4. [DOI] [PubMed] [Google Scholar]
- 51.Ingle H, Nasim A, Bolia A, et al. Subintimal angioplasty of isolated infragenicular vessels in lower limb ischemia: long-term results. J Endovasc Ther. 2002;9(4):411–6. doi: 10.1177/152660280200900404. [DOI] [PubMed] [Google Scholar]
- 52.Varty K, Bolia A, Naylor AR, et al. Infrapopliteal percutaneous transluminal angioplasty: a safe and successful procedure. Eur J Vasc Endovasc Surg. 1995;9(3):341–5. doi: 10.1016/s1078-5884(05)80141-8. [DOI] [PubMed] [Google Scholar]
- 53.Bolia A, Sayers RD, Thompson MM, Bell PR. Subintimal and intraluminal recanalisation of occluded crural arteries by percutaneous balloon angioplasty. Eur J Vasc Surg. 1994;8(2):214–9. doi: 10.1016/s0950-821x(05)80463-3. [DOI] [PubMed] [Google Scholar]
- 54.Muradin GS, Bosch JL, Stijnen T, Hunink MG. Balloon dilation and stent implantation for treatment of femoropopliteal arterial disease: meta-analysis. Radiology. 2001;221(1):137–45. doi: 10.1148/radiol.2211010039. [DOI] [PubMed] [Google Scholar]
- 55.Sarac TP, Huber TS, Back MR, et al. Warfarin improves the outcome of infrainguinal vein bypass grafting at high risk for failure. J Vasc Surg. 1998;28(3):446–57. doi: 10.1016/s0741-5214(98)70130-2. [DOI] [PubMed] [Google Scholar]
- 56.McDermott MM, Liu K, Ferrucci L, et al. Physical performance in peripheral arterial disease: a slower rate of decline in patients who walk more. Ann Intern Med. 2006;144(1):10–20. doi: 10.7326/0003-4819-144-1-200601030-00005. [DOI] [PubMed] [Google Scholar]
- 57.Montgomery PS, Gardner AW. The clinical utility of a six-minute walk test in peripheral arterial occlusive disease patients. J Am Geriatr Soc. 1998;46(6):706–11. doi: 10.1111/j.1532-5415.1998.tb03804.x. [DOI] [PubMed] [Google Scholar]
- 58.McHorney CA, Ware JE, Jr, Lu JF, Sherbourne CD. The MOS 36-item Short-Form Health Survey (SF–36): III. Tests of data quality, scaling assumptions, and reliability across diverse patient groups. Med Care. 1994;32(1):40–66. doi: 10.1097/00005650-199401000-00004. [DOI] [PubMed] [Google Scholar]
- 59.Morgan MB, Crayford T, Murrin B, Fraser SC. Developing the Vascular Quality of Life Questionnaire: a new disease-specific quality of life measure for use in lower limb ischemia. J Vasc Surg. 2001;33(4):679–87. doi: 10.1067/mva.2001.112326. [DOI] [PubMed] [Google Scholar]
- 60.Feezor RJ, Baker HV, Mindrinos M, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19(3):247–54. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 61.Laudanski K, Miller-Graziano C, Xiao W, et al. Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc Natl Acad Sci U S A. 2006;103:15564–15569. doi: 10.1073/pnas.0607028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moe TK, Ziliang J, Barathi A, Beuerman RW. Differential expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta actin and hypoxanthine phosphoribosyltransferase (HPRT) in postnatal rabbit sclera. Curr Eye Res. 2001;23(1):44–50. doi: 10.1076/ceyr.23.1.44.5420. [DOI] [PubMed] [Google Scholar]
- 63.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holter NS, Mitra M, Maritan A, et al. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci U S A. 2000;97(15):8409–14. doi: 10.1073/pnas.150242097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doniger SW, Salomonis N, Dahlquist KD, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dahlquist KD, Salomonis N, Vranizan K, et al. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 67.Rajagopalan D, Agarwal P. Inferring pathways from gene lists using a literature-derived network of biological relationships. Bioinformatics. 2005;21(6):788–93. doi: 10.1093/bioinformatics/bti069. [DOI] [PubMed] [Google Scholar]
- 68.Tavazoie S, Hughes JD, Campbell MJ, et al. Systematic determination of genetic network architecture. Nat Genet. 1999;22(3):281–5. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 69.Storey JD, Xiao W, Leek JT, et al. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102(36):12837–42. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spellman PT, Sherlock G, Zhang MQ, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hitchcock D, Casella G, Booth J. Improved Estimation of Dissimilarities by Presmoothing Functional Data. J Am Stat Assoc. 2006;101(473):211–222. [Google Scholar]
- 72.Ramsay JO, Spellman BW. Functional Data Analysis. New York: Springer-Verlag; 1997. [Google Scholar]
- 73.Ramsay JO, Spellman BW. Applied Functional Data Analysis: Methods and Case Studies. New York: Springer-Verlag; 2002. [Google Scholar]
- 74.Luan Y, Li H. Clustering of time-course gene expression data using a mixed-effects model with B-splines. Bioinformatics. 2003;19(4):474–82. doi: 10.1093/bioinformatics/btg014. [DOI] [PubMed] [Google Scholar]
- 75.Fong YM, Marano MA, Moldawer LL, et al. The acute splanchnic and peripheral tissue metabolic response to endotoxin in humans. J Clin Invest. 1990;85(6):1896–904. doi: 10.1172/JCI114651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feezor RJ, Baker HV, Xiao W, et al. Genomic and proteomic determinants of outcome in patients undergoing thoracoabdominal aortic aneurysm repair. J Immunol. 2004;172(11):7103–9. doi: 10.4049/jimmunol.172.11.7103. [DOI] [PubMed] [Google Scholar]


