Abstract
The sequencing of the Strongylocentrotus purpuratus genome provides a unique opportunity to investigate the function and evolution of neural genes. The neurobiology of sea urchins is of particular interest because they have a close phylogenetic relationship with chordates, yet a distinctive pentaradiate body plan and unusual neural organization. Orthologues of transcription factors that regulate neurogenesis in other animals have been identified and several are expressed in neurogenic domains before gastrulation indicating that they may operate near the top of a conserved neural gene regulatory network. A family of genes encoding voltage-gated ion channels is present but, surprisingly, genes encoding gap junction proteins (connexins and pannexins) appear to be absent. Genes required for synapse formation and function have been identified and genes for synthesis and transport of neurotransmitters are present. There is a large family of G-protein-coupled receptors, including 874 rhodopsin-type receptors, 28 metabotropic glutamate-like receptors and a remarkably expanded group of 161 secretin receptor-like proteins. Absence of cannabinoid, lysophospholipid and melanocortin receptors indicates that this group may be unique to chordates. There are at least 37 putative G-protein coupled peptide receptors and precursors for several neuropeptides and peptide hormones have been identified, including SALMFamides, NGFFFamide, a vasotocin-like peptide, glycoprotein hormones, and insulin/insulin-like growth factors. Identification of a neurotrophin-like gene and Trk receptor in sea urchin indicates that this neural signaling system is not unique to chordates. Several hundred chemoreceptor genes have been predicted using several approaches, a number similar to that for other animals. Intriguingly, genes encoding homologues of rhodopsin, Pax6 and several other key mammalian retinal transcription factors are expressed in tube feet, suggesting tube feet function as photosensory organs. Analysis of the sea urchin genome presents a unique perspective on the evolutionary history of deuterostome nervous systems and reveals new approaches to investigate the development and neurobiology of sea urchins.
Introduction
Neural organization is a defining feature of animal body plans that is often emphasized in analyses of the relationships among major taxa. The arrangement of the nervous system in echinoderms is a feature that distinguishes them from other deuterostomes (chordates and hemichordates). Echinoderm nervous systems are dispersed, but they are not a simple nerve net. The adult is not cephalized, yet the radial nerves are segmentally organized. The larva has a nervous system, but it appears to be independent of the nervous system of the adult. Identifying neural genes in the sea urchin genome has permitted an unprecedented glimpse into how this poorly understood nervous system develops and functions. This genomic approach circumvents many of the technical problems that have hampered conventional neurobiological approaches in echinoderms. Moreover, the list of putative neural genes identified in the genome provides a unique resource for developing hypotheses on gene function and numerous opportunities for new approaches to understanding neural development and function in sea urchins.
Neural development and organization
Neurons of the larval nervous system appear as neuroblasts in the thickened ectoderm of the animal plate and in the presumptive ciliary band of the late gastrula (Bisgrove and Burke, 1986; Yaguchi et al. 2000; Nakajima et al. 2004) (Figs 1, 2). A phase of neurite projection follows during which the tracts of axons at the base of the ciliary bands are formed. Neurons continue to be added throughout larval development and the fully formed, eight-armed pluteus has a complex array of sensory neurons, interneurons, tracts of axons and ganglia that are closely associated with the larval ciliary bands and larval muscles (Bisgrove and Burke, 1986; Beer et al. 2001). The larval nervous system has been described using ultrastructure, histochemistry, antibodies to neurotransmitters, tissue-specific monoclonal antibodies and neuron-specific, in situ hybridization probes (Burke, 1978; Bisgrove and Burke, 1986, 1987; Beer et al. 2001, Yaguchi and Katow, 2003; Nakajima et al. 2004)(Fig. 2). Neurons and neurites are principally associated with the ciliary bands that surround the oral field and form at the rim of the larval mouth (Beer et al. 2001; Nakajima et al. 2004). Several clusters of neurons with associated neuropil are organized as ganglia. The largest of these is the apical organ, a bilaterally symmetric patch of neurons at the anterior end of the larva between the preoral arms. In the early larva, the apical organ is composed of 4–6 bilaterally positioned sensory cells containing serotonin, a central cluster of 10–12 neurons and several non-neural supporting cells. The neurons associated with the ciliary band are arrayed at intervals along the length of the ciliary band on the side of the band closest to the larval mouth. These appear to be primary sensory cells that contribute axons to a central tract that lies at the base of the ciliated cells and appears to run the length of the ciliary band. The lateral ganglia are two clusters of neurons, on the left and right sides of the larva that project axons or dendrites beneath the epidermis throughout the posterior end of the larva (Nakajima et al. 2004). The larval nervous system grows and becomes more complex as the larva increases in size (Fig. 2, Beer et al. 2001).
Figure 1.
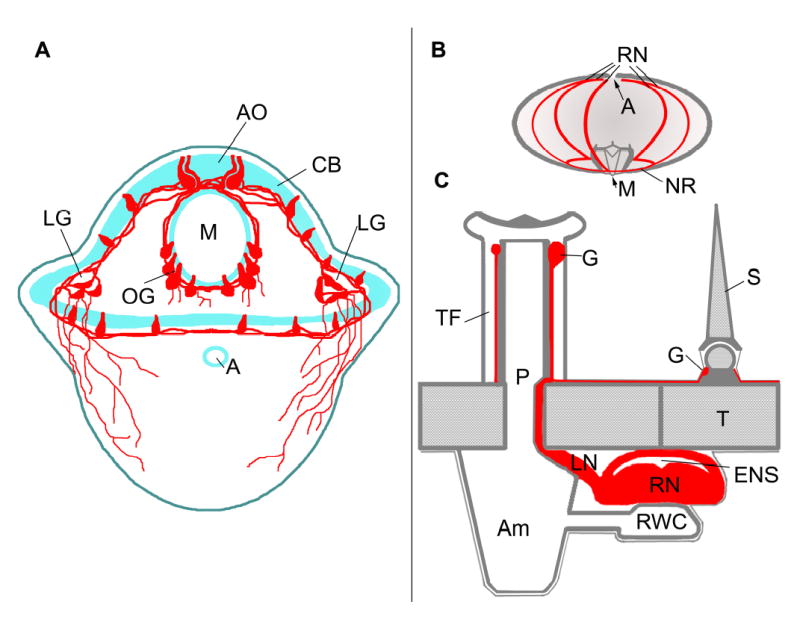
Diagrams of larval and adult nervous systems. A. Larval nervous sytem of an early pluteus. The nervous system is associated with the ciliary band (CB) surrounding the oral epidermis and the larval mouth (M). The apical organ (AO) is at the foremost end of the larva and there are oral ganglia (OG) in the lower lips of the larval mouth and paired lateral ganglia (LG) between the larval arms. Neurites project beneath the aboral epidermis. B. Lateral perspective of an adult sea urchin showing the 5 radial nerves (RN), anus (A) and mouth (M). The radial nerves are joined by commissures forming a nerve ring (NR) around the base of the lantern apparatus. C. Cross-section of adult radial nerve and test. The radial nerve (RN) and epineural sinus (ENS) lie against the inner surface of the test (T). Each segment of the radial nerve gives rise to a lateral nerve (LN) that projects through a pore (P) to connect to the base of a tube foot (TF). The water vascular system, comprised of a radial water canal (RWC) and ampulla (Am), connects to the lumen of the tube foot and overlies the radial nerve and radial hemal sinus (not shown). Each tube foot has a terminal ganglion (G) and ring of neuropil. Spines (S) all have a ganglion (G) and ring of neuropil at their base that connect to the radial nerve. Neural tissues are in red.
Figure 2.
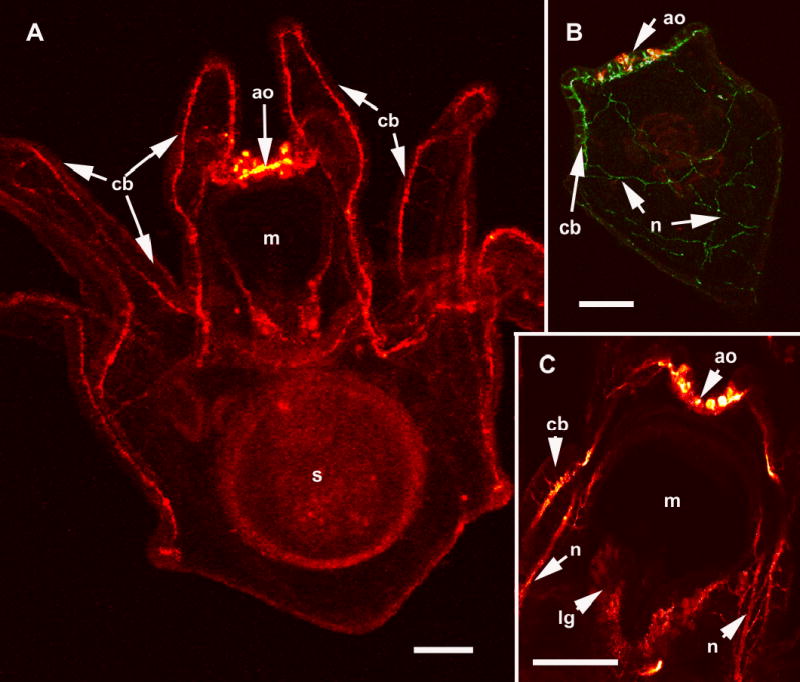
The larval nervous system of S. purpuratus revealed with immunolocalization of anti-synaptotagmin and anti-serotonin. A. Confocal image of an eight arm pluteus larva showing the tracts of axons that underlie the ciliated cells of the ciliary band (cb), anti-synaptotagmin. The apical organ (ao) is located on the oral hood at the foremost end of the larva and is thought to be a sensory organ. The larval mouth (m) is also innervated and nerves surround the esophagus, a tube connecting the mouth and the stomach (s). B. In the more familiar 4 arm pluteus larva the apical organ (ao) is double labelled with anti-serotonin (red) and the ciliary band neurons are labelled with anti-synaptotagmin (green). The neural cells bodies of the ciliary bands (cb) are arrayed at intervals along the oral side of the ciliated cells. The neurons in the ciliary bands project neurites to the posterior end of the larva (n). C. A confocal image (anti-synaptotagmin, red) showing details of the mouth (m) region. The lip ganglion (lg) contains serotonergic and non-serotonergic neurons. In addition to the ciliary band neurons and axon tracts (cb), there are bundles of neurites (n) of unknown function projecting from the apical organ, under the oral ectoderm. Bars = 25 μm
How the larval nervous system functions has been deduced from neuroanatomy and behavior. The ciliated cells of the ciliary band and musculature of the esophagus and mouth appear to be the principal effectors (Strathmann, 1975). It is assumed that coordinated reversals of cilia, reversals of peristalsis in the esophagus and contraction of the muscles that open the mouth and flex the arms are neurally controlled (Bisgrove and Burke, 1986). Mackie et al. (1969) recorded electrical activity from ciliary bands coincident with ciliary reversals, but there have been no neurophysiological investigations of larval nerves.
The oral half of the adult body develops as a rudiment on the left side of the larva, arising from a disc of ectoderm from the larval oral field that invaginates and lies against a mesodermal epithelium. This mesoderm forms a toroid and becomes the water vascular system while the overlying ectoderm makes five radially arrayed neural infoldings (Hyman, 1955 after von Ubisch, 1913). The lumen of each of the infoldings becomes an epineural sinus and the epithelium at the base of each infolding differentiates as the neural epithelium of the radial nerves (Fig. 1). Initially a single segment forms, but additional segments are added as ambulacral tube feet appear (Fig. 3). In many species metamorphosis takes place while the adult rudiment has only a single segment. The neurons of the appendages (tube feet, spines, sphaeridia, and pedicellariae, Fig. 4) appear to develop within the appendages. This is most apparent in pedicellariae that arise from separate placode-like ectodermal invaginations of the surface of the larva (Burke, 1980). The larval and adult nervous systems appear to have no direct connections and during metamorphosis the larval nervous system is largely lost (Chia and Burke, 1978).
Figure 3.

The adult nervous system of S. purpuratus. Segmental radial nerves in adult echinoderms are known only from ophiouroids (Cobb, 1987). The adult sea urchin radial nerve appears to have a similar organization. A. Dissected radial nerve showing the segmental organization. Each segment spans the width of the nerve and a single nerve bundle projects from each segment. The segments are arranged so that alternating segments project nerve bundles to alternating sides of the nerve. A single segment is outlined (arrows). The patterned pigmentation is in the cellular layer. Bar = 1 cm B. Cross-section of a radial nerve showing that the cellular layer (c) is on the surface of the nerve and extensive neuropil (n), comprised of closely apposed neurites beneath the cellular layer. Bar = 500 μm C. The segmentation of the nervous system is revealed by whole mount of a nerve in which the lipophilic tracer DiI was injected into the lateral projections of two segments and neurites were allowed to back-fill overnight. The backfilling procedure identifies a similar cluster of nerve cell bodies in both segments (n) indicating the segments have identical organization and are repeated elements. Neurites also project between segments in longitudinal tracts that are predominantly on the lateral edges of the nerves. The darker pigmentation pattern (p) of the tissue provides orientation. Bar = 200 μm D. The adult nervous system arises in the developing adult rudiment on the side of the larval body. In this immunofluorescent preparation with anti-synaptotagmin the oral surface of the rudiment is uppermost and the larval nervous system (lns) is out of focus behind the specimen. Nerves appear in the developing appendages, spines (s) and tube feet (tf) simultaneously with the appearance of neurons in the radial nerves. Bar = 50 μm E. In confocal images of later rudiments, connecting neurites from the radial nerves to appendages appear (arrow heads). Bar = 25 μm F. The radial nerves (rn) first appear as a single segment and the neuronal cell bodies distinguish the radial nerves from the nerve ring (nr), which is simply a commissure of neurites. As the juvenile grows, additional segments are added to the radial nerves. Bar = 50 μm
Figure 4.
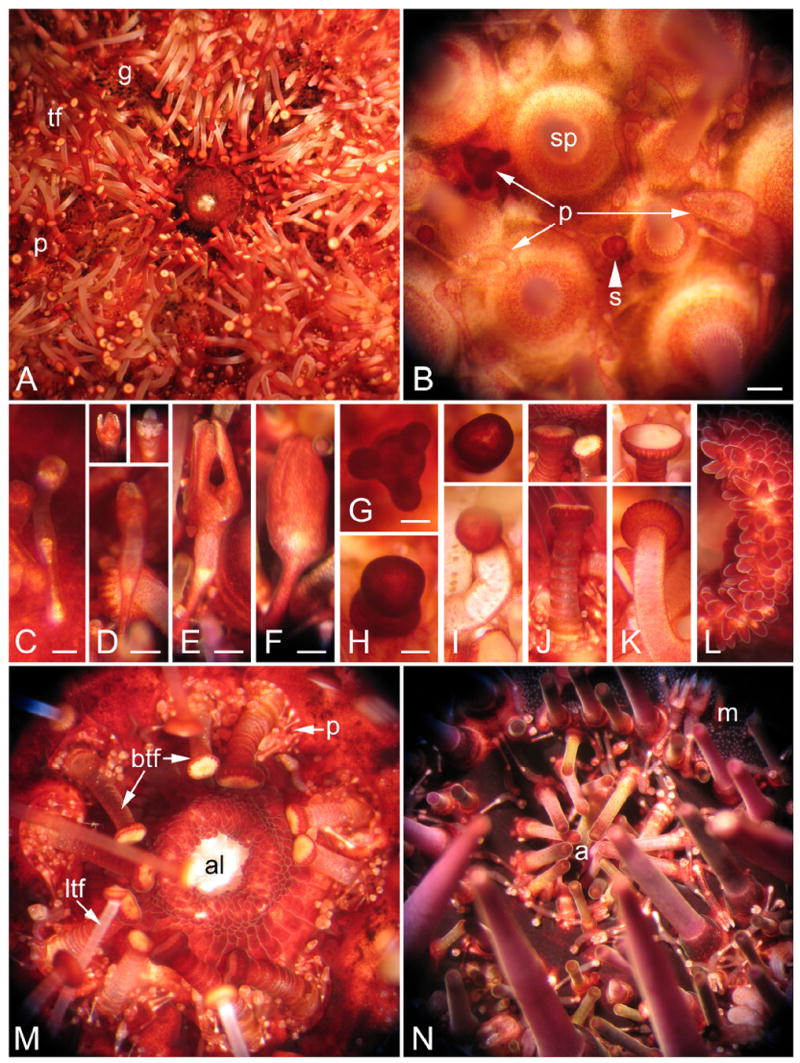
Appendages of adult S. purpuratus. Appendage types are classified according to Hyman (1955). A. Oral view, showing the mouth and large numbers of tube feet (tf), pedicellariae (p), and gills (g). B. Lateral surface of the test, showing spines (sp) extending upward from tubercles, pedicellariae (p), and sphaeridium (s). Bar = 1mm. C. Triphyllous pedicellaria; tiny but numerous. Bar = 0.25mm. D. Ophiocephalous or trifoliate pedicellaria, with three blunt jaws (insets) that do not meet. Bar = 0.5mm. E. Tridentate pedicellaria; the largest and very numerous. Bar = 0.5mm. F. Globiferous pedicelleria containing poison sacs. Bar = 0.5mm. G. Claviform pedicellaria, which may arise from the loss of jaws from globiferous pedicellariae. Bar = 0.5mm. H. Sphaeridium. Bar = 0.25mm. I. Tube foot with bulbous end (inset). J. Buccal tubefoot from near the mouth; has a shorter, stouter stalk than locomotory tubefoot, and modified sucker (inset); a cavity surrounding the base of each stalk contains numerous tiny pedicellariae. K. Locomotory tube foot having typical cupped sucker (inset). L. Edge of the mouth (left) with many lobes reminiscent of taste buds on the vertebrate tongue. M. Oral side, including the peristome, showing the teeth of Aristotle’s lantern (al) protruding from the lips of the mouth, buccal tube feet (btf) on stalks stouter than locomotory tube feet (ltf), and many tiny pedicellariae (p). N. Aboral surface, showing many spines and pedicellariae (p) surrounding the periproct and anus (a) and gonopores (not shown). The single madreporic (m) plate defines an axis of polarity and plane of bilateral symmetry.
The adult echinoid nervous system is comprised of 5 radial nerve cords, which are joined at their base by commissures that form a ring surrounding the mouth (Cobb, 1970; Cavey and Markel, 1994) (Fig. 1). Tube feet, spines and pedicellariae have ganglia and a complement of sensory and motor neurons (Fig. 1, 4). The viscera are well innervated (Garcia-Arraras et al. 2001) and there are neurons in almost all tissues. Each segment of the radial nerve cords contains a layer of neurons overlying an extensive neuropil (Fig. 3). Neurites project laterally within each segment and longitudinally to adjacent segments. Each segment gives rise to a single nerve bundle that projects laterally to a tube foot and adjacent appendages.
Neural organization is similar to the organization of the internet, an interconnected system of smaller local networks. Sensory input within tube feet, spines and pedicellariae project to ganglia within the appendages that integrate and control local motor responses (Cobb, 1987). Each of these nodes is interconnected to adjacent nodes and to the radial nerves. The radial nerves appear to function in overall control and coordination, but appendages have independent sensory-interneuron-motor reflex arcs and for the most part lack direct motor and sensory input from a central nervous system. Cobb (1987) emphasized that this is a highly organized, interconnected set of independent reflex pathways.
The echinoderm nervous system may be the least well studied of all those in the major metazoan phyla. Although morphological studies provide the essential neuroanatomy of the echinoid nervous system, conduction pathways are unknown because only a few neurophysiological studies using wick or metal filled electrodes have been done (Pentreath and Cobb, 1972; Binyon, 1972). The development of the echinoid nervous system is equally mysterious. Embryonic neurogenesis giving rise to larval nerves is accessible to experimental study (Yaguchi et al. 2006), but the descriptions of adult nervous system development are fragmentary and based on histological studies (Bury, 1896; von Ubisch, 1913; MacBride, 1903). Thus, while the sea urchin is used as a modern biomedical model, we have only fragmentary understanding of its neurobiology.
Analysis of the neural gene set of the sea urchin genome holds great promise for achieving a better understanding of relationships within the deuterostomes. Furthermore, a better understanding of the sea urchin nervous system permits a more incisive analysis of the features of neural development and organization shared by deuterostomes that distinguish them from other bilaterians. Identification of neural genes in the sea urchin provides a wealth of comparative data that indicates how the metazoan gene set has been adapted to construct this puzzling organism.
Neural Development
Neural specification
Understanding the structure and composition of gene regulatory networks that specify neurons is critical for learning how the sea urchin nervous system develops. Although little is currently known about neural specification in either larvae or adults, echinoderms are in an informative phylogenetic position and have the potential to contribute to our understanding of neural evolution. Here, our objective is to analyze the S. purpuratus genome to define neurogenic gene regulatory networks in the larva, which we predict is likely to include core metazoan elements.
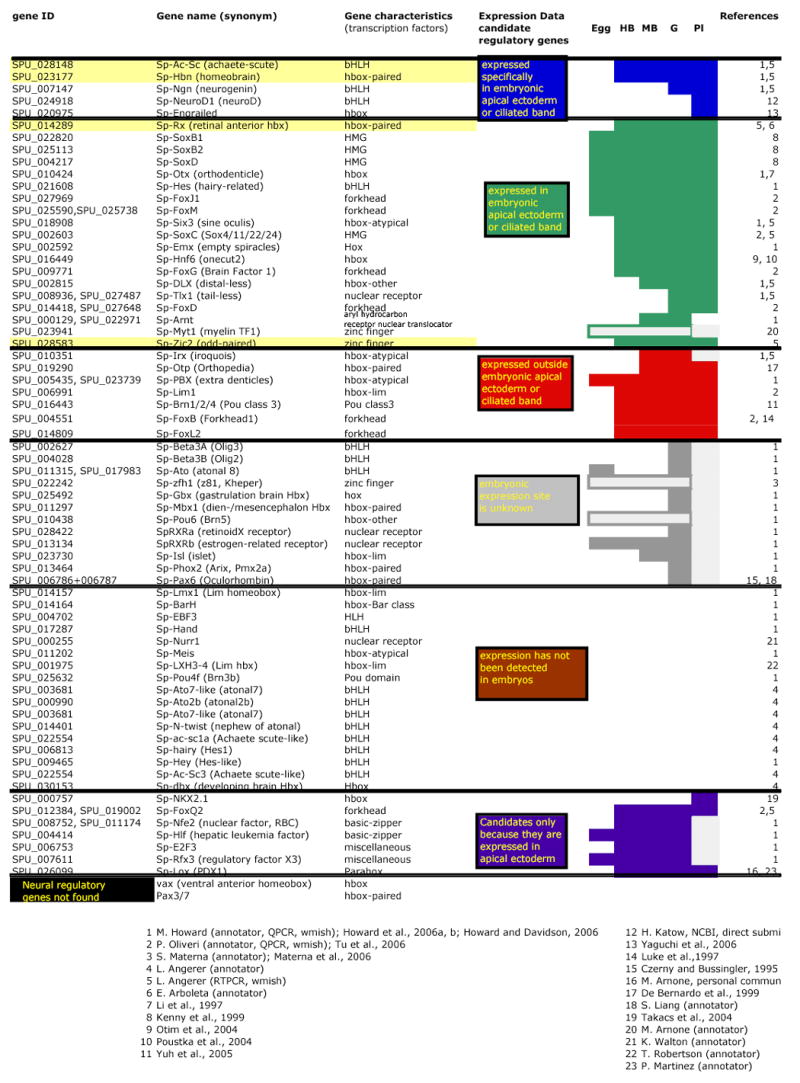
Searches of the genome have identified homologues of a number of genes known to encode transcription factors that are expressed in neural tissues in other animals (genomic methods are described in Sodergren et al. 2006). PCR, in situ hybridization, or microarray analyses (Samanta et al. 2006) indicate that many of these genes are expressed during early development. Expression of several of these genes begins before gastrulation, the time when neuroblasts are first detected. We have categorized candidate neurogenic genes by what is known about their temporal and spatial patterns of expression during embryogenesis (Table 1, colored boxes). Whereas this list includes many genes, it represents only a starting point since more candidate genes will undoubtedly be found as experimentation and analysis progress.
Table 1.
|
One group comprises genes expressed exclusively in the neurogenic ectoderm at the animal pole (Table 1, blue). Two genes in this group begin to be expressed between the hatching blastula and early mesenchyme blastula stages, before the onset of morphogenesis. Transcripts from each of these genes accumulate in a nearly contiguous patch at the animal pole (Fig. 5A, C). Two of these, Sp-Ac-Sc (achaete-scute) and Sp-Hbn (homeobrain), are homologues of genes known to be involved in early neural specification in other systems: Achaete-scute is a basic-HLH atonal class factor whose function in neurogenesis is conserved in cnidarians, flies and vertebrates (Bertrand et al. 2002). In Drosophila embryos, Hbn, which encodes a paired-class transcription factor, is required for development of the brain (Davis et al. 2003) and is expressed in the presumptive brain, at the cellular blastoderm stage (Walldorf et al. 2000). Other sea urchin genes expressed exclusively in animal plate ectoderm encode the well-known proneural, atonal class factors, neurogenin and neuroD, and are expressed later in development, apparently exclusively in neural cells (LMA, unpublished; H. Katow, NCBI submission).
Figure 5.
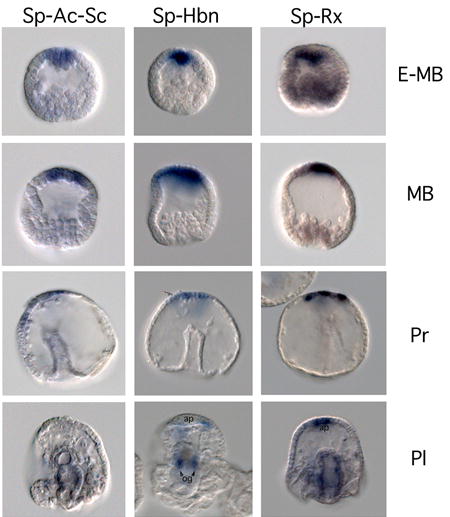
Sp-Sc-Ac (achaete-scute), Sp-Hbn (homeobrain), Sp-Rx (retinal anterior homeobox) transcripts accumulate in the apical ectoderm at blastula stage and in a subset of cells of the nervous system in the pluteus larva. Hybridization probes are shown in columns and stages in rows; lateral views of embryos have animal plate ectoderm at the top. Sp-Rx RNA accumulates in animal plate ectoderm and to lower levels in vegetal cells of blastulae (E-MB), is restricted to animal plate ectoderm in mesenchyme blastulae (MB), and is in scatted cells of the animal plate (ap) and around the gut in plutei (Pl). Sp-Ac is expressed in scattered cells of the animal plate ectoderm at prism stage (Pr). Sp-Hbn is expressed in oral ganglia (og) and scattered cells near the animal plate of plutei. Whole mount in situ hyrbridizations were carried out according to Minokawa et al. (2004). Embryos were photographed with DIC optics.
Genes in a second group are also expressed in the animal plate ectoderm, but expression is not confined to this region (Table 1, green). The onset of expression refers to the whole embryo and in most cases expression in the animal plate is later than in other tissues. This group includes genes widely expressed in the ectoderm (SoxB1 and B2, Kenny et al. 1999) or throughout most of the embryo (Otx, Li et al. 1997), but which are critical factors in neural development in other animals (Sasai, 2001, Boyl et al. 2001). Also in this group is the gene encoding Sp-Rx (retinal anterior homeobox), another paired-class transcription factor. In vertebrates, Rx is expressed in the early anterior neural plate at the gastrula stage (Furukawa et al. 1997) and subsequently in the retina where it is required for eye development (Mathers et al. 1997). The early expression domain of Sp-Rx is nearly identical to that of Sp-Ac-Sc and Sp-Hbn (Fig. 5E). As in Drosophila, the expression patterns of these three genes establish that a neurogenic territory is specified after the period of rapid cell division and before morphogenetic movements. Thus Sp-Rx, Sp-Hbn and Sp-Ac-Sc likely operate near the top of a neuronal gene regulatory network. As development proceeds, expression of these genes becomes confined either to a subset of cells in the apical plate of the prism-stage embryo (Sp-Ac-Sc; Fig. 5B; Sp-Rx; Fig. 5F), or to neurons in the oral ganglia and adjacent to the ciliary band of the pluteus larva (Sp-Hbn; Fig. 5D).
Additional candidate genes that function in neural development in other animals are not detectable in the larval nervous system (Table 1, red). However, they may function in later neural development. Genes in this group are similar in sequence to known neural factors, but either their spatial patterns of expression are not yet known (gray) or not detectable in the embryo (brown). Interestingly, additional proneural-like genes are found among those not detectably expressed during the first two days of embryogenesis, the period covered by the microarray analysis (Samanta et al. 2006), raising the possibility that they regulate later stages of neural development in either the larva or adult. A few genes have been included as neural candidates because they are expressed in the embryonic neurogenic region, although their homologues have not yet been shown to be expressed specifically in neural tissue (purple group). One of these (NK2.1) is not required for initial differentiation of embryonic neurons (Takacs et al. 2004; K. Peterson, personal communication), but a subsequent role in the nervous system remains possible. Finally, by the criterion of reciprocal BLAST searches of the genome, ventral anterior homeobox (vax) and Pax3/7 have not been conclusively identified.
As expected, this survey has shown that the sea urchin genome contains many homologues of genes involved in neurogenesis in deuterostomes and protostomes. The neurogenic functions of several of these genes are ancient because they are conserved in cnidarians. In fact, the genes encoding transcription factors that are expressed in early phases of neurogenesis, (Sp-Rx and Sp-Hbn, and Sp-Ac-Sc; Fig. 5A, C, E) are similar to those expressed at early stages of the neural program in Hydra that include the paired-class protein genes, prdla and prdlb (Miljkovic-Licina et al. 2004) and the achaete-scute homologue, CnASH I (Grens et al. 1995). This set of conserved early neurogenic factors may also include Zic, a Drosophila odd-paired homologue. The Hydra homologue, HyZic, is expressed in neural stem cells and is involved with the proliferation of neuroblasts (Lindgrens et al. 2004); the ascidian homologue, HrZic, functions in early specification of some neuronal precursors (Wada and Saiga, 2002); and the Xenopus homologue is thought to function as a link between the neural inducer, chordin, and the neurogenic gene, neurogenin (Mizuseki et al. 1998). A sea urchin Zic protein, Sp-Zic2, also a homologue of odd paired, has been shown by microarray analysis to be expressed in the embryo between egg and gastrula stages. Reciprocal BLAST identifies Sp-Zic2 as homologous to odd paired and its expression begins during early blastula stage (LMA, unpublished). This conserved collection of genes, paired-class, achaete-scute and Zic (Table 1, yellow) appears to be expressed at the same points in the neurogenic programs of a diverse set of organisms. This supports the hypothesis they are core components of a metazoan neurogenic gene regulatory network and its expression begins in the animal plate ectoderm of early blastulae.
Insight into the evolution of the nervous system may come from detailed phylogenetic comparisons of sea urchin, cnidarian and protostome neurogenic gene sets leading to discovery of ancestral, deuterostome-specific genes. Shared components and pathways have the potential of revealing how metazoan neurogenic gene regulatory networks have been shaped by evolution to produce the vertebrate nervous system.
Signaling that specifies neurons
Cellular interactions mediated by ligands and receptors are known to specify and pattern the nervous systems of metazoans. The genome of S. purpuratus encodes ligands and receptors for all of the major signaling molecules that have previously been demonstrated to have a role in neural specification and patterning (Croce et al. 2006a; Beane et al. 2006; Walton et al. 2006; Lapraz et al. 2006). Many of these genes are expressed during early cleavage, the time when such specification must occur. Some are expressed in neurogenic domains (Croce et al. 2006b) and there is evidence that early specification events have an impact on the abundance and patterning of larval nerves (Yaguchi et al. 2006). Access to genomic data will undoubtedly reveal approaches to investigating the signaling that underlies neural specification in sea urchin embryos. An understanding of the signaling mechanisms specifying neural tissues in urchins will provide critical data for comparison with this area of research that is well-studied in Drosophila, C.elegans, and vertebrates.
Axon Guidance
Neurogenesis in sea urchin embryos involves elongation of neurites that have terminal growth cones and the neurites form precise patterns in which neurons and effector cells are interconnected in a predictable pattern (Nakajima et al. 2004). The implication of these observations is that axon guidance mechanisms function during neurogenesis. The genome contains orthologues of all the deuterostome axon guidance molecules. There is a single B-type Eph receptor and a single transmembrane, B type eprhin ligand. An orthologue of semaphorin and numerous plexin receptors have been identified (Whittaker et al. 2006). There are predictions for orthologues of Netrin and the axon repulsion receptor Unc-5. A Slit orthologue and its receptor Robo have also been identified. In addition, the neural adhesion molecule L1 (Neurofascin) is present. Preliminary data indicate Sp-Eph is expressed in the ciliary bands of the larva and in adult radial nerves (RDB, preliminary data) and L1 appears to be expressed in larval nerves (D. McClay, personal communication). The small number of paralogues may permit incisive determinations of how this set of metazoan axon guidance molecules functions in neurogenesis in sea urchins.
Synapse formation
Echinoderm synaptic morphology is reported to be unusual in that morphological studies have failed to identify direct synaptic contacts and pre- and post-synaptic specializations. Pre-synaptic terminals appear as swellings, but they do not appear to directly contact post-synaptic membranes (Cobb and Laverack, 1967; Cobb and Pentreath, 1976, 1977; Pentreath and Cobb, 1972). In some situations, it has been proposed that substantial distances separate neurons and effector cells (Florey and Cahill, 1977). The genome encodes several genes that in other metazoans mediate the processes of synapse formation. Orthologues for neuroligin, β- neurexin, agrin, MUSK and thrombospondin have been identified by reciprocal BLAST searches, PFAM and Smart searches (Whittaker et al. 2006). In vertebrates β-neurexin binds neuroligin, which appears to function in recruiting post-synaptic components (Washbourne et al. 2004). Agrin is a nerve derived, heparan sulphate proteoglycan extracellular matrix component that interacts with MUSK to aggregate acetycholine receptors in post synaptic terminals (Gautam et al. 1999). Echinoids appear to have these molecules that are specialized for roles in synapse formation in vertebrates, but it is not known if they are involved in synapse formation in urchins. EST’s encoding Agrin (SPU_0 22633) and thrombospondin (SPU_017370) have been identified in primary mesenchyme libraries suggesting a more generalized role in cell adhesion.
Electrical signaling
The ability of neurons to generate and transmit electrical signals is dependent on expression of voltage-gated ion channels (VGICs) that are permeable to ions including potassium (K+), calcium (Ca++) and sodium (Na+). The proteins that form the ion pore of VGICs (alpha subunit) have sequence similarity consistent with a common evolutionary ancestry and members of this family have also been identified in prokaryotes (Hille, 2001). Relatively little is known about the electrophysiological properties of neurons in echinoderms, although there is evidence for calcium-based action potentials (Cobb and Moore, 1989). No VGIC molecules have previously been identified in echinoderms, but our analysis of the sea urchin genome has uncovered 11 K+ channels, 4 Ca++ channels and 1 Na+ channel (Table S-1). Voltage-gated sodium channels in animals probably evolved from T-type calcium channels prior to the radiation of the bilateria because they share more sequence similarity with T-type calcium channel alpha subunits than with other VGICs and are present in protostomes and deuterostomes. However, subtle changes in the sequences of voltage-gated ion channels can switch ion selectivity and therefore it is possible that the single putative Na+ channel in S. purpuratus actually functions as a calcium channel. Experimental studies are required to address this issue.
Gap junction proteins
Gap junctions are intercellular channels derived from multimeric channel proteins that attach head to head to form a pore that permits the passage of small molecules between adjacent cells. Gap junctions provide ionic or electrical coupling between neurons and in vertebrates gap junction proteins are expressed in all tissues. Two unrelated families of proteins are known to form gap junctions: connexins, which have only been found in chordates, and pannexins, which include innexins and are found throughout metazoans (Panchin, 2005). Searches with BLAST, pfam and IPRSCAN have failed to find representative genes for any of these proteins in the urchin genome. Gap junctions have distinctive ultrastructural characteristics and several morphological studies have searched systematically for gap junctions without success (Cobb, 1987; Cavey and Wood, 1991; Florey and Cahill, 1977; 1980). Thus, sea urchins appear to lack gap junction proteins, which implies that their neurons lack ionic coupling and must rely on chemical synapses for communication. Some classes of cnidarians appear also not to have gap junction proteins, suggesting that this condition may have arisen more than once in metazoan evolution (Mackie et al. 1984; Mire et al. 2000).
Neurotransmitter release
Neurotransmitter release at nerve terminals is mediated by calcium-dependent fusion of synaptic vesicles (SV) with the plasma membrane. Synaptic vesicle trafficking has been studied in a few model organisms: rodents, Drosophila, and C. elegans (Sudhof, 2004; Richmond and Brodie, 2002). In the SV trafficking cycle, vesicles are filled with neurotransmitters, dock at the active zone, and are primed to become calcium sensitive. Arrival of action potentials in the axon terminal causes voltage-gated calcium channels to open and trigger fusion of SVs with the plasma membrane, releasing neurotransmitter into the synaptic cleft. Following fusion, empty SVs are recycled and refilled for another round of neurotransmitter release.
Regulated secretion (exocytosis) appears to proceed through common mechanisms in eukaryotes, mediated by highly conserved proteins; some exist as single copy genes and others belong to multigene families (Jahn, 2003). These proteins are generally conserved in the S. purpuratus genome, with numbers of isoforms that are similar to other invertebrate metazoans (Table S-2). There are no published studies on the SV cycle in S. purpuratus, however, homologues of several critical proteins have been characterized in studies on the role of calcium-triggered exocytosis in egg maturation and fertilization (Avery et al. 1997; Conner et al. 1997; Leguia et al. 2004; Schulz et al. 1998; Tahara et al. 1998; Whalley et al. 2004).
Membrane attachment
The Rab GTPases are a multigene family that mediates targeting of intracellular vesicles to target membranes (Pereira-Leal and Seabra, 2001). S. purpuratus has 33 Rab genes, similar to C. elegans and D. melanogaster (29) and fewer than human (~50). Rab3 is the major isoform bound to SVs. The S. purpuratus homologue has 47% identity to vertebrate and mammalian Rab3 (Table S-2). The GTP-bound form of Rab3 binds to specific effectors in the active zone: Unc-13 and RIMs (Rab3-interacting molecules). S. purpuratus Unc-13 is smaller (898aa) than its homologues in zebrafish (1121aa) or mouse (1797aa). These proteins have highly conserved (50–80% identity) C2 domains at their amino- and carboxy-termini. The size difference is due to expansion of the central region of Unc-13 in mouse and zebrafish. In vertebrates, RIMs are large, multidomain proteins (~1500 aa) with a Rab3 binding site near the amino-terminus. The single S. purpuratus RIM homologue (Sp-RIM2) is a small protein (305 aa) similar to the C-terminal region of mammalian RIM2 (55% identity) and lacking the Rab3 binding site (Table S-2). These differences in Rab3 effectors suggest that the mechanism of SV docking to the active zone may differ between echinoderms and vertebrates.
The exocyst complex is another highly conserved Rab effector, consisting of eight subunits (Sp-Exoc1-8). It mediates insertion of proteins into the plasma membrane, and possibly formation of synaptic junctions (Clandinin, 2005).
SNARE and SM proteins
The SNAREs are small, membrane-anchored proteins with one or two 70-residue SNARE domains. Genetic studies indicate that SNAREs are required for both intracellular vesicle trafficking and secretion in eukaryotes (Sudhof, 2004; Jahn, 2003 ). The four classes of SNARE domains, Qa, Qb, Qc and R, form a stable alpha helical bundle (core complex) comprised of 3–4 SNAREs (some SNAREs contain both a Qb and a Qc domain) anchored in the vesicle and target membranes. S. purpuratus has at least 17 SNAREs including eight syntaxins (Stx 1, 5, 6, 8, 12, 16, 17,18) (Table S-2). Syntaxins are highly conserved among S. purpuratus, vertebrates (Danio) and mammals, having 30–70% identity over their entire length. It is interesting to note that S. purpuratus is apparently missing a node from the mammalian syntaxin family tree comprising syntaxins 2, 3, 4 and 11 (Teng, 2001 ). Syntaxins 2–4 are located in the plasma membrane and mediate transport of proteins to the cell surface and protein secretion.
In SV fusion, VAMP1 (vesicle-associated membrane protein)/synaptobrevin (R-SNARE) on the vesicle, forms the core complex with SNAP25 (Qb, Qc-SNARE) and syntaxin 1 (Qa-SNARE) on the target membrane. S. purpuratus appears to have single-copy genes coding for VAMP1, SNAP25, and syntaxin 1 homologues (Table S-2).
SM proteins bind to SNAREs and may regulate core complex assembly (Sudhof, 2004; Jahn, 2003). Genetic studies in rodents indicate that these proteins are required for SV fusion. S. purpuratus has a single gene encoding a homologue of the mammalian Sec1/munc18 family, Sp-Stxbp with 60% identity to vertebrate and mammalian Stxbp1.
Following vesicle fusion with the target membrane, the SNARE complex is disassembled through the action of the NSF ATPase (N-ethyl maleimide-sensitive factor) and soluble NSF-attachment proteins (SNAPs). S. purpuratus has single genes encoding NSF (Sp-Nsf) and alphaSNAP (Sp-Napa) homologues with >60% identity the vertebrate and mammalian proteins.
The expression patterns of these SV-specific SNAREs and associated proteins in echinoderm neural tissue have not yet been described. However, these proteins are also expressed in echinoderm eggs and early embryos where they are localized to the cortical region of the cell and may mediate exocytosis required for early embryogenesis (Conner et al. 1997; Leguia et al. 2004; Tahara et al. 1998 ).
Synaptotagmins
Synaptotagmins (Syts) are a multiprotein family that have attracted considerable interest as potential sensors for calcium-triggered neurotransmitter release (Sudhof, 2002). Synaptotagmins are anchored in vesicle and target membranes at their amino-terminus, and have two tandem C2 domains (termed C2A and C2B) which bind to phospholipid in the presence of calcium. Membrane fusion may be triggered by binding of the C2 domains to the target membranes, in cooperation with the SNARE complex (Arac et al. 2006). Synaptotagmins bind to SNAREs in vitro and co localize with SNAREs and SM proteins in vivo.
There are 17 Syt isoforms in human and rodents, expressed mainly in neural tissue (a few isoforms are non-neuronal). Genetic studies indicate that Syts 1 and 2 are required for the fast, synchronous component of neurotransmitter release (Sudhof, 2002; Jahn, 2003; Sudhof, 2004 ). Other Syts may mediate the slow, asynchronous component of release, and secretion from neurendocrine cells. Some isoforms (Syts 8, 12–16) lack calcium-binding activity; their functions are unknown.
S. purpuratus has 14 synaptotagmin isoforms (Table S-2). These proteins have tandem C2 domains with greater sequence similarity to the C2A-C2B domains of Syts than to other C2 domain proteins (> 30% identity, Expect value < 1e-15). We have adopted a provisional nomenclature for S. purpuratus syts, based on the most similar mammalian isoform. Nine isoforms exhibit a high degree of similarity to a particular mammalian isoform (>50% identity, Expect value < 1e-50) and may be considered to be orthologues: Syt1-1, Syt5, Syt6-1, Syt7, Syt9, Syt12, Syt14, Syt15-1, Syt17. The other isoforms: Syt1-2, Syt4, Syt6-2, Syt15-2 have similar degrees of similarity to two or more mammalian syts and are thus difficult to classify according to the mammalian tree. Three of these isoforms have a tandem duplication of the N-terminal 1/2 (Syt1-2, Syt12) or whole C2A domain (Syt15-1). Such duplications have not been described in syts from other species. This may reflect problems with the current genomic assembly; hopefully, cDNA cloning of these genes will resolve this issue.
A pairwise comparison of the predicted C2A-C2B domains, of the 14 S. purpuratus isoforms indicates that Syts 1-1, 5, 6-1, 7, and 9 comprise a closely related group (Fig. 6). These are all orthologues of mammalian syts known to have calcium-phospholipid binding activity and to be calcium sensors for regulated exocytosis (Chenna et al 2003). The others, including orthologues of mammalian syts which lack this activty (Syts 12, 14, 17) are more distantly related (Fig. 6).
Figure 6.
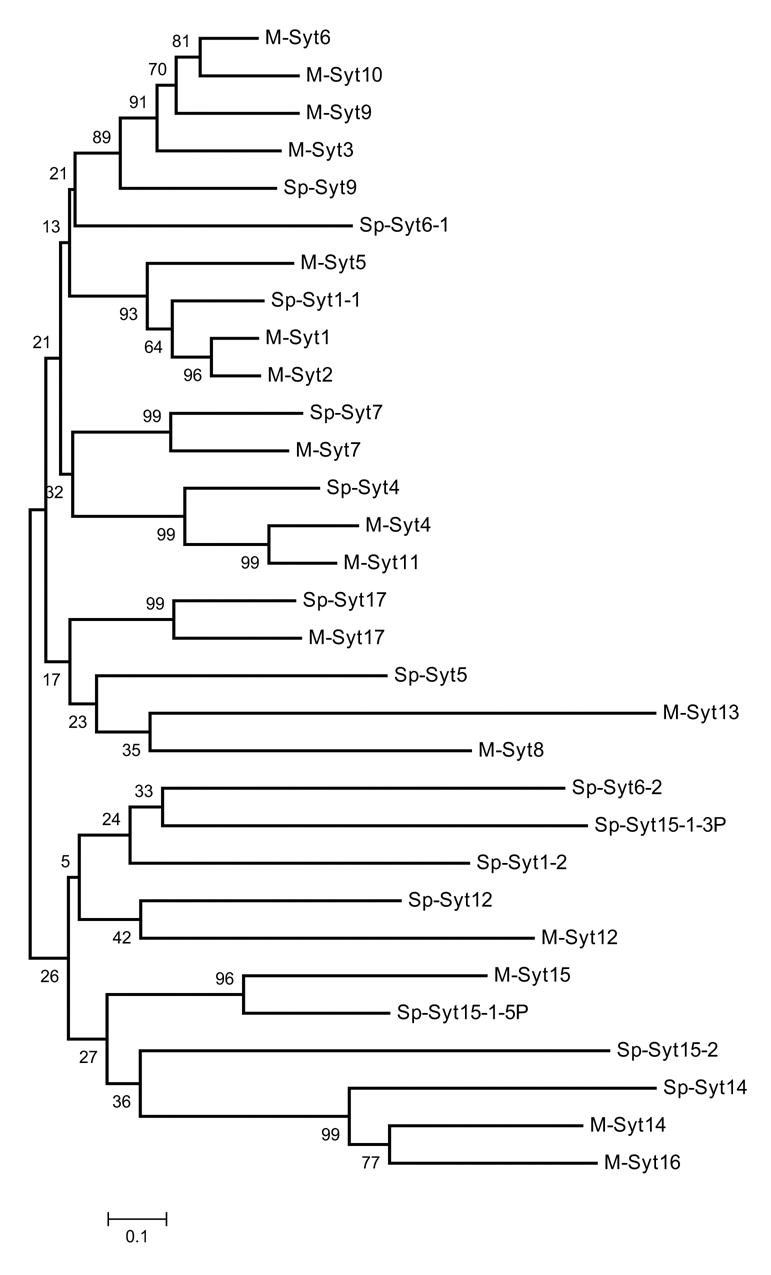
Neighbor joining tree of the 14 synaptotagmin isoforms in S. purpuratus. Alignments of the C2A-C2B domains were made using ClustalW, numbers indicate bootstrap values (1000 replicates). For this analyis, the extra N-terminal C2 domain in SPU_16698 (Sp-Syt15-1-5P) and the two C-terminal C2 domains in SPU_28132 (Sp-Syt15-1-3P), were treated as different genes rather than alleles. The predicted amino acid sequence for Sp-Syt1-1 derived from cDNA (AAB67801) was used instead of SPU_005854, as this gene model is missing part of the C2B region.
Syt1-1 (also termed Synaptotagmin B) appears to be the orthologue of mammalian Syt1, the calcium sensor for SV fusion. Both Syt1-1 mRNA and protein are expressed in the early pluteus (72–96 hrs.) in the apical organ, the ciliary band, and the adoral ciliary band surrounding the mouth, in cells resembling neurons and tracts of neurites, and also in adult neural tissues, consistent with a role in neurotransmitter release (Burke et al. 2006).
Neurotransmitters, transporters and receptors
Early studies of echinoid neurotransmitters used biochemical methods to detect neurotransmitters extracted from tissues and this provided good evidence that acetylcholine and some associated enzymes are present (Pentreath and Cottrell, 1968). Applications of neurotransmitters and inhibitors and histochemical methods have provided inconclusive information on other putative transmitters (Binyon, 1972; Cobb, 1987). Immunocytochemical methods have been helpful, but they have not been systematically applied. Thus, the full spectrum of neurotransmitters employed by echinoids is not known (Welsh, 1966; Cobb, 1987). We have analyzed the sea urchin genome for orthologues of proteins involved in the biosynthesis, reception or inactivation of neurotransmitters in other animals (Table S-3).
Serotonin is synthesized from tryptophan by tryptophan hydroxylase (TPH) and aromatic amino acid decarboxylase (AADC). Sea urchin TPH is known from Hemicentrotus pulcherrimus, (Yaguchi and Katow, 2003) and there is a single TPH homologue in the Stronglylocentrotus genome that is almost identical to HpTPH (93 % amino acid identity). We identified 1 AADC gene from the S.purpuratus genome and 1 serotonin transporter gene that is most similar to serotonin transporters of other invertebrates (Table S-3). Melatonin is derived from serotonin in a pathway that includes Serotonin N-acetyltransferase and acetylserotonin O-methyltransferase. However, neither gene has been found in either ascidians or sea urchins, suggesting that this may be a pathway unique to vertebrates.
Monoamine neurotransmitters, such as serotonin and dopamine are inactivated/degraded in the synaptic cleft and pre-synaptic terminal by the activites of monoamine oxydase (MOA) and catechol O-metyltransferase (COMT). Several MOAs have been identified in the sea urchin genome, and they include both types of MOA-a and MOA-b (Table S-3). One COMT was also found in the genome.
Serotonin receptors are categorized into 7 types and 18 subtypes based on their amino acid sequences and responses to agonists and antagonists (Adayev et al. 2005). To date, invertebrate genomes have revealed only a few types of serotonin receptors. In the sea urchin genome, 4 serotonin receptors were found searching with extracellular and cytoplasmic domains of all known types of serotonin receptor (Fig. 7A). Although the BLAST results are strong for all of these predictions, in neighbor joining trees of aligned sequences clear relationships can not be readily resolved. Type-3 serotonin receptors differ from other serotonin receptors in not being G-protein coupled receptors. Urchins appear not to have type 3 receptors, suggesting that these might be a chordate innovation (Table S-3).
Figure 7.
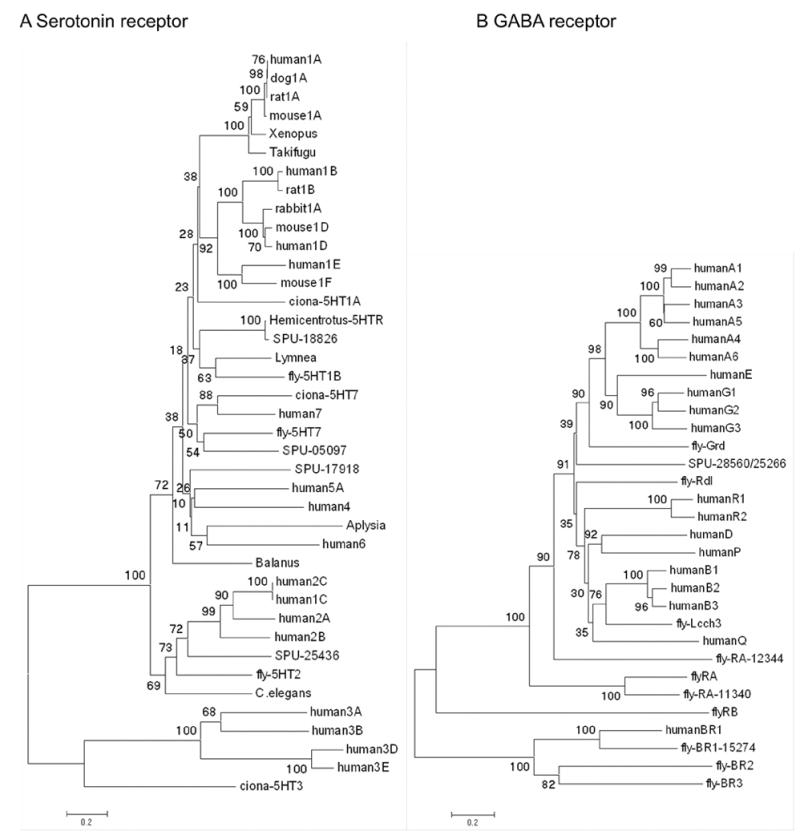
Neighbor joining trees based on alignment of full length sequences and predictions for serotonin receptors indicate there are 4 sea urchin homologues that can be associated with known families of vertebrate and invertebrate receptors. B Neighbor joining tree based on alignments of full length cDNA sequences or predictions for GABA receptors. Numbers indicate bootstrap values (1000 replicates).
The catecholamine dopamine is synthesized from tyrosine in a pathway including tyrosine hydroxylase (TH) and AADC. We have identified both genes in the sea urchin genome (Table S-3). In the Drosophila genome, there are two types of dopamine receptor, whereas we found 7 putative dopamine receptors in the S. purpuratus genome. In a phylogenetic analysis, the sea urchin and Drosophila genes are most similar to the human D1-type dopamine receptor.
Noradrenaline is derived from dopamine by dopamine ß-hydroxylase (DBH). Phenylethanolamine N-methyltransferase (PNMT) catalyzes the conversion of noradrenaline to adrenaline. A DBH-like gene is present in the sea urchin genome but PNMT genes have not been identified in either the tunicate or the sea urchin genome, suggesting that adrenaline may be specific to vertebrates. We identified 1 adrenergic receptor from the sea urchin genome that clusters weakly with Ciona and human adrenergic receptors (Table S-3; Fig. S-1).
GABA is synthesized from glutamate by glutamic acid decarboxylase (GAD). Drosophila and human genomes have two types of GAD and there is a single GAD gene in the Ciona genome. In the sea urchin genome, we found 1 GAD that is most similar to Drosophila GAD1 (Table S-3). We found 1 ionotropic (A –type) GABA receptor gene in the sea urchin genome, whereas Drosophila and humans have several. In phylogenetic analysis, the sea urchin GABA receptor is most similar to the human GABA receptor type E (Fig. 7B)
Histamine is synthesized from histidine by histidine decarboxylase (HDC). HDC genes are known from Drosophila and humans, but in ascidian genomes HDC and histamine receptors have not been identified (Dehal et al. 2002). There are two HDC gene predictions in the sea urchin genome and three histamine receptors (Table S-3). Phylogenetic analysis of the receptors suggests that two of these form a clade with human HRH3. Another appears to be most similar to a Drosophila histamine receptor. Thus, histamine appears to have a bilaterian origin with subsequent loss in ascidians.
Drosophila, ascidians, and humans have a single choline acetyltransferase (ChAT) and a single acetylcholinesterase (AChE). In the sea urchin genome, we found 1 ChAT gene, and several AChE genes (Table S-3). Vesicular acetylcholine transporter was also found in the sea urchin genome as in Drosophila and human.
Acetylcholine receptors are classified into two major groups, muscarinic (mAChR) and nicotinic (nAChR). In humans, there are 5 types of mAChR receptors (M1–M5). We identified four mAChR in the sea urchin genome (Table S-3). Phylogenetic analysis shows two of these form 1 group with human mAChR-M4. The other two position weakly within the same clade. The sea urchin genome also contains 12 genes for nAChR. In phylogenetic analyses, four of these form clades with human nAChRs, and the other 4 group with non-vertebrate clades. The remainder has unresolved affinities (Fig. S-1).
Glycine is known to function as a neurotransmitter through the glycine receptor (GLR). Humans have 4 A-type and 1 B-type glycine receptors. In the sea urchin genome, we found two glycine receptors (Table S-3). One is similar to human GLRA1 and the other has no clear affinity to vertebrate glycine receptors.
These gene predictions indicate that sea urchins are likely to use a broad range of neurotransmitters. However, there is no evidence that they are able to use either adrenalin or melatonin, as is the case for Ciona (Dehal et al. 2002). These data indicate that although the organization of the echinoid nervous system differs from that of other deuterostomes we can anticipate that the neurons are similar with respect to synthesis and perception of neurotransmitters.
Neuromodulators
Adenosine, cannabinoids, lysophospholipids and melanocortins
The G-protein coupled receptors of classical neurotransmitters and neuromodulators, such as acetylcholine, catecholamines and serotonin are rhodopsin-like receptors known as α-type receptors (Fredriksson et al. 2003). A sub-group of mammalian α-type receptors includes adenosine, cannabinoid, lysophospholipid and melanocortin receptors. Orthologues of adenosine receptors are present in Drosophila melanogaster and other protostomes, indicating a bilaterian ancestry of adenosine receptors. Consistent with this, we have identified an adenosine receptor gene in S. purpuratus (SPU_008789). Orthologues of mammalian cannabinoid, lysophospholipid and melanocortin receptors are not present in Drosophila and C. elegans (Elphick and Egertová, 2001, 2005). However, the Ciona genome contains an orthologue of mammalian CB1/CB2-type cannabinoid receptors and genes encoding receptors related to mammalian lysophospholipid receptors (Elphick et al. 2003; MRE, unpublished). Orthologues of mammalian CB1/CB2 cannabinoid receptors, lysophospholipid receptors and melanocortin receptors were not identified in the S. purpuratus genome, suggesting that this group of receptors is unique to chordates. However, cannabinoid binding sites have been detected on sea urchin sperm and endocannabinoids may prevent of polyspermy by blocking the acrosome reaction (Chang et al. 1993; Schuel et al. 1994). Further studies are required to determine the identity of the putative cannabinoid receptor detected on sea urchin sperm.
Nitric Oxide
Nitric oxide (NO) modulates many physiological processes in animals. NO is produced by nitric oxide synthase (NOS) and often operates by stimulating the soluble guanylyl cyclase (sGC) receptor to produce cGMP. NO causes SGC/cGMP-dependent relaxation of the cardiac stomach in the starfish Asterias rubens (Elphick and Melarange, 1998) and subsequent studies indicate that NO acts as a general muscle relaxant in echinoderms (Elphick and Melarange, 2001; Melarange and Elphick, 2003). Metamorphosis of the urchin Lythechinus pictus is negatively regulated by NO and cGMP (Bishop and Brandhorst, 2001). Immunostaining indicates that NOS is expressed in the larval nervous system in the apical organ, oral ganglion, gut and arms of plutei and a few ciliated neuronal cell bodies in the ventral transverse ciliary band that appear during larval development (Bishop and Brandhorst, 2001; and unpublished). These latter cells have projections to the apical organ that stain for NOS and microsurgical experiments are consistent with the hypothesis that these cells mediate the response of the larva to chemical cues for metamorphosis.
Analysis of the S. purpuratus genome reveals the presence of a gene that encodes an orthologue of the NOS proteins that have been identified in other metazoans (Table S-4). However, there are also 4 genes encoding NOS-like proteins and further studies are needed to determine the functions of these proteins. A family of genes encoding putative sGC subunits are present in S. purpuratus, which include 1 α subunit, 2 β1 subunits, 1 β2 subunit and several sGC-like proteins (Table S-4).
Neuropeptides, peptide hormones and G-protein coupled receptors
Secreted peptides generated by cleavage of larger precursor proteins are important neuroendocrine signaling molecules throughout the metazoans (Strand, 1999). There is evidence for a neuroendocrine system in echinoderms and several neuropeptides have been identified in starfish and sea cucumbers. However, neuropeptides have not been identified in sea urchins and it is not known if peptide hormones are produced by non-neural endocrine organs.
Most neuropeptides and many peptide hormones exert their effects by activating G-protein coupled receptors (GPCRs). To obtain insight into the potential diversity of peptide signaling, we searched the S. purpuratus genome for orthologues of neuropeptide receptors. There are five families of GPCRs (Glutamate, Rhodopsin, Adhesion, Frizzled, and Secretin) and most peptide receptors belong to the rhodopsin or secretin families (Fredriksson et al. 2003). There are 37 S. purpuratus G-protein coupled peptide receptors in the rhodopsin family (Table 2). In a similar analysis of the Drosophila genome 32 putative G-protein coupled peptide receptors were identified (Hewes and Taghert, 2001). The identities of the peptides that activate many of these Drosophila GPCRs have subsequently been identified (Cazzamali and Grimmelikhuijzen, 2002). Based on orthology with human GPCRs, we predict genes for a vasopressin/oxytocin receptor and a cholecystokinin/gastrin receptor (Table 2). The family of secretin receptors is exceptionally large with 161 members compared to only 15 in the human genome. Although genes that are related to the human corticotrophin-releasing hormone (CRH) and calcitonin receptors were identified, most sea urchin receptors in this family are not closely related to known vertebrate receptors. The expanded secretin family of GPCRs may be unique to sea urchins.
Although 37 sea urchin GPCRs related to human neuropeptide or peptide hormone receptors were identified (Table 2), it was more difficult to identify genes encoding precursors of the putative peptide ligands for these receptors. However, we have identified a gene encoding a peptide (echinotocin, Table 2) related to vasopressin/oxytocin/vasotocin peptides of vertebrates and protostomes (Van Kesteren et al. 1992). Echinotocin may act as the ligand for a sea urchin orthologue of human vasopressin and oxytocin receptors.
Glycoprotein hormones occur throughout bilaterians and include the mammalian hormones luteinizing hormone, follicle stimulating hormone, and thyroid stimulating hormone (Hsu et al. 2002; Luo et al. 2005). The sea urchin genome contains members of this family of hormones and GPCRs related to human glycoprotein hormone receptors (Table 2). Putative sea urchin glycoprotein hormones are of particular interest because they may function as gonadotropins. Gonadotropins in echinoderms were first discovered in extracts of starfish nerve cords nearly 50 years ago (Chaet and McConnaughy, 1959), but their molecular identity remains elusive. Two other members of the glycoprotein hormone family in S. purpuratus appear to be orthologues of the alpha and beta subunits of the insect hormone bursicon (Table 2; Luo et al. 2005).
The sequences of several neuropeptides have been determined in starfish and sea cucumbers. The first to be identified in echinoderms were SALMFamides, which act as muscle relaxants (Elphick et al. 1991; Elphick and Melarange, 2001). Two related peptides, S1 and S2, were isolated from Asterias and subsequently other SALMFamides were found in holothurians. A SALMFamide precursor gene in S. purpuratus contains two exons encoding a signal peptide and seven putative SALMFamides (Elphick and Thorndyke, 2005). Although a number of other echinoderm neuropeptides have been identified in the sea cucumber Stichopus japonicus, based on a muscle activity assay (Iwakoshi et al. 1995; Ohtani et al. 1999), homologues of most of these could not be identified in the sea urchin genome. One predicted gene encoding two copies of a putative neuropeptide (NGFFFamide) does appear to be related to a Stichopus peptide (NGIWYamide), which causes muscle contraction (Inoue et al. 1999). A promising approach for further studies of many of the putative neuroendocrine peptides will be to determine if they have activity in standard bioassays using echinoid tissues.
Growth factors and growth factor receptors
Growth factors that bind to non-G-protein coupled receptors include insulin, neurotrophins and the ependymins. From a structural and functional perspective, the distinction between peptide hormones and peptide/protein growth factors is arbitrary. Although often products of the nervous system, several are also expressed in non-neural tissues.
Insulin and IGF family
The insulin peptide family is one of the best-known neuroendocrine systems of chordates. In vertebrates, insulin (INS) is a pancreatic endocrine hormone with a role in regulating of carbohydrate metabolism. Other members of this family include the insulin-like growth factors (IGFs), relaxins (RLNs) and insulin-like factors (INSLs) (Chan and Steiner 2000, Conlon 2000; 2001, Reinecke and Collett 1998). IGFs are ubiquitously expressed and regulate cell proliferation and plasticity (Reinecke and Collett 1998). RLN is a mammalian placental hormone important in angiogenesis and softening of ligaments during parturition (Wilkinson et al. 2005). Mature RLN and INS are derived from single chain precursors in which B- and A domains are linked by a connecting C-peptide. During processing, the amino-terminal signal sequence is removed and the intervening C-peptide is cleaved leaving B and A domains linked by disulphide bridges. In contrast, IGFs not only retain the C-peptide but also contain D- and E domains. This family of peptides interacts with tyrosine kinase receptors (INS/IGFs) or GPCRs (INSLs/RLNs) and there are binding proteins responsible for their transport. Invertebrate insulin-like peptides constitute large, divergent multi-gene families that share the basic insulin structure, including the conserved cysteines essential for formation of inter- and intra-chain disulphide bonds. Most invertebrate insulin-like peptides are expressed in the nervous system and act as neurohormonal growth regulators (Smit et al. 1998).
The S. purpuratus genome has two genes encoding members of the insulin family with paired cysteines, a propeptide containing the amino-terminal signal sequence and B, C and A domains (Table S-5; Fig. S-2). Sp-IGF1 has characteristics of both the INSLNs and IGFs, including dibasic processing sites at the carboxy- and amino-termini of B and A domains, that may allow removal of the C domain, as in vertebrate insulin and relaxin. However, the A domain extends to D and E domains, as in the IGFs. In contrast, Sp-IGF2, is more IGF-like as predicted cleavage sites suggest that the C-peptide is not processed for removal. Gene predictions also exist for appropriate processing enzymes, receptors, and binding proteins for the Sp-IGFs (Lapraz et al 2006).
Both amphioxus and Ciona intestinalis have genes encoding proteins similar to Sp-IGF1 and Sp-IGF2. Amphi ILP has been variously interpreted as being derived from a common ancestor of both INSLN/IGF or derived from an ancestral IGF (Chan et al. 1990). However, like Sp-IGF1, Amphi ILP has potential processing sites at the borders of B-C and C-A domains so the relationship is not straightforward. In Ciona Ci INS-L2, shares with Sp-IGF1 processing sites for removal of the C domain while Ci-INS-L1 and 3 are likely processed in a more IGF-like fashion and thus resemble Sp-IGF2 (Olinski et al. 2006) (Table S-5). The S. purpuratus INSLN and IGFs appear more similar to chordate peptides than to invertebrate INSLN and IGF.
Previous work suggests that Sp-IGFs may be expressed in the sea urchin larval gut (dePablo et al. 1988) and in adult starfish gut (Wilson and Falkmer, 1965). Microarray data confirms expression in the sea urchin embryo (Samanta et al. 2006).
Neurotrophins and Trk receptors
Neurotrophins regulate survival, growth and plasticity of neurons. Neurotrophins have two transmembrane-receptor systems: the Trk receptor tyrosine kinases and the common p75NTR. In tetrapods, neurotrophins and Trks include nerve growth factor (NGF) for which TrkA is a receptor, brain derived neurotrophic factor (BDNF) and neurotrophin-4/5 (NT-4/5) for which TrkB is a receptor, and neurotrophin-3 (NT-3) for which TrkC is a receptor. Phylogenetic analysis suggests that the four neurotrophins and three Trks in extant vertebrates are each derived from an ancestral neurotrophin and Trk gene that was duplicated in concert with two whole genome duplications, on the lineage leading to the gnathostomes (Hallböök, 1999).
The neurotrophin/Trk signaling system has been proposed to be a vertebrate innovation (Satou et al. 2003) because no genes encoding neurotrophophins, Trks or p75 receptors have been found in the Ciona intestinalis genome (Dehal et al. 2002). However, the recent discovery of a Trk receptor in amphioxus established that Trk receptors originated prior to the cephalochordate/vertebrate split (Benito-Gutierrez et al. 2005). Furthermore, there appears to be a molluscan Trk-like receptor (LTrk) (van Kesteren et al. 1998), raising the possibility that this system is much older, present in the common ancestor to deuterostomes and protostomes. Analysis of the S. purpuratus genome provides further support for this idea because neurotrophin-, Trk- and p75NTR genes are bona fide orthologues of vertebrate genes, as shown by phylogenetic analysis (Fig. 8). Sp-NT protein is encoded within 1 exon, as are its vertebrate counterparts, and contains all the cysteines necessary to form a characteristic neurotrophin cysteine knot. Sp-Trk has a potential signal peptide and a canonical vertebrate domain structure (Fig. 9). The extracellular region of Sp-Trk shares with vertebrate Trk receptors two immunoglobulin (Ig)-like domains following a leucine-rich repeat. The most carboxy-terminal Ig domain contains conserved asparagine residues with structural roles for ligand-receptor interactions. On the cytoplasmic side, Sp-Trk includes a tyrosine kinase (TK) domain belonging to class II tyrosine kinase receptors (Prosite pattern PS00239 [DVYstdYYR]). Sp-NT does not cluster with any 1 vertebrate neurotrophin, implying a pro-orthologous ancestor (Fig. 8).
Figure 8.

Neighbor joining tree of ClustalW aligned mature neurotrophin protein sequences, numbers indicate bootstrap values (1000 replicates).
Figure 9.
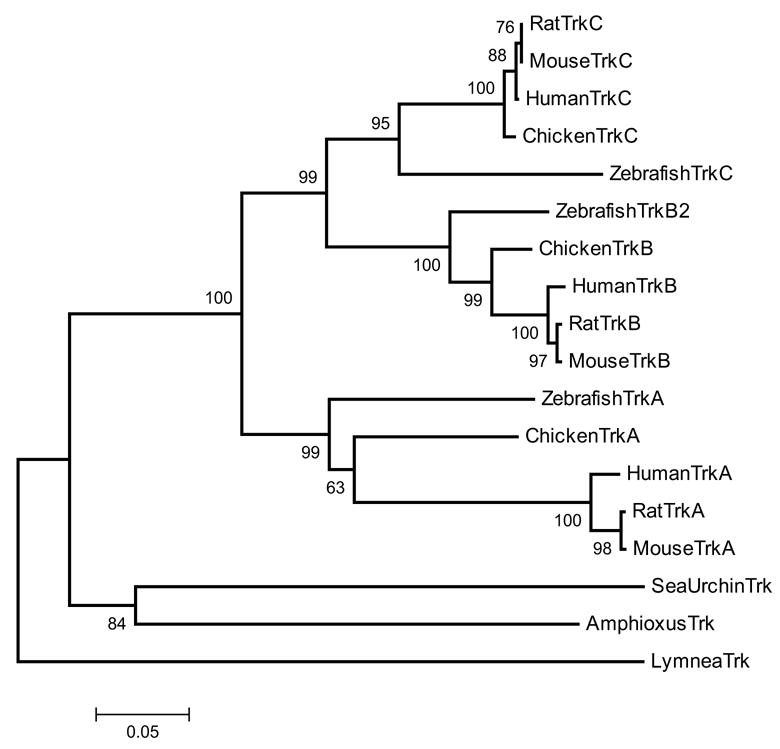
Neighbor joining tree of the Trk receptor protein sequences with 500 bootstrap replicates.
The neurotrophin/Trk system in S. purpuratus suggests that neurotrophin/Trk is a shared deuterostome feature. An important question is whether the urchin neurotrophin system is involved in neurotrophic regulation in a similar manner to the vertebrates.
Ependymins
Ependymin is a secreted glycoprotein implicated in a variety of cellular actions that are hallmarks of the complex neurotrophic factor family of proteins (Shashoua, 1991). Evidence links ependymin to consolidation of long term memory, neuronal regeneration, learning, and neuronal growth (Shashoua, 1985; Piront and Schmidt, 1988; Schmidt and Shashoua, 1988; Shashoua and Hesse, 1989; Shashoua, 1991; Schmidt et al. 1992). Ependymins have been postulated to play a role in establishing specific cell contacts (Hoffmann and Schwarz, 1996; Nimmrich et al. 2001). Recently, short peptides from goldfish ependymins were shown to function like neurotrophins (NGF and BDNF) during neuronal survival, proliferation and axon guidance (Gaiddon et al. 1996; Marsh and Palfrey, 1996; Adams et al. 2003; Shashoua et al. 2001). Ependymins, like neurotrophins, were proposed to be vertebrate-specific molecules. Recently, an ependymin-related gene was found in two holothurian species and in the sea urchin Lytechinus variegatus (Suarez-Castillo et al. 2004).
Predicted S. purpuratus ependymin genes encode peptides that contain four conserved cysteines crucial for dimeration, the strictly conserved residues D59, L107, P117, G129 and W140, and have sequence similarity to vertebrate ependymin-related proteins (Fig. 10). Sp-Epen is expressed during early larval development (Poustka et al. 2003) and transcripts have been localized to the intestine, esophagus, and gonads of Lytechinus variegates and in regenerating intestine of sea cucumbers (Suarez-Castillo et al. 2004). The expression of ependymin in these tissues may be neural since in fish, it is mainly expressed in the brain.
Figure 10.
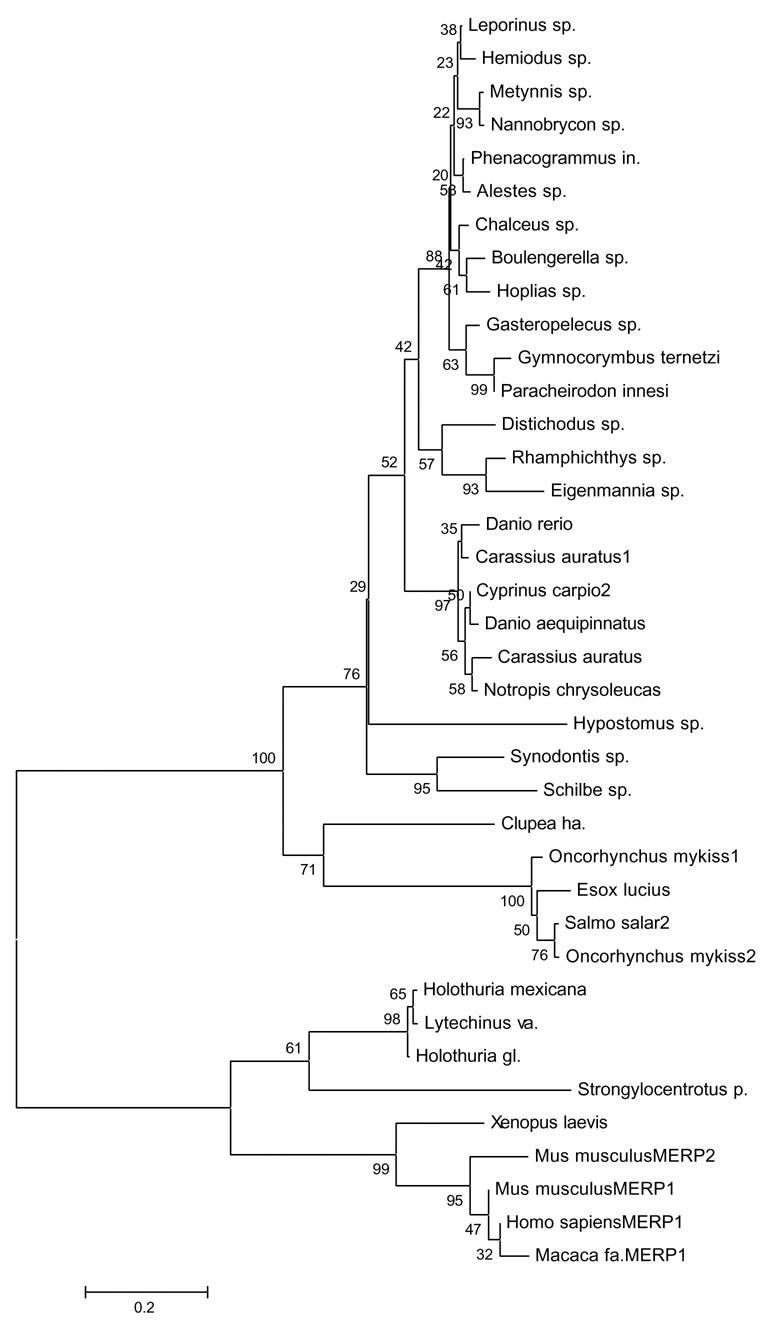
Neighbor joining tree of ClustalW aligned vertebrate and echinoderm ependymin-related protein sequence, numbers indicate bootstrap values (1000 replicates).
Other endocrine systems
Classic endocrine pathways in vertebrates and invertebrates include the steroid and thyroid nuclear receptors and their ligands, such as estrogens, progestogens, androgens, thyroxine and tri-iodothyronine in chordates and ecdysteroids in protostomes. Nuclear hormone receptors are transcriptional regulators involved in development, differentiation and endocrine hormone activity (Mangelsdorf et al. 1995). Studies of steroid hormones in echinoderms have focused on their roles in the gonads (Janer et al. 2005, Wasson and Watts 2000), and recently on environmental aspects (Roepke et al. 2005). Only 1 member of the steroid/thyroid hormone receptor family has previously been characterized in S. purpuratus (Chan et al. 1992), but analysis of the urchin genome has now revealed genes for estrogen, androgen, glucocorticoid, and retinoic acid receptors. A more complete analysis of nuclear receptors and transcription factors may be found in Howard-Ashby et al. (2006).
The chordate thyroid gland is a compact mass of endocrine cells derived from the pharynx. At least some thyroid functions are conserved throughout deuterostomes. In fishes, endocrine cells are scattered ventrally around the aorta, and in tunicates, ancestral thyroidal tissue is a subset of endostyle cells. A thyroidal system is present in tunicates, although thyroglobulin itself has not been identified (Campbell et al. 2004). In echinoderms thyroxine plays a role in metamorphosis (Heyland and Hodin 2004, Heyland et al. 2004). The sea urchin genome contains predictions for several thyroidal proteins including a sodium iodide transporter, a thyroid peroxidase for activating and coupling iodide to thyroglobulin, thyroglobulin and thyroid hormone nuclear receptors. Expression of the putative urchin thyroidal system remains to be determined. The key role of thyroid hormones in chordate development and metamorphosis raises the intriguing possibility that these regulators may play a similar role in echinoderms.
Sensory biology and behavior
Adult sea urchins detect and integrate a range of external stimuli using several sensory systems. They can discriminate among different foods; their spines respond to touch; they are probably thermotactic; and some are negatively geotactic (Lindahl and Runnström, 1929). Spines, tube feet, and pedicellariae are extensively innervated, presumably for both motor and sensory purposes. The test and spines have a ciliated epithelium including interspersed cells with sensory hairs suggested to have a mechanosensory role (Hyman, 1955). Sphaeridia on the surface of their tests are thought to function as balance organs (Hyman, 1955; Fell and Pawson, 1966).
Echinoderm larvae also have a range of behaviors dependent upon sensory systems. Plutei use cilia to sweep currents of water over their bodies and redirect food particles toward the mouth where it is swallowed using coordinated muscular activity (Strathmann, 1975). Plutei quickly change their direction of swimming and move their arms in an evasive response to chemicals or touch. Plutei can maintain a fixed orientation with their arms up and exhibit phototactic and geotactic responses, including an ability to influence their position in the water column (Yoshida, 1966). Echinoderm larvae competent to metamorphose often select a substrate based on chemical cues prior to settling and initiating metamorphosis (Cameron and Hinegardner, 1974, Burke, 1983; Bishop and Brandhorst, 2001).
Chemosensory receptors
Although the behavior of urchin larvae and adults indicates that they respond to chemicals, olfactory organs have not been identified with any certainty. Chemoreceptors are likely included in the large family of GCPRs. Because of rapid divergence, genes encoding chemoreceptors are difficult to identify (Clyne et al. 1999). We used two approaches to identify putative chemoreceptors genes. In the first, termed the 7TM/PFAM/C.elegans search, we identified GLEAN3 predictions that contain the 7tm_1 Pfam domain (rhodopsin-like domain). Genes with significant similarity (e-value <= 1e−20) to any non-chemoreceptor in C. elegans were excluded. This yielded 678 putative chemoreceptor genes (Table S-6). Many of these genes form tandem clusters, a property shared by chemoreceptor genes in all sequenced eukaryotic genomes. For example, 12 genes in a large tandem gene cluster consist of two sub-clusters, with a gap of 120 kb between them. Genes within each sub-cluster share considerable similarity, suggesting repetitive tandem duplication has occurred (Fig. 11). The majority of these predicted genes lack introns, a property shared by essentially all chemoreceptor genes in vertebrates, but not those in the protostomes, C. elegans or Drosophila. Nevertheless, some of these putative echinoid chemoreceptors are similar to known chemoreceptors in protostomes, such as C. elegans. Raible et al. (2006) have also performed a 7TM screen and identified 979 putative chemoreceptors after selecting full, non-overlapping domains at an e-value cutoff of 0.001. The sets of 678 and 979 putative chemoreceptors genes each contain many closely related genes that are arranged in clusters and many of the genes lack introns. Raible et al. (2006) have gone on to demonstrate that some of these genes are expressed in larvae and adult tissues including tube feet.
Figure 11.
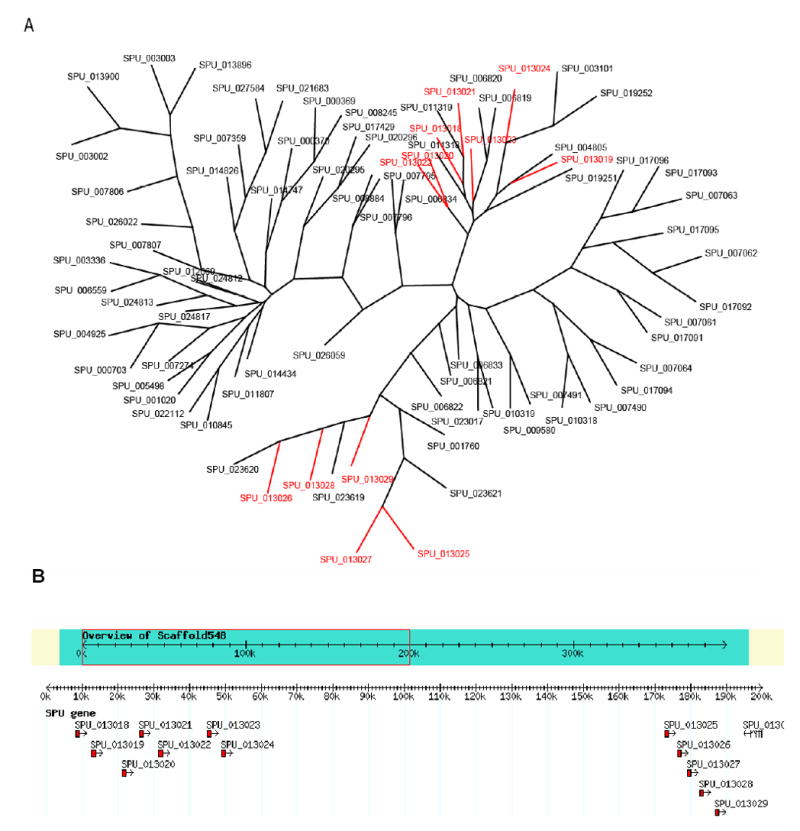
Putative sea urchin chemosensory genes form clusters in genome. A. Genes that show significant similarity (e-value <= e-100) to the putative chemosensory gene G13018 were pooled and aligned with ClustalW (Thompson et al 1994). Alignment in the PHYLIP format was then analyzed in sequence with seqboot, protdist, neighbor, consense (PHYLIP, Felsenstein, 1988) with 100 bootstrap iterations. The resulting tree was then viewed (treeview) and edited (Adobe Illustrator). Genes coded in red are physically clustered on a single scaffold. B. An example of the gene clustering found within putative chemosensory genes. Individual gene predictions that appear to be paralogous are found on the same scaffold. Genes in the cluster on the left are shown in the upper right of the tree in A, while those clustered on the right are shown on the lower left.
The second approach combined two strategies – an odorant-specific motif-based search and searches with three hidden Markov models (profile HMMs). The motif, LxxxxxxRxxAlxxPL, occurs in most zebrafish and mouse odorant receptors (ORs) (Alioto and Ngai, 2005) and was found in 43 GLEAN3 models. Blastp searches identified eleven additional closely related (≥70% identity) protein sequences that lack the motif. Five genes were subsequently removed from the set because they appear to have a non-homologous LxxxxxxRxxAIxxPL sequence or were ≥70% identical to a sequence with a non-homologous motif. However, not all mouse or zebrafish ORs possess the motif described above. To expand the list of candidate sea urchin chemoreceptors, we constructed profile hidden Markov models using HMMER (http://hmmer.wustl.edu) to query GLEAN3 peptides. The three profile HMMs used were based upon alignments of 12 sea lamprey odorant receptors (Berghard and Dryer, 1998), 16 fish T1R taste receptors (Ishimaru et al., 2005), and 239 fish odorant receptors (Alioto and Ngai, 2005). The lamprey odorant profile identified 91 sea urchin genes, the fish T1R profile identified 15 sea urchin genes and the fish odorant receptor profile identified 64 sea urchin genes (e-values ≤ 10−10) (Table S-7). Together the motif- and HMM-based searches identified 192 genes encoding putative sea urchin chemoreceptors. Of these, 74 can also be found in the set of 678 generated in the 7TM/PFAM/C.elegans search. The several hundred putative urchin chemoreceptors predicted by our approaches and by Raible et al (2006) are similar to the numbers of chemoreceptors identified in many other animals, suggesting an elaborate chemosensory system operates in sea urchins.
Precursors of sensory placodes
The visual, olfactory, acoustic, and gustatory systems of the peripheral nervous system of vertebrates develop from cells of the neural crest and several pairs of neurogenic cranial placodes that initiate as patches of thickened ectoderm (reviewed by Baker and Bronner-Fraser, 2001). Gans and Northcutt (1983) proposed that the neural crest and cranial placodes are distinctive innovations of vertebrate embryos that allowed for the extensive remodeling of the head during early vertebrate evolution. They also speculated that placodes evolved from a diffuse ectodermal nerve plexus similar to that in echinoderms and hemichordates (Lowe et al., 2003). However, whether the chordate ancestor had placodes or neural crest is controversial. On the one hand, marker gene expression and anatomical features suggest that embryos of the ascidian Ciona intestinalis (Mazet et al. 2005; Mazet and Shimeld, 2005) and the larvacean Oikopleura dioica (Bassham and Postlethwait, 2005) possess placode-like ectodermal specializations that form sensory organs. Furthermore, migratory neural crest-like cells have been detected by Jeffery et al. (2004) in ascidian embryos. However, Holland and Holland (2001) argue that the sensory organ primordia derived from ectoderm in amphioxus are not placodes, but may have evolved into placodes and the neural crest. Schlosser (2005) agrees that sensory primordia in urochordates and cephalochordates are not the integrated placodes characteristic of vertebrates. These considerations raise the interesting question of whether basal deuterostomes, like echinoderms, have primordial patches of ectodermal cells resembling placodes that give rise to sensory organs.
To address this question, we used RT-PCR to assess the expression of members of gene families that are associated with placode development in vertebrates (Table 3). We compared expression levels of genes in sea urchin gastrula-stage embryos, early larvae, late larvae containing rudiments, adult radial nerves and adult tube feet. All the genes were expressed, often differentially. Transcript levels for Sp-Eya1, Sp-Phox2, Sp-Pax1-9, Sp-EBF3, and Sp-Pitx3 were relatively high in adult radial nerves compared to tube feet or larvae. Sp-PaxB was expressed at very high levels in all samples tested. The high level of expression in gastrulae presumably reflects involvement of PaxB with late histone gene regulation (Czerny et al. 1997). These observations indicate that genes collectively associated with placode formation are expressed in urchins when sensory organ primordia may be forming. It will be informative to determine the spatial patterns of expression of these genes in larvae and adults, particularly in the tube feet and pedicellariae that are derived from placode-like structures.
Table 2.
Gene predictions for G-protein coupled peptide receptors and peptide precursors
| Rhodopsin-type G-protein coupled peptide receptors | |
|---|---|
| Vasopressin / Oxytocin / Gonadotropin-releasing hormone-like receptors | SPU_021291, SPU_028207, SPU_001850, SPU_026012 |
| Vasopressin / Oxytocin receptors | SPU_021290 |
| Endothelin / Bombesin-like / Gastrin- Releasing Peptide / Neuromedin-B receptors | SPU_009901, SPU_027116, ,SPU_010606, SPU_011360, SPU_021160, SPU_008003 |
| Neuromedin U / Neurotensin / Growth hormone secretagogue / Ghrelin receptors | SPU_004603, SPU_015957, SPU_016898 |
| Cholecystokinin / Gastrin receptors | SPU_026458 |
| Orexin receptors | SPU_023223, SPU_012799 |
| Thyrotropin-releasing hormone receptor | SPU_010167 |
| Tachykinin receptors | SPU_015140, SPU_015139 |
| Galanin receptors | SPU_024231, SPU_022317 |
| RFamide receptor (GPR54) | SPU_021219, SPU_007422, SPU_012318, SPU_022376, SPU_022468 |
| RFamide receptor (GPR103) | SPU_006927, SPU_024924, |
| Prolactin-releasing hormone receptor | SPU_020376 |
| Leucine-rich-repeat-containing receptors 7 & 8 | SPU_012382, SPU_005162, SPU_012383 |
| Gonadotropin receptors | SPU_027375, SPU_000995, SPU_004389 |
| Neuropeptide and peptide hormone precursors | |
| Echinotocin | SPU_006899 |
| SALMFamides | SPU_021565 |
| NGFFFamide | SPU_030074 |
| Glycoprotein hormone β subunit | SPU_004405, SPU_005842, SPU_011451 |
| Bursicon-α | SPU_003984 |
| Bursicon-β | SPU_017707 |
Photoreception
It is clear from their behavior that echinoderms are photosensitive. Although most echinoderms lack obvious light-sensitive organs, they nevertheless respond to light, photoperiod and lunar cycles (Boolootian, 1966; Yoshida, 1966). S. purpuratus adults are negatively phototactic (Giese and Farmanfarmaian, 1963) and photoperiod influences their reproductive cycle (Boolootian, 1963). Most urchins will quickly initiate a righting reaction to keep their oral surface in contact with the substratum and this behavior can be influenced by light (Reese, 1966; Yoshida, 1966). Many urchin species will select opaque rather than transparent objects to cover themselves in response to lighting from above (Millott, 1966; Yoshida, 1966).
Although sea urchins lack conventional eyes, they probably evolved from organisms that collected light through an aperture and focused it with a lens onto photoreceptor cells specialized to convert photons into neural signals (Fernald, 2000). All echinoderms display dermal photosensitivity based on diffuse dermal light receptors (Yoshida, 1966). Both surfaces of the calcitic endoskeleton (the test) and the pigmented radial nerves are photosensitive as are tube feet, which exhibit phototropic behavior and reflexive responses to light (Yoshida, 1966).
The translucent echinoderm test lined with photosensitive tissue provides an opportunity for lenses and filters to evolve. Single calcite crystals in brittle star skeletal ossicles can serve as microlenses in specialized photosensory organs that may collectively function as a compound eye (Aizenberg et al., 2001). A diffuse photoreceptor system with directional sensitivity may perceive images, albeit with limited resolution. Woodley (1982) suggested that the opaque spines of urchins might restrict the direction and angle of light reaching the photosensitive test, with the diffuse photoreception system of the test becoming a large compound eye. Consistent with this prediction, Blevins and Johnsen (2004) reported that urchins could locate and move toward dark targets and locate hiding places, apparently using their spatial vision.
Molecular evidence for photosensory organs in echinoderms is sparse, but the case is strongest for tube feet. There is a terminal nerve ring and a ganglion in each tube foot. As well, tube feet also connect directly with the radial nerves underneath the test. The buccal tube feet surrounding the mouth have modified suckers and are thought to be sensory appendages (Fig. 4; Hyman, 1955). The most intriguing observation is that cells adjacent to the longitudinal muscle fibers express a homologue of Pax6 (Czerny and Busslinger, 1995), a critical transcriptional regulator of vertebrate and invertebrate eye development (Gehring, 2005).
Retinal gene homologues
To investigate the cellular and genetic basis of echinoderm photosensitivity, we searched the S. purpuratus genome for genes homologous to known mammalian retinal genes because the retina is the neural tissue responsible for phototransduction and for making connections with other neurons required for visual responses. The first approach was based on 228 Math5-dependent mouse genes identified by microarray analysis, which have roles in specifying retinal cell fates (Mu et al., 2005). Math5 is a mouse homologue of Drosophila atonal, a gene encoding a proneural bHLH transcription factor required for development of sensory organs and retinal ganglion cells (Wang et al. 2001; Brown et al. 2001). We used 228 protein sequences to BLAST search the GLEAN3 gene predictions at a threshold of e−20, and found 117 putative homologues. In a second approach we searched for candidate genes encoding transcription factors known to play important roles in mammalian eye and retinal development (Hatakeyama and Kageyama, 2004; Mu and Klein, 2004).
These searches identified 20 genes encoding transcription factors (Table 4). Some of these are involved in early retinal patterning and specification events include Pax6 (Gehring, 2005), three members of the Six gene family (Zhu et al., 2002) (Donner and Maas, 2004), and Rx, a homeodomain gene essential for vertebrate eye development (Bailey et al., 2004). Genes involved in the later events of cell type specification and differentiation include three members of the atonal family, NeuroD, POU4f and POU6f homologues, several homeodomain-containing genes, and genes for zinc finger transcription factors. Sequence alignments suggest that there are single genes equally similar to mouse POU4F1 or POU4f2, NeuroD1 or NeuroD4, and Six3 or Six6 (Fig. S-3).
Table 3.
Expression of Placode Gene Markers. Expression for each gene is based on a qRT-PCR assay and is shown as the percentage of the level of expression of Sp-ubiquitin in that sample, based on average of two determinations for each sample.
| Predicted gene | Gene Name or Synonym | Domain Structure or Orthology | Gastrula | Early Larva | Late Larva | Adult Tube Foot | Adult Radial Nerve |
|---|---|---|---|---|---|---|---|
| SPU_002815 | Sp-Dlx | homeobox TF | 0.04 | 0.02 | 0.01 | ND | ND |
| SPU_013869 | Sp-Eya1 | tyrosine phosphatase | 2.10 | 0.65 | 0.09 | 0.55 | 17.19 |
| SPU_013464 | Sp-Phox2 | fork head TF | 0.02 | 0.17 | 0.23 | 0.86 | 13.92 |
| SPU_004702 | Sp-EBF3 | COE family TF | 2.12 | 1.48 | 0.89 | 0.24 | 8.11 |
| SPU_014461 (contains SPU_024163) | Sp-Pitx1 (primer set1)_ | homeobox TF | 0.03 | 0.16 | 0.78 | 0.02 | 0.04 |
| SPU_014461 (contains SPU_024163) | Sp-Pitx1 (primer set2) | homeobox TF | 0.52 | 0.25 | 1.73 | 0.08 | 0.02 |
| SPU_004599 | Sp-Pitx2 | homeobox TF | 0.14 | 0.34 | 0.42 | 0.01 | 0.17 |
| SPU_006159 (contains SPU_004598) | Sp-Pitx3 | homeobox TF | 0.01 | 0.01 | 0.91 | 0.01 | 1.90 |
| SPU_023894 | Sp-FoxI | fork head TF | 0.68 | 0.39 | 0.89 | 0.00 | 0.02 |
| SPU_027334 | Sp-PaxA | paired box TF | 0.02 | 0.15 | 0.10 | 0.07 | 0.04 |
| SPU_018351 | Sp-PaxB | paired box TF | 138.99 | 18.95 | 6.34 | 4.80 | 5.89 |
| SPU_006683 | Sp-Pax1-9 | paired box TF | 2.38 | 3.54 | 2.46 | 1.62 | 25.35 |
| SPU_014539 | Sp-Pax2-5-8 | paired box TF | 7.83 | 0.70 | 0.55 | 0.16 | 0.06 |
In addition, we searched for genes encoding photo opsins because they are critical components in phototransduction. Four opsin homologues were identified with many signature features indicative of a prototypical photopigment (Table 4; Fig. S-4). These include a highly conserved lysine (K398) that binds 11-cis-retinal (Wang et al., 1980), which absorbs photons causing a protein conformational change essential for phototransduction (Hargrave et al., 1984), a conserved aromatic amino acid (Y170) in the third trans-membrane domain that stabilizes the 11-cis-retinal attachment and two critical cysteines (C167 and C245), essential for correct tertiary structure (Karnik et al., 1988). Three of the four S. purpuratus homologues contain a conserved arginine-tyrosine (R192-Y193) required for opsin-mediated G-protein activation, although only one has the preceding conserved acidic amino acid. Raible et al. (2006) independently report finding two additional opsin homologues in the S. purpuratus genome, including rhabdomeric and ciliary opsins. Genes encoding a serine/threonine phosphatase (phosphatase 2A) and a tyrosine phosphatase (phosphatase 4) associated with phototransduction events downstream of opsin were also identified as Math5-dependent homologues in the urchin genome (Selke et al., 1998; Ensslen and Brady-Kalnay, 2004), as well as genes associated with neural transmission (a voltage-gated potassium channel) and with neuronal microtubules (Tubb2/Tubb3, Tubb4) (Table 4).
The fact that the sea urchin genome contains many genes encoding homologues of retinal transcription factors, opsins, and phototransduction proteins provides strong evidence for a phototransduction system. Because tube feet are excellent candidates to have such systems, we tested whether other retinal gene homologues were expressed in this tissue using real time RT-PCR (Table3, S-8 for primers). In addition to Pax6, Sp-NeuroD, Sp-Blimp1, Sp-Six3, Sp-BarH1, Sp-Rx and Sp-Ato are all expressed in tube feet at levels ranging from 0.0015% to 2% of levels for Sp-ubiquitin transcripts (Table 4). Detectable PCR products were not obtained for Sp-Ato2b, Sp-Ato7, and an Sp-opsin (SPU_027634). However, there are 3 Sp-opsins that are expressed in tube feet. Raible et al. (2006) also report that opsin genes are expressed in this tissue as well as in growing larvae. Although we have not shown that expression is specific to tube foot neurons, or that the genes are expressed together in the same cells, the results support the hypothesis that tube feet contain cells that express many of the genes expressed in mammalian retinas.
We also tested whether the set of retinal genes expressed in adult tube feet are also expressed during embryogenesis (Table 4). Two opsins, Sp-Opsins and Sp-BarH1 are expressed only in tube feet, whereas 1 Sp-opsin, SpPou4f1, and Sp-NeuroD are expressed in 72 h embryos and tube feet but not in 24 h embryos. Furthermore, the relative expression levels were quite different among the three samples. For example, Sp-Blimp1 and Sp-Six3 are expressed at relatively high levels in embryos while the same genes are expressed at lower relative levels in tube feet. Raible et al (2006) report that opsin genes are expressed in growing larvae and adult tube feet. These results suggest that the retinal gene set may be used for distinct purposes in different cells.
Circadian genes
Diurnal rhythms are widespread in metazoans, and usually linked to daily and seasonal cycles that are a feature of feeding, reproduction, migration (Merrow et al. 2005). Circadian rhythms are not well known in echinoids although photoperiodic activity has been recorded for both feeding and reproductive behaviors (Rosenberg and Lundberg 2004, Coppard and Campbell 2005) and temporal variability in larval settlement is also noted (Hereu et al. 2004).
Studies in flies and fish show that the gene regulatory network that controls circadian rhythms includes transcription factors and inhibitory and enhancing pathways. The central clock genes of this network include Period (Per), the first clock gene identified (Konopka and Benzer, 1971), Cryptochrome (Cry), Clock (CLK), Timeless (Tim) and Brain and muscle Arnt-like protein (BMAL) (Glaser and Stanewsky, 2005; Hirayama and Sassone-Corsi, 2005). Per proteins appear to be a central component of the circadian clock across the animal kingdom and together with Cry and Tim proteins are fundamental to circadian behavior pathways.
Analysis of the urchin genome using reciprocal BLAST searches has revealed the presence of at least 4 genes including Sp-Clock, Sp-CRY, Sp-Timeless1 and Sp-Timeless2 that may belong to a Clock gene network (Table S-9). There is also evidence for urchin genes encoding other transcription factors that may be part of a Clock network, such as Sp-Arnts and Sp-Arnt-like genes (Table S-9). At present, we know little about the temporal or spatial expression patterns of these key genes although the tiling data (Samanta et al., 2006) suggests that except for one of the Arnt-like genes, all are expressed in embryos. It is clear that one of the most exciting challenges of sea urchin genomics will be an analysis of this putative network.
Conclusions
Our analysis of the genome has provided a rich source of information that has opened several completely new lines of investigation into the neural development and neurobiology of sea urchins. It appears that sea urchins share with other metazoans the gene regulatory programs that specify embryonic neurons. The experimental dissection of the sea urchin neurogenic network is now possible. How this network is deployed will prove particularly important in resolving how the distinctive pentaradial, yet segmented, echinoid nervous system relates to that of other deuterostomes and how it was derived from the deuterostome ancestor. The unusual synaptic connections between neurons and between neurons and effectors in echinoids can now be approached with molecular and cellular studies. In addition, investigations of the mechanisms underlying embryonic axon guidance and neural adhesion appear tractable. The array of neuromodulators, neuropeptides and growth factors that have been identified in the genome suggests that the echinoid nervous system fully utilizes these modes of communication and regulation. Investigation of the physiological roles of these molecules will provide fascinating insights into the evolution of chemical signaling systems of animals. The expression of a set of retinal genes in tube feet, which are non-ocular structures, provides a novel framework for understanding how organisms perceive light. Similarly, the characterization of chemoreceptors will provide novel tools for an analysis of sea urchin sensory modalities. Analysis of the sea urchin genome has facilitated a revitalization of sea urchin neurobiology that will improve on its status of being among the least studied metazoan nervous systems.
The list of neural gene predictions provides a resource for developing hypotheses of gene function in the nervous system. Most of the gene predictions will need to be verified with full length cDNAs and expression studies and experimental perturbations will be necessary to determine if gene functions are conserved. The separate larval and adult phases of sea urchin life history provide two distinct body plans with what appear to be separate nervous systems. A more complete analysis of the development and organization of echinoderm nervous systems is central to the problem of fitting these two body plans into the evolutionary history of deuterostomes as well as helping to resolve longstanding questions of deuterostome ancestry.
Supplementary Material
Table 4.
S. purpuratus homologues of mammalian retinal genes. Expression values are from qRT-PCR using the procedures of Revilla-i-Domingo et al. (2004). RNA from 500 embryos (24h, 72h) or from 5 tube feet was used as template. ND, not detected
| Predicted gene | Mouse Gene Name (S. purpuratus Gene name) | Gene Function (Domain Class) | Expressionb 24h/72h/tube foot 104 |
|---|---|---|---|
| SPU_000990 | Atonal (Sp-Ato2b) | bHLH transcription factor | ND/ND/ND |
| SPU_003681 | Atonal7 | bHLH transcription factor | ND/ND/ND |
| SPU_011351/ | Atonal | bHLH transcription factor | 3.2/16.8/2.3 |
| SPU_014289 | Rx | Homeobox transcription | 2.5/7.4/0.43 |
| SPU_010438 | Pou6f2 | POU domain transcription | |
| SPU_025632 | Pou4f1/Pou4f2 | POU domain transcription | ND/1.9/3.1 |
| SPU_004702 | Ebf1/Ebf3/Olf-1 | Zinc finger transcription factor | |
| SPU_000237 | Islet1 | LIM homeobox transcription | 0.95/4.6/3.1 |
| SPU_017627 | Gli1 | Zinc finger transcription factor | |
| SPU_010351 | Irx2/Irx5 | Homeobox transcription | |
| SPU_024918 | NeuroD1/NeuroD4 | bHLH transcription factor | ND/1.8/5.0 |
| SPU_017348 | Atbf1 | Zinc finger homeobox | |
| SPU_027235 | Blimp1 | Zinc finger transcription factor | 46.6/25.0/3.5 |
| SPU_006786+ SPU_006787 | Pax6 | Paired class homeobox transcription factor | 1.2/6.3/20.8 |
| SPU_017379 | Six1/Six2 | Homeobox transcription | |
| SPU_018908 | Six3/Six6 | Homeobox transcription | 35.8/17.6/1.6 |
| SPU_017380 | Six4/Six5 | Homeobox transcription | |
| SPU_007599 | GLASS 2 | Zinc finger transcription factor | |
| SPU_012645 | GfI1 | Zinc finger transcription factor | |
| SPU_014164 | BarH1 | Homeobox transcription | ND/ND/8.3 |
| SPU_005569 | Rhodopsin (Sp-opsin1) | Photo pigment | ND/4.0/0.60 |
| SPU_006737 | Rhodopsin (Sp-opsin5 | Photo pigment | ND/ND/0.15 |
| SPU_027634 | Rhodopsin (Sp-opsin3.1) | Photo pigment | ND/ND/ND |
| SPU_022851 | Melanopsin (Sp-opsin4) | Photo pigment | ND/ND/5.6 |
| SPU_000178 | Phosphatase 2a | Photo transduction | |
| SPU_008502 | Phosphatase 4 (receptor | Photo transduction | |
| SPU_019810 | Notch | Notch signaling | |
| SPU_000092 | K voltage-gated channel | Nerve transmission | |
| SPU_000062 | Tubb2/Tubb3/Tubb4 | Tubulin cytoskeleton | |
| SPU_009708 | Trim2 | Not assigned |
Acknowledgments
RDB receives support from NSERC and CIHR; LMA is supported by the intramural program of the National Institute for Dental and Craniofacial Research, NIH; KW, FH, RO, MCT receive support from Swedish Science Research Council, Network of Excellence Marine Genomics Europe (GOCE-04-505403); EU RTN FP5 Trophic Neurogenome HPRN-ct-2002-00263, and the Royal Swedish Academy of Sciences; MRE was supported by BBSRC (UK) grant S19916; WHK receives support from National Institute of Child Health and Human Development (HD22619), The National Eye Institute (EY11930), and the Robert A. Welch Foundation; BPB, NC and JST receive support from NSERC. GWH is supported by the intramural program of the National Institute of Child Health and Human Development, NIH
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams D, Hasson B, Boyer-Boiteau A, El-Khishin A, Shashoua V. A peptide fragment of ependymin neurotrophic factor uses protein kinase C and the mitogen-activated protein kinase pathway to activate c-Jun N-terminal kinase and a functional AP-1 containing c-Jun and c-Fos proteins in mouse NB2a cells. J Neurosci Res. 2003;72:405–416. doi: 10.1002/jnr.10590. [DOI] [PubMed] [Google Scholar]
- Adayev T, Ranasinghe B, Banerjee P. Transmembrane signaling in the brain by serotonin, a key regulator of physiology and emotion. Bioscience Reports. 2005;25:363–385. doi: 10.1007/s10540-005-2896-3. [DOI] [PubMed] [Google Scholar]
- Aizenberg J, Tkachenko A, Weiner S, Addadi L, Hendler G. Calcitic microlenses as part of the photoreceptor system in brittlestars. Nature. 2001;412:819–822. doi: 10.1038/35090573. [DOI] [PubMed] [Google Scholar]
- Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173–187. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac D, Chen X, Khant HA, Ubach J, Ludtke SJ, Kikkawa M, Johnson AE, Chiu W, Sudhof TC, Rizo J. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–17. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- Avery J, Hodel A, Whitaker M. In vitro exocytosis in sea urchin eggs requires a synaptobrevin-related protein. Journal of Cell Science. 1997;110:1555–1561. doi: 10.1242/jcs.110.14.1555. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Baker CVH, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Developmental Biology. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Bassham S, Postlethwait JH. The evolutionary history of placodes: a molecular genetic investigation of the larvacean urochordate Oikopleura dioica. Development. 2005;132:4259–4272. doi: 10.1242/dev.01973. [DOI] [PubMed] [Google Scholar]
- Beane WS, Voronina E, Wessel G, McClay D. GTP-binding proteins and their regulators in S. purpuratus: GTPases and teh prediction of reduced complexity in early deuterostomes. Dev Biol 2006 [Google Scholar]
- Beer AJ, Moss C, Thorndyke M. Development of serotonin-like and SALMFamide-like immunoreactivity in the nervous system of the sea urchin Psammechinus miliaris. Biological Bulletin. 2001;200:268–280. doi: 10.2307/1543509. [DOI] [PubMed] [Google Scholar]
- Benito-Gutierrez E, Nake C, Llovera M, Comella JX, Garcia-Fernandez J. The single AmphiTrk receptor highlights increased complexity of neurotrophin signaling in vertebrates and suggests an early role in developing sensory neuroepidermal cells. Development. 2005;132:2191–202. doi: 10.1242/dev.01803. [DOI] [PubMed] [Google Scholar]
- Berghard A, Dryer L. A novel family of ancient vertebrate odorant receptors. Journal of Neurobiology. 1998;37:383–392. [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nature Reviews Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Binyon J. Physiology of Echinoderms. Pergammon Press; Oxford: 1972. [Google Scholar]
- Bisgrove BW, Burke RD. Development of serotonergic neurons in embryos of the sea urchin Strongylocentrotus purpuratus. Development Growth & Differentiation. 1986;28:569–574. doi: 10.1111/j.1440-169X.1986.00569.x. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Burke RD. Development of the nervous system of the pluteus larva of Strongylocentrotus droebachiensis. Cell and Tissue Research. 1987;248:335–343. doi: 10.1007/BF00222422. [DOI] [PubMed] [Google Scholar]
- Bishop CD, Brandhorst BP. The role of NO/cGMP and HSP90 in regulating metamorphosis of the sea urchin Lytechinus pictus. Developmental Biology. 2001;235:251–251. [Google Scholar]
- Blevins E, Johnsen S. Spatial vision in the echinoid genus Echinometra. J Exp Biol. 2004;207:4249–4253. doi: 10.1242/jeb.01286. [DOI] [PubMed] [Google Scholar]
- Boolootian RA. Response of the testes of purpurple sea urchins to variations in temperature and light. Nature. 1963;197:403. [Google Scholar]
- Boolootian RA. Reproductive Physiology. In: Boolootian RA, editor. Physiology of Echinodermata. Interscience; New York: 1966. pp. 561–614. [Google Scholar]
- Boyl PP, Signore M, Annino A, Barbera JPM, Acampora D, Simeone A. Otx genes in the development and evolution of the vertebrate brain. International Journal of Developmental Neuroscience. 2001;19:353–363. doi: 10.1016/s0736-5748(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RD. The structure of the nervous system of the pluteus larva of Strongylocentrotus purpuratus. Cell and Tissue Research. 1978;191:233–247. doi: 10.1007/BF00222422. [DOI] [PubMed] [Google Scholar]
- Burke RD. Development of pedicellariae in the pluteus larva of Lytechinus pictus. Canadian Journal of Zoology. 1980;58:1674–1682. [Google Scholar]
- Burke RD. Neural Control of metamorphosis in Dendraster excentricus. Biological Bulletin. 1983;164:176–188. [Google Scholar]
- Burke RD, Osborne L, Wang D, Murabe N, Yaguchi S, Nakajima Y. Neuron-specific expression of a synaptotagmin gene in the sea urchin Strongylocentrotus purpuratus. J Comp Neurol. 2006;496:244–51. doi: 10.1002/cne.20939. [DOI] [PubMed] [Google Scholar]
- Bury H. The metamorphosis of echinoderms. Quarterly Journal of Microscopy. 1896;38:45–135. [Google Scholar]
- Cameron RA, Hinegardner RT. Initiation of Metamorphosis in Laboratory Cultured Sea-Urchins. Biological Bulletin. 1974;146:335–342. doi: 10.2307/1540409. [DOI] [PubMed] [Google Scholar]
- Campbell RK, Satoh N, Degnan BM. Piecing together evolution of the vertebrate endocrine system. Trends Genet. 2004;20:359–366. doi: 10.1016/j.tig.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Cavey MJ, Wood RL. Organization of the adluminal and retractor cells in the coelomic lining from the tube foot of a phanerozonain starfish, Luidia foliata. Canadian Journal of Zoology-Revue Canadienne de Zoologie. 1991;69:911–923. [Google Scholar]
- Cavey MJ, Markel K. Echinoidea. In: Harrison FW, Chia FS, editors. Microscopic Anatomy of Invertebrates. Vol. 14. Wiley-Liss; New York: 1994. pp. 345–400. [Google Scholar]
- Cazzamali G, Saxild NPE, Grimmelikhuijzen CJP. Molecular cloning and functional expression of a Drosophila corazonin receptor. Biochemical and Biophysical Research Communications. 2002;298:31–36. doi: 10.1016/s0006-291x(02)02398-7. [DOI] [PubMed] [Google Scholar]
- Chaet AB, McConnaughy RA. Physiologic activity of nerve extracts. Biological Bulletin. 1959;117:407. [Google Scholar]
- Chan SJ, Cao QP, Steiner DF. Evolution of the insulin superfamily: cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. 1990;87:9319–9323. doi: 10.1073/pnas.87.23.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SM, Naidu X, Niemeyer CC, Bone JR, Flytzanis CN. SpCOUP-TF: A sea urchin member of the steroid/thyroid hormone receptor family. Proc Natl Acad Sci USA. 1992;89:10568–10572. doi: 10.1073/pnas.89.22.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SJ, Steiner DF. Insulin through the ages: Phylogeny of a growth Promoting and Metabolic Regulatory Hormone. Am Zool. 2000;40:213–222. [Google Scholar]
- Chang MC, Berkery D, Schuel R, Laychock SG, Zimmerman AM, Zimmerman S, Schuel H. Evidence for a Cannabinoid Receptor in Sea-Urchin Sperm and Its Role in Blockade of the Acrosome Reaction. Molecular Reproduction and Development. 1993;36:507–516. doi: 10.1002/mrd.1080360416. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia FS, Burke RD. Echinoderm metamorphosis: Fate of larval structures. In: Chia FS, Rice ME, editors. Settlement and Metamorphosis of Marine Invertebrate Larvae. Elsevier-North Holland; New York: 1978. pp. 219–234. [Google Scholar]
- Clandinin TR. Surprising twists to exocyst function. Neuron. 2005;46:164–166. doi: 10.1016/j.neuron.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim JH, Carlson JR. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Cobb JLS, Laverack MS. Neuromuscular systems in echinoderms. Symp zool Soc Lond. 1967;20:25–51. [Google Scholar]
- Cobb JLS. The significance of the radial nerve cords in Asteroids and Echinoids. Z Zellforsch. 1970;108:457–474. doi: 10.1007/BF00339653. [DOI] [PubMed] [Google Scholar]
- Cobb JLS, Pentreath VW. The identification of chemcial synapses in echinoderm nervous tissue. Thalass Jugoslavica. 1976;12:81–85. [Google Scholar]
- Cobb JLS, Pentreath VW. Anatomical studies of simple invertebrate synapses using stage rotation electron microscopy and densitometry. Tissue and Cell. 1977;9:125–135. doi: 10.1016/0040-8166(77)90054-4. [DOI] [PubMed] [Google Scholar]
- Cobb JLS. Neurobiology of Echinodermata. In: Ali MA, editor. Nervous Systems of Invertebrates. Plenum; New York: 1987. pp. 483–525. [Google Scholar]
- Cobb JLS, Moore A. Studies on the integration of sensory information by the nervous system of the brittlestar. Ophiura ophiura Mar Behav Physiol. 1989;14:211–222. [Google Scholar]
- Conlon JM. Molecular Evolution of Insulin in Non-Mammalian Vertebrates. Am Zool. 2000;40:200–212. [Google Scholar]
- Conlon JM. Evolution of the insulin molecule: insights into structure-activity and phylogenetic relationships. Peptides. 2001;22:1183–1193. doi: 10.1016/s0196-9781(01)00423-5. [DOI] [PubMed] [Google Scholar]
- Conner S, Leaf D, Wessel G. Members of the SNARE hypothesis are associated with cortical granule exocytosis in the sea urchin egg. Mol Reprod Dev. 1997;48:106–18. doi: 10.1002/(SICI)1098-2795(199709)48:1<106::AID-MRD13>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Coppard SE, Campbell AC. Lunar periodicities of diadematid echinoids breeding in Fiji. Coral Reefs. 2005;24:324–332. [Google Scholar]
- Croce JC, Wu S-Y, Byrum C, Xu R, Duloquin L, Wikramanayake AH, Gache C, McClay D. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006a doi: 10.1016/j.ydbio.2006.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce J, Duloquin L, Lhomond G, McClay DR, Gache C. Frizzled5/8 is required in secondary mesenchyme cells to initiate archenteron invagination during sea urchin development. Development. 2006b;133:547–557. doi: 10.1242/dev.02218. [DOI] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-Binding and Transactivation Properties of Pax-6 - 3 Amino-Acids in the Paired Domain Are Responsible for the Different Sequence Recognition of Pax-6 and Bsap (Pax-5) Molecular and Cellular Biology. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Bouchard M, Kozmik Z, Busslinger M. The characterization of novel Pax genes of the sea urchin and Drosophila reveal an ancient evolutionary origin of the Pax2/5/8 subfamily. 1997;67:179–192. doi: 10.1016/s0925-4773(97)00119-6. [DOI] [PubMed] [Google Scholar]
- Davis RJ, Tavsanli BC, Dittrich C, Walldorf U, Mardon G. Drosophila retinal homeobox (drx) is not required for establishment of the visual system, but is required for brain and clypeus development. Developmental Biology. 2003;259:272–287. doi: 10.1016/s0012-1606(03)00201-x. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, de Tomaso A, Davidson B, di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- DePablo F, Chamber SA, Ota A. Insulin-related molecules and insulin effects in the sea urchin embryo. Dev Biol. 1988;130:304–310. doi: 10.1016/0012-1606(88)90436-8. [DOI] [PubMed] [Google Scholar]
- Di Bernardo M, Castagnetti S, Bellomonte D, Oliveri P, Melfi R, Palla F, Spinelli G. Spatially restricted expression of PlOtp, a Paracentrotus lividus Orthopedia-related homeobox gene, is correlated with oral ectodermal patterning and skeletal morphogenesis in late-cleavage sea urchin embryos. Development. 1999;126:2171–2179. doi: 10.1242/dev.126.10.2171. [DOI] [PubMed] [Google Scholar]
- Donner AL, Maas RL. Conservation and non-conservation of genetic pathways in eye specification. International Journal of Developmental Biology. 2004;48:743–753. doi: 10.1387/ijdb.041877ad. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Price DA, Lee TD, Thorndyke MC. The SALMFamides - a new family of neuropeptides isolated from an Echinoderm. Proceedings of the Royal Society of London Series B-Biological Sciences. 1991;243:121–127. doi: 10.1098/rspb.1991.0020. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Melarange R. Nitric oxide function in an echinoderm. Biological Bulletin. 1998;194:260–266. doi: 10.2307/1543096. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Egertova M. The neurobiology and evolution of cannabinoid signalling. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2001;356:381–408. doi: 10.1098/rstb.2000.0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR, Melarange R. Neural control of muscle relaxation in echinoderms. Journal of Experimental Biology. 2001;204:875–885. doi: 10.1242/jeb.204.5.875. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Satou Y, Satoh N. The invertebrate ancestry of endocannabinoid signalling: an orthologue of vertebrate cannabinoid receptors in the urochordate Ciona intestinalis. Gene. 2003;302:95–101. doi: 10.1016/s0378-1119(02)01094-6. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Thorndyke MC. Molecular characterisation of SALMFamide neuropeptides in sea urchins. Journal of Experimental Biology. 2005;208:4273–4282. doi: 10.1242/jeb.01910. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Egertova M. The phylogenetic distribution and evolutionary origins of endocannabinoid signalling. Handb Exp Pharmacol. 2005;168:283–297. doi: 10.1007/3-540-26573-2_9. [DOI] [PubMed] [Google Scholar]
- Ensslen SE, Brady-Kalnay SM. PTP mu signaling via PKC delta is instructive for retinal ganglion cell guidance. Molecular and Cellular Neuroscience. 2004;25:558–571. doi: 10.1016/j.mcn.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Fell HB, Pawson DL. The general biology of echinoderms. In: Boolootian RA, editor. Physiology of Echinoderms. Interscience; New York: 1966. pp. 1–48. [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fernald RD. Evolution of eyes. Current Opinion in Neurobiology. 2000;10:444–450. doi: 10.1016/s0959-4388(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Florey E, Cahill MA. Ultrastructure of sea urchin tube feet. Cell and Tissue Research. 1977;177:195–214. doi: 10.1007/BF00221081. [DOI] [PubMed] [Google Scholar]
- Florey E, Cahill MA. Cholinergic motor control of sea urchin tube feet: evidence for chemical transmission without synapses. J Exp Biol. 1980;88:281–292. doi: 10.1242/jeb.88.1.281. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin L, Schioth HB. The G-protein coupled receptors in the human genome form five main families: phylogenetic analysis. Molecular Pharmacology. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C, Loeffler J, Larmet Y. Brain-derived neurotrophic factor stimulates AP-1 and cyclic AMP-responsive element dependent transcriptional activity in central nervous system neurons. J Neurochem. 1996;66:2279–2286. doi: 10.1046/j.1471-4159.1996.66062279.x. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural Crest and the Origin of Vertebrates - a New Head. Science. 1983;220:268–274. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Garcia-Arraras JE, Rojas-Soto M, Jimenez LB, Diaz-Miranda L. The enteric nervous system of echinoderms: Unexpected complexity revealed by neurochemical analysis. Journal of Experimental Biology. 2001;204:865–873. doi: 10.1242/jeb.204.5.865. [DOI] [PubMed] [Google Scholar]
- Gautam M, DeChiara TM, Glass DJ, Yancopoulos GD, Sanes JR. Distinct phenotypes of mutant mice lacking agrin, MuSK, or rapsyn. Developmental Brain Research. 1999;114:171–178. doi: 10.1016/s0165-3806(99)00013-9. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. Journal of Heredity. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- Giese AC, Farmanfarmanian A. Resistance of the purple sea urchin to osmotic stress. Biological Bulletin. 1963;124:182–192. [Google Scholar]
- Glaser FT, Stanewsky R. Temperature synchronization of the Drosophila circadian clock. Current Biology. 2005;15:1352–1363. doi: 10.1016/j.cub.2005.06.056. [DOI] [PubMed] [Google Scholar]
- Grens A, Mason E, Marsh JL, Bode HR. Evolutionary conservation of a cell fate specification gene: the Hydra achaete-scute homologue has proneural activity in Drosophila. Development. 1995;121:4027–4035. doi: 10.1242/dev.121.12.4027. [DOI] [PubMed] [Google Scholar]
- Gutman GA, Chandy KG, Grissmer S, Lazdunksi M, Mckinnon D, Pardo LA, Robertson GA, Rudy B, Sanguinetti MC, Stuhmer W, Sand W, Wang X. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage gated potassium channels. Pharmac Rev. 2005;57:473–508. doi: 10.1124/pr.57.4.10. [DOI] [PubMed] [Google Scholar]
- Hallböök F. Evolution of the vertebrate neurotrophin and Trk receptor gene families. Curr Opin Neurobiol. 1999;9:616–21. doi: 10.1016/S0959-4388(99)00011-2. [DOI] [PubMed] [Google Scholar]
- Hargrave PA, McDowell JH, Felmann RJ, Atkinson PH, Rao JK, Argos P. Rhodopsin's protein and carbohydrate structure: selected aspects. Vision Research. 1984;24:1487–1499. doi: 10.1016/0042-6989(84)90311-0. [DOI] [PubMed] [Google Scholar]
- Hargrave PA, McDowell JH, Felmann RJ, Atkinson PH, Rao JK, Argos P. Rhodopsin's protein and carbohydrate structure: selected aspects. Vision Research. 1984;24:1487–1499. doi: 10.1016/0042-6989(84)90311-0. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Seminars in Cell & Developmental Biology. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hereu B, Zabala M, Linares C, Sala E. Temporal and spatial variability in settlement of the sea urchin Paracentrotus lividus in the NW Mediterranean. Marine Biology. 2004;144:1011–1018. [Google Scholar]
- Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Research. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyland A, Hodin J. Heterochronic developmental shift caused by thyroid hormone in larval sand dollars and its implications for phenotypic plasticity and the evolution of nonfeeding development. Evolution Int J Org Evolution. 2004;58:524–538. [PubMed] [Google Scholar]
- Heyland A, Reitzel AM, Hodin J. Thyroid hormones determine developmental mode in sand dollars (Echinodermata: Echinoidea) Evol Dev. 2004;6:382–392. doi: 10.1111/j.1525-142X.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. Sinauer; New York: 2001. [Google Scholar]
- Hirayama J, Sassone-Corsi P. Structural and functional features of transcription factors controlling the circadian clock. Current Opinion in Genetics & Development. 2005;15:548–556. doi: 10.1016/j.gde.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hoffmann W, Schwarz H. Ependymins: meningeal-derived extracellular matrix proteins at the blood-brain barrier. Int Rev Cytol. 1996;165:121–158. doi: 10.1016/s0074-7696(08)62221-4. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Holland ND. Evolution of neural crest and placodes: amphioxus as a model for the ancestral vertebrate? Journal of Anatomy. 2001;199:85–98. doi: 10.1046/j.1469-7580.2001.19910085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna S, Brown T, Chen L, Cameron A, Davidson EH. The identification and characterization of transcription factor families in early Strongylocentrotus purpuratus development. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna S, Brown T, Chen L, Cameron A, Davidson EH. Utilization of forkhead transcription factors in the early development of the sea urchin. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Nakabayashi K, Bhalla A. Evolution of glycoprotein hormone subunit genes in bilateral metazoa: Identification of two novel human glycoprotein hormone subunit family genes, GPA2 and GPB5. Molecular Endocrinology. 2002;16:1538–1551. doi: 10.1210/mend.16.7.0871. [DOI] [PubMed] [Google Scholar]
- Hyman LH. The Invertebrates: Echinodermata. McGraw-Hill; New York: 1955. [Google Scholar]
- Inoue M, Birenheide R, Koizumi O, Kobayakawa Y, Muneoka Y, Motokawa T. Localization of the neuropeptide NGIWYamide in the holothurian nervous system and its effects on muscular contraction. Proceedings of the Royal Society of London Series B-Biological Sciences. 1999;266:993–1000. [Google Scholar]
- Ishimaru Y, Okada S, Naito H, Nagai T, Yasuoka A, Matsumoto I, Abe K. Two families of candidate taste receptors in fishes. Mechanisms of Development. 2005;122:1310–1321. doi: 10.1016/j.mod.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Iwakoshi E, Ohtani M, Takahashi T, Moneoka Y, Ikeda T, Fujita T, Minakata H, Nomoto H. Comparative aspects of structure and action of bioactive peptides isolated from the sea cucumber Stichopus japonicus. In: Ohno M, editor. Peptide Chemistry 1994. Protein Research Foundation; Osaka: 1995. pp. 261–264. [Google Scholar]
- Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–33. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Janer G, LeBlanc GA, Porte C. A comparative study on androgen metabolism in three invertebrate species. Gen Comp Endocrinol. 2005;143 doi: 10.1016/j.ygcen.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–699. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- Karnik SS, Sakmar TP, Chen HB, Khorana HG. Cysteine Residue-110 and Residue-187 Are Essential for the Formation of Correct Structure in Bovine Rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny AP, Kozkowski DJ, Oleksyn DW, Angerer LM, Angerer RC. SpSoxB1, a maternally encoded transcription factor asymmetrically distributed among early sea urchin blastomeres. Development. 1999;126:5473–5483. doi: 10.1242/dev.126.23.5473. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock Mutants of Drosophila-Melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:2112–2120. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapraz F, Rottinger E, Duboc V, Range R, Duloquin L, Walton K, Wu S-Y, Bradham C, Loza MA, Wilson K, Poustka A, Angerer LM, McClay D. Genes for receptors tyrosine kinases and TGFB signaling pathways encoded in the sea urchin genome. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguia M, Wessel GM. Selective expression of a sec1/munc18 member in sea urchin eggs and embryos. Gene Expr Patterns. 2004;4:645–57. doi: 10.1016/j.modgep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Li XT, Chuang CK, Mao CA, Angerer LM, Klein WH. Two Otx proteins generated from multiple transcripts of a single gene in Strongylocentrotus purpuratus. Developmental Biology. 1997;187:253–266. doi: 10.1006/dbio.1997.8610. [DOI] [PubMed] [Google Scholar]
- Lindahl PE, Runnstrom J. Variation und Okologie von Psammechinus miliaris. Acta Zoologica. 1929;10:401–484. [Google Scholar]
- Lindgens D, Holstein TW, Technau U. Hyzic, the Hydra homolog of the zic/odd-paired gene, is involved in the early specification of the sensory nematocytes. Development. 2004;131:191–201. doi: 10.1242/dev.00903. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- Luke NH, Killian CE, Livingston BT. Spfkh1 encodes a transcription factor implicated in gut formation during sea urchin development. Development Growth & Differentiation. 1997;39:285–294. doi: 10.1046/j.1440-169x.1997.t01-2-00004.x. [DOI] [PubMed] [Google Scholar]
- Luo CW, Pisarska MD, Hsueh AJW. Identification of a stanniocalcin paralog, stanniocalcin-2, in fish and the paracrine actions of stanniocalcin-2 in the mammalian ovary. Endocrinology. 2005;146:469–476. doi: 10.1210/en.2004-1197. [DOI] [PubMed] [Google Scholar]
- MacBride EW. The development of Echinus esculentus, together with some points on the development of E. miliaris and E.acutus. Philosophical Transactions of the Royal Society, London series B. 1903;195:285–325. [Google Scholar]
- Mackie GO, Spencer AN, Strathmann R. Electrical Activity associated with Ciliary Reversal in an Echinoderm Larva. 1969;223:1384–1385. [Google Scholar]
- Mackie GO, Anderson PAV, Singla CL. Apparent Absence of Gap-Junctions in 2 Classes of Cnidaria. Biological Bulletin. 1984;167:120–123. [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schiitq G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evan RM. The Nuclear Receptor Superfamily: The Second Decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh H, Palfrey HC. Neurotrophin-3 and brain-derived neurotrophic factor activate multiple signal transduction events but are not survival factors for hippocampal pyramidal neurons. J Neurochem. 1996;67:952–963. doi: 10.1046/j.1471-4159.1996.67030952.x. [DOI] [PubMed] [Google Scholar]
- Materna S, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc fingers of the Strongylocentrotus purpuratus genome. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Mazet F, Shimeld SM. Molecular evidence from ascidians for the evolutionary origin of vertebrate cranial sensory placodes. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution. 2005;304B:340–346. doi: 10.1002/jez.b.21054. [DOI] [PubMed] [Google Scholar]
- Mazet F, Hutt JA, Milloz J, Millard J, Graham A, Shimeld SM. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Developmental Biology. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Melarange R, Elphick MR. Comparative analysis of nitric oxide and SALMFamide neuropeptides as general muscle relaxants in starfish. Journal of Experimental Biology. 2003;206:893–899. doi: 10.1242/jeb.00197. [DOI] [PubMed] [Google Scholar]
- Merrow M, Spoelstra K, Roenneberg T. The circadian cycle: daily rhythms from behaviour to genes - First in the Cycles Review Series. Embo Reports. 2005;6:930–935. doi: 10.1038/sj.embor.7400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljkovic-Licina M, Gauchat D, Galliot B. Neuronal evolution: analysis of regulatory genes in a first-evolved nervous system, the hydra nervous system. Biosystems. 2004;76:75–87. doi: 10.1016/j.biosystems.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Millott N. Coordination of spine movements in echinoids. In: Boolootian RA, editor. Physiology of Echinoderms. Interscience; New York: 1966. pp. 187–220. [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expression Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Mire P, Nasse J, Venable-Thibodeaux S. Gap junctional communication in the vibration-sensitive response of sea anemones. Hearing Research. 2000;144:109–123. doi: 10.1016/s0378-5955(00)00047-2. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mu XQ, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Seminars in Cell & Developmental Biology. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Mu XQ, Fu XY, Sin HX, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Developmental Biology. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Kaneko H, Murray G, Burke RD. Divergent patterns of neural development in larval echinoids and asteroids. Evolution & Development. 2004;6:95–104. doi: 10.1111/j.1525-142x.2004.04011.x. [DOI] [PubMed] [Google Scholar]
- Nimmrich I, Erdmann S, Melchers U, Chtarbova S, Finke U, Hentsch S, Hoffmann I, Oertel M, Hoffmann W, Muller O. The novel ependymin related gene UCC1 is highly expressed in colorectal tumor cells. Cancer Lett. 2001;165:71–79. doi: 10.1016/s0304-3835(01)00390-1. [DOI] [PubMed] [Google Scholar]
- Ohtani M, Iwakoshi E, Muneoka Y, Minakata H, Nomoto H. Isolation and characterization of bioactive peptides from the sea cucumber Stichopus japonicus. In: Shimonishi Y, editor. Peptide Science - Present and Future. Springer; Berlin: 1999. pp. 419–420. [Google Scholar]
- Olinski RP, Dahlberg C, Thorndyke M, Hallböök F. Three insulin-relaxin-like genes in Ciona intestinalis. Peptides. 2006 doi: 10.1016/j.peptides.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Otim O, Amore G, Minokawa T, McClay DR, Davidson EH. SpHnf6, a transcription factor that executes multiple functions in sea urchin embryogenesis. Developmental Biology. 2004;273:226–243. doi: 10.1016/j.ydbio.2004.05.033. [DOI] [PubMed] [Google Scholar]
- Panchin YV. Evolution of gap junction proteins - the pannexin alternative. Journal of Experimental Biology. 2005;208:1415–1419. doi: 10.1242/jeb.01547. [DOI] [PubMed] [Google Scholar]
- Pentreath VW, Cottrell GA. Acetylcholine and Cholinesterase in Radial Nerve of Asterias Rubens. Comparative Biochemistry and Physiology. 1968;27:775. doi: 10.1016/0010-406x(68)90617-8. [DOI] [PubMed] [Google Scholar]
- Pentreath VW, Cobb JLS. Neurobiology of echinodermata. Biological Reviews of the Cambridge Philosophical Society. 1972;47:363–392. doi: 10.1111/j.1469-185x.1972.tb00977.x. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Piront M, Schmidt R. Inhibition of long-term memory formation by anti-ependymin antisera after active shock-avoidance learning in goldfish. Brain Res. 1988;442:53–62. doi: 10.1016/0006-8993(88)91431-x. [DOI] [PubMed] [Google Scholar]
- Poustka A, Groth D, Hennig S, Thamm S, Cameron A, Beck A, Reinhardt R, Herwig R, Panopoulou G, Lehrach H. Generation, annotation, evolutionary analysis, and database integration of 20,000 unique sea urchin EST clusters. Genome Res. 2003;13:2736–2746. doi: 10.1101/gr.1674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka AJ, Kuhn A, Radosavljevic V, Wellenreuther R, Lehrach H, Panopoulou G. On the origin of the chordate central nervous system: expression of onecut in the sea urchin embryo. Evolution & Development. 2004;6:227–236. doi: 10.1111/j.1525-142X.2004.04028.x. [DOI] [PubMed] [Google Scholar]
- Raible F, Tessmar-Raible K, Arboleda E, Kaller T, Bork P, Arendt D, Arnone MI. Opsins and clusters of sensory G-protein coupled receptors in the sea urchin genome. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Reese ES. The complex behavior of echinoderms. In: Boolootian RA, editor. Physiology of Echinoderms. Interscience; New York: 1966. pp. 15–218. [Google Scholar]
- Reinecke M, Collett C. The phylogeny of the insulin-like growth factors. Int Rev Cytol. 1998;183:1–94. doi: 10.1016/s0074-7696(08)60142-4. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: a cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Developmental Biology. 2004;274:438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Broadie KS. The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans. Curr Opin Neurobiol. 2002;12:499–507. doi: 10.1016/s0959-4388(02)00360-4. [DOI] [PubMed] [Google Scholar]
- Roepke TA, Snyder MJ, Cherr GN. Estradiol and endocrine disrupting compounds adversely affect development of sea urchin embryos at environmentally relevant concentrations. Aquat Toxicol. 2005;71:155–173. doi: 10.1016/j.aquatox.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Lundberg L. Photoperiodic activity pattern in the brittle star Amphiura filiformis. Marine Biology. 2004;145:651–656. [Google Scholar]
- Samanta MP, Tongprasit W, Istrail S, Cameron A, Tu Q, Davidson EH. A high-resolution transcriptome map of the sea urchin embryo. Dev Biol. 2006 doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- Sasai Y. Regulation of neural determination by evolutionarily conserved signals: anti-BMP factors and what next? Current Opinion in Neurobiology. 2001;11:22–26. doi: 10.1016/s0959-4388(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Satou Y, Sasakura Y, Yamada L, Imai K, Satoh N, Degnan B. A genomewide survey of developmentally relevant genes in Ciona intestinalis. V. Genes for receptor tyrosine kinase pathway and notch signaling pathway. Dev Genes Evol. 2003;213:254–263. doi: 10.1007/s00427-003-0317-9. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Evolutionary origins of vertebrate placodes: Insights from developmental studies and from comparisons with other deuterostomes. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution. 2005;304B:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Shashoua V. Antibodies to ependymin block the sharpening of the regenerating retinotectal projection in goldfish. Brain Res. 1988;446:269–84. doi: 10.1016/0006-8993(88)90886-4. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Rother S, Schlingensiepen K, Brysch W. Neuronal plasticity depending on a glycoprotein synthesized in goldfish Leptomeninx. Prog Brain Res. 1992;91:7–12. doi: 10.1016/s0079-6123(08)62309-2. [DOI] [PubMed] [Google Scholar]
- Schuel H, Goldstein E, Mechoulam R, Zimmerman AM, Zimmerman S. Anandamide (Arachidonylethanolamide), a Brain Cannabinoid Receptor Agonist, Reduces Sperm Fertilizing-Capacity in Sea-Urchins by Inhibiting the Acrosome Reaction. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7678–7682. doi: 10.1073/pnas.91.16.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JR, Sasaki JD, Vacquier VD. Increased association of synaptosome-associated protein of 25 kDa with syntaxin and vesicle-associated membrane protein following acrosomal exocytosis of sea urchin sperm. J Biol Chem. 1998;273:24355–9. doi: 10.1074/jbc.273.38.24355. [DOI] [PubMed] [Google Scholar]
- Selke D, Anton H, Klumpp S. Serine/threonine protein phosphatases type 1, 2A and 2C in vertebrate retinae. Acta Anatomica. 1998;162:151–156. doi: 10.1159/000046480. [DOI] [PubMed] [Google Scholar]
- Shashoua V. The role of brain extracellular proteins in neuroplasticity and learning. Cell Mol Neurobiol. 1985;5:183–207. doi: 10.1007/BF00711092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashoua V, Hesse G. Classical conditioning leads to changes in extracellular concentrations of ependymin in goldfish brain. Brain Res. 1989;484:333–339. doi: 10.1016/0006-8993(89)90377-6. [DOI] [PubMed] [Google Scholar]
- Shashoua V. Ependymin, a brain extracellular glycoprotein, and CNS plasticity. Ann N Y Acad Sci. 1991;627:94–114. doi: 10.1111/j.1749-6632.1991.tb25916.x. [DOI] [PubMed] [Google Scholar]
- Shashoua V, Adams D, Boyer-Boiteau A. CMX-8933, a peptide fragment of the glycoprotein ependymin, promotes activation of AP-1 transcription factor in mouse neuroblastoma and rat cortical cell cultures. Neurosci Lett. 2001;312:103–107. doi: 10.1016/s0304-3940(01)02119-x. [DOI] [PubMed] [Google Scholar]
- Smit AB, van Kesteren RE, Li KW, Van Minnen J, Spijker S, Van Heerikhuizen H, Geraerts WP. Towards understanding the role of insulin in the brain: lessons from insulin-related signaling systems in the invertebrate brain. Prog Neurobiol. 1998;54:35–54. doi: 10.1016/s0301-0082(97)00063-4. [DOI] [PubMed] [Google Scholar]
- Smith JE. Echinodermata. In: Bullock TH, Horridge GA, editors. Structure and Function in the Nervous Systems of Invertebrates. W.H. Freeman and Co; London: 1965. pp. 1519–1558. [Google Scholar]
- Sodergren E, Shen U, Song X, Zhang L, Weinstock G, Gibbs R. Shedding genetic light on Aristotle's lantern. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Strand FL. Neuropeptides. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- Strathmann RR. Larval feeding in echinoderms. American Zoologist. 1975;15:717–730. [Google Scholar]
- Suarez-Castillo E, Medina-Ortiz W, Roig-Lopez J, Garcia-Arraras J. Ependymin, a gene involved in regeneration and neuroplasticity in vertebrates, is overexpressed during regeneration in the echinoderm Holothuria glaberrima. Gene. 2004;334:133–143. doi: 10.1016/j.gene.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Synaptotagmins: why so many? J Biol Chem. 2002;277:7629–32. doi: 10.1074/jbc.R100052200. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–47. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Tahara M, Coorssen JR, Timmers K, Blank PS, Whalley T, Scheller R, Zimmerberg J. Calcium can disrupt the SNARE protein complex on sea urchin egg secretory vesicles without irreversibly blocking fusion. J Biol Chem. 1998;273:33667–73. doi: 10.1074/jbc.273.50.33667. [DOI] [PubMed] [Google Scholar]
- Takacs CM, Amore G, Oliveri P, Poustka AJ, Wang D, Burke RD, Peterson KJ. Expression of an NK2 homeodomain gene in the apical ectoderm defines a new territory in the early sea urchin embryo. Developmental Biology. 2004;269:152–164. doi: 10.1016/j.ydbio.2004.01.023. [DOI] [PubMed] [Google Scholar]
- Teng FYH, Wang Y, Tang BL. The syntaxins. Genome Biol. 2001;2:3012.1–3012.7. doi: 10.1186/gb-2001-2-11-reviews3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W - Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorndyke MC, Crawford BD, Burke RD. Localization of a SALMFamide neuropeptide in the larval nervous system of the sand dollar, Dendraster excentricus. Acta Zoologica. 1992;73:207–212. [Google Scholar]
- Tu Q, Davidson EH, Oliveri P. A high-resolution transcriptome map of the sea urchin embryo. Dev Biol 2006 [Google Scholar]
- van Kesteren RE, Smit AB, Dirks RW, Dewith ND, Geraerts WPM, Joosse J. Evolution of the Vasopressin Oxytocin Superfamily - Characterization of a Cdna-Encoding a Vasopressin-Related Precursor, Preproconopressin, from the Mollusk Lymnaea stagnalis. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4593–4597. doi: 10.1073/pnas.89.10.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kesteren RE, Fainzilber M, Hauser G, van Minnen J, Vreugdenhil E, Smit AB, Ibanez CF, Geraerts WPM, Bulloch AGM. Early evolutionary origin of the neurotrophin receptor family. Embo Journal. 1998;17:2534–2542. doi: 10.1093/emboj/17.9.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ubisch L. Die Entwicklung con Strongylocentrotus lividus. Ztschr Wiss Zool. 1913;106:409–488. [Google Scholar]
- Wada S, Toyoda R, Yamamoto H, Saiga H. Ascidian otx gene Hroth activates transcription of the brain-specific gene HrTRP. Developmental Dynamics. 2002;225:46–53. doi: 10.1002/dvdy.10135. [DOI] [PubMed] [Google Scholar]
- Walldorf U, Kiewe A, Wickert M, Ronshaugen M, McGinnis W. Homeobrain, a novel paired-like homeobox gene is expressed in the Drosophila brain. Mechanisms of Development. 2000;96:141–144. doi: 10.1016/s0925-4773(00)00380-4. [DOI] [PubMed] [Google Scholar]
- Walton K, Croce JC, Glenn TD, Wu S-Y, McClay D. Genomics and expression profiles of the Hedgehog and Notch signaling pathways in sea urchin development. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JK, Mcdowell JH, Hargrave PA. Site of Attachment of 11-Cis-Retinal in Bovine Rhodopsin. Biochemistry. 1980;19:5111–5117. doi: 10.1021/bi00563a027. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun DT, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes & Development. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Dityatev A, Scheiffele P, Biederer T, Weiner JA, Christopherson KS, El-Husseini A. Cell adhesion molecules in synapse formation. Journal of Neuroscience. 2004;24:9244–9249. doi: 10.1523/JNEUROSCI.3339-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson KM, Watts SA. Progesterone metabolism in the ovaries and testes of the echinoid Lytechinus variegatus Lamarck (Echinodermata) Comp Biochem Physiol C Toxicol Pharmacol. 2000;127:263–272. doi: 10.1016/s0742-8413(00)00153-5. [DOI] [PubMed] [Google Scholar]
- Welsh JH. Neurohumors and neurosecretion. In: Boolootian RA, editor. Physiology of Echinoderms. Wiley; New York: 1966. p. 822. [Google Scholar]
- Whalley T, Timmers K, Coorssen J, Bezrukov L, Kingsley DH, Zimmerberg J. Membrane fusion of secretory vesicles of the sea urchin egg in the absence of NSF. J Cell Sci. 2004;117:2345–56. doi: 10.1242/jcs.01077. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Bergeron KF, Whittle JA, Brandhorst BP, Burke RD, Hynes RO. The echinoderm adhesome. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson TN, Speed TP, Tregear GW, Bathgate RA. Evolution of the relaxin-like peptide family. Bmc Evolutionary Biology. 2005;5 doi: 10.1186/1471-2148-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Falkmer S. Starfish Insulin. Can J Biochem. 1965;43:1615–1624. doi: 10.1139/o65-178. [DOI] [PubMed] [Google Scholar]
- Woodley JD. Photosensitivity in Diadema antillarum: does it show scototaxis? In: Lawrence JM, editor. The International Echinoderm Conference. Balkema; Rotterdam: 1982. p. 61. [Google Scholar]
- Yaguchi S, Kanoh K, Amemiya S, Katow H. Initial analysis of immunochemical cell surface properties, location and formation of the serotonergic apical ganglion in sea urchin embryos. Development Growth & Differentiation. 2000;42:479–488. doi: 10.1046/j.1440-169x.2000.00535.x. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Katow H. Expression of Tryptophan 5-hydroxylase gene during sea urchin neurogenesis and role of serotonergic nervous system in larval behavior. Journal of Comparative Neurology. 2003;466:219–229. doi: 10.1002/cne.10865. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD. Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development. 2006;133:2337–2346. doi: 10.1242/dev.02396. [DOI] [PubMed] [Google Scholar]
- Yoshida M. Photosensitivity. In: Boolootian RA, editor. Physiology of Echinoderms. Interscience; New York: 1966. pp. 435–464. [Google Scholar]
- Yuh CH, Dorman ER, Davidson EH. Brn1/2/4, the predicted midgut regulator of the endo16 gene of the sea urchin embryo. Developmental Biology. 2005;281:286–298. doi: 10.1016/j.ydbio.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Zhu CQC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


