Abstract
Most field isolates of the swine pathogen Actinobacillus pleuropneumoniae form tenacious biofilms on abiotic surfaces in vitro. We purified matrix polysaccharides from biofilms produced by A. pleuropneumoniae field isolates IA1 and IA5 (serotypes 1 and 5, respectively), and determined their chemical structures by using NMR spectroscopy. Both strains produced matrix polysaccharides consisting of linear chains of N-acetyl-D-glucosamine (GlcNAc) residues in β(1,6) linkage (poly-β-1,6-GlcNAc or PGA). A small percentage of the GlcNAc residues in each polysaccharide were N-deacetylated. These structures were nearly identical to those of biofilm matrix polysaccharides produced by Escherichia coli, Staphylococcus aureus and S. epidermidis. PCR analyses indicated that a gene encoding the PGA-specific glycoside transferase enzyme PgaC was present on the chromosome of 15 out of 15 A. pleuropneumoniae reference strains (serotypes 1-12) and 76 out of 77 A. pleuropneumoniae field isolates (serotypes 1, 5 and 7). A pgaC mutant of strain IA5 failed to form biofilms in vitro, as did wild-type strains IA1 and IA5 when grown in broth supplemented with the PGA-hydrolyzing enzyme dispersin B. Treatment of IA5 biofilms with dispersin B rendered them more sensitive to killing by ampicillin. Our findings suggest that PGA functions as a major biofilm adhesin in A. pleuropneumoniae. Biofilm formation may have relevance to the colonization and pathogenesis of A. pleuropneumoniae in pigs.
Keywords: Crystal violet, Dispersin B, dspB, pgaABCD
1. Introduction
Surface-associated colonies of bacteria known as biofilms play a role in the pathogenesis of many chronic infections [1]. Bacterial cells in a biofilm are encased in a self-synthesized, extracellular hydrogel matrix that holds the cells together in a mass and firmly attaches the bacterial mass to the underlying surface [2]. This matrix, also referred to as the slime layer, glycocalyx, or extracellular polymeric substance (EPS) matrix, can comprise up to 90 % of the biofilm biomass [3]. In addition to its structural role, the EPS matrix provides biofilm cells with a protected microenvironment containing dissolved nutrients, secreted enzymes, DNA, and bacteriophages. The EPS matrix also contributes to the increased resistance to antibiotics and host defenses exhibited by biofilm cells [4]. Polysaccharide is a major component of the EPS matrix in most bacterial biofilms [2].
Actinobacillus pleuropneumoniae is a member of the Pasteurellaceae, a family of Gram-negative bacteria that includes many important human and animal pathogens. A. pleuropneumoniae colonizes the lungs of pigs and causes the severe and contagious respiratory disease swine pleuropneumonia [5]. Most field isolates of A. pleuropneumoniae form tenacious biofilms on abiotic surfaces in vitro [6]. A. pleuropneumoniae biofilms contain a hexosamine-rich polysaccharide that is functionally and genetically related to extracellular polysaccharide adhesins produced by Escherichia coli, Staphylococcus aureus and S. epidermidis [7]. These polysaccharides, usually referred to as PGA, PNAG or PIA (polysaccharide intercellular adhesin), consist of linear chains of N-acetyl-D-glucosamine (GlcNAc) residues in β(1,6) linkage (hereafter referred to as PGA). Various forms of PGA appear to differ in their molecular weight, in the degree of N-deacetylation of the GlcNAc residues, and in the presence of O-succinate substituents [8–11]. PGA has been shown to play a role in abiotic surface attachment and intercellular adhesion [12–15], protection from host innate defenses including phagocytosis and antimicrobial peptides [16], and virulence [17]. PGA appears to be essential for A. pleuropneumoniae biofilm formation in vitro because biofilms treated with a PGA-hydrolyzing enzyme were efficiently detached from surfaces [7].
The purpose of the present study was to gain better insight into the structural and functional role of PGA in A. pleuropneumoniae biofilm formation. We purified PGA polysaccharides from two biofilm-producing field isolates of A. pleuropneumoniae and determined their chemical structures by using NMR spectroscopy. We also investigated the phylogenetic distribution of PGA biosynthetic genes among 92 A. pleuropneumoniae reference strains and field isolates by using a PCR assay. Finally, we investigated the role of PGA in A. pleuropneumoniae biofilm formation in vitro by using both a PGA mutant strain and a PGA-degrading enzyme. In this report we present the structure of A. pleuropneumoniae PGA along with evidence that PGA mediates intercellular adhesion, biofilm formation and antibiotic resistance in phylogenetically diverse A. pleuropneumoniae strains.
2. Results
2.1 Purification of A. pleuropneumoniae PGA
PGA was purified from two biofilm-positive A. pleuropneumoniae field isolates, IA1 and IA5 (serotypes 1 and 5, respectively). Extraction of IA1 and IA5 biofilms with saline was not sufficient to release PGA from the cells. Sonication, however, liberated large amounts of PGA, which eluted as a single peak on a gel filtration column (Fig. 1). Based on the elution profile, the molecular weight of A. pleuropneumoniae PGA was similar to that of staphylococcal PGA (>20 kDa; [10]). A. pleuropneumoniae PGA was partially soluble in water. Complete solubility of PGA was achieved by solubilizing the samples in a small volume of 5M HCl. GLC analysis revealed that glucosamine was the only monosaccharide component of A. pleuropneumoniae PGA.
Fig. 1.
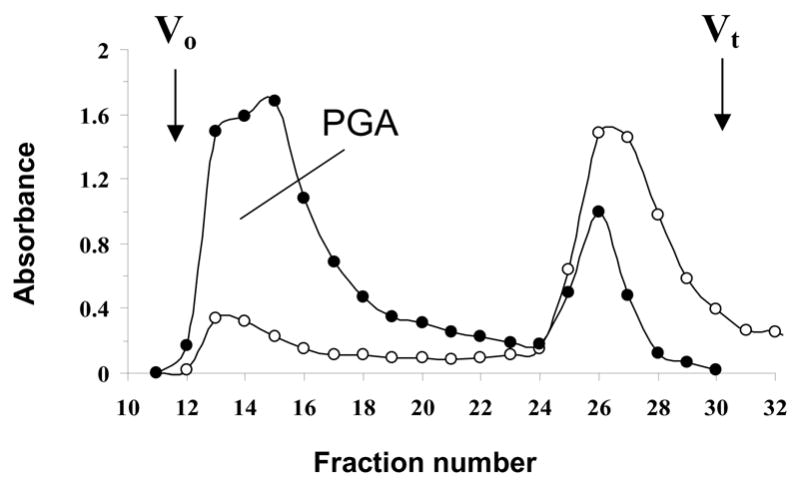
Elution profile of a crude extracellular extract of A. pleuropneumoniae IA1 biofilm cells on a Sephacryl S-300 column irrigated with water. Aliquots (200 μl) of each 5-ml fraction were assayed for neutral sugars (●, A485) and aminosugars (○, A530). Void (Vo) and total (Vt) volumes of the column are indicated with arrows.
2.2 A. pleuropneumoniae PGA is a β(1,6)-linked GlcNAc polymer
Purified PGA polysaccharides were analyzed by 1H-NMR spectroscopy (Fig. 2). The 1H-NMR spectra of PGA from A. pleuropneumoniae IA1 and IA5 (Fig. 2, spectra 1 and 2, respectively) were nearly identical to each other and to that of PGA purified from S. epidermidis strain RP62A (Fig. 2, spectrum 3). In addition, all of the major signals of the β(1,6)-linked GlcNAc residues had shift assignments that were nearly identical to those reported for 1H-NMR spectra of PGA isolated from various other staphylococcal strains and from E. coli [8–11,15,18]. The presence of minor peaks, including one at 2.7 ppm (H2 of GlcNH2), may be due to partial N-deacetylation of GlcNAc residues [8,9]. These results indicate that A. pleuropneumoniae PGA is a linear polymer with the structure →6)-β-GlcNAc-(1→.
Fig. 2.
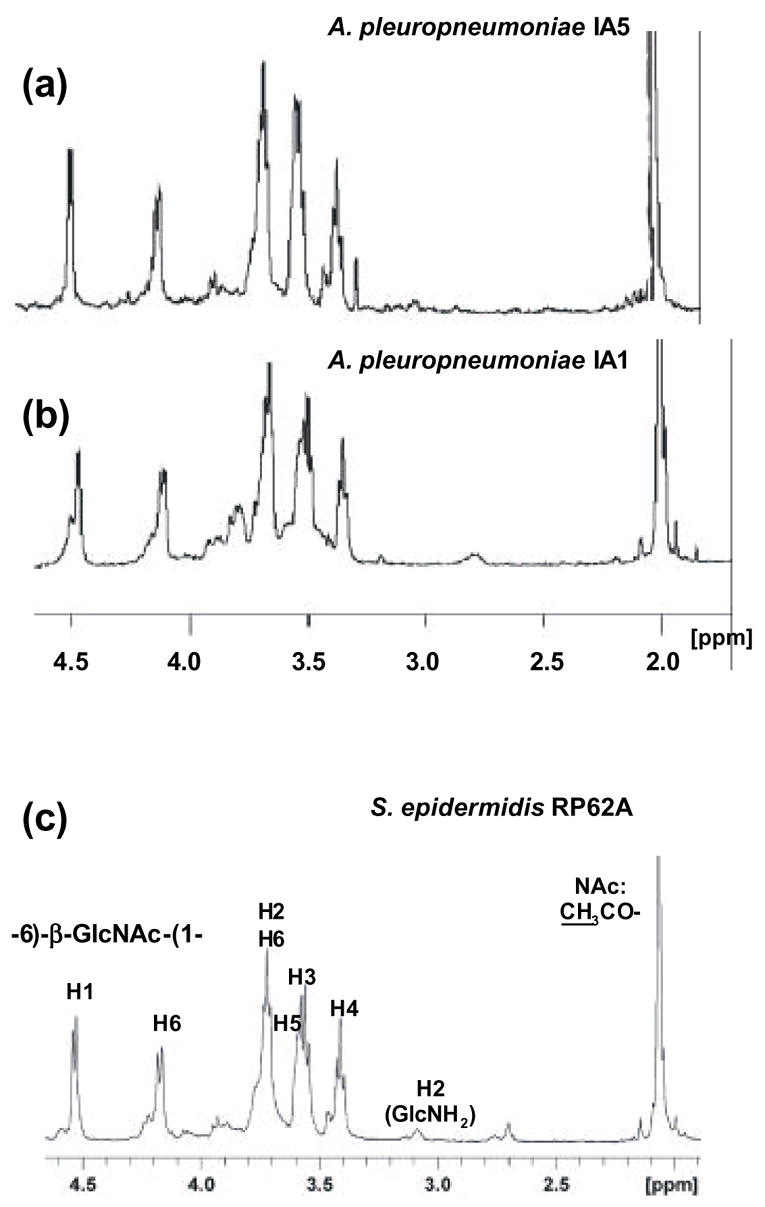
1H NMR spectra of PGA from of A. pleuropneumoniae strains IA5 (a), A. pleuropneumoniae strain IA1 (b), and PGA from S. epidermidis RP62A (c). The spectrum in panel (c) is from [8]. Peak assignments are according to Mack et al. [9].
The chemical structure of A. pleuropneumoniae PGA was confirmed by 2D-NMR spectroscopy (Fig. 3). 1H and 13C chemical shifts of both GlcNH2 and GlcNAc residues (Table 2) closely corresponded to those reported for PGA purified from S. aureus strain MN8m [8]. PGA from strain IA1 showed more heterogeneity than PGA from IA5 due to a higher degree of N-deacetylation, which was evident by the relative intensity of the peak at 2.7 ppm. According to the NMR data, ~25% of the total glucosamine residues in this IA1 PGA preparation were N-deacetylated. This result was in good agreement with the value obtained colorimetrically using the Smith and Gilkerson method (22 ± 2%). The degree of N-deacetylation in PGA from strain IA5 varied from 1–16%, depending on the PGA preparation. No evidence for partial O-succinate substitution of A. pleuropneumoniae PGA was found.
Fig. 3.
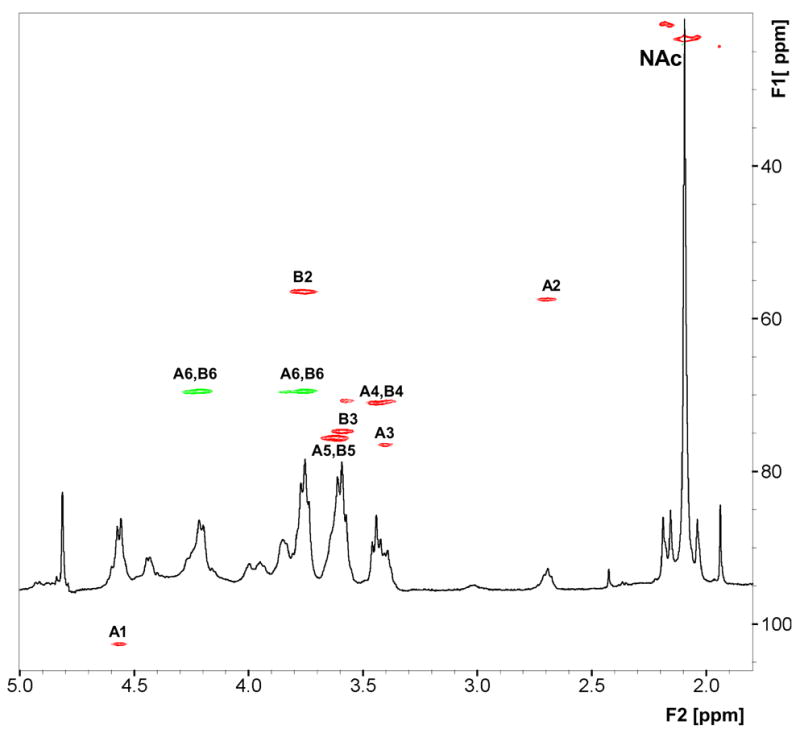
Partial 500-MHz heteronuclear 1H-13C chemical shift correlation (HSQC) spectrum of A. pleuropneumoniae IA1 PGA in D2O at 25°C. Correlation peaks are labeled with residue identification of β-GlcNH2 (A) and β-GlcNAc (B), as in [9].
Table 2.
1H and 13C-NMR chemical shifts of A. pleuropneumoniae IA1 PGA. NAc:CH3 signals were at 2.09 (1H) and 23.3 (13C) ppm.
| Residue | Nucleus | 1 | 2 | 3 | 4 | 5 | 6a | 6b |
|---|---|---|---|---|---|---|---|---|
| β-GlcNH2 A | 1H | 4.54 | 2.69 | 3.40 | 3.39 | 3.62 | 3.75 | 4.21 |
| 13C | 102.3 | 57.4 | 76.5 | 70.8 | 75.6 | 69.4 | ||
| β-GlcNAc B | 1H | 4.54 | 3.76 | 3.58 | 3.44 | 3.62 | 3.75 | 4.21 |
| 13C | 102.3 | 56.4 | 74.7 | 71.0 | 75.6 | 69.4 |
2.3 PGA biosynthetic genes are widespread among A. pleuropneumoniae strains
The A. pleuropneumoniae pgaC gene encodes an integral membrane glycoside transferase enzyme (PgaC) that catalyzes the polymerization of PGA from UDP-GlcNAc monomers [7]. We used a PCR assay to identify pgaC in 15 A. pleuropneumoniae reference strains and 77 A. pleuropneumoniae field isolates (Table 1). Genomic DNA isolated from all 15 reference strains (serotypes 1–12) and from 76 out of 77 field isolates (serotypes 1, 5 and 7) produced PCR products of the expected size when amplified with pgaC-specific PCR primers (data not shown).
Table 1.
A. pleuropneumoniae strains.
| Strain | Relevant characteristicsa | Biofilm phenotypeb | pgaC genotypeb | Source or referencec |
|---|---|---|---|---|
| Reference strains | ||||
| 27088 | Serotype 1A | − | + | ATCC |
| ISU158 | Serotype 1B | − | + | [35] |
| 27089 | Serotype 2 | − | + | ATCC |
| 27080 | Serotype 3 | − | + | ATCC |
| 33378 | Serotype 4 | − | + | ATCC |
| ISU178 | Serotype 5 | − | + | [35] |
| K-17 | Serotype 5A | − | + | [35] |
| L20 | Serotype 5B | + | + | [35] |
| 33590 | Serotype 6 | − | + | ATCC |
| ISU63 | Serotype 7 | − | + | [35] |
| 405 | Serotype 8 | − | + | [35] |
| CVJ1326 | Serotype 9 | − | + | [35] |
| 13038 | Serotype 10 | − | + | [35] |
| 56513 | Serotype 11 | + | + | [35] |
| 1096 | Serotype 12 | − | + | [35] |
| Field isolates | ||||
| IA1 | Serotype 1 | + | + | [7] |
| IA5 | Serotype 5 | + | + | [7] |
| Strain collection | 77 field isolatesd | (53) | (99) | [6] |
| Mutant strains | ||||
| IA5N | Spontaneous Nalr mutant of IA5 | + | + | This study |
| EI1001 | IA5N (pgaC::nadV) | − | − | This study |
| EI1005 | IA5N (pgaC::nadV) | − | − | This study |
Nalr, nalidixic acid resistant
Numbers in parentheses indicate the percentage of positive strains. Biofilm phenotypes of reference strains and filed isolates are from [6].
ATCC, American Type Culture Collection
Serotypes 1, 5 and 7
2.4 An A. pleuropneumoniae pgaC mutant is deficient in PGA production and biofilm formation
We used natural transformation to construct an isogenic mutant of A. pleuropneumoniae IA5N that contained Haemophilus ducreyi nadV inserted into the middle of pgaC on the chromosome. This mutant strain (named EI1001) produced white colonies on Congo red agar, as opposed to the red colonies produced by wild-type strain IA5. Congo red binding has been correlated with PGA production in other bacteria [7,19]. EI1001 cells also released significantly less total hexosamine when treated with the PGA-hydrolyzing enzyme dispersin B compared to the amount released by wild-type cells (<1 μMole of total hexosamine per mg of wet cells for pgaC mutant IA1001 versus ~100 μMoles/mg for wild-type strain IA5). Strain EI1001 was also completely deficient in biofilm formation when tested in a microtiter plate assay (Fig. 4A). We attempted to genetically complement the pgaC mutation in EI1001 with pVK93, a broad-host-range plasmid that contains the A. pleuropneumoniae pgaCD genes located downstream from an IPTG-inducible tac promoter [7]. Plasmid pVK93 was previously shown to complement pgaC mutations in A. actinomycetemcomitans and E. coli [7]. Although pVK93 replicated in A. pleuropneumoniae, it did not restore the ability of strain EI1001 to synthesize PGA or to produce biofilms. This result could be due to poor expression of the pVK93 tac promoter in A. pleuropneumoniae [20]. To confirm that the biofilm-negative phenotype in strain EI1001 was due to the nadV insertion and not to mutations in other locations on the chromosome, we isolated genomic DNA from EI1001 and used it to transform A. pleuropneumoniae IA5N to NAD-independence. The resulting transformant (EI1005) was also deficient in biofilm formation (Fig. 4A). These results are consistent with the hypothesis that pgaC expression is essential for PGA production and biofilm formation in A. pleuropneumoniae.
Fig. 4.
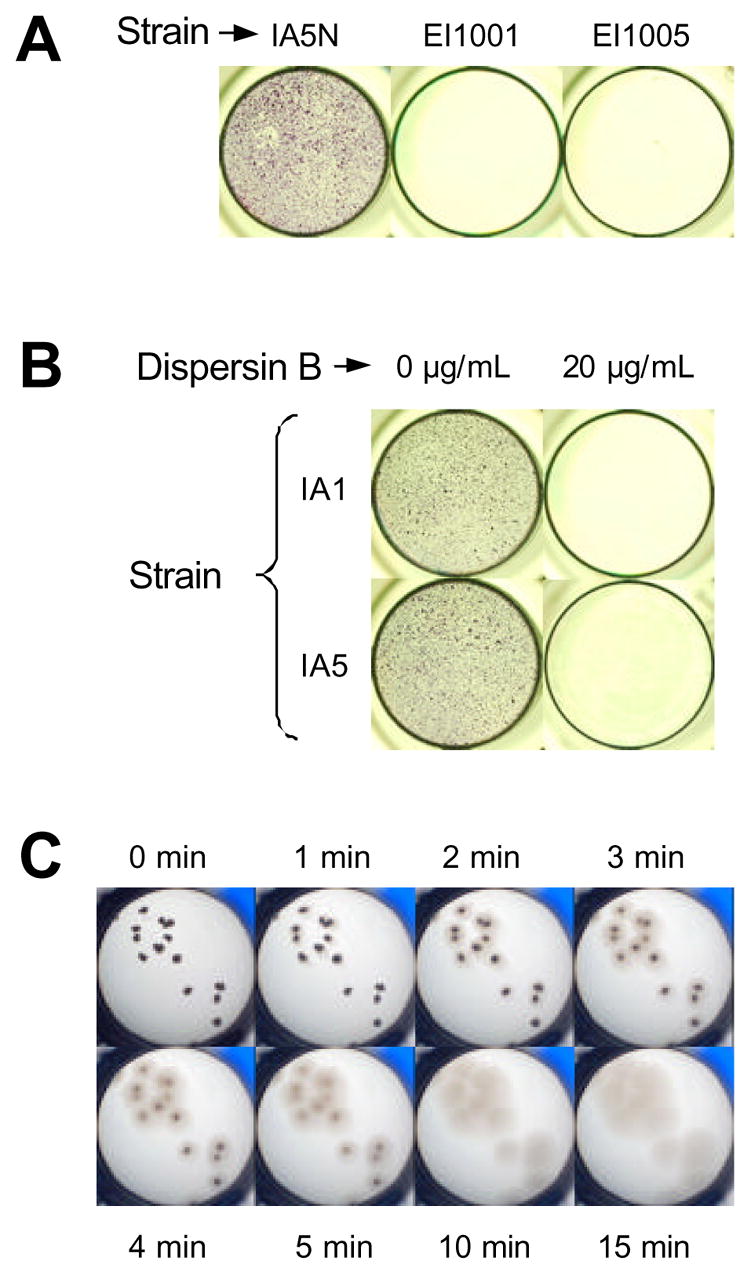
Growth and detachment of A. pleuropneumoniae biofilms in 96-well microtiter plates. (A) Growth of biofilm-forming strain IA5N and isogenic pgaC mutants EI1001 and EI1005 in MHB. (B) Growth of wild-type strains IA1 and IA5 in MHB containing 0 or 20 μg/ml of dispersin B. Biofilms in panels A and B were stained with crystal violet. (C) Detachment of IA5 biofilm colonies by dispersin B. A well containing a small number of biofilm colonies (dark spots) was aspirated and filled with 100 μL of dispersin B (20 μg/mL in PBS) and photographed at the indicated times.
2.5 Depolymerization of A. pleuropneumoniae PGA inhibits biofilm formation and increases antibiotic sensitivity
To confirm that PGA mediates A. pleuropneumoniae biofilm formation, we grew wild-type strains IA1 and IA5 in 96-well microtiter plates in broth containing 0 or 20 μg/ml of dispersin B. The presence of dispersin B in the broth completely inhibited biofilm formation by both strains (Fig. 2B). Concentrations of 2 and 0.2 μg/mL of dispersin B also completely inhibited A. pleuropneumoniae biofilm formation (data not shown). Fig. 2C shows live, 24-h old biofilm colonies of strain IA5 that were treated with dispersin B. The biofilm colonies began to dissolve almost immediately after addition of the enzyme and were completely detached by gentle rinsing after 2 min of treatment. Control experiments showed that dispersin B dispersed IA5 biofilm into uniformly turbid cell suspensions but did not lyse the cells (data not shown). This phenotype was similar to the detachment phenotype exhibited by E. coli and S. epidermidis biofilms treated with dispersin B [21,22].
We also measured the effect of PGA depolymerization on the sensitivity of IA5 biofilms to killing by ampicillin (Fig. 5). Biofilms were grown in polystyrene microcentrifuge tubes for 18 h. The broth was then removed and replaced with fresh broth containing 0, 10 or 100 μg/mL of ampicillin and 0 or 20 μg/mL of dispersin B. After 4 h the number of CFUs per tube was determined. Biofilms treated with ampicillin exhibited a significant dose-dependent decrease in the number of CFUs/tube when compared to biofilms treated with unsupplemented broth (P < 0.02). Also, biofilms treated with ampicillin plus dispersin B exhibited a significant decrease in the numbers of CFUs/tube when compared to biofilms treated with ampicillin alone (P < 0.02). There was not a significant difference between the CFUs/tube in tubes treated with 100 μg/mL of ampicillin and in tubes treated with 10 μg/mL of ampicillin plus dispersin B, indicating that A. pleuropneumoniae biofilm cells were resistant to approximately 10-fold higher concentrations of ampicillin than were planktonic cells.
Fig. 5.
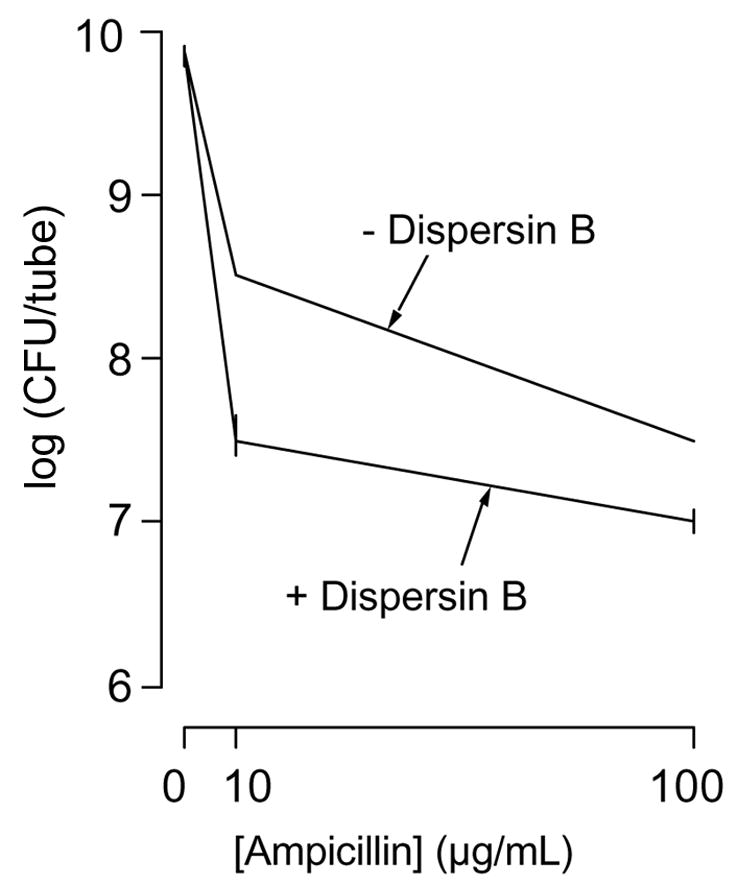
Killing of A. pleuropneumoniae biofilms by ampicillin. Biofilms were grown for 18 h in MHB and then for 4 additional h in MHB with 0, 10 or 100 μg/ml of ampicillin and 0 or 20 μg/ml of dispersin B. CFUs were determined by dilution plating. Values indicate the mean number of CFUs/tube. Error bars indicate range.
3. Discussion
PGA polysaccharides were isolated from biofilms produced by two field isolates of A. pleuropneumoniae, IA1 and IA5. Structural analyses indicated that A. pleuropneumoniae PGA is a linear polymer of GlcNAc residues in β(1,6) linkage (Fig. 2 and 3). Approximately 1–25% of the GlcNAc residues were N-deacetylated depending on the strain and PGA preparation. This structure was identical to that reported for staphylococcal PGA [9], except that A. pleuropneumoniae PGA did not contain O-succinate substitutions. Varying degrees of N-deacetylation in different staphylococcal PGA preparations have been reported: 20% in S. epidermidis RP62A [9]; 43% in S. aureus MN8m [8]; and 2–20% in clinical staphylococcal strains [11]. Although E. coli PGA was shown to be a linear polymer of β(1,6)-linked GlcNAc residues [15], no NMR or chemical evidence for partial N-deacetylation of E. coli PGA was reported.
In S. epidermidis, N-deacetylation of PGA is catalyzed by IcaB, a protein encoded by the staphylococcal PGA biosynthetic operon (icaADBC) [23]. A S. epidermidis icaB mutant that produced fully-acetylated PGA exhibited reduced biofilm formation, immune evasion, and virulence, which suggests that N-deacetylation is crucial for PGA function [24]. In addition, N-deacetylated PGA was shown to be the major epitope in the human antibody response to S. aureus in cystic fibrosis patients [25], and antiserum raised against N-deacetylated PGA, but not against fully-acetylated PGA, was effective at clearing S. aureus in a mouse bacteremia model [26]. Interestingly, the PGA biosynthetic operon of A. pleuropneumoniae (pgaABCD) encodes a protein (PgaB) that exhibits homology to IcaB and to several other polysaccharide N-deacetylases [7]. The N-terminal half of the PgaB polypeptide comprises the deacetylase domain and the C-terminal half comprises a novel domain of unknown function. These observations suggest that PgaB may be a bifunctional PGA-specific N-deacetylase, and that N-deacetylation may be required for biological activity of PGA in A. pleuropneumoniae.
We found that the PGA biosynthetic gene pgaC was present in 15 out of 15 A. pleuropneumoniae reference strains and 76 out of 77 A. pleuropneumoniae field isolates. Previous studies showed that only 2 of these 15 reference strains and 41 of these 77 field isolates exhibited biofilm formation when tested in polystyrene tube and microtiter plate assays [6]. One explanation for this discrepancy may involve the fact that A. pleuropneumoniae strains can undergo a spontaneous and irreversible transition from a biofilm-positive to a biofilm-negative phenotype upon subculture in the laboratory [6]. The genetic mechanism of this phenotypic switch is unknown. However, the closely related bacterium A. actinomycetemcomitans undergoes a similar irreversible switch from a biofilm-positive to a biofilm-negative phenotype upon continuous subculture [27], and this phenotypic switch in A. actinomycetemcomitans involves the down-regulation of PGA production (J.B. Kaplan, unpublished). A. pleuropneumoniae strains that exhibit a biofilm-negative phenotype may have undergone a similar down-regulation of PGA production. This hypothesis is supported by the fact that the frequency of the biofilm-negative phenotype is higher among reference strains, which are likely to have been subcultured more frequently than field isolates.
Our findings suggest that PGA functions as a major biofilm matrix adhesin in A. pleuropneumoniae. PGA appears to be essential for A. pleuropneumoniae biofilm formation because PGA mutants were completely deficient in biofilm formation (Fig. 4A), and depolymerization of PGA by dispersin B inhibited biofilm formation by wild-type strains (Fig. 4B) and caused the rapid detachment and disaggregation of preformed wild-type biofilms (Fig. 4C). Pretreatment of A. pleuropneumoniae biofilms with dispersin B rendered them more sensitive to killing by ampicillin (Fig. 5), indicating that A. pleuropneumoniae biofilm cells exhibit increased resistance to antibiotics compared to the resistance exhibited by planktonic cells.
4. Materials and methods
4.1 Bacterial strains, media and growth conditions
The A. pleuropneumoniae strains used in this study are listed in Table 1. Strain IA5N was isolated as a spontaneous nalidixic acid-resistant mutant of strain IA5 after plating of dense suspensions of cell on TSA containing nalidixic acid. Strain IA5N exhibits the same PGA production and biofilm formation phenotypes as strain IA5. Bacteria were grown in Mueller-Hinton broth (MHB) or on Trypticase Soy agar (TSA) or Brain Heart Infusion (BHI) agar. MHB and TSA were supplemented with 6 g of yeast extract and 8 g of glucose per liter. Except where indicated, all media were also supplemented with 10 mg of NAD per liter. For Congo red binding assays, TSA plates were further supplemented with 0.1 % (w/v) of Congo red dye. Strains were passaged twice weekly on TSA. All cultures were incubated statically at 37 °C.
4.2 Preparation of inocula
A 100-mm-diam tissue-culture-treated polystyrene Petri dish (Corning no. 430167) containing 20 mL of broth was inoculated with a single colony from a TSA plate and incubated for 24 h. The broth was discarded and the biofilm was rinsed with fresh broth, scraped from the surface with a cell scraper, and resuspended in 3 mL of fresh broth. The cells were vortexed for 15 s and then incubated statically for 5–10 min to allow the clumps to settle. The upper layer was transferred to a new tube and vortexed briefly. The resulting inoculum contained >108 CFU/mL.
4.3 Large-scale biofilm cultures
Bacteria were grown in 150-mm-diam tissue-culture-treated polystyrene petri dishes (Corning no. 430199) or in 3 L borosilicate glass (Pyrex) Erlenmeyer flasks containing MHB (60 mL for dishes and 400 mL for flasks). For each purification, 10 dishes or 3 flasks were employed. Culture vessels were inoculated with a 0.2 % volume of inoculum and incubated 18–24 h. Biofilms were washed with 0.9 % (w/v) NaCl, detached from the surface using a cell scraper (for dishes) or 6-mm-diam glass beads (for flasks), and harvested by centrifugation (2,200 g, 4 °C, 15 min). Cell pellets were processed immediately after harvesting.
4.4 Purification of A. pleuropneumoniae PGA
The cell pellet (approx. 1.5 g of wet cells) was resuspended in 40 mL of 0.9 % NaCl. The suspension was sonicated twice for 30 s on ice using an Ikasonic sonicator (IKA Labortechnik) set to 30 % amplitude and 50 % duty cycle. The sonicate was clarified twice by centrifugation (2,200 g, 4 °C, 15 min, then 8,700 g, 4 °C, 10 min), and the supernatant was concentrated using an Amicon 10-kDa cutoff ultrafiltration cell. TCA was added to a concentration of 5 % and the precipitate was removed by centrifugation (8,700 g, 4 °C, 10 min). The deproteinated extract was fractionated on a Sephacryl S-300 column (1-cm × 90-cm; Pharmacia) irrigated with water. Fractions (5 mL) were assayed colorimetrically for aldoses [28] and amino sugars [29]. Amino sugar-rich fractions corresponding to PGA were pooled and lyophilized. The average yield was approximately 4 mg of PGA per g of wet cells.
4.5 Structural analyses
For NMR analysis, 3 mg of lyophilized PGA was dissolved in 30 μL of 20 % (w/v) DCl/D2O and the total volume was adjusted to 1 mL with D2O. NMR spectra were recorded at 25 °C in D2O with a Varian Unity Inova 500 MHz spectrometer using acetone as internal reference (1H, δ 2.225 ppm, 13C, δ 31.5 ppm). Varian standard programs COSY and HSQC were used. The degree of N-acetylation of PGA was estimated from the ratio of GlcNAc/GlcNH2 CH-2 signal intensities in the HSQC spectrum. N-deacetylation of PGA was also assessed colorimetrically by dissolving lyophilized PGA samples in 5 M HCl, adjusting the concentration to 0.5 M HCl, and then measuring the amount of total hexosamine and nonacetylated hexosamine as described by Smith and Gilkerson [30], with and without heating at 110 °C for 2 h, respectively. Absorbance (A260, A485, A530 and A650) was measured using 1-cm semi-micro cuvettes (Kartell) and a Helios β spectrophotometer (Unicam).
4.6 Other carbohydrate techniques
Polysaccharides were converted to alditol acetates by conventional methods following hydrolysis with 4 M trifluoroacetic acid at 120 °C for 2 h. Monosaccharides were identified by GLC with a Shimatzu GC-14 gas chromatograph equipped with a flame ionization detector and a Zebron ZB-5 capillary column (30-m × 0.25-mm, Phenomenex) with hydrogen as a carrier gas and an initial programmed temperature of 170 °C for 3 min followed by an increase to 260 °C at a rate of 5 °C/min. The amount of total hexosamine released from dispersin B-treated cells was measured as previously described [7].
4.7 PGA-specific PCR assay
A small loopful of cells from a 24-h old BHI agar plate (approx. 5 mg) was transferred to a microcentrifuge tube containing 100 μL of sterile water. Cells were disrupted by vortex agitation and then incubated at 100°C for 10 min to lyse the cells. Tubes were centrifuged for 5 min to pellet cell debris and the supernatant was transferred to a new tube. A total of 5 μL of lysate was amplified by PCR using primers 5-TTTATTGGGCTTTTGCA-3 and 5-CTCCAATACAGCCAAGG-3, which hybridize to bp 234–256 and 876–896, respectively, in 1,236 bp A. pleuropneumoniae pgaC coding region (bp 1-1,236; GenBank accession no. AY618480). The PCR conditions and the sequences of 16 S rRNA-specific PCR primers used as a control were described previously [31]. PCR products were electrophoresed on agarose gels and visualized by staining with ethidium bromide. The expected sizes of the PCR products were 663 bp for the pgaC-specific primers and 478 bp for the 16 S rRNA-specific primers.
4.8 Construction of PGA mutant strains
Genomic DNA isolated from A. pleuropneumoniae IA5 was amplified by PCR using primers 5-GTCACGAATTCTTATTGGGCTTTTGCAGGGC-3 and 5-CTCAGGGTACCTTCGGACACTTGTAAGCGTCC-3, which hybridize to bp 57–76 and 529–549, respectively, in A. pleuropneumoniae pgaC (accession no. AY618480). The resulting PCR product (525 bp) was digested with EcoRI and KpnI (recognition sequences underlined) and ligated into the EcoRI/KpnI sites of pC18KnadV, which contains the H. ducreyi nadV gene with a constitutive Tn903 kanamycin promoter ligated into the PstI site of pUC18 [32]. The nadV gene of pC18KnadV confers NAD independence in A. pleuropneumoniae [33]. The resulting plasmid (pVK208) contained the 5′ half of A. pleuropneumoniae pgaC located upstream from nadV. The 3′ half of A. pleuropneumoniae pgaC was amplified by PCR using primers 5-GACTCGCATGCCGCAATGTTTAATGGGAACC-3 and 5-CTGAGAAGCTTCCAATACAGCCAAGGATACC-3, which hybridize to bp 581–600 and 1,118–1,137, respectively, in A. pleuropneumoniae pgaC (accession no. AY618480). The PCR product (579 bp) was digested with SphI and HindIII and ligated into the SphI/HindIII sites of pVK208. The resulting plasmid (pVK215) contained the nearly complete pgaC coding region interrupted by a 1.6 kb nadV gene that was transcribed in the same orientation as pgaC. Plasmid pVK215 DNA (linearized with ScaI) was used to transform strain IA5N to NAD independence using natural transformation [34]. Transformant were selected on TSA without NAD. Six transformants were obtained, all of which contained nadV inserted into pgaC on the chromosome as determined by PCR using primers that hybridize to nadV and to DNA sequences outside the homologous regions in pVK215. For genetic complementation, plasmid pVK93, which contains the A. pleuropneumoniae pgaCD genes downstream from an IPTG-inducible tac promoter, was conjugated from E. coli strain C600 into the mutant strains using the helper plasmid pRK21761 as previously described [7]. Transconjugants were plated on TSA containing 3 μg/mL of chloramphenicol and 20 μg/mL of nalidixic acid.
4.9 Biofilm growth and detachment assays
Biofilms were grown in 96-well tissue-culture-treated polystyrene microtiter plates (Falcon no. 324662). Wells were filled with 100 μL of inoculum (diluted 1:1,000 in MHB) and incubated for 12–16 h. For genetic complementation experiments, broth was supplemented with 3 μg/mL of chloramphenicol and 1 μg/mL of IPTG. For inhibition studies, media were supplemented with 0.2–20 μg of A. actinomycetemcomitans dispersin B per mL (ca. 103 units per mg of protein; [7]). For detachment studies, bacteria were grown in broth without dispersin B and the wells were then washed with phosphate buffered saline (PBS) and filled with 100 μL of PBS containing 0 or 20 μg/mL of dispersin B. After incubation at 37 °C for 5 min, the wells were rinsed with water and stained for 1 min with 100 μL of crystal violet solution (Fisher no. 23255960). Wells were then rinsed, dried and photographed. All assays were performed in duplicate or triplicate wells, which exhibited minimal variation. All assays were performed on at least three separate occasions with similar results.
4.10 Biofilm killing assay
Biofilms were grown in 1.5-mL polypropylene microcentrifuge tubes (Sarstedt). Tubes were filled with 200 μL of inoculum (diluted 1:1,000 in fresh broth). After 16 h the broth was aspirated and replaced with fresh broth containing 0, 10 or 100 μg/mL of ampicillin and 0 or 20 μg/mL of dispersin B. After 4 h the cells were pelleted and rinsed with sterile PBS three times to remove the ampicillin. Cell pellets were resuspended in 200 μL of PBS containing 20 μg/mL of dispersin B and incubated for 5 min to disaggregate the cells. Tubes were vortexed briefly and the number of CFUs/tube was determined by dilution plating. Killing assays were performed in duplicate tubes and on two separate occasions with similar results. The significance of differences between groups was determined by a one-way ANOVA test.
Acknowledgments
We thank Robin Kastenmayer (Michigan State University) for providing plasmid pC18KnadV; David Furgang (New Jersey Dental School) for help with the statistical analyses; and Leann MacLean (National Research Council), Rhiannon LaVeque (Michigan State University), and Jason Vladimer and Arianita Mulahu (New Jersey Dental School) for technical assistance. This work was supported by grants DE15124 and DE16291 from the United States Public Health Service, grant 2002-35204-11622 from the United States Department of Agriculture, the Center for Microbial Pathogenesis at Michigan State University, and the Regional Council of Nord-Pas-de-Calais (France).
Abbreviations
- BHI
Brain Heart Infusion
- EPS
extracellular polymeric substance
- GlcNAc
N-acetyl-D-glucosamine
- GlcNH2
glucosamine
- MHB
Mueller-Hinton broth
- PGA
poly-β-1,6-N-acetyl-D-glucosamine
- PBS
phosphate buffered saline
- TSA
Tryptic Soy agar
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland IW. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 3.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–6. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Haesebrouck F, Chiers K, Van Oberbeke I, Ducatelle R. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet Microbiol. 1997;58:239–49. doi: 10.1016/s0378-1135(97)00162-4. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan JB, Mulks MH. Biofilm formation is prevalent among field isolates of Actinobacillus pleuropneumoniae. Vet Microbiol. 2005;108:89–94. doi: 10.1016/j.vetmic.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–20. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce JG, Abeygunawardana C, Xu Q, Cook JC, Hepler R, Przysiecki CT, et al. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res. 2003;338:903–22. doi: 10.1016/s0008-6215(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 9.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–83. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maira-Litrán T, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, Goldmann DA, et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–40. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadovskaya I, Chaignon P, Kogan G, Chokr A, Vinogradov E, Jabbouri S. Carbohydrate-containing components produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol Med Microbiol. 2006;47:75–82. doi: 10.1111/j.1574-695X.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- 12.Agladze K, Wang X, Romeo T. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J Bacteriol. 2005;187:8237–46. doi: 10.1128/JB.187.24.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–91. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 14.McKenney D, Hubner J, Muller E, Wang Y, Goldmann DA, Pier GB. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun. 1998;66:4711–20. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–34. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–75. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 17.Kropec A, Maira-Litrán T, Jefferson KK, Grout M, Cramton SE, Götz F, et al. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun. 2005;73:6868–76. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain Staphylococcus epidermidis RP62A. Infect Immun. 2005;73:3007–17. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39:2151–6. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West SE, Romero MJ, Regassa LB, Zielinski NA, Welch RA. Construction of Actinobacillus pleuropneumoniae-Escherichia coli shuttle vectors: expression of antibiotic-resistance genes. Gene. 1995;160:81–6. doi: 10.1016/0378-1119(95)00236-y. [DOI] [PubMed] [Google Scholar]
- 21.Itoh Y, Wang X, Hinnebusch BJ, Preston JF, 3rd, Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–7. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004;48:2633–6. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Götz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–93. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 24.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–6. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 25.Kelly-Quintos C, Kropec A, Briggs S, Ordonez CL, Goldmann DA, Pier GB. The role of epitope specificity in the human opsonic antibody response to the staphylococcal surface polysaccharide poly N-acetyl glucosamine. J Infect Dis. 2005;192:2012–9. doi: 10.1086/497604. [DOI] [PubMed] [Google Scholar]
- 26.Maira-Litrán T, Kropec A, Goldmann DA, Pier GB. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or N-deacetylated staphylococcal poly-N-acetyl-beta-(1-6)-glucosamine. Infect Immun. 2005;73:6752–62. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fine DH, Furgang D, Schreiner HC, Goncharoff P, Charlesworth J, Ghazwan G, et al. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145:1335–47. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 28.Dubois M, Gilles KA, Hamilton JF, Rebers PA, Smyth F. Colorimetric methods for determination of sugars and related substances. Anal Biochem. 1956;28:350–6. [Google Scholar]
- 29.Enghofer E, Kress H. An evaluation of the Morgan-Elson assay for 2-amino-2-deoxy sugars. Carbohydr Res. 1979;76:233–8. doi: 10.1016/0008-6215(79)80022-1. [DOI] [PubMed] [Google Scholar]
- 30.Smith RL, Gilkerson E. Quantitation of glycosaminoglycan hexosamine using 3-methyl-2-benzothiazolone hydrazone hydrochloride. Anal Biochem. 1979;98:478–80. doi: 10.1016/0003-2697(79)90170-2. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan JB, Schreiner HC, Furgang D, Fine DH. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J Clin Microbiol. 2002;40:1181–7. doi: 10.1128/JCM.40.4.1181-1187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kastenmayer RJ. PhD Thesis. Michigan State University; 2002. Characterization of the genes of Actinobacillus pleuropneumoniae involved in oxidative stress and pathogenesis. [Google Scholar]
- 33.Martin PR, Shea RJ, Mulks MH. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol. 2001;183:1168–74. doi: 10.1128/JB.183.4.1168-1174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosse JT, Nash JH, Kroll JS, Langford PR. Harnessing natural transformation in Actinobacillus pleuropneumoniae: a simple method for allelic replacements. FEMS Microbiol Lett. 2004;233:277–81. doi: 10.1016/j.femsle.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 35.Cruz WT, Nedialkov YA, Thacker BJ, Mulks MH. Molecular characterization of a common 48-kilodalton outer membrane protein of Actinobacillus pleuropneumoniae. Infect Immun. 1996;64:83–90. doi: 10.1128/iai.64.1.83-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


