Abstract
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide, accounting for an estimated 600,000 deaths annually. Aberrant methylation, consisting of DNA hypomethylation and/or promoter gene CpG hypermethylation, is implicated in the development of a variety of solid tumors, including HCC. We analyzed the global levels of DNA methylation as well as the methylation status of 105 putative tumor suppressor genes and found that the extent of genome-wide hypomethylation and CpG hypermethylation correlates with biological features and clinical outcome of HCC patients. We identified activation of Ras and downstream Ras effectors (ERK, AKT, and RAL) due to epigenetic silencing of inhibitors of the Ras pathway in all HCC. Further, selective inactivation of SPRY1 and -2, DAB2, and SOCS4 and -5 genes and inhibitors of angiogenesis (BNIP3, BNIP3L, IGFBP3, and EGLN2) was associated with poor prognosis. Importantly, several epigenetically silenced putative tumor suppressor genes found in HCC were also inactivated in the nontumorous liver. Our results assign both therapeutic and chemopreventive significance to methylation patterns in human HCC and open the possibility of using molecular targets, including those identified in this study, to effectively inhibit HCC development and progression.
Introduction
Human hepatocellular carcinoma (HCC) is one of the most common solid tumors, rated fifth in incidence and the third in mortality worldwide (1). Although the major etiological agents have been identified, the molecular pathogenesis of HCC remains unclear (2). Deregulation of a number of oncogenes such as c-MYC, cyclin D1, and β-catenin as well as of tumor suppressor genes including P16INK4A, P53, E-cadherin, DLC-1, and pRb is observed at different frequencies in HCC (2). However, the specific genes and the molecular pathways that play pivotal roles in liver tumor development have not been identified (2).
Aberrant DNA methylation occurs commonly in human cancers in the form of genome-wide hypomethylation and regional hypermethylation (3–8). Global DNA hypomethylation (also known as demethylation) is associated with activation of protooncogenes, such as c-JUN, c-MYC, and c-Ha-Ras, and generation of genomic instability. Hypermethylation on CpG islands located in the promoter regions of tumor suppressor genes results in transcriptional silencing and genomic instability (3–11). CpG hypermethylation (also known as de novo methylation) acts as an alternative and/or complementary mechanism to gene mutations causing gene inactivation, and it is now recognized as an important mechanism in carcinogenesis. Although the mechanism(s) responsible for de novo methylation in cancer are poorly understood, it has been hypothesized that epigenetic silencing depends on activation of a number of proteins known as DNA methyltransferases (DNMTs) that possess de novo methylation activity. The importance of DNMTs in CpG methylation was substantiated by the observation that genetic disruption of both DNMT1 and DNMT3b genes in HCT116 cell line nearly eliminated methyltransferase activity (12). However, more recent findings indicate that HCT116 cells retain a truncated, biologically active form of DNMT1 and maintain 80% of their genomic methylation (13, 14). Further reduction of DNMT1 levels by an siRNA approach resulted in decreased cell viability, increased apoptosis, enhanced genomic instability, checkpoint defects, and abrogation of replicative capacity. These data show that DNMT1 is required for cell survival and suggest that DNTM1 has additional functions that are independent of its methyltransferase activity (13, 14). Concomitant overexpression of DNMT1, -3A, and -3b has been found in various tumors including HCC (15, 16). However, no changes in the expression of DNMTs were found in other neoplasms, such as colorectal cancer, suggesting the existence of alternative mechanisms (5). In HCC, a novel DNMT3b splice variant, known as DNMT3b4, is overexpressed (17). DNMT3b4 lacks DNMT activity and competes with DNMT3b3 for targeting of pericentromeric satellite regions in HCC, resulting in DNA hypomethylation of these regions and induction of chromosomal instability, further linking aberrant methylation and generation of genomic alterations (16).
It is now well accepted that methylation changes occur early and ubiquitously in cancer development. The case has been made that tumor cell heterogeneity is due, in part, to epigenetic variation in progenitor cells and that epigenetic plasticity together with genetic lesions drive tumor progression (18). The early emergence of epigenetic changes in cancer development offers an opportunity to explore the impact of these changes on the mechanism(s) of tumor development; in particular, to what extent these early epigenetic changes are retained in the fully developed tumors and/or subclasses of tumors. We have used human liver cancer to address these issues.
A growing number of genes undergoing aberrant CpG island hypermethylation in HCC have been discovered, suggesting that de novo methylation is an important mechanism underlying malignant transformation in the liver (19–22). However, most of the previous studies have focused on a single or a limited number of genes, and few have attempted to analyze the methylation status of multiple genes in HCC and associated chronic liver diseases. In addition, the functional consequence(s) of global DNA hypomethylation and CpG island hypermethylation in human liver cancer has not been investigated to date. Furthermore, to our knowledge no comprehensive analysis of CpG island hypermethylation involving activation of signaling pathways in HCC has been performed.
Our recent study on global gene expression profiles revealed that human HCC harbor common molecular features that differ greatly from those of nontumorous surrounding tissues. Also, human HCC can be subdivided into 2 broad but distinct subclasses that are associated with length of patient survival (23). These data suggest that molecular mechanisms both common for all HCC and specific for the 2 HCC subclasses may exist. This notion is further supported by recent data from our laboratory showing that frequent and concomitant aberrant methylation of Ras (RASSF1A, NORE1A) and Jak/Stat (CIS, SOCS1, -2, -3; and SHP1) inhibitors in human HCC resulted in persistent activation of the Ras and Jak/Stat pathways (24).
In the present study, we determined the levels and the functional significance of global DNA hypomethylation and CpG hypermethylation in normal livers, HCC, and matching nonneoplastic surrounding liver tissues. Subsequently, we investigated the promoter methylation status of 105 putative tumor suppressor genes, which we correlated to the biologic features of HCC and the survival data from the same patients. In addition, we analyzed molecular pathways activated following the silencing of tumor suppressor genes via promoter hypermethylation. The results indicate that the extent of global DNA hypomethylation and CpG hypermethylation correlates with biologic features and clinical outcomes of HCC. Also, we have identified molecular pathways that are both commonly activated in all HCC and those that are specific for the survival subclasses of HCC. Furthermore, we show that a number of the putative tumor suppressor genes that are epigenetically silenced in HCC are already inactivated in the surrounding nontumorous part of the liver. Our data suggest that aberrant methylation is a major event in both early and late stages of liver malignant transformation and might constitute a critical target for cancer risk assessment, treatment, and chemoprevention of HCC.
Results
Genome-wide hypomethylation correlates with HCC progression and genomic instability.
Previously, 2 distinct subclasses of HCC (subclasses A and B) were identified by gene expression profile analysis (23). Survival of subclass A HCC patients was significantly lower than that of subclass B HCC patients (23). In this study we assessed the level of global DNA hypomethylation in these 2 predefined subclasses and compared it with that found in matching nonneoplastic liver tissues and normal livers. As a measure of genome-wide hypomethylation (Figure 1A), the incorporation of [3H]dCTP into nontumorous surrounding livers was significantly higher than that in normal livers (P = 4.1 × 10–6). No differences were detected between nonneoplastic surrounding livers from the 2 subclasses of HCC. Genomic hypomethylation was significantly increased in HCC of both subclasses A and B when compared with matched nonneoplastic surrounding livers (P = 6.7 × 10–10) and was higher in subclass A than subclass B (P = 3.6 × 10–7) HCC.
Figure 1. Analysis of DNA hypomethylation, degree of genomic instability, and CpG hypermethylation in normal livers (NL), nonneoplastic surrounding liver tissues (ST), and HCC with shorter (subclass A) and longer (subclass B) survival rate.
(A) As a measure of genome-wide hypomethylation, the incorporation of [3H]dCTP into DNA was assessed after treatment with HpaII endonuclease, cutting only nonmethylated sites. In nontumorous surrounding livers, global DNA hypomethylation was significantly higher than in normal livers. Genomic hypomethylation progressively increased from nonneoplastic surrounding livers to HCC. Liver tumors with poor prognosis displayed the highest levels of DNA hypomethylation. (B) Degree of genomic alterations as detected by RAPD analysis. The level of genomic alterations was significantly higher in subclass A than in subclass B HCC (P = 7.8 × 10–8). (C) Correlation between DNA hypomethylation and genomic alterations. r2 = 0.945; P < 0.001. (D) To determine CpG regional hypermethylation, DNA was pretreated with BssHII endonuclease. Since BssHII cuts only unmethylated sites, the lower [3H]dCTP incorporation indicates fewer unmethylated (cleaved) CpG sites and, therefore, a higher de novo methylation. CpG methylation levels were highest in HCC, particular in tumors of subclass A.
Since hypomethylation contributes to carcinogenesis by promoting genomic instability (5, 9–11), we determined the level of genomic alterations by comparing each tumor with corresponding nonneoplastic liver tissue using the random amplified polymorphic DNA (RAPD) method. RAPD was selected because it allows the quantitative evaluation of a wide number of genetic alterations, ranging from large deletions and genomic rearrangements to single nucleotide mutations without prior information about loci (25, 26). The results of RAPD analysis showed that HCC of subclass A exhibited significantly higher levels of genomic instability (Figure 1B) than the subclass B tumors (P = 7.8 × 10–8). Statistical analysis showed that correlation between levels of genomic alterations and DNA hypomethylation was highly significant (Figure 1C; r2 = 0.945; P < 0.001).
CpG methylation positively correlates with HCC development and progression.
To assess levels of CpG methylation, DNA from normal liver samples, HCC, and surrounding nonneoplastic tissues was subjected to BssHII digestion (Figure 1D). CpG islands are normally unmethylated in genomic DNA. Therefore, the decrease in [3H]dCTP incorporation as compared with that in normal livers is consistent with the presence of de novo methylation. The most pronounced decrease in [3H]dCTP uptake and therefore the highest level of CpG methylation was found in subclass A HCC (P = 4.0 × 10–4 compared with subclass B tumors). Like genomic hypomethylation, CpG hypermethylation was correlated to the number of genomic alterations (r2 = 0.3395; P < 0.0007). Thus, HCC development and progression are characterized by progressively increased levels of global DNA hypomethylation, regional CpG hypermethylation, and genomic instability.
Tumor suppressor genes are differentially methylated during HCC development and progression.
We investigated the promoter status of 105 putative tumor suppressor genes by methylation-specific PCR analysis (Figure 2 and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI31457DS1). The genes were selected based on their capacity to modulate signaling pathways (Ras, Jak/Stat, Wingless/Wnt, and RELN) and/or biologic features of the tumors (proliferation, apoptosis, angiogenesis, invasion, DNA repair, immune response, and detoxification). Normal livers showed the absence of promoter methylation for all genes examined. Based on promoter methylation frequency, the tumor suppressor genes can be divided in 4 different categories: (a) rarely or not methylated; (b) methylated at comparable frequency in HCC and the matching nonneoplastic surrounding livers; (c) highly methylated in all HCC; and (d) highly methylated only in subclass A HCC (Figure 2 and Supplemental Table 1). At least 1 of the genes involved in inhibition of Ras (ARHI, CLU, DAB2, hDAB2IP, HIN-1, HRASL, LOX, NORE1A, PAR4, RASSF1A, RASSF2, RASSF3, RASSF4, RIG, RRP22, and SPRY 2 and -4), Jak/Stat (ARHI, CIS, SHP1, PIAS-1, PIAS-γ, SOCS1, -2, and -3, SYK, and GRIM-19), and Wnt/β-catenin (APC, E-cadherin, γ-catenin, SFRP1, -2, -4, and -5, DKK-1 and -3, WIF-1, and HDPR1) pathways was affected by de novo methylation in all HCC. Significantly, a number of these genes were also highly methylated in the surrounding nontumorous liver (Supplemental Table 1). In contrast, inactivation of at least 1 of the genes implicated in the RELN pathway (DAB1, reelin) was detected only in HCC of subclass A but very rarely in subclass B tumors, implying their role in liver tumor aggressiveness and progression. This is consistent with the observation that suppression of the RELN pathway enhances the invasiveness of pancreatic cancer cells and is associated with significantly shorter survival of pancreatic cancer patients (27).
Figure 2. Summary of promoter hypermethylation frequency in subclass A and B HCC as detected by methylation-specific PCR.
Black and gray boxes indicate the presence or absence of promoter hypermethylation, respectively. Note the significantly higher number of genes affected by methylation in subclass A HCC. Only genes that were affected by promoter methylation in at least 1 sample are shown.
To accurately quantify the degree of promoter hypermethylation, we selected 30 genes and subjected them to combined bisulfite restriction analysis (COBRA; Supplemental Table 2). Again, mean methylation density for all genes was significantly higher in HCC when compared with surrounding nonneoplastic liver tissues (Supplemental Table 2). Furthermore, 20 of 30 (66.6%) genes displayed significantly higher methylation density in subclass A HCC when compared with subclass B tumors. Three genes (LOX, MAD2, and RASSF2) exhibited higher methylation in subclass B HCC, and 7 genes exhibited similar hypermethylation in the 2 subclasses of HCC. Additional quantitative RT-PCR analysis of 8 genes showed that levels of gene expression inversely correlated with the degree of promoter methylation (Supplemental Figure 1).
Next, we assessed the prognostic value of promoter hypermethylation in hepatocarcinogenesis by determining the methylation index in HCC of subclasses A and B (Figure 3A). The mean methylation frequency was significantly higher in subclass A than subclass B HCC (P = 3.6 × 10–9). A bimodal distribution for number of genes methylated in each case was also consistent with the existence of 2 subclasses of HCC with distinct hypermethylation profiles (Supplemental Figure 2). Furthermore, an inverse correlation was found between the methylation index and the length of patient survival (r2 = 0.7387; P < 0.0001) (Figure 3B). Taken together, these data indicate that levels of gene promoter methylation progressively increase during HCC development and progression and suggest a prognostic significance for the methylation index in human HCC.
Figure 3. Methylation index in HCC from patients with shorter (subclass A) and longer (subclass B) survival.
Methylation index (or mean frequency of methylation) was defined as the ratio between the numbers of methylated genes to total number of examined genes in each sample. (A) Methylation index was highest in subclass A HCC. (B) Methylation index negatively correlated with the length of patient’s survival (r2 = 0.7387; P < 0.0001).
Epigenetic silencing of multiple tumor suppressor genes maintains activation of the Ras pathway.
A major finding of our analysis was the concurrent hypermethylation of multiple inhibitors of the Ras pathway. Among them, several inhibitors were equally methylated in the 2 subclasses of HCC, whereas ARHI, DAB2, hDAB2IP, NORE1A, PAR4, RRP22, and SPRY2 and -4 were almost exclusively methylated and silenced in HCC of subclass A, suggesting their role in liver tumor progression (Figure 2 and Supplemental Table 1). Consistent with methylation data, Ras was significantly more active in HCC than in surrounding (P = 2.25 × 10–8) or normal livers (P = 1.4 × 10–7) (Figure 4A). HCC of subclass A displayed higher levels of Ras activation than subclass B HCC (P = 0.03). Similar results were obtained when determining ERK and AKT activation in the same collection of samples (Figure 4, B and C). Intriguingly, activation of Ral A was detected mostly at the neoplastic stage, with subclass A HCC showing the highest levels of Ral A activity (Figure 4D).
Figure 4. Activation of Ras and Ras downstream effectors in normal livers, nonneoplastic surrounding tissues, and HCC.
Activity of Ras (A), ERK (B), AKT (C), and Ral A (D) progressively increased from nontumorous surrounding livers to HCC, reaching the highest levels in subclass A HCC. Of note, the Ras downstream effector Ral A was active mostly in subclass A HCC. Proteins levels of total Ras (Pan Ras), ERK, AKT, and Ral A are shown to ensure equal protein loading. MBP, myelin basic protein; pGSK-3β, phospho–glycogen synthase kinase–3β.
To rule out the possibility that Ras activation was due to somatic mutations in 1 or more Ras members, we assessed the mutation frequency in H-Ras, K-Ras, and N-Ras genes as well as in EGFR (an upstream modulator of Ras) and downstream effectors (A-RAF, B-RAF, and RAF-1). No mutations were detected in any of the examined genes (data not shown). Subsequent Western blot analysis showed that RAF-1 expression was higher in subclass A HCC, whereas no significant differences were detected in A-RAF and B-RAF levels in the 2 subclasses of HCC, as wells as normal and nontumorous surrounding livers (Supplemental Figure 3A). Furthermore, mutations in the catalytic subunit p110α of phosphatidylinositol 3-kinase (PIK3CA), an upstream regulator of AKT, were detected in 50% (15 of 30) of HCC of subclass A but only in 10% (3 of 30) of HCC of subclass B. Mutations were confined to the residues H1047 (11 of 18; 61.1%), E545 (6 of 18; 33.3%), and E542 (1 of 18; 5.5%), which result in gain of enzymatic function and oncogenic activity of PIK3CA (28). The highest levels of PIK3CA protein were detected in the subclass A HCC, independent of PIK3CA somatic mutations (Supplemental Figure 3A). The high frequency of PIK3CA mutations and/or its upregulation in subclass A HCC might be at least partly responsible for the Akt hyperactivation found in HCC with poor prognosis.
It is well documented that Ras is a downstream effector of many growth factors and cytokines, including EGFR (29). Also, using a comparative functional genomics analysis, we have recently shown that HCC from transgenic mice coexpressing TGF-α, a ligand for EGFR, and c-Myc is a good model for subclass A HCC (30). Indeed, we found that both total and phosphorylated levels of EGFR progressively increased from surrounding livers to subclass A HCC (Figure 5A), indicating that Ras activation might be partly due to increased EGFR signaling. Levels of EGFR inversely correlated with those of EGFR inhibitors SOCS4 and -5 (31, 32), which were suppressed by methylation in most of subclass A HCC. In contrast, SOCS4 and-5 were generally upregulated in subclass B HCC (Figure 5A, Supplemental Table 1, and Supplemental Figure 4). Accordingly, levels of EGFR bound to SOCS5 and thus targeted for degradation were highest in subclass B HCC and lowest in subclass A HCC (Figure 5A).
Figure 5. Representative Western blot and immunoprecipitation analyses of Ras inhibitors and Ras downstream effectors in normal livers, nonneoplastic surrounding tissues, and HCC.
(A) Downregulation of EGFR inhibitors SOCS4 and SOCS5 resulted in increased EGFR levels, EGFR activation, and low EGFR/SOCS5 complexes in subclass A HCC. (B) Downregulation of SPRY1, -2, and -4 led to FGF receptor (FGFR) upregulation and increased FGF-mediated Ras signaling activation, as shown by a decrease in SPRY1/GRB2 and SPRY2/GRB2 complexes and increase in FRS2/GRB2 complexes in subclass A HCC. (C) In contrast, subclass B HCC showed elevated levels of both total (t) and membranous (m) SPRY1 and -2, leading to SPRY1 and -2 activation by tyrosine phosphorylation, increase in SPRY1/GRB2 and SPRY2/GRB2 complexes, and decrease in FRS2/GRB2 complexes (C). (D) DAB2 was downregulated in subclass A HCC, with consequent upregulation of c-Fos, Src, and ILK.
We also found a progressive increase in expression of FGF receptor (FGFR) which was paralleled by downregulation of SPRY2 and SPRY4 via promoter methylation (Figure 2, Figure 5B, Supplemental Table 1, and Supplemental Figure 4). In addition, levels of SPRY1, another inhibitor of the FGF-driven Ras/ERK activation, were lowest in subclass A HCC, although unrelated to promoter hypermethylation (Figure 2, Figure 5B, and Supplemental Table 1). Levels of activated SPRY1 and SPRY2 by tyrosine phosphorylation (33) were lowest in subclass A and highest in subclass B HCC (Figure 5C). Membranous localization of SPRY1 and SPRY2, another marker of SPRY1 and SPRY2 activation, was detected in tumors and to a lesser extent in surrounding liver tissues and was associated with an increase in SPRY1/GRB2 and SPRY2/GRB2 complexes and a decrease in FRS2/GRB2 complexes (Figure 5, B and C). In contrast, downregulation of SPRY1 and SPRY2 in subclass A HCC was paralleled by low SPRY1/GRB2 and SPRY2/GRB2 complexes and increased FRS2/GRB2 complexes (Figure 5C).
Similarly, levels of DAB2, another Ras inhibitor, were lowest in HCC of subclass A, in accordance with promoter methylation data (Figure 2, Figure 5D, and Supplemental Figure 4). In addition, the frequency of promoter hypermethylation at DAB2 downstream effectors hDAB2IP and GATA factors 4, 5, and 6 was significantly higher in subclass A HCC (Figure 2 and Supplemental Table 2). In particular, the hDAB2IP gene was affected by hypermethylation in all subclass A HCC, whereas at least 1 of GATA factors 4, 5, and 6 was hypermethylated in all HCC, with shorter survival length.
DAB2 suppresses the Ras cascade via binding to GRB2, thus disrupting the binding between GRB2 and SOS1 (34). As expected, subclass A HCC exhibited low DAB2/GRB2 and high SOS1/GRB2 complexes, whereas subclass B HCC showed high levels of both DAB2 and GRB2/DAB2 complexes and low SOS1/GRB2 complexes (Figure 5D and Supplemental Figure 4). As a consequence, the levels of c-Fos, ILK, and activated c-Src, which are downregulated by DAB2 (35, 36), were higher in the subclass A HCC (Figure 5D).
Selective pressure toward Ras activation in hepatocarcinogenesis was further substantiated by the progressive induction of other upstream inducers of Ras. The expression of PGDFRα and -β, VEGFR2, and RET was considerably higher in subclass A than in subclass B HCC (Supplemental Figure 3B). Thus, concomitant suppression of multiple Ras inhibitors and upregulation of Ras upstream inducers underlie unrestrained activation of Ras cascade during HCC development and progression.
Promotion of Ras-induced angiogenesis in human HCC.
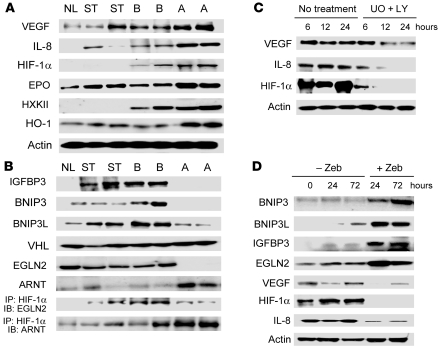
Previously, it has been shown that activation of the Ras pathway contributes to tumor angiogenesis by transcriptional activation of VEGF, HIF-1α, and IL-8 target genes (37, 38). Expression levels of these proteins as well as of HIF-1α targets, including erythropoietin (EPO), hexokinase II (HXKII), and heme oxygenase–1 (HO-1), progressively increased from normal livers to subclass A HCC (Figure 6A). In addition, the latter tumors displayed significantly more frequent promoter hypermethylation and decreased levels of HIF-1α proapoptotic targets, including BNIP3, BNIP3L, and IGFBP3 (Figure 2, Figure 6B, Supplemental Figure 4, and Supplemental Table 1). However, we could not detect the presence of the HIF-1α785 splice variant, shown to be specifically induced via Raf/MEK/ERK axis (39), in any of the samples analyzed by RT-PCR (data not shown), indicating that HIF-1α785 does not play a role in HCC angiogenesis.
Figure 6. Representative Western blot and immunoprecipitation (IP) analyses of Ras proangiogenic targets (A) and inhibitors of Ras-driven angiogenesis (B).
Upregulation of HIF-1α, VEGF, and IL-8 (A) was associated with high HIF-1α/ARNT complexes, low levels of HIF-1α inhibitors (IGFBP3, BNIP3, BNIP3L, and EGLN2), and low EGLN2/HIF-1α complexes in subclass A HCC (B). (C) Suppression of Ras downstream effectors MEK (UO126 [UO]) and AKT (LY294002 [LY]) led to downregulation of HIF-1α, VEGF, and IL-8 in Focus hepatoma cell lines. (D) Reactivation of HIF-1α inhibitors and EGLN2 by Zebularine (Zeb) reduced HIF-1α, VEGF, and IL-8 expression in Focus hepatoma cell lines. Equivalent results were obtained in SNU-387 and 7703 hepatoma cell lines (data not shown).
Since HIF-1α stability is regulated by various posttranslational modifications (40), we assessed the levels of HIF-1α regulators VHL and EGLN2. VHL slightly increased concomitantly with HIF-1α induction from nonneoplastic to neoplastic lesions, presumably in an attempt to counteract HIF-1α activity. In contrast, loss or reduction of EGLN2 expression was detected only in HCC of subclass A, in accordance with promoter methylation data (Figure 6B). As a consequence, very low levels of EGLN2/HIF-1α complexes were found in subclass A HCC (Figure 6B). Furthermore, subclass A showed increased levels of the HIF-1α stabilizer ARNT (HIF-1β), leading to increased HIF-1α/ARNT nuclear complexes (Figure 6B). The relevance of Ras proangiogenic effector upregulation in HCC was further demonstrated by higher microvessel density (MVD) in HCC of subclass A (189.70 ± 8.12 versus 115.23 ± 7.70 in subclass B HCC; P = 1.43 × 10–7).
To more directly assess the role played by Ras activation in HCC angiogenesis, 3 HCC cell lines (SNU-387, 7703, and Focus) with elevated levels of HIF-1α, VEGF, and IL-8, were treated with MEK/ERK (UO126) and AKT (LY294002) inhibitors. Each inhibitor was able to reduce the amount of proangiogenic proteins described above (data not shown), but their combination led to almost complete inhibition of VEGF, HIF-1α, and IL-8 (Figure 6C) in all 3 hepatoma cell lines. Similarly, treatment of the same cell lines with the demethylating agent Zebularine caused a marked upregulation of BNIP3, BNIP3L, IGBP3, and EGLN2 proteins concomitantly with downregulation of VEGF, HIF-1α, and IL-8 (Figure 6D), indicating a crucial role for Ras pathway in human HCC angiogenesis.
Discussion
The results of this study demonstrate that both DNA hypomethylation and regional CpG hypermethylation are dominant events during human HCC development and progression. Importantly, no significant association between methylation patterns and specific etiologic agents (i.e., HBV, HCV, ethanol, etc.) has been detected in the present investigation, further substantiating the conclusion that aberrant methylation is a ubiquitous phenomenon in hepatocarcinogenesis. Current evidence suggests that hypomethylation might promote malignant transformation via multiple mechanisms, including chromosomal instability, activation of protooncogenes, reactivation of transposable elements, and loss of imprinting (3–11). Generation of genomic instability by hypomethylation seems to depend on the propensity of hypomethylated DNA to recombine with a high frequency, resulting in a variety of chromosomal rearrangements in regions containing hypomethylated DNA (5, 9–11). Here, we show that levels of DNA hypomethylation directly correlate with those of genomic alterations in human HCC. The degree of DNA hypomethylation progressively increased from nonneoplastic livers to fully malignant HCC, indicating that genomic hypomethylation is an important prognostic factor in HCC, as reported for brain, breast, and ovarian cancer (41–43).
Regional CpG hypermethylation was also enhanced during the course of HCC disease. The more aggressive subclass A HCC displayed a very high frequency of gene promoter hypermethylation, a condition similar to the CpG island methylator phenotype (CIMP) described in human colorectal cancer (44, 45) but absent in breast tumors (46). However, in contrast to CIMP+ colorectal tumors, which have mutated B-RAF and show a high rate of microsatellite instability, HCC are characterized by wild-type B-RAF and gross chromosomal alterations, as detected in this study by RAPD analysis and by others using cytogenetic and microsatellite analyses (47).
Extensive study of tumor suppressor gene promoters showed that CpG methylation was frequently detected both in surrounding nontumorous livers and HCC. Numerous tumor suppressor genes commonly silenced by hypermethylation in other solid tumors were not affected in HCC, indicating that their suppression might be tissue specific. However, this finding does not exclude the possibility that these genes may be inactivated in HCC via nonepigenetic mechanisms. Interestingly, LOX, MAD2, and RASSF2 genes were preferentially inactivated in subclass B HCC. The reason for selective hypermethylation of their promoters in HCC with better prognosis is not clear. In this regard, it is tempting to hypothesize that inactivation of LOX, RASSF2, and MAD2 is required during HCC development, whereas reactivation of these genes during HCC progression might provide growth and invasive advantages to tumor cells. According to this hypothesis, inactivation of E-cadherin is predominantly detected in HCC with no vascular invasion, whereas increased E-cadherin expression is characteristic for liver tumors with marked vascular invasion (48). Similarly, MAD2 overexpression is a frequent event in aggressive tumors with elevated genomic instability and poor prognosis, where MAD2 upregulation is associated with defective mitosis leading to aneuploidy (49). In addition, although LOX methylation and downregulation have been detected in gastric tumors (50), LOX upregulation can confer migration and invasive advantages to tumor cells during hypoxia-induced metastasis (51, 52). These data suggest that LOX, MAD2, and presumably RASSF2 might play diverse and seemingly paradoxical roles in HCC, reflecting specific requirements during different stages of tumor progression.
A major question addressed in the present study concerns the prognostic value of the CpG methylation index in human HCC. Our data indicate that the methylation index might be useful in predicting survival of patients with HCC. In particular, we identified a number of candidate genes whose promoter methylation is highly associated with liver tumor development and/or progression. Future studies using a selected number of these genes might be helpful in predicting the life expectancy of the patients as well as the therapeutic approach.
Among other important findings of our analysis is the concomitant inactivation of several inhibitors of the Jak/Stat, Wnt/β-catenin, and Ras pathways in HCC as well as a high methylation frequency of many of the same inhibitors found in the surrounding nontumorous liver. These data indicate that unrestrained activation of Jak/Stat, Wnt/β-catenin, and Ras cascades is required for liver cancer development. We hypothesize that the methylation-controlled gene suppression starts at an early stage (i.e., chronic hepatitis and/or dysplastic stage) of hepatocarcinogenesis, similar to what has been described for other cancers (53). In combination with mutations and genomic instability, it provides a milieu for the selection of simultaneous suppression of numerous regulatory genes. Accordingly, we found the concurrent suppression of multiple inhibitors of the Ras/ERK axis, including SPRY1, SPRY2, SPRY4, and DAB2. Furthermore, inactivation of another sprouty family member, SPRED-1, has recently been described in HCC, and SPRED-1 levels were inversely correlated with the incidence of tumor invasion and metastasis (54). In accordance with this hypothesis, activation of Ras and downstream effectors of Ras (Raf/ERK, AKT, and Ral) was associated with downregulation of multiple inhibitors in a stage-dependent and cumulative manner. Promoter hypermethylation of CNK1 and HRASLS was an early event but was also present in HCC, whereas silencing of DAB2, GATA 4, -5, and -6, hDAB2IP, NORE1A, SPRY2 and -4, and SOCS-4 and -5 occurred almost exclusively in HCC, and most frequently in subclass A. Thus, it seems that the magnitude of Ras signaling is defined by the number of inhibitors affected by promoter hypermethylation. Furthermore, since Ras inhibitors regulate numerous molecular pathways, their suppression may lead to persistent activation of signaling cascades other than the Ras pathway. Indeed, we show that deregulation of DAB2 was accompanied by higher levels of c-Fos, ILK, and activated c-Src in HCC of subclass A.
Furthermore, subclass A HCC exhibit a remarkable hyperactivation of Ras due to induction of numerous upstream Ras inducers, including PDGFR, VEGFR2, and RET; although further studies are needed to determine whether the activation of these kinases is caused by the silencing of their inhibitors, these findings may have important implications for the development of new therapeutic approaches for human HCC. Recent evidence indicates a strong anticancer activity of the multikinase inhibitor sorafenib via suppression of RAF, PDGFR, VEGFR2, and RET in a variety of preclinical tumor models, including HCC (55). Our results support the value of the ongoing phase III clinical trials using sorafenib for HCC patients (55).
The role of Ras oncogenes in driving cellular transformation, proliferation, and survival is well established. However, the contribution of Ras signaling to tumor angiogenesis remains poorly characterized. Here, we demonstrate that a number of proangiogenic proteins, including VEGF, HIF-1α, and IL-8, are Ras targets in liver cancer. These proteins are predominantly upregulated in subclass A HCC, consistent with the notion that angiogenesis is associated with tumor progression. In particular, upregulation of Ras proangiogenic proteins in tumors with poor prognosis was the result of multiple, apparently independent alterations, such as promoter hypermethylation of genes involved in downregulation of HIF-1α (EGLN2) and HIF-1α–induced apoptosis (BNIP3, BNIP3L, IGFBP3) as well as strong induction of the HIF-1α stabilizer ARNT. Thus, liver tumors with poor prognosis are characterized by selective pressure toward accumulating alterations in genes regulating angiogenesis. Importantly, treatment of HCC cell lines with the demethylating agent Zebularine suppressed Ras proangiogenic targets and reactivated antiangiogenic genes, including BNIP3, BNIP3L, IGFBP3, and EGLN2, suggesting that Zebularine might be an effective therapeutic agent against HCC-driven angiogenesis. These results further strengthen and substantiate the recent finding that demethylating agents posses a strong angiostatic activity (56).
In summary, the present work underlines the functional importance and the prognostic value of genomic hypomethylation and regional CpG hypermethylation in human hepatocarcinogenesis. Therapeutic strategies able to modify the methylation status of liver cancer cells and to target either Ras upstream inducers (55) or downstream effectors (e.g., RAF-1, ERK, AKT, Ral) may be highly beneficial in the treatment of human HCC. Finally, we propose that aberrant methylation is a major event during early stages of malignant transformation as well as in the progression of HCC and therefore might represent a critical target for risk assessment, chemoprevention, and treatment of HCC.
Methods
Human tissue samples and preparation of DNA.
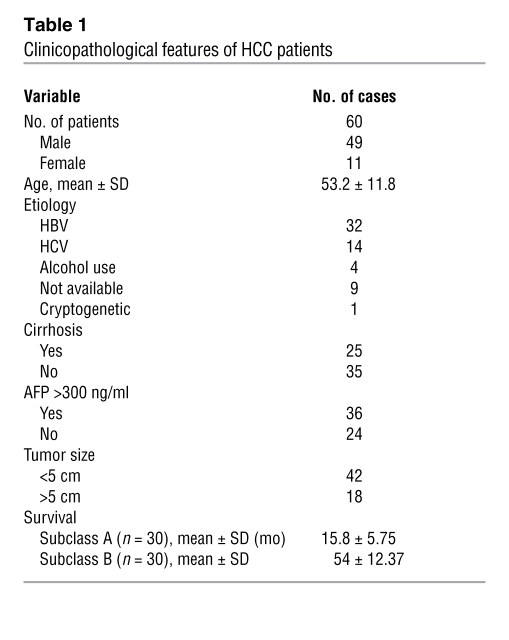
A total of 10 normal livers, 60 HCC, and matched surrounding nontumor liver tissues were used. Clinicopathological features of the patients in this study are shown in Table 1. HCC were divided in 2 subclasses based on patient’s survival length as previously reported (23). In brief, HCC of subclasses A or B were characterized by a shorter (<3 years) or longer (>3 years) survival length following liver partial resection, respectively. Liver samples were kindly provided by Z. Sung (National Laboratory of Molecular Oncology, Cancer Institute, Beijing, China) and the Liver Tissue Procurement and Distribution System (LTPDS), which was funded by NIH contract no. N01-DK-9-2310. Institutional Review Board approval was obtained at participating hospitals in the LTPDS (University of Minnesota, Minneapolis, Minnesota, USA, and University of Pittsburgh, Pittsburgh, Pennsylvania, USA) and the NIH. High-molecular-weight DNA was isolated from frozen tissues as described previously (57).
Table 1 .
Clinicopathological features of HCC patients
Assessment of global and regional DNA methylation status.
Two micrograms of genomic DNA from 6 normal livers, 30 HCC (15 subclass A and 15 subclass B), and matched nonneoplastic surrounding livers were pretreated with either HpaII or BssHII endonucleases (to assess the degree of DNA hypomethylation or CPG hypermethylation, respectively) and subjected to the cytosine extension assay as described previously (58). MspI, an isoschizomer of HpaII insensitive to methylation status, was used as a control for efficiency of enzyme digestion. Relative incorporation of [3H]dCTP was compared between samples. The assay was repeated at least 3 times, and the data expressed as mean ± SEM.
RAPD analysis.
Fifteen GC-rich arbitrary primers (D1Byu1-D9Byu1; ref. 26) were used to score genomic alterations in 30 HCC (15 subclass A and 15 subclass B) and respective nonneoplastic surrounding livers. RAPD reaction was performed as described previously (26). Differences were scored in case of change in the intensity of a band, absence of a band, or appearance of a new band in HCC when compared with corresponding nontumorous livers.
Methylation-specific PCR, methylation index, and COBRA.
Genomic DNA from 10 normal livers, 60 HCC, and matching surrounding livers was modified using the EZ DNA Methylation Kit (Zymo Research). CpGenome Universal Methylated DNA and CpG Universal Unmethylated DNA (Chemicon International) were used as positive and negative control for each reaction, respectively. Primers used in this study were as previously published (19–22, 27, 46, 48, 50, 56, 59–69), and the identity of PCR products was verified by genomic bisulfite sequencing. A total of 30 ng of DNA was amplified in a reaction volume of 15 μl consisting of 0.4 μM each primer, 200 μM each dNTP, 10× PCR buffer, 5× GC-RICH solution, and 0.4 U of FastStart Taq DNA Polymerase (Roche Applied Science). Methylation index was assessed by determining the ratio of methylated genes to the number of genes examined. Quantitation of promoter methylation density of 30 genes was determined using the combined bisulfite restriction analysis (COBRA) as previously described (70). COBRA consists of a standard sodium bisulfite PCR treatment followed by restriction digestion and quantitation (70).
Western blot and immunoprecipitation analyses.
Five normal livers, 38 HCC (20 subclass A and 18 subclass B), and matching surrounding livers were used. Hepatic tissue samples were processed as previously described (71). To assess SPRY1 and SPRY2 subcellular localization, membranous fractions were obtained using the Mem-PER Eukaryotic Membrane Protein Extraction Kit (Pierce Biotechnology) following the manufacturer’s protocol. For immunoprecipitation studies, a total of 500 mg of normal livers (n = 5), HCC (n = 28; 14 subclass A and 14 subclass B), and corresponding surrounding nonneoplastic liver samples were homogenized and processed as previously reported (72).
Real-time RT-PCR analysis.
Primers for SOCS4, SOCS5, SPRY2, SPRY4, DAB2, IGFBP3, BNIP3, BNIP3L, EGLN2, RASSF1A, SOCS2, HRASLS, LOX, MAD2, ARHI, DAPK, CASP8, and ribonucleic acid ribosomal 18S (RNR-18; used as internal control) were chosen by the TaqMan Gene Expression Assays (Applied Biosystems). PCR reactions were performed with 75–300 ng of cDNA, using an ABI Prism 7000 and TaqMan Universal PCR Master Mix (Applied Biosystems) following the manufacturer’s instructions. Quantitative values were calculated using PE Biosystems Analysis software. Data are expressed as the mean ± SD of N target (NT); NT = 2–ΔCt, where the ΔCt value of the sample was calculated by subtracting the average Ct value of the target gene from the average Ct value of the 18S gene.
Ras, ERK, AKT, and Ral activation assay.
Ras, MAPK/ERK, AKT, and Ral activation was determined on 8 normal livers, 22 HCC (11 subclass A and 11 subclass B), and matched surrounding livers by using the Ras (Chemicon International), MAPK/ERK, AKT (Cell Signaling Technology), and Ral (Upstate USA Inc.) kinase assay kits, respectively, following the manufacturers’ protocols. The bands on the photos were scanned, and the densities of the peaks were calculated in arbitrary units using ImageQuaNT 5.1 software (Molecular Dynamics). Values are expressed as mean ± SEM.
Mutation analyses.
Mutations at H-Ras, K-Ras, N-Ras, A-Raf, B-Raf, Raf-1, EGFR, and PIK3CA were assessed in 10 normal livers, 60 HCC, and respective surrounding nontumor liver tissues as previously described (73–76).
Hepatoma cell line treatments.
The SNU-387, 7703, and Focus human hepatoma cell lines were maintained as monolayer cultures in either DMEM or RPMI medium supplemented with 10% FBS as described previously (77). Cell lines were plated at a density of 2.0 × 106 cells in 10-cm dishes and treated with 20 mM UO126 (MEK inhibitor), 50 μM LY294002 (AKT inhibitor), or both for 6, 12, and 24 hours and with Zebularine (200 μM) for 24–72 hours. Zebularine was provided by the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute.
Evaluation of MVD.
Fifteen HCC per group were subjected to immunostaining with mouse monoclonal anti-CD34 antibody (Vector Laboratories). HCC were screened at low power (×40) to identify the areas of highest MVD. The 4 highest MVD areas for each tumor were photographed at high power (×200) and the size of each area standardized using ImageJ software (http://rsb.info.nih.gov/ij/). MVD was expressed as the percentage (mean ± SEM) of the total CD34-stained spots per section area (0.94 mm2).
Statistics.
The 2-tailed Student’s t test was used to evaluate statistical significance. Pearson’s correlation testing was used to assess the correlation between DNA hypomethylation and genomic alterations by PRISM 2.00 (GraphPad Software). P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We are grateful to M. Goggins for providing the primers for DAB1 methylation studies. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Nonstandard abbreviations used: COBRA, combined bisulfite restriction analysis; DNMT, DNA methyltransferase; HCC, hepatocellular carcinoma(s); MVD, microvessel density; RAPD, random amplified polymorphic DNA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2713–2722 (2007). doi:10.1172/JCI31457
References
- 1.Bruix J., Boix L., Sala M., Llovet J.M. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson S.S., Grisham J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 3.Baylin S.B. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin. Cancer Biol. 2002;12:331–337. doi: 10.1016/s1044-579x(02)00053-6. [DOI] [PubMed] [Google Scholar]
- 4.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. . 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson A.S., Power B.E., Molloy P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–16. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg A.P. The epigenetics of cancer etiology. Semin. Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M. DNA methylation and cancer therapy: new developments and expectations. Curr. Opin. Oncol. . 2005;17:55–60. doi: 10.1097/01.cco.0000147383.04709.10. [DOI] [PubMed] [Google Scholar]
- 8.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 9.Chen R.Z., Pettersson U., Beard C., Jackson-Grusby L., Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 10.Gaudet F., et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 11.Kim M., Trinh B.N., Long T.I., Oghamian S., Laird P.W. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 2004;32:5742–5749. doi: 10.1093/nar/gkh912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee I., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 13.Egger G., et al. Identification of DNMT1 hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown K.D., Robertson K.D. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. . 2007;39:289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 15.Robertson K.D., et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito Y., et al. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. . 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y., et al. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinberg A.P., Ohlsson R., Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 19.Lee S., et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am. J. Pathol. 2003;163:1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang B., Guo M., Herman J.G., Clark D.P. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am. J. Pathol. 2003;163:1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J., et al. Methylation profiles of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer. 2002;2:29. doi: 10.1186/1471-2407-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J., et al. Methylation profiling of twenty four genes and the concordant methylation behaviors of nineteen genes that may contribute to hepatocellular carcinogenesis. Cell Res. 2003;13:319–333. doi: 10.1038/sj.cr.7290177. [DOI] [PubMed] [Google Scholar]
- 23.Lee J.S., et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 24.Calvisi D.F., et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2005;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Atienzar F.A., Jha A.N. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat. Res. . 2006;613:76–102. doi: 10.1016/j.mrrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Calvisi D.F., Factor V.M., Ladu S., Conner E.A., Thorgeirsson S.S. Disruption of beta-catenin pathway or genomic instability define two distinct categories of liver cancer in transgenic mice. Gastroenterology. . 2004;126:1374–1386. doi: 10.1053/j.gastro.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Sato N., Fukushima N., Chang R., Matsubayashi H., Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–565. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Bader A.G., Kang S., Zhao L., Vogt P.K. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 29.Repasky G.A., Chenette E.J., Der C.J. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell. Biol. 2004;14:639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.S., et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 31.Kario E., et al. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J. Biol. Chem. 2005;280:7038–7048. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- 32.Nicholson S.E., et al. Suppressor of cytokine signaling 5 is a potential negative regulator of epidermal growth factor signaling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2328–2333. doi: 10.1073/pnas.0409675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H.J., Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat. Rev. Mol. Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., Scholes J., Hsieh J.T. Characterization of a novel negative regulator (DOC-2/DAB2) of c-Src in normal prostatic epithelium and cancer. J. Biol. Chem. 2003;278:6936–6941. doi: 10.1074/jbc.M210628200. [DOI] [PubMed] [Google Scholar]
- 35.He J., Smith E.R., Xu X.X. DAB-2 exerts its tumor suppressor activity by uncoupling c-Fos expression and MAP kinase activation. J. Biol. Chem. 2001;276:26814–26818. doi: 10.1074/jbc.M101820200. [DOI] [PubMed] [Google Scholar]
- 36.Wang S.C., et al. DOC-2/hDab-2 inhibits ILK activity and induces anoikis in breast cancer cells through an Akt-independent pathway. Oncogene. 2001;20:6960–6964. doi: 10.1038/sj.onc.1204873. [DOI] [PubMed] [Google Scholar]
- 37.Kranenburg O., Gebbink M.F., Voest E.E. Stimulation of angiogenesis by Ras proteins. Biochim. Biophys. Acta. 2004;1654:23–37. doi: 10.1016/j.bbcan.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Sparmann A., Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 39.Lim J.H., et al. Ras-dependent induction of HIF-1α785 via the Raf/MEK/ERK pathway: a novel mechanism of Ras-mediated tumor promotion. . Oncogene. 2004;23:9427–9431. doi: 10.1038/sj.onc.1208003. [DOI] [PubMed] [Google Scholar]
- 40.Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 41.Soares J., et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- 42.Uhlmann K., et al. Distinct methylation profiles of glioma subtypes. Int. J. Cancer. 2003;106:52–59. doi: 10.1002/ijc.11175. [DOI] [PubMed] [Google Scholar]
- 43.Widschwendter M., et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 44.Toyota M., et al. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisenberger D.J., et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 46.Bae Y.K., et al. Hypermethylation in histologically distinct classes of breast cancer. Clin. Cancer Res. 2004;10:5998–6005. doi: 10.1158/1078-0432.CCR-04-0667. [DOI] [PubMed] [Google Scholar]
- 47.Piao Z., Kim H., Malkoshyan S., Park C. Frequent chromosomal instability but not microsatellite instability in hepatocellular carcinomas. Int. J. Oncol. . 2000;17:507–512. doi: 10.3892/ijo.17.3.507. [DOI] [PubMed] [Google Scholar]
- 48.Wei Y., et al. Altered expression of E-cadherin in hepatocellular carcinoma: correlations with genetic alterations, beta-catenin expression, and clinical features. Hepatology. 2002;36:692–701. doi: 10.1053/jhep.2002.35342. [DOI] [PubMed] [Google Scholar]
- 49.Hernando E., et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 50.Kaneda A., et al. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410–6415. doi: 10.1158/0008-5472.CAN-04-1543. [DOI] [PubMed] [Google Scholar]
- 51.Kirschmann D.A., et al. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002;62:4478–4483. [PubMed] [Google Scholar]
- 52.Erler J.T., et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 53.Herman J.G. Epigenetic changes in cancer and preneoplasia. Cold Spring Harb. Symp. Quant. Biol. 2005;70:329–333. doi: 10.1101/sqb.2005.70.036. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida T., et al. Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene. 2006;25:6056–6066. doi: 10.1038/sj.onc.1209635. [DOI] [PubMed] [Google Scholar]
- 55.Liu L., et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 56.Hellebrekers D.M., et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol. Cancer Ther. 2006;5:467–475. doi: 10.1158/1535-7163.MCT-05-0417. [DOI] [PubMed] [Google Scholar]
- 57.Laird P.W., et al. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pogribny I., Yi P., James S.J. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem. Biophys. Res. Commun. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki H., et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki H., et al. Distinct methylation pattern and microsatellite instability in sporadic gastric cancer. Int. J. Cancer. 1999;83:309–313. doi: 10.1002/(sici)1097-0215(19991029)83:3<309::aid-ijc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland K.D., et al. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726–7733. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- 62.McKie A.B., et al. Epigenetic inactivation of the human sprouty2 (hSPRY2) homologue in prostate cancer. Oncogene. 2005;24:2166–2174. doi: 10.1038/sj.onc.1208371. [DOI] [PubMed] [Google Scholar]
- 63.Lu R., Au W.C., Yeow W.S., Hageman N., Pitha P.M. Regulation of the promoter activity of interferon regulatory factor-7 gene: activation by interferon and silencing by hypermethylation. J. Biol.Chem. 2000;275:31805–31812. doi: 10.1074/jbc.M005288200. [DOI] [PubMed] [Google Scholar]
- 64.Dammann R., et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 65.Irimia M., Fraga M.F., Sanchez-Cespedes M., Esteller M. CpG island promoter hypermethylation of the Ras-effector gene NORE1A occurs in the context of a wild-type K-ras in lung cancer. Oncogene. 2004;23:8695–8699. doi: 10.1038/sj.onc.1207914. [DOI] [PubMed] [Google Scholar]
- 66.Elam C., et al. RRP22 is a farnesylated, nucleolar, Ras-related protein with tumor suppressor potential. Cancer Res. 2005;65:3117–3125. doi: 10.1158/0008-5472.CAN-04-0749. [DOI] [PubMed] [Google Scholar]
- 67.Moreno-Bueno G., et al. Inactivation of the candidate tumor suppressor Par-4 in endometrial cancer. Cancer Res. 2007;67:1927–1934. doi: 10.1158/0008-5472.CAN-06-2687. [DOI] [PubMed] [Google Scholar]
- 68.Bae Y.K., et al. Hypermethylation in histologically distinct classes of breast cancer. Clin. Cancer Res. 2004;10:5998–6005. doi: 10.1158/1078-0432.CCR-04-0667. [DOI] [PubMed] [Google Scholar]
- 69.Du Y., et al. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res. 2001;61:8094–8099. [PubMed] [Google Scholar]
- 70.Xiong Z., Laird P.W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calvisi D.F., Factor V.M., Loi R., Thorgeirsson S.S. Activation of beta-catenin during hepatocarcinogenesis in transgenic mouse models: relationship to phenotype and tumor grade. Cancer Res. . 2001;61:2085–2091. [PubMed] [Google Scholar]
- 72.Santoni-Rugiu E., Jensen M.R., Thorgeirsson S.S. Disruption of the pRb/E2F pathway and inhibition of apoptosis are major oncogenic events in liver constitutively expressing c-myc and transforming growth factor alpha. Cancer Res. . 1998;58:123–134. [PubMed] [Google Scholar]
- 73.Jiang W., Kahn S.M., Guillem J.G., Lu S.H., Weinstein I.B. Rapid detection of ras oncogenes in human tumors: applications to colon, esophageal, and gastric cancer. Oncogene. 1989;4:923–928. [PubMed] [Google Scholar]
- 74.Fransen K., et al. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–533. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- 75.Kosaka T., et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. . 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 76.Samuels Y., et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 77.Lee J.S., Thorgeirsson S.S. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002;35:1134–1143. doi: 10.1053/jhep.2002.33165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.