Abstract
S6K1 has emerged as a critical signaling component in the development of insulin resistance through phosphorylation and inhibition of IRS-1 function. This effect can be triggered directly by nutrients such as amino acids or by insulin through a homeostatic negative-feedback loop. However, the role of S6K1 in mediating IRS-1 phosphorylation in a physiological setting of nutrient overload is unresolved. Here we show that S6K1 directly phosphorylates IRS-1 Ser-1101 in vitro in the C-terminal domain of the protein and that mutation of this site largely blocks the ability of amino acids to suppress IRS-1 tyrosine and Akt phosphorylation. Consistent with this finding, phosphorylation of IRS-1 Ser-1101 is increased in the liver of obese db/db and wild-type, but not S6K1−/−, mice maintained on a high-fat diet and is blocked by siRNA knockdown of S6K1 protein. Finally, infusion of amino acids in humans leads to the concomitant activation of S6K1, phosphorylation of IRS-1 Ser-1101, a reduction in IRS-1 function, and insulin resistance in skeletal muscle. These findings indicate that nutrient- and hormonal-dependent activation of S6K1 causes insulin resistance in mice and humans, in part, by mediating IRS-1 Ser-1101 phosphorylation.
Keywords: mTOR, nutrient sensing, amino acids, insulin signaling
Maintenance of blood glucose levels involves a complex interplay between peripheral tissues and pancreatic β cells (1). The impaired ability of insulin to trigger downstream metabolic actions, defined as insulin resistance, is closely associated with the development of type 2 diabetes (2). Multiple mechanisms have been proposed as a molecular basis underlying the induction of insulin resistance (2, 3), of which phosphorylation of IRS-1 on serine residues has emerged as a key event (4). Indeed, a large number of protein kinases have been shown to cause serine phosphorylation of IRS-1, including JNK (5), IKK (6), ERK (7), PKC-ζ (8, 9), PKC-θ (10), mTOR (11), and, more recently, S6K1 (12, 13). Phosphorylation of IRS-1 by serine/threonine kinases is known to cause inhibition of insulin signaling by negating IRS-1's ability to serve as a mediator of insulin receptor tyrosine kinase signals (4).
Nutrient overload represents a defining cause of insulin resistance (14), and recent evidence suggests that the mTOR/raptor/GβL (mTOR Complex 1) signaling complex, an effector of nutrient and hormonal signals (15–17), and its downstream target, S6K1, are critical components in this response (13, 18, 19). Activation of this pathway by insulin or amino acids has been shown to cause increased serine phosphorylation of IRS-1, disruption of phosphatidylinositol 3-kinase (PI3-kinase) signaling, and inhibition of glucose transport in L6 myocytes and 3T3-L1 adipocytes (20–23). Moreover, injection of rapamycin in mice was found to reduce serine phosphorylation of IRS-1 and prolong insulin-stimulated PI3-kinase activity in skeletal muscle (24). Further insights into the molecular mechanism linking S6K1 activation to IRS-1 serine phosphorylation was revealed by studies of the TSC1–TSC2 tumor suppressor complex, a component of the canonical PI3-kinase signaling pathway, which acts to suppress insulin-induced mTOR Complex1 signaling and S6K1 activation (13). Disruption of this complex in mouse embryonic fibroblasts leads to constitutive S6K1 activation, leading to IRS-1 phosphorylation and degradation, as well as inhibition of IRS-1 transcription (12, 25). The negative effects observed on IRS-1 function in TSC1–TSC2-deficient cells are much more pronounced than those observed in models of insulin resistance (12, 25) potentially because of the constitutive activation of S6K1. Indeed, the physiological importance of S6K1 in the settings of nutrient overload may be distinct from that of loss of TSC1–TSC2. S6K1's critical role in such settings is reflected by its elevated levels of activation in the liver, adipose, and muscle of obese animals (26, 27) and the fact that S6K1-deficient mice are protected against diet-induced obesity and insulin resistance (27).
Although the role of S6K1 in modulating insulin action is clear (13, 18, 19), it remains to be established whether S6K1 directly mediates IRS-1 phosphorylation in physiological settings of nutrient overload. Rapamycin treatment has been shown to inhibit phosphorylation of IRS-1 on Ser-307, Ser-312, Ser-612, and Ser-636/639 (human notation of IRS-1 Ser residues) (11, 12, 23, 24, 27, 28). Of these residues, mTOR has been shown to directly catalyze the phosphorylation of Ser-636/639 (11), whereas S6K1 was shown to phosphorylate Ser-307 (mouse Ser-302) in vitro (12). S6K1−/− mice showed decreased phosphorylation of IRS-1 on both Ser-312 (mouse Ser-307) and Ser-636/639 (27), although neither of these sites reside in a S6K1 recognition motif. Here we have identified Ser-1101 as an S6K1-mediated phosphorylation site in IRS-1. We further found that phosphorylation of Ser-1101 is increased upon nutrient overload and in the obese setting, and it also participates in suppressing insulin action through inhibition of PI3-kinase/Akt signaling, which leads to insulin resistance.
Results
Increased Amino Acid Availability Causes a Rapamycin-Sensitive Inhibition of Insulin Signaling.
Nutrient sensing through the mTOR Complex 1 signaling pathway is known to cause inhibition of insulin signaling in a number of cell types (13). Using L6 muscle cells, we found that supplementation of amino acids to the incubation medium significantly increased S6K1 kinase activity, which was linked to hyperphosphorylation of IRS-1 on serine and threonine residues as evidenced by decreased mobility of the protein on SDS/PAGE and impaired activation of PI3-kinase when compared with amino acid-deprived cells [supporting information (SI) Fig. 6 A–C]. Importantly, these signaling defects were linked to a defective stimulation of glucose transport by insulin (SI Fig. 6D). In contrast, inhibition of mTOR Complex 1 signaling by rapamycin abrogated S6K1 activity and restored IRS-1 migration behavior, PI3-kinase activity, and glucose uptake in amino acid-treated cells (SI Fig. 6 E–H). These findings indicate that amino acid availability promotes an insulin-resistant state in L6 myocytes, which is mediated by rapamycin-sensitive IRS-1 serine and threonine phosphorylation.
S6K1 Phosphorylates IRS-1 at Ser-1101.
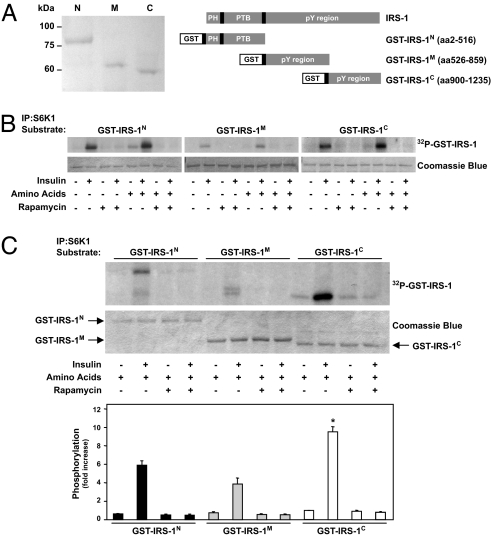
To address the ability of S6K1 to directly phosphorylate IRS-1, GST-fusion proteins encoding specific domains of IRS-1 were tested as S6K1 substrates in an in vitro kinase assay (Fig. 1A). The results of independent experiments show that S6K1 immunopurified from L6 muscle cells has the ability to phosphorylate the N-, middle- and C-terminal domains of IRS-1 (Fig. 1B). Moreover, the magnitude of S6K1-mediated phosphorylation of IRS-1 in each experiment depended on insulin and amino acid stimulation, and this response was abolished by rapamycin treatment (Fig. 1B). To determine the relative preference of S6K1 for each of the specific IRS-1 fragments, the kinase reaction products from the same experiments were resolved on a common SDS/PAGE, and 32P incorporation was normalized for the amount of GST-IRS-1 used for each assay. Although it may not reflect the affinity of S6K1 for specific phosphorylation sites in vivo, we found that, in an in vitro setting, S6K1 preferred GST-IRS-1C (10-fold above basal; P < 0.01), compared with either GST-IRS-1N or GST-IRS-1M (6- and 4-fold above basal, respectively; P < 0.01) (Fig. 1C).
Fig. 1.
S6K1 phosphorylates IRS-1. (A) Serum-deprived L6 cells were incubated in an amino acid-deprived or amino acid-containing medium for 1 h and stimulated or not with 100 nM insulin for the last 30 min of incubation as indicated, and then 0.01% DMSO vehicle or 25 nM rapamycin was added during the 1-h incubation. (B and C) Kinase activity of S6K1 was determined by using GST-fusion proteins encoding various domains of IRS-1 (as shown in A). Controls for substrate amount (GST-IRS-1) were analyzed by SDS/PAGE, followed by Coomassie blue staining. Autoradiograms are representative of at least three independent experiments. The bars in C Lower represent the means ± SE of three independent determinations of S6K1 activity by using GST-IRS-1 as a substrate. *, P < 0.01 vs. corresponding S6K1 assay by using GST-IRS-1N or GST-IRS-1M as a substrate.
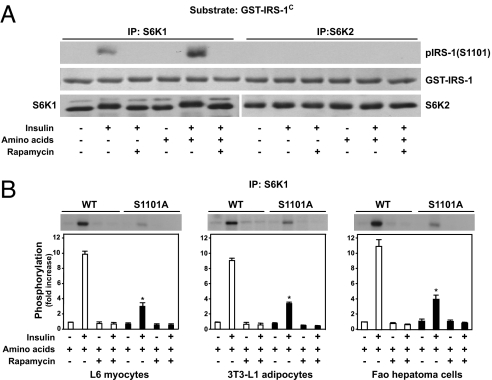
To identify potential S6K1-mediated phosphorylation sites within the C-terminal domain of IRS-1, we searched for the known S6K1 substrate recognition motif, RxRxxSx (29), and identified Ser-1101 (1) as the unique sequence having this motif. To test the ability of S6K1 to mediate phosphorylation of Ser-1101 in vitro, the GST-IRS-1C fragment was incubated with S6K1 derived from L6 muscle cells treated with insulin in the absence or presence of rapamycin, and the extent of Ser-1101 phosphorylation was determined with a phosphospecific antibody. The results show that S6K1 derived from insulin-treated cells increases Ser-1101 phosphorylation, and that this effect is potentiated by the presence of amino acids and is totally blunted by pretreatment with rapamycin (25 nM) (Fig. 2A). Parallel experiments revealed that Ser-1101 was not phosphorylated by S6K2 immunoprecipitated from the same cells (Fig. 2A), although it is active toward 40S ribosomal protein S6 (data not shown and ref. 30). We also found that neither S6K2 nor mTOR were present in S6K1 immunoprecipitates from L6 cells, consistent with a specific involvement of S6K1 versus S6K2 in mediating IRS-1 Ser-1101 phosphorylation (SI Fig. 7). Importantly, a serine-to-alanine mutation of this site strongly diminished 32P incorporation into GST-IRS-1C S1101A by S6K1 immunoprecipitated from either insulin-stimulated L6 myocytes, 3T3-L1 adipocytes, or Fao hepatoma cells (Fig. 2B), suggesting that Ser-1101 is the major S6K1-mediated phosphorylation site in the C-terminal domain of IRS-1. The fact that phosphorylation still takes place in the GST-IRS-1C S1101A peptide raised the possibility of alternative S6K1 phosphorylation sites, albeit with weak recognition motifs. We previously reported that PKC-θ also phosphorylates IRS-1 Ser-1101 (10). However, neither PKC-θ, δ, z, nor γ are present in S6K1 immunoprecipitates (SI Fig. 7), ruling out the potential contribution from these kinases in S6K1-mediated IRS-1 Ser-1101 phosphorylation.
Fig. 2.
S6K1 phosphorylates IRS-1 at Ser-1101. Serum-deprived L6 cells were incubated in an amino acid-deprived or amino acid-containing medium for 1 h and stimulated or not with 100 nM insulin for the last 30 min of incubation as indicated, and then 0.01% DMSO vehicle or 25 nM rapamycin was added during the 1-h incubation. (A) Kinase activity (by using cold ATP only) of S6K1 and S6K2 was determined by using GST-IRS-1C as a substrate, and then the reaction product was analyzed by SDS/PAGE by using an antibody that detects IRS-1 only when phosphorylated at Ser-1101. Controls for substrate amount (GST-IRS-1C) and immunoprecipitated kinases (S6K1 or S6K2) were analyzed by SDS/PAGE, followed by Coomassie blue staining and Western blot with specific antibodies, respectively. (B) Kinase activity of S6K1 was determined by in vitro kinase assay by using GST-IRS-1C or GST-IRS-1C (S1101A) as substrates. The means ± SE from three individual experiments are shown for B. *, P < 0.01 vs. corresponding S6K1 assay by using wild-type GST-IRS-1C as a substrate.
Phosphorylation of IRS-1 at Ser-1101 Depends on Nutrient Availability and Causes Inhibition of Insulin Signaling.
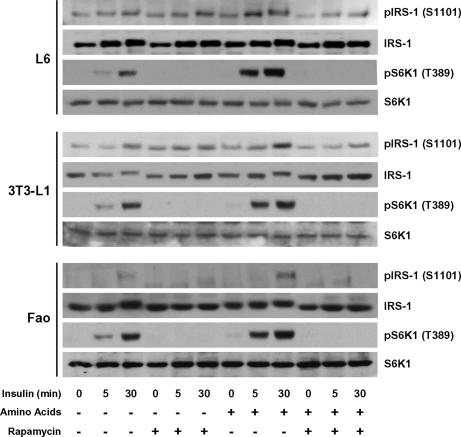
Because amino acids augment insulin-induced hyperphosphorylation of IRS-1 on serine and threonine residues (SI Fig. 6B), we next determined whether this effect was reflected in specific cell models by increased IRS-1 Ser-1101 phosphorylation. The results demonstrate that insulin-induced IRS-1 Ser-1101 and S6K1 Thr-389 phosphorylation were both increased when L6 muscle cells were incubated in an amino acid-enriched medium, an effect that was suppressed by rapamycin (Fig. 3Top). Similar results were obtained in 3T3-L1 adipocytes (Fig. 3 Middle) and Fao hepatoma cells (Fig. 3 Bottom). Thus, the phosphorylation of IRS-1 at Ser-1101 and the activation status of S6K1 in insulin target cells closely mirrored the data obtained in the in vitro assays, in which S6K1 was found to mediate phosphorylation of IRS-1 at Ser-1101 in a rapamycin-sensitive manner.
Fig. 3.
Phosphorylation of IRS-1 at Ser-1101 depends on nutrient availability. Serum-deprived L6 muscle cells, 3T3-L1 adipocytes, or Fao hepatoma cells were incubated either in an amino acid-containing or amino acid-deprived medium for 1 h and stimulated or not with 100 nM insulin as indicated, and then 0.01% DMSO vehicle or 25 nM rapamycin was added during the 1-h incubation. Phosphorylation of IRS-1 and S6K1 was determined by using antibodies that recognized IRS-1 and S6K1 only when phosphorylated at Ser-1101 and Thr-389, respectively. Stripped membranes were reprobed for total IRS-1 and S6K1. Results are of one representative experiment repeated at least three times.
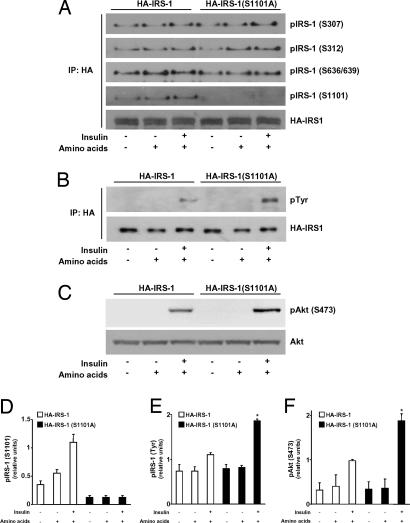
To assess the functional consequences of IRS-1 Ser-1101 phosphorylation on insulin signaling, we next measured the ability of insulin to induce tyrosine phosphorylation of either an HA epitope-tagged wild-type IRS-1 (HA-IRS-1) or an IRS-1 S1101A (HA-IRS-1 S1101A) mutant after their transfection in CHO cells. Incubation of CHO cells with amino acids and insulin increased the phosphorylation of HA-IRS-1 on Ser-1101, but not that of the HA-IRS-1 S1101A mutant (Fig. 4 A and D). Importantly, phosphorylation of IRS-1 at Ser-307, Ser-312, and Ser-636/639 (equivalent to mouse Ser-302, Ser-307, and Ser-632/635) appeared to be unaffected by the S1101A mutation of HA-IRS-1 (Fig. 4A and SI Fig. 8). We further observed that insulin-induced tyrosine phosphorylation of HA-IRS-1 S1101A in the presence of amino acids was significantly elevated, compared with that of wild-type HA-IRS-1 (Fig. 4 B and E). These findings indicate that phosphorylation of Ser-1101 by S6K1 is inhibitory to insulin receptor signaling. Consistent with this conclusion, we found that Akt Ser-473 phosphorylation, a downstream effector of PI3-kinase, was enhanced in cells expressing the S1101A-mutated form of IRS-1, compared with those expressing its wild-type counterpart (Fig. 4 C and F).
Fig. 4.
Phosphorylation of IRS-1 at Ser-1101 causes inhibition of insulin signaling. Serum-deprived CHO cells expressing either HA-IRS-1 or HA-IRS-1 (S1101A) were incubated either in an amino acid-deprived or amino acid-containing medium for 1 h and stimulated or not with 100 nM insulin for the last 30 min of incubation as indicated. (A and B) Phosphorylation of IRS-1 was determined in anti-HA immunoprecipitates by using phosphospecific antibodies against IRS-1 Ser-307, Ser-312, Ser-636/639, and Ser-1101 (A) and phosphotyrosine (B). Levels of HA-IRS-1 recovered in anti-HA immunoprecipitates also are shown. (C) Phosphorylation and expression of Akt were determined in whole-cell extracts by using antibodies against phospho-Akt Ser-473 (Upper) and Akt (Lower). Results are of one representative experiment repeated at least three times. (D–F) Quantification of IRS-1 Ser-1101 (D), IRS-1 Tyr (E), and Akt Ser-473 (F) phosphorylation. *, P < 0.05 vs. corresponding CHO cells expressing wild-type HA-IRS-1.
Increased IRS-1 Ser-1101 Phosphorylation in Obesity- and Nutrient-Induced Insulin Resistance in Mice and Humans: The Role of S6K1.
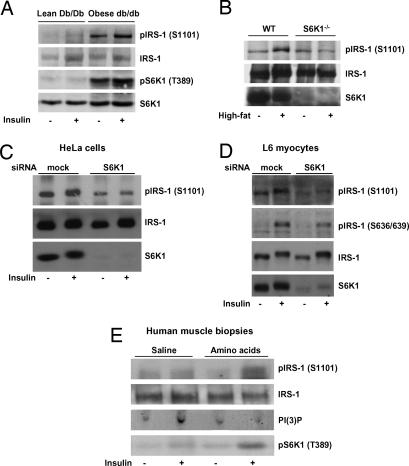
We previously showed that S6K1 Thr-389 phosphorylation in the liver of diet-induced obese rats is markedly elevated in the fasting state and that this response is accelerated in the presence of insulin, peaking 5 min after injection (26). Here we determined whether genetically induced obesity in mice is associated with S6K1 activation and increased phosphorylation of IRS-1 on Ser-1101. As shown in Fig. 5A, S6K1 Thr-389 and IRS-1 Ser-1101 phosphorylation were barely detectable in the liver of lean Db/Db mice. Acute (5-min) insulin injection slightly increased S6K1 activation and IRS-1 Ser-1101 phosphorylation in these lean mice. In marked contrast, S6K1 activation and IRS-1 Ser-1101 phosphorylation were sharply increased in the liver of obese diabetic db/db mice, and this finding was further enhanced by acute insulin stimulation (Fig. 5A). To further evaluate the role of S6K1 in mediating IRS-1 Ser-1101 phosphorylation in the animal, wild-type and S6K1−/− mice were placed on a normal (chow) or high-fat diet for 14 weeks. We recently showed that a high-fat diet induces marked insulin resistance in wild-type, but not in S6K1−/−, mice (27). Here we find that development of insulin resistance in animals placed on a high-fat diet is associated with increased IRS-1 Ser-1101 phosphorylation in wild-type, but not S6K1−/−, mice (Fig. 5B). These latter findings are consistent with our previous observations that S6K1 is constitutively activated in the liver of obese rodents (26, 27) and therefore contributes to IRS-1 Ser-1101 hyperphosphorylation in this insulin-resistant setting.
Fig. 5.
Increased IRS-1 Ser-1101 phosphorylation in obesity- and nutrient-induced insulin resistance in mice and humans: role of S6K1. (A) Fourteen-week-old lean (Db/Db) and obese diabetic (db/db) mice were acutely (5 min) injected i.v. with either saline or insulin at 3.8 units/kg before the liver was collected. (B) Ten-week-old wild-type or S6K1−/− mice were maintained on a normal chow (−) or high-fat (+) diet for 14 weeks before the liver was collected. (A and B) Results are of one representative experiment repeated in at least three different animals for each group. (C and D) Lowering S6K1 by siRNA reduces IRS-1 Ser-1101 phosphorylation. Serum-deprived HeLa cells (C) or L6 myocytes (D) were stimulated with amino acids with or without 200 nM insulin for 30 min. (E) Human skeletal muscle biopsies were obtained during saline and amino acid infusion. Biopsies were sampled 120 min before insulin infusion (−) and after 30 min of prandial-like hyperinsulinemia (+). S6K1 and IRS-1 expression, as well as IRS-1 Ser-636/639 or Ser-1101 phosphorylation, were detected by Western blotting analysis. Results are of one representative experiment repeated in at least five paired subjects.
Using siRNA-mediated gene silencing, we previously demonstrated that S6K1 increases IRS-1 Ser-312 and Ser-636/639 phosphorylation in a cell-autonomous manner (27). Therefore, we tested whether this finding also was the case for IRS-1 Ser-1101. Reduction of S6K1 levels with siRNAs in HeLa cells (Fig. 5C) and L6 myocytes (Fig. 5D) resulted in a lower IRS-1 Ser-1101 phosphorylation in the absence or presence of insulin stimulation. In addition, we showed that silencing of S6K1 partially reduced phosphorylation of IRS-1 at Ser-636/639 in L6 muscle cells (Fig. 5D), which is in agreement with our previous observation in HeLa cells (27). Thus, removal of S6K1 reduced IRS-1 Ser-1101 phosphorylation in a cell-autonomous manner.
Finally, we tested the hypothesis that nutrient-induced insulin resistance also was linked to S6K1-mediated impairment of insulin signaling by IRS-1 Ser-1101 phosphorylation in humans. Paired healthy subjects were infused with saline or amino acids, and muscle biopsies were obtained before and after insulin stimulation. Using this experimental paradigm, we previously reported that the induction of insulin resistance by amino acids in human was associated with the hyperactivation of S6K1 (31, 32). Here we show that the activation status of S6K1 as shown by increased Thr-389 phosphorylation was tightly linked to IRS-1 Ser-1101 phosphorylation and insulin resistance in human skeletal muscle (Fig. 5E). Indeed, although insulin alone slightly increased phosphorylation of IRS-1 at Ser-1101, a combined infusion of amino acids and insulin resulted in a dramatic increase in the phosphorylation of IRS-1 at Ser-1101 and severely blunted the ability of IRS-1-associated PI3-kinase to catalyze the in vitro formation of PI3-phosphate, all of which are consistent with a hyperphosphorylated state of S6K1 in these subjects (Fig. 5E).
Discussion
The mTOR Complex 1–S6K1 signaling axis emerged as a central integration point for multiple inputs, including amino acids and hormones (15–17). First recognized as important modulators of translational events, mTOR Complex 1 and its downstream effector, S6K1, are now considered instrumental to the operation of a negative-feedback loop that acts on IRS-1 (13, 18, 19, 22, 27). We proposed the existence of this negative feedback mechanism based on the observation that inhibition of insulin-stimulated PI3-kinase activity by amino acids was rapidly and temporally related to the activation of S6K1 and IRS-1 serine phosphorylation, a process found to be fully abrogated by the mTOR Complex 1/S6K1 inhibitor, rapamycin (13, 22). Evidence implicating S6K1 as a key element of this negative-feedback loop first emerged in studies in Drosophila, where it was demonstrated that the Drosophila orthologue of S6K1, dS6K, was a negative regulator of dAkt signaling (33, 34). Using biochemical and genetic approaches, recent studies demonstrated the involvement of S6K1 in this inhibitory feedback loop toward the IRS-1–PI3-kinase–Akt pathway in mammalian cells (13, 18, 19, 27). Moreover, S6K1 was found to be highly active in the liver, muscle, and adipose tissue of obese animals, an effect that is paralleled by increased serine phosphorylation of IRS-1 and the absence of Akt activation (26, 27). The underlining mechanism by which S6K1 mediates the nutrient-dependent and rapamycin-sensitive phosphorylation of IRS-1 has become a critical issue in insulin signaling.
To address this question, we designed an S6K1 in vitro kinase assay in which GST-fusion proteins encoding specific domains of IRS-1 were used as substrates. We found that S6K1 purified from insulin- and amino acid-treated cells possesses the ability to phosphorylate multiple residues located within distinct domains of IRS-1. Further analysis of S6K1-mediated phosphorylation of the N-terminal domain of IRS-1 by mass spectrometry and Western blotting identified Ser-307 as a direct target of the kinase (F. Tremblay, S. Brûlé, J. Hunter, and A. Marette, unpublished data). These results are in agreement with the finding of Harrington et al. (12), who found that IRS-1 was phosphorylated in vitro by S6K and that mutation of IRS-1 at Ser-302 (S302A) (equivalent to human Ser-307) blunted phosphorylation of its N-terminal portion. Analysis of the middle portion of IRS-1 (GST-IRS-1M) revealed immunoreactivity to an antibody that recognizes IRS-1 when phosphorylated at Ser-636/639 (data not shown). This finding is not surprising given studies showing that nutrient- and hormonal-dependent phosphorylation of these sites is enhanced in obese mice (27), blocked by rapamycin (23, 24, 26), and found to be severely reduced in adipose tissue of S6K1−/− mice (27), as well as after siRNA-mediated knockdown of S6K1 in HeLa cells and L6 myocytes (Fig. 5 C and D). Consistent with these findings, Shah and Hunter (35) found that Ser-636/639 phosphorylation, although not directly mediated by S6K1, depends on S6K1 phosphorylation of IRS-1 in vivo. Employing a rapamycin-resistant form of S6K1, they showed that, in the presense of rapamycin, IRS-1 Ser-636/639 phosphorylation was unaffected, arguing that the kinase-regulating phosphorylation of this site was distinct from mTOR (35). However, others showed that mTOR/raptor directly phosphorylate IRS-1 at Ser-636/639, and that rapamycin inhibits this response even in NIH 3T3 cells expressing a rapamycin-resistant form of S6K1 (36). It also should be noted that overexpression of a kinase-dead mutant of S6K1 blunted IRS-1 Ser-636/639 phosphorylation in 293HEK cells (36), supporting the notion that S6K1, through second-site phosphorylation, can mediate phosphorylation of this site. Clearly, more studies are needed to clarify the mechanisms by which S6K1 facilitates Ser-636/639 phosphorylation by mTOR/raptor and/or another proline-directed kinase.
The major finding of the present study is the identification of a S6K1 phosphorylation site in IRS-1 located at its C-terminal extremity, Ser-1101. Experiments carried out in the skeletal muscle, liver, and adipose cells indicate that phosphorylation of this site depends on insulin stimulation and was modulated by the presence of amino acids. Moreover, we showed that phosphorylation of IRS-1 at Ser-1101 and the activation status of S6K1 in L6 myocytes closely mirrored the in vitro assay findings, in which S6K1 was found to mediate phosphorylation of IRS-1 at Ser-1101 in a rapamycin-sensitive manner. Consistent with this finding, serine-to-alanine mutation of this site markedly reduced phosphorylation of the IRS-1C fragment, in agreement with this residue being the principal site of S6K1 phosphorylation. Mutational analysis further revealed the importance of this site in the development of insulin resistance in vitro because cells expressing a Ser-1101-unphosphorylatable form of IRS-1 showed enhanced insulin-induced tyrosine phosphorylation of IRS-1 and Akt Ser-473 phosphorylation. The mechanism by which phosphorylation of Ser-1101 interferes with its tyrosine phosphorylation remains to be clarified, but one could speculate that it causes a steric hindrance on one or more of many potential tyrosine phosphorylation sites on the C-terminal region of IRS-1, potentially antagonizing the PI3-kinase binding (37). Importantly, improved insulin signaling to Akt in cells expressing the mutant form of IRS-1 (S1101A) occurred despite unchanged phosphorylation at Ser-307, Ser-312, and Ser-636/639, residues known to be associated with insulin resistance (5, 11, 12, 24). These results suggest that phosphorylation of IRS-1 at Ser-1101 by S6K1 contributes to the induction of insulin resistance under conditions in which nutrients are available in excess.
S6K was recently shown by Harrington et al. (12) to phosphorylate an IRS-1 fragment (IRS-1899−1235) encompassing residues 899–1235 of murine IRS-1. However, the in vitro phosphorylation of IRS-1899−1235 was weak compared with the ability of S6K to phosphorylate the IRS-1 fragment (IRS-1108−516) containing Ser-302. In comparing these findings, it is important to note that Harrington et al. used recombinant S6K2 purified from insect cells for the in vitro kinase assays, whereas we used S6K1 immunoprecipitated from insulin- and amino acid-stimulated mammalian cells. We found that S6K1, but not S6K2, mediates phosphorylation at Ser-1101. Given the large numbers of serine and threonine residues within GST-IRS-1C, it is conceivable that S6K1 and S6K2 preferentially target distinct residues within IRS-1.
High-fat feeding in rodents leads to the development of insulin resistance by promoting excessive storage of triglycerides not only in the adipose tissue, but also in the liver and skeletal muscle (38–40). There also is evidence for increased availability of amino acids in skeletal muscle of high-fat fed rats (41). We recently observed that S6K1 was overactivated in the fat, liver, and muscle of obese animals (26, 27) and that deletion of S6K1 in mice protected these animals against diet-induced obesity and the development of insulin resistance (13, 27). We now provide evidence that phosphorylation of IRS-1 at Ser-1101 is increased in the liver of animals rendered obese by increased fat intake, compared with lean, low-fat diet-fed mice. The pathophysiological role of S6K1 in this process is evidenced by the observation that S6K1-deficient mice show no increase in the phosphorylation of IRS-1 at Ser-1101 when fed a high-fat diet, a finding consistent with their improved insulin sensitivity (27). That these effects are mediated in a cell-autonomous manner was shown by the finding of blunted IRS-1 Ser-1101 phosphorylation after S6K1 removal by RNA interference.
We previously reported that phosphorylation of Ser-1101 was induced by “classical” mediators of insulin resistance, such as free fatty acids and TNF-α (10). We further found that PKC-θ was a prominent IRS-1 serine kinase, among other members of the PKC family, and that phosphorylation of IRS-1 at Ser-1101 was reduced in PKC-θ−/− mice (10). These results underscore the importance of Ser-1101 as an IRS-1 phosphorylation site that integrates stress/obesity signals through PKC-θ and nutrient/hormonal inputs by S6K1 to create a state of insulin resistance. In contrast, a recent study failed to observe elevated Ser-1101 phosphorylation in TSC1–TSC2-deficient mouse embryo fibroblasts, where loss of function of the TSC1–TSC2 complex gives rises to a marked and unregulated hyperactivation of the mTOR–S6K1 pathway (35). However, mTOR and S6K1 are constitutively activated in this model, leading to a dramatic hyperphosphorylation of IRS-1 on other serine residues and marked IRS-1 degradation (12, 25, 35). This finding is clearly different from the comparatively modest and regulated overactivation of S6K1 in classical models of insulin resistance, such as the liver and muscle from dietary and genetic models of obesity (Fig. 5 A and B) (26, 27) or in muscle of human subjects infused with amino acids (Fig. 5E). Indeed, in those models, we find no evidence for IRS-1 degradation despite increased phosphorylation of Ser-1101, suggesting that Ser-1101 phosphorylation by S6K1 is an early event in the activation of the negative-feedback loop and potentially not essential for proteasomal degradation of IRS-1.
As in mice, overconsumption of nutrients in humans promotes insulin resistance (13, 14). Early studies also have shown that plasma concentrations of amino acids, particularly branched-chain amino acids, are elevated in obese insulin-resistant subjects (13, 42, 43). Similar to our observation in L6 myocytes, increased amino acid availability in humans during physiological hyperinsulinemia impairs glucose uptake in skeletal muscle (31). In the present report, we explored the possibility that amino acid-induced insulin resistance is linked with S6K1-mediated phosphorylation of IRS-1 at Ser-1101. We found that, although infusion of insulin alone barely increases S6K1 activity, a combination of insulin and amino acids leads to a strong activation of S6K1 and increased phosphorylation of IRS-1 at Ser-1101. These data strongly support our previous results showing that S6K1 is promoting a state of insulin resistance in humans when nutrients are present in excess (32), consistent with our previous findings in mice (27). Whether increased activation of S6K1 is a common feature of human obesity and insulin resistance is currently unknown, but the use of phosphospecific antibodies against IRS-1 (Ser-1101) and S6K1 (Thr-389) as diagnostic tools might prove to be useful to predict and design therapeutic treatments.
In summary, the present study argues that increased amino acid availability leads to insulin resistance in part through promoting S6K1-mediated phosphorylation of IRS-1 at Ser-1101. More importantly, our data strongly suggest that phosphorylation of IRS-1 Ser-1101 by S6K1 is an important causal element involved in the development of insulin resistance in both animals and humans during nutrient satiation.
Methods
GST-Fusion Proteins.
GST-fusion proteins containing rat IRS-1 fragments, GST-IRS-1N (amino acids 2–516), GST-IRS-1M (amino acids 526–859), and GST-IRS-1C (amino acids 900–1235) have been previously described (44). The S1101A mutation was generated by using pGEX-2T/GST-IRS-1 (amino acids 900–1235) as a template and 5′-GAGGCACAGCGCCGAGACCT-TCTCG-3′ and 5′-CGAGAAGGTCTCGGCGCTGTGCCTC-3′ as mutagenic primers. Inserted mutation was confirmed by sequencing.
Antibodies.
Polyclonal antibodies for immunoprecipitation of S6K1, S6K2, and IRS-1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal antibodies for immunoblotting of IRS-1 and S6K1 were obtained from Upstate Biotechnology (Lake Placid, NY). Antibodies against phosphotyrosine, IRS-1 (Ser-307, Ser-312, Ser-636/639, and Ser-1101), S6K1 (Thr-389), and Akt (Ser-473) were purchased from Cell Signaling Technology.
Cell Culture and Treatment.
L6 skeletal muscle cells were cultured in α-MEM with 10% FBS until they reached 70% confluency. To induce differentiation, cells were further cultured in α-MEM containing 2% FBS for 7 days. The 3T3-L1 adipocytes were grown and differentiated as previously described (23). Rat hepatoma Fao cells were grown and maintained in RPMI medium 1640 supplemented with 10% FBS. CHO IR/IRS-1 cells were cultured in DMEM containing 10% FBS and transfected with wild-type HA-IRS-1 or HA-IRS-1 (S1101A) as previously described (10).
RNA Interference.
RNAi duplexes corresponding to human S6K1 (5′-AAGGGGGCTATGGAAAGGTTT-3′) were purified, annealed, and transfected into HeLa cells by using oligofectamine (Invitrogen, Carlsbad, CA). Controls were treated in the same manner, except no siRNAs were transfected. After 60 h, cells were deprived of serum overnight and then were deprived of amino acids for 2 h. At this point, cells were stimulated with amino acids with or without 200 nM insulin for 30 min. The effect of RNAi on S6K1 expression was measured by Western blot analysis.
Mouse Studies.
Animal studies were approved by the Animal Care and Handling Committees of Laval University and the Friedrich Miescher Institute for Biomedical Research (Basel, Switzerland). S6K1−/− mice were generated as previously described (45). At 10 weeks of age, male mice were assigned to either a normal chow diet (KLIBA-NAFAG, diet 3807) or a high-fat diet (Research Diets, diet D12492) for 14 weeks. Liver from 6-h fasted mice were collected and frozen in liquid nitrogen until analysis. Lean Db/Db and obese db/db mice were fasted for 5 h before tail vein injection with saline or insulin (3.8 units/kg). Five min after the injection, the liver was removed and frozen in liquid nitrogen until analysis as previously described (26).
Human Subject and Muscle Biopsies.
Healthy male volunteers were studied twice during infusion of amino acids and control saline infusion in random order as previously described (31).
PI3-Kinase Assay.
PI3-kinase activity was measured in IRS-1 immunoprecipitates as previously described (22).
S6K1/2 Kinase Assay.
S6K1 activity was measured by using S6K1 substrate as previously described (23). In some experiments, S6K1 and S6K2 kinase activities also were measured by using 5 μg of GST-IRS-1C as a substrate.
Statistical Analysis.
The effects of amino acids, insulin, and rapamycin were compared by ANOVA, followed by a least-square means determination. Differences were considered to be statistically significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Attila Brehm and Dr. Elisabeth Bernroider for the human study protocol, Dr. Luce Dombrowski for help with the biochemical analysis of the muscle biopsies, and Patrice Dallaire and Sandra Favret for their expert help with the maintenance and care of db/db and Db/Db mice. This work was supported by a Canadian Institutes of Health Research Investigator award and grant (to A.M.), a National Institutes of Health (NIH) grant (to G.T.), NIH Grant R01 DK73802 (to G.T. and K.M.), a Novartis Institute for Biomedical Research grant (to G.T.), Austrian Science Foundation Grant P15656, a European Foundation for the Study of Diabetes Novo Nordisk Type 2 diabetes grant, the Herzfelder Family Trust (to M.R.), an Austrian Diabetes Association grant (to M.K.), a Canadian Institutes of Health Research studentship (to F.T.), and a Fonds de la Recherche en Santé du Québec National Researcher grant (to A.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706517104/DC1.
References
- 1.Accili D. Diabetes. 2004;53:1633–1642. doi: 10.2337/diabetes.53.7.1633. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd PR, Kahn BB. N Engl J Med. 1999;341:248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 3.Virkamaki A, Ueki K, Kahn CR. J Clin Invest. 1999;103:931–943. doi: 10.1172/JCI6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zick Y. Trends Cell Biol. 2001;11:437–441. doi: 10.1016/s0962-8924(01)02129-8. [DOI] [PubMed] [Google Scholar]
- 5.Aguirre V, Uchida T, Yenush L, Davis R, White MF. J Biol Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- 6.Gao Z, Hwang D, Bataille F, Lefevre M, York D, Quon MJ, Ye J. J Biol Chem. 2002;277:48115–48121. doi: 10.1074/jbc.M209459200. [DOI] [PubMed] [Google Scholar]
- 7.De Fea K, Roth RA. J Biol Chem. 1997;272:31400–31406. doi: 10.1074/jbc.272.50.31400. [DOI] [PubMed] [Google Scholar]
- 8.Liu YF, Paz K, Herschkovitz A, Alt A, Tennenbaum T, Sampson SR, Ohba M, Kuroki T, LeRoith D, Zick Y. J Biol Chem. 2001;276:14459–14465. doi: 10.1074/jbc.M007281200. [DOI] [PubMed] [Google Scholar]
- 9.Ravichandran LV, Esposito DL, Chen J, Quon MJ. J Biol Chem. 2001;276:3543–3549. doi: 10.1074/jbc.M007231200. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Soos TJ, Li X, Wu J, Degennaro M, Sun X, Littman DR, Birnbaum MJ, Polakiewicz RD. J Biol Chem. 2004;279:45304–45307. doi: 10.1074/jbc.C400186200. [DOI] [PubMed] [Google Scholar]
- 11.Ozes ON, Akca H, Mayo LD, Gustin JA, Maehama T, Dixon JE, Donner DB. Proc Natl Acad Sci USA. 2001;98:4640–4645. doi: 10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Um SH, D'Alessio D, Thomas G. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Krebs M, Roden M. Curr Med Chem. 2004;11:901–908. doi: 10.2174/0929867043455620. [DOI] [PubMed] [Google Scholar]
- 15.Hay N, Sonenberg N. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 16.Jaeschke A, Dennis PB, Thomas G. Curr Top Microbiol Immunol. 2004;279:283–298. doi: 10.1007/978-3-642-18930-2_17. [DOI] [PubMed] [Google Scholar]
- 17.Shah OJ, Anthony JC, Kimball SR, Jefferson LS. Am J Physiol. 2000;279:E715–E729. doi: 10.1152/ajpendo.2000.279.4.E715. [DOI] [PubMed] [Google Scholar]
- 18.Harrington LS, Findlay GM, Lamb RF. Trends Biochem Sci. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Shah OJ, Hunter T. Cell Cycle. 2005;4:46–51. doi: 10.4161/cc.4.1.1343. [DOI] [PubMed] [Google Scholar]
- 20.Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, Olefsky JM, Kobayashi M. Mol Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 21.Takano A, Usui I, Haruta T, Kawahara J, Uno T, Iwata M, Kobayashi M. Mol Cell Biol. 2001;21:5050–5062. doi: 10.1128/MCB.21.15.5050-5062.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tremblay F, Marette A. J Biol Chem. 2001;276:38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- 23.Tremblay F, Gagnon A, Veilleux A, Sorisky A, Marette A. Endocrinology. 2005;146:1328–1337. doi: 10.1210/en.2004-0777. [DOI] [PubMed] [Google Scholar]
- 24.Gual P, Gremeaux T, Gonzalez T, Marchand-Brustel Y, Tanti JF. Diabetologia. 2003;46:1532–1542. doi: 10.1007/s00125-003-1223-4. [DOI] [PubMed] [Google Scholar]
- 25.Shah OJ, Wang Z, Hunter T. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 26.Khamzina L, Veilleux A, Bergeron S, Marette A. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- 27.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 28.Carlson CJ, White MF, Rondinone CM. Biochem Biophys Res Commun. 2004;316:533–539. doi: 10.1016/j.bbrc.2004.02.082. [DOI] [PubMed] [Google Scholar]
- 29.Flotow H, Thomas G. J Biol Chem. 1992;267:3074–3078. [PubMed] [Google Scholar]
- 30.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krebs M, Krssak M, Bernroider E, Anderwald C, Brehm A, Meyerspeer M, Nowotny P, Roth E, Waldhausl W, Roden M. Diabetes. 2002;51:599–605. doi: 10.2337/diabetes.51.3.599. [DOI] [PubMed] [Google Scholar]
- 32.Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, Nowotny P, Waldhausl W, Marette A, Roden M. Diabetes. 2005;54:2674–2684. doi: 10.2337/diabetes.54.9.2674. [DOI] [PubMed] [Google Scholar]
- 33.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Genes Dev. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radimerski T, Montagne J, Rintelen F, Stocker H, van der KJ, Downes CP, Hafen E, Thomas G. Nat Cell Biol. 2002;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- 35.Shah OJ, Hunter T. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzatsos A, Kandror KV. Mol Cell Biol. 2006;26:63–76. doi: 10.1128/MCB.26.1.63-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White MF. Am J Physiol. 2002;283:E413–E422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- 38.Kraegen EW, Clark PW, Jenkins AB, Daley EA, Chisholm DJ, Storlien LH. Diabetes. 1991;40:1397–1403. doi: 10.2337/diab.40.11.1397. [DOI] [PubMed] [Google Scholar]
- 39.Storlien LH, James DE, Burleigh KM, Chisholm DJ, Kraegen EW. Am J Physiol. 1986;251:E576–E583. doi: 10.1152/ajpendo.1986.251.5.E576. [DOI] [PubMed] [Google Scholar]
- 40.Storlien LH, Jenkins AB, Chisholm DJ, Pascoe WS, Khouri S, Kraegen EW. Diabetes. 1991;40:280–289. doi: 10.2337/diab.40.2.280. [DOI] [PubMed] [Google Scholar]
- 41.Herrero MC, Remesar X, Blade C, Arola L. Int J Obes Relat Metab Disord. 1997;21:698–703. doi: 10.1038/sj.ijo.0800464. [DOI] [PubMed] [Google Scholar]
- 42.Felig P, Marliss E, Cahill GF., Jr N Engl J Med. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- 43.Felig P. Annu Rev Biochem. 1975;44:933–955. doi: 10.1146/annurev.bi.44.070175.004441. [DOI] [PubMed] [Google Scholar]
- 44.Qiao LY, Goldberg JL, Russell JC, Sun XJ. J Biol Chem. 1999;274:10625–10632. doi: 10.1074/jbc.274.15.10625. [DOI] [PubMed] [Google Scholar]
- 45.Pende M, Kozma SC, Jaquet M, Oorschot V, Burcelin R, Marchand-Brustel Y, Klumperman J, Thorens B, Thomas G. Nature. 2000;408:994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.