Abstract
GTP cyclohydrolase 1 (GCH1) is rate limiting in the provision of the cofactor tetrahydrobiopterin for biosynthesis of catecholamines and NO. We asked whether common genetic variation at GCH1 alters transmitter synthesis and predisposes to disease. Here we undertook a systematic search for polymorphisms in GCH1, then tested variants’ contributions to NO and catecholamine release as well as autonomic function in twin pairs. Renal NO and neopterin excretions were significantly heritable, as were baroreceptor coupling (heart rate response to BP fluctuation) and pulse interval (1/heart rate). Common GCH1 variant C+243T in the 3′-untranslated region (3′-UTRs) predicted NO excretion, as well as autonomic traits: baroreceptor coupling, maximum pulse interval, and pulse interval variability, though not catecholamine secretion. In individuals with the most extreme BP values in the population, C+243T affected both diastolic and systolic BP, principally in females. In functional studies, C+243T decreased reporter expression in transfected 3′-UTRs plasmids. We conclude that human NO secretion traits are heritable, displaying joint genetic determination with autonomic activity by functional polymorphism at GCH1. Our results document novel pathophysiological links between a key biosynthetic locus and NO metabolism and suggest new strategies for approaching the mechanism, diagnosis, and treatment of risk predictors for cardiovascular diseases such as hypertension.
Introduction
GTP cyclohydrolase I (GCH1) catalyzes the first and rate-limiting step in tetrahydrobiopterin (BH4) biosynthesis (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI31093DS1). BH4 is an essential cofactor for several transmitter biosynthetic pathway enzymes, including tyrosine hydroxylase (TH) and NOSs, as well as phenylalanine hydroxylase and tryptophan hydroxylase. Studies in vitro indicate that BH4 levels are tightly linked to hydroxylation of l-tyrosine (to L-DOPA) and BH4 bioavailability modulates catecholamine synthesis (1).
The human GCH1 locus experiences rare natural inactivating mutations with profound consequences: such mutations cause 2 disorders: autosomal dominant hereditary progressive dystonia/DOPA-responsive dystonia (HPD/DRD) and autosomal recessive GCH1-deficient hyperphenylalaninemia (HPA). Biochemical characterizations of these diseases demonstrate lower cerebrospinal fluid levels of BH4, neopterin, and homovanillic acid, with low levels of TH activity in the striatum (2). A mouse model for dominantly inherited GTP cyclohydrolase deficiency also showed low brain levels of BH4, catecholamines, serotonin, and their metabolites together with low levels of TH activity within the striatum (3).
The gaseous transmitter NO, derived from 3 NOS isoforms, plays a key role in vascular tone, and alterations in NO production influence endothelium-dependent vasodilation and BP. Previous studies in cultured cells showed that GCH1 inhibition lowers BH4 levels, leading to decreased NO production (4, 5). Exogenous BH4 in the spontaneously (genetic) hypertensive rat can prevent development of elevated BP (6).
Does more common genetic variation at GCH1 also cause enzymatic activity changes of NOS or TH in humans? Might such genetic variation alter endothelial or autonomic activity and thus predispose to cardiovascular disease? To answer these questions, we explored common allelic variation at the GCH1 locus. To probe the impact of GCH1 variation on disease pathways, we resequenced approximately 1.2 kbp of 5′ promoter as well as all 6 exons and adjacent intronic regions for rare and common variants in DNA from 64 ethnically diverse subjects as well as 171 pairs of twins. The twins were phenotyped for biochemical and physiological traits likely to be influenced by variation in GCH1, including transmitters, baroreceptor indices, and BP. The study tested whether GCH1 single nucleotide polymorphisms (SNPs) influenced human endothelial or autonomic activity. Twin pairs enabled us to study whether GCH1 allelic variation contributed to heritable control of the circulation.
Results
GCH1 genomics: systematic human polymorphism discovery
To identify genetic variants in GCH1 that might alter its function, we resequenced all 6 exons and adjacent intronic regions, as well as approximately 1.2 kbp of 5′ promoter adjacent to exon 1 (Supplemental Table 1) in DNA from 64 unrelated human subjects. Later we resequenced GCH1 from an additional 171 pairs of twins collected as part of our cardiorenal phenotypic studies.
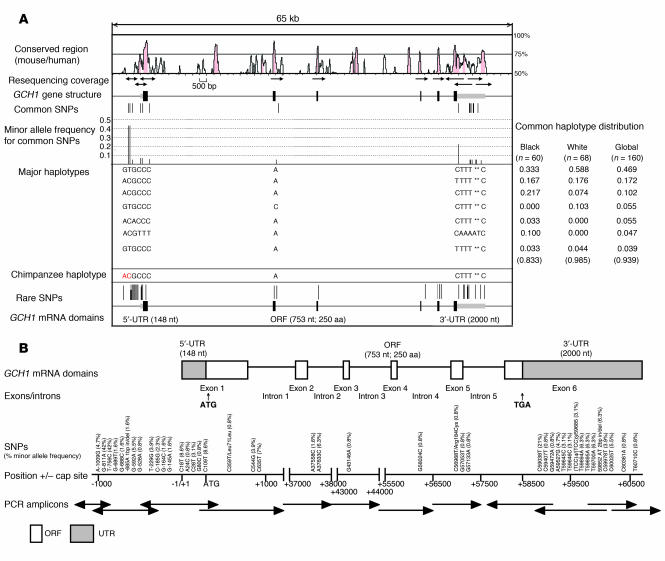
Figure 1A shows sequences conserved between mouse and human GCH1 at the local genomic region resequenced on human chromosome 14q22.1–q22.2. Thirty-nine biallelic SNPs, 2 short (1- to 2-bp) insertion/deletions, and 1 trinucleotide repeat/microsatellite were identified in our initial sample of 64 subjects. Among the 39 SNPs, 11 were located in the approximately 1.2-kbp proximal promoter, 2 in coding regions (1 synonymous, 1 nonsynonymous; both rare), 18 in the untranslated regions (UTRs), and 8 in exon-adjacent intronic regions. Twelve SNPs were common (minor allele frequency ≥5%), including 3 in the GCH1 proximal promoter, 7 in the UTRs, and 2 in the introns. One of the 2 insertion/deletions was located in the proximal promoter, while 1 was in the 3′-UTRs (relatively common, at 6.3%). The trinucleotide repeat/microsatellite located in the 3′-UTRs was rare.
Figure 1. Polymorphism at GCH1: distribution across the gene.
(A) Local genomic region. GCH1 resequencing strategy and identified variants. Sequences conserved between mouse and human GCH1 were visualized with VISTA (http://genome.lbl.gov/vista/index.shtml). Location of common (upper) and rare (lower) SNPs relative to exons and conserved noncoding sequences is indicated by position. Red lines represent nonsynonymous SNPs, while black rods represent synonymous SNPs. Nucleotides in red in the chimpanzee haplotype indicate the minor allele in the human sequence. Computationally reconstructed haplotypes are indicated, along with their relative frequencies in ethnogeographic groups within our sample population. Nucleotide deletions in haplotype sequences are indicated by an asterisk. n = 42 variants were discovered; n = 13 were common (≥5%), while n = 29 were rare (<5%). (B) Functional domains and coding region SNPs. The distribution of variants across GCH1 exons and functional protein domains is illustrated. ATG, translational start codon; Cap, transcriptional initiation site; ORF, open reading frame.
Applying the HAP algorithm (7) to 12 common (minor allele frequency >5%) variants and 1 common insertion/deletion across GCH1, we inferred 12 haplotypes (Figure 1A). The 3 most common haplotypes accounted for 74.3% of chromosomes examined: the most common haplotype (GTGCCCACTTT**C, where ** is the 2-bp AT insertion in exon 6) was frequent (at 58.8%) in the white population but less common (at 33.3%) in samples from black individuals. The third most common haplotype (most likely ancestral, ACGCCCACTTT**C) was more common in black than white subjects (21.7% versus 7.4%).
Figure 1B illustrates GCH1 functional domains and coding region SNPs in the resequenced 64 individuals. Both of the 2 coding region polymorphisms were unusual (minor allele frequency <0.6%), and only 1 specified an amino acid substitution (Arg184Cys).
Supplemental Table 1 presents GCH1 SNP results in the original discovery sample (all 64 individuals unrelated): 39 SNPs, 2 insertion/deletions, and 1 trinucleotide repeat/microsatellite were identified. Of these, 12 were relatively common variants: G–811A at 42.2%, T–756C at 42.2%, G–592A at 5.5%, C18T at 8.6%, C109T at 8.6%, C635T at 7.0%, A37633C at 6.3%, C59038T at 21.1%, T59694A at 6.3%, T59695A at 6.3%, T59700A at 6.3%, the AT 2-bp insertion/deletion (59,892–59,893) at 6.3%, and C60025T at 5.5%. Among the 39 SNPs, 9 were purine/purine (A/G) transitions, and 14 were pyrimidine/pyrimidine (C/T) transitions, while 16 were purine/pyrimidine (R/Y) transversions.
SNP genotype frequencies differed substantially by ethnicity: greater numbers of relatively common (minor allele frequency >5%) SNPs were identified in black (35 such SNPs) than whites individuals (13 such SNPs) (Supplemental Table 1). GCH1 contains a single trinucleotide repeat (microsatellite) polymorphism: (TCC)n at 3–4 copies of the repeat [(TCC)3–4] within the 3′-UTRs; (TCC)4/(TCC)3 heterozygosity was the only diploid genotype observed in these subjects.
Linkage disequilibrium across the GCH1 locus
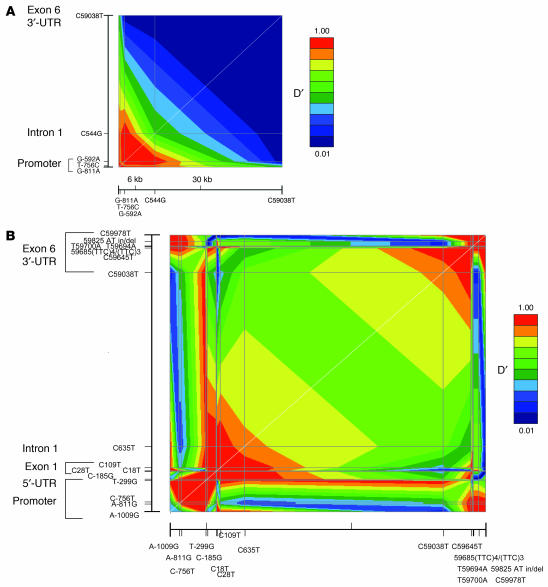
Pairwise linkage disequilibrium (LD) between common SNPs across GCH1 was quantified as parameter D′, scaled from 0 to 1. Figure 2 shows a Graphical Overview of Linkage Disequilibrium (GOLD) plot across GCH1 in subjects of European (Figure 2A) or sub-Saharan African (Figure 2B) ancestry, revealing 2 blocks of particularly high LD (D′ > 0.9) at the 5′ (promoter to intron 1) and 3′ (exon 6) regions of the gene in black individuals. Of note, common polymorphism C59038T in the 3′-UTRs of exon 6 displayed virtually no LD with other, more 5′ variants in subjects of European ancestry (Figure 2A).
Figure 2. Human GCH1 polymorphisms: patterns of LD across the entire GCH1 locus.
Graphical Overview of LD (GOLD) plots of point-by-point linkage disequilibrium (LD). The white diagonal is the line of identity (y = x). (A) European (white) ancestry. Five SNPs with high minor allele frequencies (>5%) spanning the GCH1 locus, and proceeding approximately 1.2 kbp upstream (5′; promoter region). This LD plot was constructed using data from n = 198 individuals (1 MZ twin/pair, both DZ twins/pair, and parents). (B) Sub-Saharan African (black, African-American) ancestry. Sixteen SNPs with high minor allele frequencies (>5%) spanning the GCH1 locus and proceeding approximately 1.2 kbp upstream (5′; promoter region). This LD plot was constructed using data from n = 30 unrelated individuals. in/del, insertion/deletion.
GCH1 haplotypes
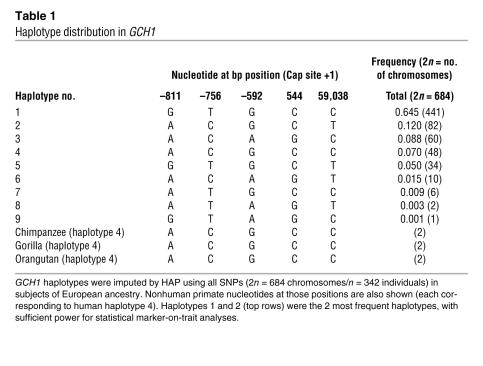
Table 1 describes the GCH1 haplotype distribution in white twins. GCH1 haplotypes were imputed by HAP using all 5 common SNPs across the GCH1 locus in subjects of European ancestry (2n = 684 chromosomes, n = 342 individuals). Nine haplotypes were inferred, with the 2 most common accounting for 76.5% of chromosomes examined. The most common haplotype overall (no. 1, GTGCC) alone accounted for 64.5% of chromosomes.
Twin phenotypes: descriptive statistics
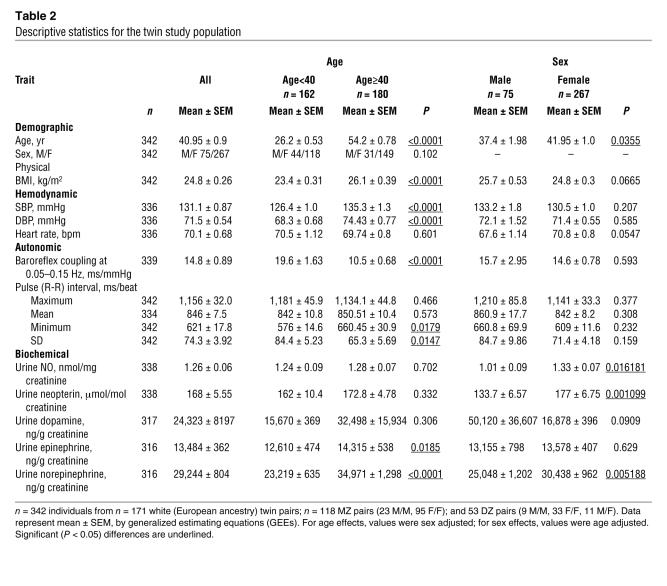
Table 2 describes the twin subject population (n = 342 individuals). Females (n = 264) had higher renal excretion of NO (P = 0.016), neopterin (P = 0.001), and norepinephrine (P = 0.0052) than males (n = 74).
Older subjects (age ≥40 years; n = 180) had higher BMI (P < 0.0001), higher basal systolic BP (SBP) (P < 0.0001) and diastolic BP (DBP) (P < 0.0001), as well as higher minimum pulse interval (higher maximum heart rate; P = 0.0179) and pulse interval variability (SD, P = 0.0147) than younger subjects (<40 years; n = 162). Older subjects also had lower baroreceptor coupling (P < 0.0001). Urinary epinephrine (P = 0.0185) and norepinephrine (P < 0.0001) levels were increased in older subjects.
Heritability in twins
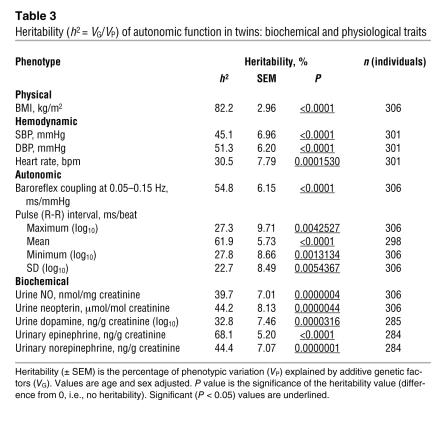
Renal/urinary NO and neopterin excretions were significantly heritable (at h2 = 39.7 ± 7.01% and h2 = 44.2 ± 8.13% [where h2 is heritability]; both P < 0.0001) (Table 3). Observed values for h2 of body mass index, basal BP, heart rate, and urinary catecholamines also displayed significant heritability, consistent with our previous reports (8). Heritability was substantial for vital signs, with SBP (h2 = 45.1% ± 7.0%; P < 0.0001) and DBP (h2 = 51.3% ± 6.2%; P = <0.0001) more heritable than heart rate (h2 = 30.5% ± 7.8%; P = 0.00015). Pulse interval (1/heart rate) values were also heritable, whether expressed as maximum values, minimum values, or variability. Heritability was significant for baroreceptor coupling, at h2 = 54.8% ± 6.15% (P < 0.0001).
Table 3 .
Heritability (h2 = VG/VP) of autonomic function in twins: biochemical and physiological traits
Pleiotropy (shared heritability) in twins: genetic covariance of NO excretion
Since other heritable (Table 3) biochemical traits correlated with urine NO excretion (Supplemental Table 2), we tested such traits for shared genetic determination (pleiotropy) with the NO trait (Supplemental Table 2).
Genetic covariance.
The results indicate that urine NO excretion shares significant genetic determination with the correlated trait of urinary neopterin excretion (genetic covariance [ρG] = 0.493 ± 0.128; P = 0.00183).
Environmental covariance.
The results (Supplemental Table 2) also indicate that urinary NO excretion shares significant environmental codetermination with the correlated traits of urinary neopterin excretion (environmental covariance [ρE] = 0.326 ± 0.0754; P = 0.000069) and urinary epinephrine excretion (ρE = 0.266 ± 0.0801; P = 0.00165).
GCH1 associations with biochemical and physiological traits in twins
Across GCH1.
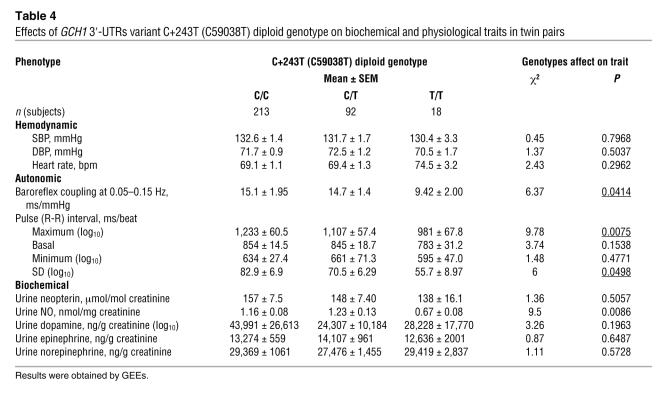
We began by testing whether any of the 5 common individual polymorphisms in GCH1 (Figures 1 and 2, and Supplemental Table 1) affected any of the biochemical traits in twins dependent upon GCH1 activity (Supplemental Figure 1): secretion levels of neopterin, NO, or catecholamines. Only GCH1 variant C59038T (in the 3′-UTRs, at C+243T) predicted any of these traits (Table 4). Of note, this 3′-UTRs variant displayed virtually no LD (D′ < 0.1) with the promoter region variants in the European ancestry sample (Figure 2A).
Table 4 .
Effects of GCH1 3′-UTRs variant C+243T (C59038T) diploid genotype on biochemical and physiological traits in twin pairs
GCH1 C59038T (3′-UTRs C+243T).
The 3′-UTRs variant predicted NO excretion (P = 0.0086), though not neopterin or catecholamine secretion (Table 4). Significant C+243T associations were also found for autonomic traits: baroreceptor coupling (P = 0.0414), maximum pulse interval (i.e., minimum heart rate; P = 0.0075), and pulse interval variability (heart rate variability; P = 0.0498). The 3′-UTRs polymorphism did not associate with basal (resting, average) BP or heart rate in these predominantly normotensive subjects (Table 4).
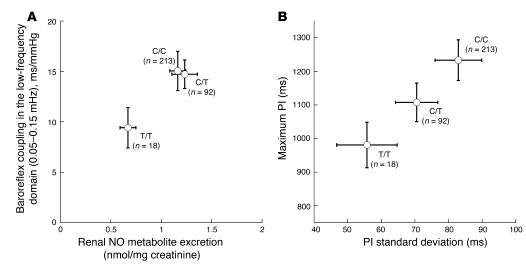
In Figure 3, A and B, these relationships are portrayed graphically. Figure 3A plots baroreflex coupling as a function of NO synthesis. C+243T minor allele homozygotes (T/T diploid genotype) displayed decreases in both NO excretion and baroreflex coupling (Figure 3A); the T allele thus seemed to act recessively on both of these traits. Figure 3B illustrates the joint effects of C+243T on maximum pulse interval as a function of heart rate variability: increasing numbers of T alleles progressively decreased values for both traits. C+243T did not affect heart rate or BP variability in the high frequency (0.15–0.4 Hz; parasympathetic) domain.
Figure 3. GCH1 3′-UTRs (C+243T) polymorphism effects on biochemical and physiological traits in twin pairs.
(A) Baroreflex coupling as a function of renal NO (NO•) production. Univariate effects of C+243T on each trait are shown. Baroreflex coupling was determined in the low-frequency domain (0.05–0.15 Hz). Renal NO production was normalized to the concentration of creatinine in the same urine sample. Univariate NO: χ2 = 9.50, P = 0.0086. Univariate baroreflex: χ2 = 6.37, P = 0. 0414. Alleles: C = 80.2%, T = 19.8%. Hardy-Weinberg equilibrium (HWE), χ2 = 1.76, P = 0.184. (B) Minimum resting heart rate as a function of heart rate variability. Univariate effects of C+243T on each trait are shown. Heart rate variability, in the resting state, was determined as pulse interval (PI) SD (ms) over 5 minutes of monitoring in the time domain. Minimum resting heart rate was determined as maximum (max) PI (ms) during the same monitoring period. Univariate PI SD: χ2 = 6.0, P = 0.0498. Univariate PI max: χ2 = 9.78, P = 0. 0075. Alleles: C = 80.2%, T = 19.8%. HWE, χ2 = 1.76, P = 0.184.
Genetic interactions of GCH1: sex and age.
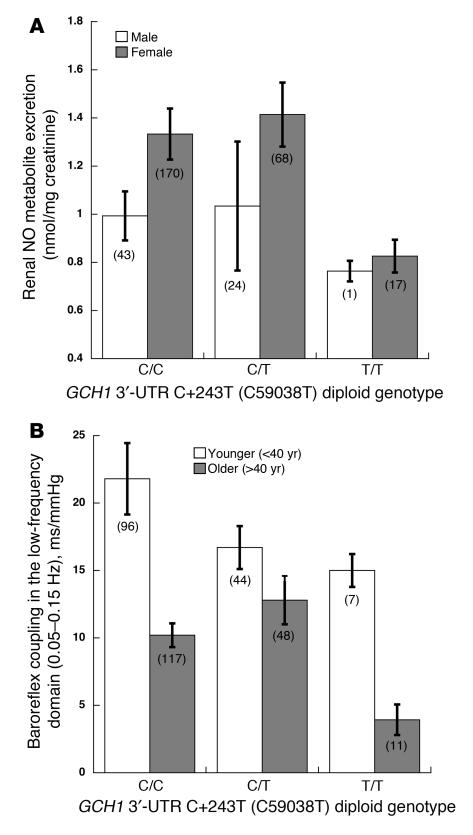
Since sex affects renal NO excretion (females with greater values than males), and age affects baroreceptor coupling (younger individuals with greater values than older individuals) (Table 2), we evaluated whether the GCH1 C+243T effects on NO or baroreceptor coupling (Figure 3A) were influenced by or interacted with sex or age. Genotype and sex interacted (P = 0.029; Figure 4A) to affect NO, with the most prominent difference between sexes in C/C (major allele) homozygotes. Genotype and age interacted (P < 0.0001; Figure 4B) to affect baroreceptor coupling, with the most prominent age effects in the homozygote classes (C/C or T/T).
Table 2 .
Descriptive statistics for the twin study population
Figure 4. GCH1 3′-UTRs C+243T genotype interactions on trait determination in twin pairs: sex and age.
Interactions were suspected based on the independent effects of sex and age on crucial traits (Table 3) and then tested by generalized estimating equations (GEEs). (A) Genotype-by-sex interaction on renal NO excretion. GEE χ2 = 9.50, P = 0.029. (B) Genotype-by-age interaction on baroreflex coupling in the low-frequency domain (0.05–0.15 Hz). Subjects were dichotomized as older or younger than 40 years. GEE χ2 = 23.4, P = 0.0001.
GCH1 haplotypes and twin traits
We approached haplotype effects by testing whether haplotype copy number (0, 1, or 2 per genome) across the GCH1 locus influenced biochemical or physiological traits in twins. This approach is feasible (i.e., may have sufficient power) for the most common haplotypes in a population (here, haplotypes 1 [GTGCC] and 2 [ACGCT]). However, copy number of neither haplotype 1 nor haplotype 2 showed significant effects on NO, catecholamines, neopterin, or any of the physiological traits (all P > 0.05).
GCH1 3′-UTRs variant C+243T and hypertension
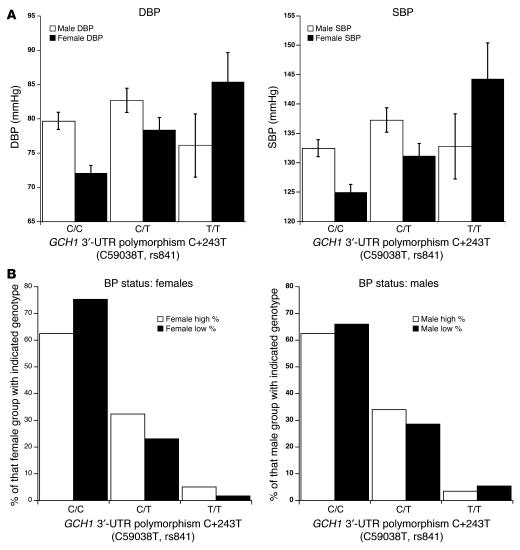
GCH1 variant C+243T was scored in n = 1,049 individuals selected from the extreme values of DBP in a large primary care population of more than 53,000 individuals (Figure 5). The C+243T diploid genotype had a significant overall effect on both DBP (P = 0.008) and SBP (P = 0.008). However, the genotype effect on DBP displayed a significant gene-by-sex interaction (P = 0.046). Indeed, when males and females were analyzed separately, females showed significant genotype effects upon both DBP (P = 0.002) and SBP (P = 0.007), while no effects were found for males alone (P = 0.22–0.24) (Figure 5A). To explore these effects in a model-free fashion, we also employed permutation (exact) tests. In 3-by-2 tables (diploid genotype by BP status), permutation revealed an effect of genotype on BP: P = 0.00258 (Figure 5B). When this analysis was confined to females, the genotype effect remained significant (P = 0.00082), while the effect was absent in males (P = 0.155).
Figure 5. Hypertension.
Intermediate phenotype–associated GCH1 variant C+243T was typed in n = 1,049 subjects selected from the most extreme DBPs (high and low) in a primary care population of more than 53,000 individuals. (A) DBP and SBP as a function of diploid genotype. Results were analyzed by 2-way ANOVA, factoring for genotype, sex, and genotype-by-sex interaction. DBP, ANOVA: genotype F = 4.81, P = 0.008; sex F = 0.150, P = 0.698; gene-by-sex F = 3.08, P = 0.046; alleles C = 81%, T= 19%. HWE: χ2 = 0.724, P = 0.395; males alone: F = 1.43, P = 0.240; females alone: F = 6.21, P = 0.002. SBP, ANOVA: genotype F = 4.84, P = 0.008; sex F = 1.49, P = 0.700; gene-by-sex F = 2.19, P = 0.112; alleles C = 81%, T = 19%. HWE: χ2 = 0.724, P = 0.395; males alone: F = 1.52, P = 0.220; females alone: F = 5.06, P = 0.007. (B) BP status (high versus low) as a function of diploid genotype. Subjects were grouped by sex and C+243T diploid genotype. Results were analyzed by permutation testing on 3-by-2 contingency tables. Gene-by-diagnosis permutation: females, P = 0.00082; males, P = 0.155.
GCH1 3′-UTRs variant C+243T: evidence of functional consequences in vitro
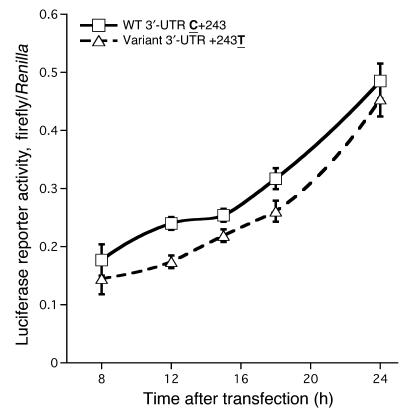
Wild-type (C+243) versus variant (+243T) 2,009-bp GCH1 3′-UTRs were ligated into reporter plasmid pGL3-Promoter (Supplemental Figure 2), downstream of the luciferase reporter open reading frame. After transfection into PC12 chromaffin cells, cellular luciferase activity accumulated in a time-dependent fashion (P < 0.001). Despite alteration by only a single base (C+243T), the two 3′-UTRs plasmids had consistently different luciferase activity across the time course of the experiment, from 8 to 24 hours (P = 0.013), with C+243 greater than +243T (Figure 6).
Figure 6. Effect of GCH1 3′-UTRs polymorphism on gene expression in vitro.
3′-UTRs expression plasmids were transfected into PC12 chromaffin cells. After continued cell growth for 8–24 hours after transfection, cells were harvested for assay of firefly luciferase as well as the cotransfected Renilla luciferase reporter pRL-CMV. Results of triplicate transfections are expressed as the ratio of firefly/Renilla luciferase activity and were analyzed by 2-way ANOVA, factoring for the effects of 3′-UTRs and time. 2-Way repeated measures ANOVA: plasmid (3′-UTRs) F = 12.04, P = 0.013; time F = 163.6, P < 0.001.
GCH1 3′-UTRs variant C+243T: RNA motifs and interspecies sequence homology
The likely ancestral allele in humans is C, based on the chimpanzee sequence. Analysis of the GCH1 3′-UTRs sequence in the local C+243T region (Regulatory RNA Motifs and Elements Finder; RegRNA, http://regrna.mbc.nctu.edu.tw/ [ref. 9]) did not reveal known RNA motifs such as A/U-rich regions or micro-RNA binding sites. RNA folding algorithms (http://www.genebee.msu.su/services/rna2_reduced.html) detected no differences in folding free energy (ΔG°=–547.1 kcal/mol, between variants), and the predicted mRNA stem/loop structures were identical.
The interspecies homology depicted in Figure 1A (top panel) indicates that C+243T lies in a region of generally conserved sequence among primate species. Figure 7 aligns the local region in species we resequenced. Although the C+243T region lies in the 3′-UTRs of the terminal exon of GCH1 in the major mRNA isoform 1 (clone NM_00016) in humans, and mRNA splicing of GCH1 displays substantial cross-species conservation (e.g., in dog and slime mold) (10), 2 additional human splice variants (NM_001024070, NM_001024071) that exclude C+243T from the mature mRNA are known in this region, and the region may be intronic in the reported isoforms of some species (mouse), or even downstream from (i.e., 3′ to) the reported terminal exon in other species (chicken, rat, zebrafish, Xenopus, roundworm, and eremoth). Cytokine induction of GCH1 transcription may also trigger its alternative splicing (10).
Figure 7. The GCH1 isoform 1 (NT_00016) 3′-UTRs spans 2,009 bp (human chromosome 14, 54378476-54380484).
The alignment shown spans 60 bp flanking the polymorphism (at position 54380242 on chromosome 14). The polymorphic base (human C+243T) is shown in uppercase bold type. Asterisk indicates that the position is identical in all primates shown. The chimpanzee, gorilla, and orangutan sequences were newly determined here.
GCH1 human phylogeny: 3′-UTRs variant C+243T
Supplemental Figure 3 illustrates a likely phylogeny for the second haplotype block within GCH1, containing the 3′-UTRs polymorphism associated with multiple phenotypes here: C+243T (C59038T). The +243T variant allele did not occur in 3 nonhuman primates (Table 2). The variation in block 2 can be grouped into 3 ancestral haplotype groups (C, D, F; defined in Supplemental Figure 3), each which differ by at least 2 SNPs. The lack of any haplotypes in the population that differ by only 1 SNP between any 2 of these ancestral groups suggests that these haplotypes existed before a putative human population bottleneck (11). The +243T variant occurs in a single haplotype (haplotype B; 27 occurrences in our resequenced subjects). Phylogenetic analysis suggests that haplotype B is derived from ancestral haplotype F, which is no longer found in the current human population.
Other genotypes in pathways controlling catecholamines or NO
To test whether our twin sample had sufficient power to detect genetic effects on transmitter biosynthesis, we also evaluated the effects of polymorphism at TH (the rate-limiting enzyme in catecholamine biosynthesis) and e-NOS (NOS3). At TH, promoter polymorphism C–824T (rs10770141) significantly predicted renal excretion of both norepinephrine (P = 0.0069) and epinephrine (P = 0.0058), while coding region polymorphism Val81Met (rs6356) did not (P > 0.1). At NOS3, coding region polymorphism Glu298Asp (rs1799983) did not predict renal excretion of norepinephrine, epinephrine, or NO (all P > 0.1). We chose this NOS3 SNP because of its influence on endothelial function (12).
Discussion
Overview
In this study, we undertook systematic polymorphism discovery at GCH1, revealing 13 common variants spanning the exons and proximal promoter. Allele frequencies and patterns of LD varied by ethnicity. In a series of twins phenotyped for autonomic and renal traits likely to be influenced by GCH1 variation, the renal excretions of neopterin, NO metabolites, and catecholamines were all highly heritable. Only NO was influenced by GCH1 polymorphism, and then only by a common variant in the 3′-UTRs (C+243T; C59038T), likely to have arisen early in the human lineage. This same polymorphism coordinately predicted not only NO but also baroreflex coupling, heart rate variability, and minimum heart rate. In the population, the 3′-UTRs variant influenced both SBP and DBP. The results are consistent with a pleiotropic cascade of events consequent upon common GCH1 polymorphism, eventuating in altered cardiovascular risk status.
Cardiovascular consequences of GCH1 polymorphism: NO, age, autonomic function, and risk predictors
GCH1 C+243T was associated with not only NO production but also baroreflex coupling (Figure 3A); in particular, T/T homozygotes displayed not only diminished NO but also decreased baroreceptor function. The influence of NO on the baroreceptor has been investigated in experimental animals: NO• acts in brainstem nuclei to inhibit sympathetic outflow while enhancing parasympathetic outflow (13). Overexpression of NOS in the rostral ventrolateral medulla causes sympathetic activity, BP, and heart rate to fall in experimental hypertension (14). Decreased baroreflex coupling in T/T homozygotes resulted in decreased overall heart rate (or pulse interval) variability (Figure 3B), eventuating in a higher minimum heart rate (maximum pulse interval; Figure 3B).
Both decreased heart rate variability (15, 16) and increased resting heart rate (17) are risk indictors for premature cardiovascular death. Our results in twin pairs document the role of heredity in these risk predictors (Table 3): baroreflex coupling at 54.8% ± 6.2% (P < 1 × 10–7), overall heart rate variability (pulse interval SD) at 22.7% ± 8.5% (P = 0.0054), and minimum heart rate (maximum pulse interval) at 27.8 ± 8.7% (P = 0013).
Finally, our genetic results define a role for a specific DNA variant (GCH1 C+243T) in a crucial pathway (Supplemental Figure 1) in triggering a pleiotropic series of effects on cardiovascular risk (Figures 3 and 5).
Heredity clearly influences human longevity (18–20), though the responsible genetic variants are not well defined in humans. Our twin results serve to quantify heritable contributions (Table 3) to risk predictors for premature mortality, such as heart rate variability and heart rate, thus refining the tractability of such traits to further genetic investigation. Of note for the consequences of C+243T on longevity, the polymorphism seemed to interact with age in determining baroreceptor coupling (Figure 4B), with the most profound age effects on the trait seen in major (C/C) or minor (T/T) allele homozygotes.
Hypertension
When subjects with the most extreme BPs in a large population were genotyped at GCH1 3′-UTRs C+243T (Figure 5A), the variant had a substantial effect upon both DBP (P = 0.008) and SBP (P = 0.008), though a gene-by-sex interaction (P = 0.046 for DBP) seemed to confine the effect to females (P = 0.002).
Critical role of GCH1 in NO and catecholamine metabolism
GCH1 encodes the first and rate-limiting enzyme in the synthesis of BH4, an essential cofactor for TH and NOS in the catecholamine and NO production pathways, respectively, thus playing a crucial role in the sympathoadrenal and endogenous nitrovasodilator systems (Supplemental Figure 1). GCH1 was isolated as the first causative gene for L-DOPA–responsive dystonia (21). Profound GCH1 deficiency in humans, as occurs after unusual inactivating mutations (Gly201Glu or Asp134Val) in homozygotes, results in widespread disturbances of neuropsychiatric function, such as autosomal recessive dystonia (22). The hph-1 mouse model of GCH1 deficiency (3) also displays reductions in levels of BH4, NO, catecholamines, serotonin, and their metabolites (23). GCH1 haplotypes also reportedly predict pain sensitivity in humans (24); we did not explore pain in our subjects.
GCH1 variants and autonomic pathways.
Since GCH1 variant C59038T (3′-UTRs, at C+243T) predicted both biochemical (Figure 3A and Supplemental Figure 1) and physiological (Figure 3, A and B) traits, the results indicate coupled genetic control of both NO secretion with baroreceptor slope coupling, as well as heart rate variability and basal heart rate (Figure 3). While we found that GCH1 genetic variation affected renal NO production (Table 4 and Figure 3A), NOS3 genetic variation (Glu298Asp) did not influence the renal NO trait (P > 0.1). These observations suggest that the rate-limiting step in NO biosynthesis may lie at the GCH1 step rather than the NOS step(s). This conclusion remains tentative, pending further data on the influence of polymorphism at other NOS loci (NOS1, NOS2).
By contrast, while GCH1 genetic variation did not seem to affect catecholamine production (Table 4), we previously found that common variation in the rate-limiting enzyme in catecholamine biosynthesis, TH, predicts interindividual variability in renal catecholamine excretion (8), an observation confirmed by our studies here on TH promoter polymorphism C–824T effects on norepinephrine and epinephrine excretion. Thus, our data are consistent with the viewpoint that TH (rather than GCH1 activity) is the rate-limiting step in the pathway toward catecholamine production (25, 26).
Sex, GCH1 polymorphism, and BP.
Our twin data (Table 2) indicate that females excrete approximately 32% more NO metabolites than males. This biochemical finding is consistent with physiological studies indicating superior endothelial function in females (27). Such biochemical and physiological differences might contribute to the lower frequency of hypertension in women (28, 29). Since both sex (Table 2) and GCH1 genotype (Table 4) influenced NO, we tested and found a significant gene-by-sex interaction in control of this trait (Figure 4A), with the most pronounced sex differences seen in major allele (C/C) homozygotes. The penetrance of profoundly inactivating mutations at GCH1 seems to be quite sex dependent, with phenotypes occurring more frequently in female carriers (30), while circulating mononuclear cell GCH1 enzymatic activity is higher in females than males (21). We did not measure GCH1 activity in these subjects. Finally, the effect of GCH1 genotype on BP in hypertension seemed to be confined to female subjects (Figure 5).
Strengths and weaknesses of this study: coupling the twin method and systematic polymorphism discovery with adrenergic and disease phenotyping
Twin phenotyping protocol.
Here we exploited the classical twin design (31, 32). Twin data offer the unique advantage of direct measurement of heritability (h2), the fraction of phenotypic variance accounted for by genetic variance, a logical estimator of the tractability of any trait to genetic investigation. Since twins are randomly sampled from the population, genetic conclusions drawn from twin studies are likely to be generalizable to the population from which they were sampled (31). Multiple autonomic phenotypes in the twins, both biochemical and physiological, allowed for inference of an integrated picture of the effects of particular genetic variants at GCH1.
Systematic polymorphism discovery.
LD mapping is an increasingly powerful tool for exploring genetic determinants of disease (33). However, the LD approach requires the fulfillment of many assumptions (34), including substantial LD between marker (surrogate) and trait (causative) alleles. Here we took another approach: systematic polymorphism discovery at a candidate genetic locus. This approach enables direct testing of marker-on-trait allelic association, rather than indirect testing relying on a hypothetical degree of LD between marker and trait alleles. Of note at the GCH1 locus, we discovered 2 blocks of LD (Figure 2) and found that the GCH1 C59038T mutation was in a different (downstream) block of LD from the promoter (upstream) block. A series of common SNPs in the GCH1 promoter (Supplemental Table 1) did not associate with the autonomic traits in this study. Given the LD structure of the GCH1 locus (Figure 2A), even quite extensive genotyping at the GCH1 locus would not have uncovered the phenotypic associations we observed (Figure 3).
Risk indicators and disease in the population.
Careful autonomic phenotyping of twins allowed us to define effects of GCH1 variation on such cardiovascular disease risk predictors as baroreflex dysfunction, reduced heart rate variability, and increased minimum heart rate (Figure 3). In turn, the availability of genomic DNA from subjects with the most extreme BPs in the population enabled us to establish a disease (hypertension) association for GCH1 3′-UTRs C+243T (Figure 5).
Sex.
Since in classical twin studies, subjects are randomly sampled from the population (31), both sexes are represented in such studies. Likewise, the availability of a large primary care population (31) enabled us to study the consequences of GCH1 variation on hypertension in both males and females. For both biochemical (Figure 4A) and physiological (Figures 3 and 4) intermediate traits, as well as the disease trait of hypertension (Figure 5), sex effects on the traits were prominent, and gene-by-sex interactions were observed (Figure 4A and Figure 5), reinforcing the value of informative sex representation in studies of cardiovascular disease (28).
Functional characterization of C+243T variant.
Finally, we demonstrated that the 3′-UTRs variant C+243T is itself functional: C→T conversion in a transfected 3′-UTRs reporter plasmid (Supplemental Figure 2) diminished gene expression in a fashion directionally appropriate to the action of the T allele in vivo to diminish NO production (Figure 6).
Caveats.
While we conducted systematic variant discovery in both black (sub-Saharan Africa) and white subjects (European ancestry) (Supplemental Table 1 and Supplemental Figure 3), the studies on physiology and disease were analyzed only in white subjects, initially to avoid the potentially spurious effects of population stratification on genetic trait associations (35). Only additional studies can determine whether the associations in white subjects are generalizable to other population groups. In studies of complex traits in the “post-genomic” era, the effects of several genetic variants may be evaluated on multiple traits, raising the specter of false positive (type I or α) statistical errors. We approached this issue in several ways: haplotyping (Table 1 and Figure 2) allowed us to evaluate several genetic variants simultaneously. Similarly, we studied the simultaneous effects of a single genetic variant upon multiple traits (Table 4 and Figures 5 and 6); since such traits are correlated, a simple Bonferroni correction would have been inappropriately conservative. We undertook permutation tests where possible (Figure 5B), documenting the independence of our findings from statistical distribution assumptions. Finally, we documented the effects of GCH1 variation on physiological traits in 2 different samples (twins and population BP extremes). Nonetheless, in the absence of full replication, any such genetic marker-on-trait results should not be characterized as definitive.
Table 1 .
Haplotype distribution in GCH1
NO.
The measurement of urinary nitrate/nitrite excretion is intended to provide an accessible index of renal NO production for consideration of its heritable determination by genetic variation at biosynthetic loci. However, urine nitrate/nitrite excretion is influenced by environment, in the form of dietary ingestion (36), as well as by heredity; our twin sample displayed substantial heritability of this trait (at h2 = 39.7% ± 7.0%; P = 4 × 10–7), despite the unrestricted diet in our protocol. Second, the source of NO in human urine is only incompletely established and may reflect production not only locally (in the kidney) but also systemically. In any event, the trait displayed substantial (P = 0.0086) determination by GCH1 genotype (Table 4 and Figure 3A).
Haplotype uncertainty.
Imputation of phase from diploid genotype data is inherently uncertain and occasionally prone to misclassification; the haplotype method we used assigns the 2 most likely haplotypes to each individual (7). While emerging haplotype methods consider uncertainty in phasing (37, 38), such methods have not yet been coupled with the computational needs of correlated twin pair statistics.
Conclusions and perspectives
Human NO production is substantially heritable (at h2 = 39.7% ± 7.0%; P = 4 × 10–7; Table 3), displaying joint genetic determination (Figures 3 and 6 and Supplemental Table 2) with autonomic activity. We further conclude that NO secretion is controlled in part by genetic variation in the pathway encoding NO synthesis, especially at the classically rate-limiting step, GCH1. The trait-associated allele in the GCH1 3′-UTRs (+243T) seems to be functionally active (Figure 6) and may have arisen early in the human lineage (Supplemental Figure 3). The results point to novel pathophysiological links between a key biosynthetic locus and NO metabolism (Figure 8 and Supplemental Figure 1) and suggest new strategies to approach the mechanism, diagnosis, and treatment of risk indicators of cardiovascular disease.
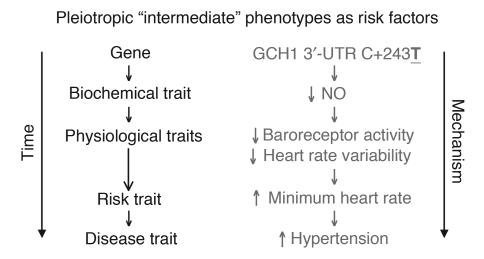
Figure 8. Intermediate phenotypes.
In the “intermediate phenotype” schema, biochemical traits (such as NO production) are postulated to be determined earlier and more proximately by genotype (such as GCH1 3′-UTRs C+243T) than are physiological traits (such as heart rate variability and resting heart rate) and, ultimately, late-penetrance disease (such as cardiovascular disease, hypertension, or sudden death). Here the concept is illustrated by the findings at GCH1 in the current study: the GCH1 3′-UTRs variant C+243T initially alters NO production, subsequently influencing baroreceptor coupling and heart variability, later minimum resting heart rate, and finally basal BP in the population.
Hypothesis schematic.
A schematic formulating our results into a global hypothesis is presented in Figure 8, outlining the role of intermediate phenotypes and the GCH1 variant C59038T (in the 3′-UTRs, at C+243T) in the determination of hypertension via the initial intermediary of NO secretion. While the pathway outlined is hypothetical, it may frame questions for future experimentation.
Implications for pathophysiology/mechanism, prediction/diagnosis, treatment.
Our results suggest that the NO pathway is centrally involved in the early pathogenesis of cardiac diseases beginning in healthy individuals, perhaps initially by altering baroreceptor function (Table 4 and Figures 3 and 6). Genetic determination of biochemical (Figure 3A) and physiological (Figure 3) traits in this pathway (Supplemental Figure 4) suggests that treatments targeting the pathway (such as BH4; ref. 39) might be beneficial in preventing later cardiac diseases if administered to subjects at specific genetic risk. Our results also raise the possibility that genetic profiling of patients with impaired endothelial or autonomic activity might yield practical pharmacogenetic predictors of patients most likely to benefit from such therapies (39). Finally, our results raise the possibility that profiling subjects for particular biosynthetic polymorphisms would provide an index of susceptibility to cardiovascular disease. This prediction awaits testing in appropriate longitudinal cohorts.
Methods
Subjects and clinical characterization
Initially, a series of n = 64 unrelated individuals was studied by resequencing of GCH1 for systematic polymorphism discovery: n = 34 of European ancestry (white) and n = 30 sub-Saharan African ancestry (black). A series of n = 171 twin pairs was also studied by resequencing the GCH1 locus.
Ethnicity was established by self-identification by the participants as well as their parents and grandparents. None of the subjects had a history of renal failure. Definitions of subject characteristics are according to previous reports from our laboratory (40). Subjects were volunteers from southern California, and each subject gave informed, written consent; the protocol was approved by the UCSD institutional review board.
Twin pairs.
We recruited a series of twin pairs, taking advantage of a large population-based twin registry in southern California (8, 41) as well as by advertisement (8). These twin individuals were all of European ancestry, to permit allelic association studies within 1 ethnicity. There were n = 342 individuals from n = 171 white (European ancestry) twin pairs; n = 118 monozygotic (MZ) pairs (23 M/M, 95 F/F); and 53 dizygotic (DZ) pairs (9 M/M, 33 F/F, 11 M/F). The twin subjects were 15–84 years old. Thirty-four of the 342 twin subjects had essential hypertension (9 male, 25 female); 30 were treated for hypertension. Seventy-four of 171 twin pairs (44%) were family history–positive for hypertension (1 or both parents), while 83 of 171 pairs (56%) were negative. Twin zygosity (MZ or DZ) was confirmed by use of more than 100 microsatellites (chromosomes 1 and 2) for self-identified DZ twins or SNP data (11–177 SNPs) as well as the TH (TCAT)n microsatellite (8) for self-reported MZ twins. The diet was the subjects’ customary unrestricted intake of nutrients and fluids.
Genomics
Genomic DNA was prepared from leukocytes in EDTA-anticoagulated blood, using PureGene extraction columns (Gentra; QIAGEN) as described previously (42). Public draft human (43) and mouse (44) genome sequences were obtained from the UCSC Genome Bioinformatics website (http://genome.ucsc.edu) and used as a scaffold for primer design and sequence alignment. The base position numbers were from NCBI GCH1 source clones NM_00016 (isoform 1), NT_0026437, and NP_000152. Promoter positions were numbered with upstream of (–) the GCH1 exon 1 start (cap) site. The following PCR primers were designed by Primer3 (45) to span approximately 1,200 bp of the proximal promoter and each of the 6 exons, as well as to include 50–100 bp of flanking intronic sequence: promoter 1, left primer 5′-GGCTTCCCTATTGGTTTGTTT-3′, right primer 5′-GTCTTTCCCCAGATGTTGGT-3′; promoter 2, left primer 5’ GAGTTCAGTTGGTGAATAGCATT-3′, right primer 5′-CCGAGTTAAGCCACCGATTA-3′; promoter 3, left primer 5′-GTGGCAGGTGCGTTTTTAAC-3′, right primer 5′-CGCAACCTGCTTAGATCACA-3′; Exon 1, left primer 5′-CCGGGCCATAAAAAGGAG-3′, right primer 5′-AGTGAGGCAACTCCGGAAA-3′; Exon 2, left primer 5′-CCTCCGTTCTCCTTCCTCTT-3′, right primer 5′-GGCTTATCCTGAGAGCCTTC-3′; Exon 3, left primer 5′-AACAGTTCCAGATGTTTTCAAGG-3′, right primer 5′-GTAGGGGACGAGAAGGAAGG-3′; Exon 4, left primer 5′-ATTTCCTCTTGCAGCCCACT-3′, right primer 5′-GAAAATGTGAAGGAATGTGCAA-3′; Exon 5, left primer 5′-AGCTGGTGTGTCTTGGCTCT-3′, right primer 5′-AGGCTCAGGGATGGAAATCT-3′; Exon 6-1, left primer 5′-CCAAACCAGCAGCTGTCTAC-3′, right primer 5′-TTCCCAATGCTCCTATGCTT-3′; Exon 6-2, left primer 5′-TACGTGCACAAAACCACTGC-3′, right primer 5′-GGCATCTACATGGATCACACA-3′; Exon 6-3, left primer 5′-TTGAGTTTCCTTTGTGTGATCC-3′, right primer 5′-CCCAATACCTCTGCCATCTT-3′; Exon 6-4, left primer 5′-CCCATCTCTGCCACTTTGAT-3′, right primer 5′-GCCCTTATGATGTCTTTGCTG-3′. Target sequences were amplified by PCR from 20 ng genomic DNA in a final volume of 25 μl, which also contained 0.1 U of Taq DNA polymerase (Applied Biosystems), 200 μM of each dNTP, 300 nM of each primer, 50 mM KCl, and 2 mM MgCl2. PCR was performed in an MJ PTC-225 thermal cycler, starting with 12 minutes of denaturation at 95°C, followed by 45 cycles at 95°C for 30 seconds, 63°C for 1 minute (annealing), and 72°C (extension) for 1 minute, and then a final extension of 8 minutes at 72°C. PCR products were treated with exonuclease I and shrimp alkaline phosphatase to remove primers and then dNTPs prior to cycle sequencing with BigDye terminators (Applied Biosystems). Sequence was determined on an ABI 3100 automated sequencer and analyzed using the Phred/Phrap/Consed suite of software to provide base quality scores (46–48). Polymorphism and heterozygosity were detected using Polyphred (49, 50) and manually confirmed. A subset of the data was cross-validated manually using base calls from Applied Biosystems software and visual inspection of trace files to identify heterozygotes. Rare SNPs were confirmed by resequencing in multiple individuals, and on the reverse strand.
Comparative genomics.
The same GCH1 regions were also resequenced in 3 nonhuman primates, with genomic DNA obtained from the Coriell Institute for Medical Research: 1 chimpanzee (NA03448A), 1 gorilla (NG05251B), and 1 orangutan (NG12256). Sequences conserved between mouse and human GCH1 were visualized with VISTA (http://genome.lbl.gov/vista/index.shtml).
Biochemical phenotyping: catecholamines and NO metabolites
Plasma and urine catecholamines were measured radioenzymatically (51). The assay uses a preconcentration step that increases sensitivity by approximately 10-fold over that of other catechol-O-methyltransferase–based (COMT-based) assays and approximately 20-fold over that of many HPLC assays, permitting accurate measurement of basal plasma epinephrine levels, which are at the limit of sensitivity for HPLC assays. Nitrate/nitrite metabolites were measured to estimate urinary NO, by the Griess spectrophotometric method at 540–570 nm, after conversion of urinary nitrate to nitrite by nitrate reductase (DE1600; R&D Systems); the interassay coefficient of variation was 3.3%–7.0%. Urine catecholamine and NO values were normalized to creatinine excretion in the same sample.
Physiological/autonomic phenotyping in vivo
Prolonged recording of BP and heart rate.
Noninvasive brachial arterial cuff BPs were obtained in seated subjects with an oscillometric device, as previously described (52). BP (in mmHg) and pulse interval (R-R interval or heart period, in ms/beat) were then recorded continuously and noninvasively for 5 minutes in seated subjects with a radial artery applanation device and dedicated sensor hardware (Colin Pilot; Colin Instruments) and software (ATLAS, WR Medical Electronics Co.; and Autonomic Nervous System, Tonometric Data Analysis [ANS-TDA], Colin Instruments), calibrated every 5 minutes against ipsilateral brachial arterial pressure with a cuff sphygmomanometer (8). Heart rate was recorded continuously with thoracic ECG electrodes to the Colin Pilot. Average, maximum, and minimum values, as well as coefficients of variation, were calculated for BP and pulse interval using the ANS-TDA software.
Baroreceptor coupling in the frequency domain.
Power spectra for SBP, DBP, and pulse (R-R) interval were obtained during the 5-minute recording, using the ANS-TDA software, by Fourier transformation of data from the time domain into the frequency domain (53). Spectral power is the variance (i.e., [SD]2) of the parameter in a particular frequency domain; e.g., for R-R interval, in milliseconds, spectral power is in milliseconds squared. The low-frequency or classical baroreceptor domain (0.05–0.15 Hz) reflects a mixture of parasympathetic and sympathetic activity. In this frequency domain, coupling of R-R interval power to SBP power (ms2/mmHg2) was computed as √([R-R power, ms2]/[SBP power/mmHg2]) = baroreflex sensitivity (ms/mmHg) in the frequency domain.
Hypertension
We studied n = 1,049 (n = 497 male, n = 552 female) white (European ancestry) subjects drawn from n = 53,078 individuals (n = 27,475 females, n = 25,538 males), recruited from a large primary care (Kaiser Permanente) population in San Diego, as previously described (54). In this primary care population, approximately 81% attended the clinic, and approximately 46% consented to participation in the study, with collection of blood for preparation of genomic DNA. From consented participants, the subjects in this study were selected, based upon measurement of DBP, to represent the highest and lowest DBP percentiles in that population. n = 509 subjects were chosen for the highest DBPs, while n = 540 subjects were chosen for the lowest DBPs. The ages of the high- and low-BP groups were not different (low BP 59 ± 0.5 years, high BP 59 ± 0.4 years [P = 0.94]); to accomplish this, low BP-individuals (SBP/DBP: 111 ± 0.6 mmHg/55 ± 0.3 mmHg) were selected from the bottom 4.8th percentile of DBP, while the high-BP group (SBP/DBP = 153 ± 0.6 mmHg/100 ± 0.8 mmHg) was selected from the top 4.9th percentile of DBP. Both SBP and DBP differed significantly between the BP extreme groups (P < 0.0001 and P < 0.0001). Forty-one percent of patients in the high-BP group were taking one or more antihypertensive medications (including 15% on diuretics and 19% on angiotensin-converting enzyme [ACE] inhibitors), while none in the low-BP group were on such treatment.
GCH1 3′-UTRs: functional consequences in vitro
The GCH1 3–UTR was amplified from a cloned bacterial artificial chromosome (BAC) target region, in BAC RP11-304L20, which spans the GCH1 gene, obtained from the BACPAC Resource at Children’s Hospital of Oakland Research Institute. The 2,009-bp 3′-UTRs was amplified with PCR primers incorporating XbaI restriction sites, facilitating ligation into the unique XbaI site in luciferase reporter plasmid pGL3-Promoter (Promega); in this plasmid, the XbaI site is just downstream (3′) of the firefly luciferase reporter open reading frame and upstream of the polyadenylation signal (pA from SV40). Correct orientation (5′→3′) of the insert was verified with asymmetric digestion by MfeI (C↓AATTG), which cuts the insert at base 88 and the vector at base 2,103 (within the pA cassette). Creation of the C+243T polymorphism was done by site-directed mutagenesis (QuikChange; Stratagene). After plasmid growth under ampicillin selection in E. coli and purification on columns (QIAGEN), supercoiled plasmid DNA was transfected into PC12 chromaffin cells using established protocols and Superfect cationic lipid reagent (QIAGEN) (55), along with the transfection efficiency control plasmid pRL-CMV, encoding the Renilla luciferase reporter under control of the CMV promoter (Promega). After transfection and cell growth over an 8- to 24-hour time course, cells were lysed for sequential measurement of the 2 luciferase enzymatic activities (firefly and Renilla) with the dual luciferase assay system (Promega) in an ultrasensitive luminometer (AutoLumat LB 953; Berthold Technologies). Luciferase results are expressed as firefly/Renilla activity ratios from triplicate transfections.
Statistics
Descriptive statistics.
Descriptive statistics (mean, SEM) were computed across all the twins, using generalized estimating equations (GEEs; PROC GENMOD) in SAS (Statistical Analysis System), creating an exchangeable correlation matrix to account for correlated trait values within each twin pair (56).
Heritability of phenotype expression in vivo.
Heritability (h2) is the fraction of phenotypic variance accounted for by genetic variance (h2 = VG/VP). Estimates of h2 were obtained using the variance component method implemented in the Sequential Oligogenic Linkage Analysis Routine (SOLAR) package (57). This method maximizes the likelihood of the estimate assuming a multivariate normal distribution of phenotypes in twin pairs (MZ versus DZ) with a mean dependent on a particular set of explanatory covariates. The null hypothesis (H0) of no heritability (h2 = 0) is tested by comparing the full model, which assumes genetic variation, and a reduced model, which assumes no genetic variation, using a likelihood ratio test. Covariates (sex and age) that were significant at P < 0.05 were retained in the heritability model. Before analyses in SOLAR, exploratory descriptive statistics were computed for each trait, and if trait values displayed excessive kurtosis, they were log10-transformed to achieve kurtosis less than 0.8.
Haplotypes and LD.
Haplotypes were inferred from unphased diploid genotypes with the likelihood-based method HAP (7) available at http://research.calit2.net/hap. We inferred the GCH1 haplotypes using 5 common (minor allele frequency >5%) SNPs discovered by resequencing n = 64 unrelated individuals (n = 128 chromosomes) chosen to span 2 diverse ethnic groups: white (European ancestry) and black (sub-Saharan African ancestry). Blocks of LD were displayed using Graphical Observation of Linkage Disequilibrium (58).
Association.
Association studies were performed for the GCH1 common alleles (minor allele frequency >5%). Each study subject was categorized according to diploid genotype at a biallelic SNP locus, or carrier status (2, 1, or 0 copies) of most frequent haplotypes.
Permutation.
To explore the effects of GCH1 3′-UTRs variant C+243T on BP status (hypertensive versus normotensive) in model-free fashion, without relying on standard asymptotic assumptions, we also employed more computationally intensive permutation (exact) tests (59) on 3-by-2 contingency tables (diploid genotype by BP status) as described at http://www.physics.csbsju.edu/stats/exact.html.
Phylogeny.
Haplotype phylogeny plots were reconstructed using an extension of the HAP algorithm (7). For common SNPs (each minor allele frequency >10%), the phylogeny over the haplotypes was computed by optimizing a probabilistic model that assumes exponential population growth after a population bottleneck. Under this assumption, any variants after the bottleneck occur in haplotypes that are also present in the modern population. Haplotype variants that have no intermediates between them (i.e., that vary by ≥2 SNPs) are assumed to originate before the bottleneck and are therefore “ancestral.” “Common” variants are single mutation or recombination events away from the ancestral variants; such events are thus likely to have happened after a major population bottleneck. The algorithm identifies “recent” haplotype variants by considering rare (<5%) haplotypes and explaining them with either a single gene conversion, mutation, or recombination event from the common haplotypes. The remaining haplotypes are clustered into ancestral groups, and any recombinants (resulting from meiotic crossover events) are identified. The algorithm is implemented as an extension to the HAP program (7), available online at http://diego.ucsd.edu/hap/html/. A P value of less than or equal to 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was funded by grants from the Department of Veterans Affairs and the NIH, including General Clinical Research Center (GCRC) grant RR00827. We appreciate the assistance of the GCRC nursing and core laboratory staff in accomplishing this study.
Footnotes
Nonstandard abbreviations used: BH4, tetrahydrobiopterin; DBP, diastolic BP; DZ, dizygotic; GCH1, GTP cyclohydrolase 1; h2, heritability; LD, linkage disequilibrium (D′); MZ, monozygotic; SBP, systolic BP; TH, tyrosine hydroxylase; UTR, untranslated region.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2658–2671 (2007). doi:10.1172/JCI31093
Lian Zhang, Fangwen Rao, and Kuixing Zhang contributed equally to this work.
References
- 1.Serova L., Nankova B., Rivkin M., Kvetnansky R., Sabban E.L. Glucocorticoids elevate GTP cyclohydrolase I mRNA levels in vivo and in PC12 cells. Brain Res. Mol. Brain Res. 1997;48:251–258. doi: 10.1016/s0169-328x(97)00098-3. [DOI] [PubMed] [Google Scholar]
- 2.Furukawa Y., et al. Dystonia with motor delay in compound heterozygotes for GTP-cyclohydrolase I gene mutations. Ann. Neurol. 1998;44:10–16. doi: 10.1002/ana.410440107. [DOI] [PubMed] [Google Scholar]
- 3.Hyland K., Gunasekara R.S., Munk-Martin T.L., Arnold L.A., Engle T. The hph-1 mouse: a model for dominantly inherited GTP-cyclohydrolase deficiency. Ann. Neurol. 2003;54(Suppl. 6):S46–S48. doi: 10.1002/ana.10695. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt K., Werner E.R., Mayer B., Wachter H., Kukovetz W.R. Tetrahydrobiopterin-dependent formation of endothelium-derived relaxing factor (nitric oxide) in aortic endothelial cells. Biochem. J. 1992;281:297–300. doi: 10.1042/bj2810297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasquez-Vivar J., et al. Superoxide generation by endothelial nitric oxide synthase: the influence of cofactors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9220–9225. doi: 10.1073/pnas.95.16.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong H.J., Hsiao G., Cheng T.H., Yen M.H. Supplemention with tetrahydrobiopterin suppresses the development of hypertension in spontaneously hypertensive rats. Hypertension. 2001;38:1044–1048. doi: 10.1161/hy1101.095331. [DOI] [PubMed] [Google Scholar]
- 7.Zaitlen N.A., et al. Inference and analysis of haplotypes from combined genotyping studies deposited in dbSNP. Genome Res. 2005;15:1594–1600. doi: 10.1101/gr.4297805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., et al. Functional allelic heterogeneity and pleiotropy of a repeat polymorphism in tyrosine hydroxylase: prediction of catecholamines and response to stress in twins. Physiol. Genomics. 2004;19:277–291. doi: 10.1152/physiolgenomics.00151.2004. [DOI] [PubMed] [Google Scholar]
- 9.Huang H.Y., Chien C.H., Jen K.H., Huang H.D. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 2006;34:W429–W434. doi: 10.1093/nar/gkl333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golderer G., et al. GTP cyclohydrolase I mRNA: novel splice variants in the slime mould Physarum polycephalum and in human monocytes (THP-1) indicate conservation of mRNA processing. Biochem. J. 2001;355:499–507. doi: 10.1042/0264-6021:3550499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halperin E., Eskin E. Haplotype reconstruction from genotype data using imperfect phylogeny. Bioinformatics. 2004;20:1842–1849. doi: 10.1093/bioinformatics/bth149. [DOI] [PubMed] [Google Scholar]
- 12.Hingorani A.D., et al. A common variant of the endothelial nitric oxide synthase (Glu298→Asp) is a major risk factor for coronary artery disease in the UK. Circulation. 1999;100:1515–1520. doi: 10.1161/01.cir.100.14.1515. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhary S., Townend J.N. Role of nitric oxide in the regulation of cardiovascular autonomic control. Clin. Sci. (Lond.). 1999;97:5–17. [PubMed] [Google Scholar]
- 14.Kishi T., et al. Cardiovascular effects of overexpression of endothelial nitric oxide synthase in the rostral ventrolateral medulla in stroke-prone spontaneously hypertensive rats. Hypertension. 2002;39:264–268. doi: 10.1161/hy0202.102701. [DOI] [PubMed] [Google Scholar]
- 15.Stein P.K., Kleiger R.E. Insights from the study of heart rate variability. Annu. Rev. Med. 1999;50:249–261. doi: 10.1146/annurev.med.50.1.249. [DOI] [PubMed] [Google Scholar]
- 16.Curtis B.M., O’Keefe J.H., Jr. Autonomic tone as a cardiovascular risk factor: the dangers of chronic fight or flight. Mayo Clin. Proc. 2002;77:45–54. doi: 10.4065/77.1.45. [DOI] [PubMed] [Google Scholar]
- 17.Palatini P., Julius S. Elevated heart rate: a major risk factor for cardiovascular disease. Clin. Exp. Hypertens. 2004;26:637–644. doi: 10.1081/ceh-200031959. [DOI] [PubMed] [Google Scholar]
- 18.Browner W.S., et al. The genetics of human longevity. Am. J. Med. 2004;117:851–860. doi: 10.1016/j.amjmed.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 19.Vijg J., Suh Y. Genetics of longevity and aging. Annu. Rev. Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- 20.Warner H.R. Longevity genes: from primitive organisms to humans. Mech. Ageing Dev. 2005;126:235–242. doi: 10.1016/j.mad.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Ichinose H., et al. Hereditary progressive dystonia with marked diurnal fluctuation caused by mutations in the GTP cyclohydrolase I gene. Nat. Genet. 1994;8:236–242. doi: 10.1038/ng1194-236. [DOI] [PubMed] [Google Scholar]
- 22.Ichinose H., Suzuki T., Inagaki H., Ohye T., Nagatsu T. Molecular genetics of dopa-responsive dystonia. Biol. Chem. 1999;380:1355–1364. doi: 10.1515/BC.1999.175. [DOI] [PubMed] [Google Scholar]
- 23.Lam A.A., Hyland K., Heales S.J. Tetrahydrobiopterin availability, nitric oxide metabolism and glutathione status in the hph-1 mouse; implications for the pathogenesis and treatment of tetrahydrobiopterin deficiency states. J. Inherit. Metab. Dis. 2007;30:256–262. doi: 10.1007/s10545-006-0502-x. [DOI] [PubMed] [Google Scholar]
- 24.Tegeder I., et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat. Med. 2006;12:1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 25.Flatmark T., Stevens R.C. Structural insight into the aromatic amino acid hydroxylases and their disease-related mutant forms. Chem. Rev. 1999;99:2137–2160. doi: 10.1021/cr980450y. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi K., Nagatsu T. Molecular genetics of tyrosine 3-monooxygenase and inherited diseases. Biochem. Biophys. Res. Commun. 2005;338:267–270. doi: 10.1016/j.bbrc.2005.07.186. [DOI] [PubMed] [Google Scholar]
- 27.Forte P., et al. Evidence for a difference in nitric oxide biosynthesis between healthy women and men. Hypertension. 1998;32:730–734. doi: 10.1161/01.hyp.32.4.730. [DOI] [PubMed] [Google Scholar]
- 28.Mendelsohn M.E., Karas R.H. Molecular and cellular basis of cardiovascular gender differences. Science. 2005;308:1583–1587. doi: 10.1126/science.1112062. [DOI] [PubMed] [Google Scholar]
- 29.Calhoun D.A., Oparil S. The sexual dimorphism of high blood pressure. Cardiol. Rev. 1998;6:356–363. doi: 10.1097/00045415-199811000-00012. [DOI] [PubMed] [Google Scholar]
- 30.Muller U., Steinberger D., Topka H. Mutations of GCH1 in Dopa-responsive dystonia. J. Neural Transm. 2002;109:321–328. doi: 10.1007/s007020200026. [DOI] [PubMed] [Google Scholar]
- 31.Boomsma D., Busjahn A., Peltonen L. Classical twin studies and beyond. Nat. Rev. Genet. 2002;3:872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- 32.MacGregor A.J., Snieder H., Schork N.J., Spector T.D. Twins. Novel uses to study complex traits and genetic diseases. Trends Genet. 2000;16:131–134. doi: 10.1016/s0168-9525(99)01946-0. [DOI] [PubMed] [Google Scholar]
- 33.Altshuler D., et al. A haplotype map of the human genome. Proc. Natl. Acad. Sci. U. S. A. 2005;37:1299–1320. [Google Scholar]
- 34.Terwilliger J.D., Hiekkalinna T. An utter refutation of the ‘Fundamental Theorem of the HapMap’. Eur. J. Hum. Genet. 2006;14:426–437. doi: 10.1038/sj.ejhg.5201583. [DOI] [PubMed] [Google Scholar]
- 35.Knowler W.C., Williams R.C., Pettitt D.J., Steinberg A.G. Gm3;5,13,14 and type 2 diabetes mellitus: an association in American Indians with genetic admixture. Am. J. Hum. Genet. 1988;43:520–526. [PMC free article] [PubMed] [Google Scholar]
- 36.Van den Brandt P.A., Willett W.C., Tannenbaum S.R. Assessment of dietary nitrate intake by a self-administered questionnaire and by overnight urinary measurement. Int. J. Epidemiol. 1989;18:852–857. doi: 10.1093/ije/18.4.852. [DOI] [PubMed] [Google Scholar]
- 37.Zaykin D.V., et al. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 38.Stram D.O., et al. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum. Hered. 2003;55:179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 39.Werner-Felmayer G., Golderer G., Werner E.R. Tetrahydrobiopterin biosynthesis, utilization and pharmacological effects. Curr. Drug Metab. 2002;3:159–173. doi: 10.2174/1389200024605073. [DOI] [PubMed] [Google Scholar]
- 40.O’Connor D.T., et al. Early decline in the catecholamine release-inhibitory peptide catestatin in humans at genetic risk of hypertension. J. Hypertens. 2002;20:1335–1345. doi: 10.1097/00004872-200207000-00020. [DOI] [PubMed] [Google Scholar]
- 41.Cockburn M., Hamilton A., Zadnick J., Cozen W., Mack T.M. The occurrence of chronic disease and other conditions in a large population-based cohort of native Californian twins. Twin Res. 2002;5:460–467. doi: 10.1375/136905202320906282. [DOI] [PubMed] [Google Scholar]
- 42.Herrmann V., et al. Beta2-adrenergic receptor polymorphisms at codon 16, cardiovascular phenotypes and essential hypertension in whites and African Americans. Am. J. Hypertens. 2000;13:1021–1026. doi: 10.1016/s0895-7061(00)01188-2. [DOI] [PubMed] [Google Scholar]
- 43.Lander E.S., et al. Initial sequencing and analysis of the human genome. Proc. Natl. Acad. Sci. U. S. A. 2001;9:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 44.Waterston R.H., et al. Initial sequencing and comparative analysis of the mouse genome. Proc. Natl. Acad. Sci. U. S. A. 2002;20:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 45.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 46.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 47.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 48.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 49.Nickerson D.A., Tobe V.O., Taylor S.L. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rieder M.J., Taylor S.L., Tobe V.O., Nickerson D.A. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy B., Ziegler M.G. A more sensitive and specific radioenzymatic assay for catecholamines. Life Sci. 1990;47:2143–2153. doi: 10.1016/0024-3205(90)90314-h. [DOI] [PubMed] [Google Scholar]
- 52.Brinton T.J., et al. Arterial compliance by cuff sphygmomanometer. Application to hypertension and early changes in subjects at genetic risk. Hypertension. 1996;28:599–603. doi: 10.1161/01.hyp.28.4.599. [DOI] [PubMed] [Google Scholar]
- 53.Appel M.L., Berger R.D., Saul J.P., Smith J.M., Cohen R.J. Beat to beat variability in cardiovascular variables: noise or music? J. Am. Coll. Cardiol. 1989;14:1139–1148. doi: 10.1016/0735-1097(89)90408-7. [DOI] [PubMed] [Google Scholar]
- 54.Waalen J., Felitti V., Gelbart T., Ho N.J., Beutler E. Prevalence of coronary heart disease associated with HFE mutations in adults attending a health appraisal center. Am. J. Med. 2002;113:472–479. doi: 10.1016/s0002-9343(02)01249-4. [DOI] [PubMed] [Google Scholar]
- 55. UCSD Chromaffin Cell & Hypertension Research. Protocols. http://medicine.ucsd.edu/hypertension/protocols.html. [Google Scholar]
- 56.Do K.A., et al. Genetic analysis of the age at menopause by using estimating equations and Bayesian random effects models. Stat. Med. 2000;19:1217–1235. doi: 10.1002/(sici)1097-0258(20000515)19:9<1217::aid-sim421>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 57.Almasy L., Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abecasis G.R., Cookson W.O. GOLD — graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 59.Clarkson D., Fan Y.A., Joe H. A remark on algorithm 643: FEXACT: an algorithm for performing Fisher’s exact test in RxC contingency tables. ACM transactions on mathematical software. 1993;19:484–488.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.