Abstract
Forkhead box O (Foxo) transcription factors govern metabolism and cellular differentiation. Unlike Foxo-dependent metabolic pathways and target genes, the mechanisms by which these proteins regulate differentiation have not been explored. Activation of Notch signaling mimics the effects of Foxo gain of function on cellular differentiation. Using muscle differentiation as a model system, we show that Foxo physically and functionally interacts with Notch by promoting corepressor clearance from the Notch effector Csl, leading to activation of Notch target genes. Inhibition of myoblast differentiation by constitutively active Foxo1 is partly rescued by inhibition of Notch signaling while Foxo1 loss of function precludes Notch inhibition of myogenesis and increases myogenic determination gene (MyoD) expression. Accordingly, conditional Foxo1 ablation in skeletal muscle results in increased formation of MyoD-containing (fast-twitch) muscle fibers and altered fiber type distribution at the expense of myogenin-containing (slow-twitch) fibers. Notch/Foxo1 cooperation may integrate environmental cues through Notch with metabolic cues through Foxo1 to regulate progenitor cell maintenance and differentiation.
Introduction
A central issue in regenerative medicine is understanding how highly specialized cell types arise from undifferentiated stem or progenitor cells (1). Germane to this issue is how biochemical signals engendered by microenvironmental and endocrine/nutritional cues are transcriptionally integrated to activate cellular differentiation processes.
The O subfamily of forkhead (Fox) proteins regulates hormonal, nutrient, and stress responses to promote cell survival and metabolism. The ability to fine-tune Foxo transcription is essential to controlling these cellular functions and is largely dependent on posttranscriptional modifications, including phosphorylation and acetylation (2). In addition to their role in terminally differentiated cells, Foxo proteins have also been implicated in myoblast (3), preadipocyte (4), and endothelial cell differentiation (5). Moreover, Foxo4 regulates vascular smooth muscle cell differentiation through interactions with myocardin (6). Foxo3 knockout mice display premature ovarian failure, consistent with a role for this gene in ovarian follicle maturation (7). The mechanisms by which Foxo proteins control cellular differentiation remain unclear, and recent conditional ablation studies are consistent with a significant degree of functional overlap among the 3 Foxo isoforms in the hematopoietic lineage (8, 9).
The Notch pathway plays an important role in neural, vascular, muscular, and endocrine differentiation during embryogenesis (10). Upon ligand-induced cleavage, the intracellular domain of the Notch receptor translocates to the nucleus, where it interacts with the DNA-binding protein Csl, changing its transcriptional properties from a suppressor to an activator of transcription (11). Csl targets include the Hairy and Enhancer of split (Hes) and Hes-related (Hey) genes. Hes1 controls gut endoderm (12), preadipocyte (13), and neurogenic differentiation (14). Active Notch signaling, or Notch1 receptor gain of function, inhibits differentiation of C2C12 and 10T/2 myoblasts by suppressing myogenic determination gene (MyoD) transcription (15–21).
It is noteworthy that Foxo1 gain of function (3–5) phenocopies Notch1 activation (13, 17, 22, 23) in every cellular differentiation context. Moreover, Foxo1 ablation (24) phenocopies Notch1 ablation (25) in mice. Despite these intriguing similarities, Foxo and Notch signal through 2 seemingly distinct mechanisms, the phosphatidylinositol 3-kinase pathway (Foxo) and the Hes/Hey pathway (Notch). In this study, we show that Foxo physically and functionally interacts with Notch by promoting corepressor clearance from Csl, thus controlling the myogenic program.
Myogenic precursors arise from mesodermal stem cells (26) and are converted into myotubes by a multistep process culminating in the expression of myogenic transcription factors of the myogenic regulatory factor (MRF) family (MyoD, myogenin, MRF4, and myogenic factor 5 [Myf5]) (27). Myogenic transcription factors heterodimerize with E proteins and promote expression of muscle-specific genes, acting in close coordination with myocyte-specific MEF2 enhancer factors (28).
Adult muscle is a heterogeneous tissue, primarily defined by its myofiber content (29). Different myosin heavy chain (MyHC) subtypes characterize different myofibers. Type I fibers express primarily slow-twitch MyHC whereas type II fibers express fast-twitch MyHC (29). The process of fiber-type specification is controlled at multiple steps. First, there appears to be heterogeneity among myogenic precursor cells, and evidence from avian embryo cross-transplantation experiments indicates that early precursors contribute primarily to slow muscle fibers and later precursors to fast fibers (29). Postnatally, fiber type specification is also affected by cell autonomous factors, including innervation and endocrine/nutritional cues (28). The Foxo coactivator Pparγ coactivator 1α (Pgc1α) plays a critical role in promoting the formation of slow-twitch fibers (30), and recent data have also implicated the Foxo deacetylase Sirt1 in this process (31). Using conditional mutagenesis in mice, we show that Foxo1’s role in suppressing MyoD-dependent myogenesis in C2C12 cells is mirrored by an increase of MyoD-containing myofibers in Foxo1-deficient skeletal muscle, consistent with a key function in myoblast lineage specification.
Results
Interaction of Foxo1 and Notch signaling in C2C12 differentiation.
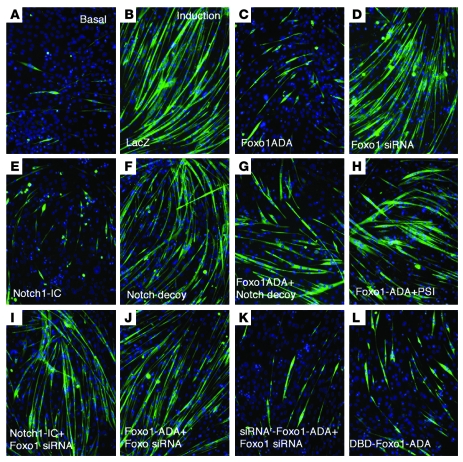
To understand whether Notch and Foxo interact to control muscle development, we used a cellular differentiation model. C2C12 cells undergo myogenic conversion and myotube fusion upon growth factor withdrawal, a process associated with Foxo1 nuclear translocation (3). Accordingly, transduction of adenovirus encoding a constitutively active Foxo1 mutant (Foxo1-ADA; a mutant Foxo1 with the following amino acid substitutions: T24A, S253D, and S316A) (4) blocked the effect of serum withdrawal to induce C2C12 differentiation, as reflected by inhibition of myoblast fusion (Figure 1, A–C). Conversely, Foxo1 inhibition by siRNA did not affect these processes (Figure 1D). Similarly, constitutively active Notch1 (Notch1-IC) phenocopied Foxo1-ADA in blocking myoblast differentiation (Figure 1E). Virtually all cells became transduced with the adenoviruses (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI32054DS1). Foxo1 siRNA effectively suppressed expression of both endogenous Foxo1 and transfected FLAG-Foxo1 (Supplemental Figure 2) in a dose-dependent manner, without affecting control proteins or other Foxo isoforms (Supplemental Figure 3). Neither Foxo1-ADA nor Notch1-IC affected C2C12 proliferation (Supplemental Figure 4).
Figure 1. Regulation of myoblast differentiation by Foxo and Notch.
C2C12 cells were immunostained with anti-myosin antibody (green) and DAPI (blue). See text for panel description. Each experiment was repeated at least 6 times. Original magnification, ×10.
We asked whether we could preempt the effect of Foxo1-ADA by inhibition of endogenous Notch signaling. To this end, we used a truncated Notch1 receptor lacking the transmembrane anchor and intracellular domain, which acts as a decoy receptor by binding Notch ligands (32, 33) (our unpublished observations). The decoy did not affect C2C12’s ability to undergo differentiation in response to growth factor withdrawal (Figure 1F) but partly rescued Foxo1-ADA inhibition of myoblast differentiation (Figure 1G). As an alternative probe to block Notch signaling, the presenilin inhibitor (PSI) compound E (34) also rescued Foxo1-ADA inhibition of myoblast differentiation (Figure 1H).
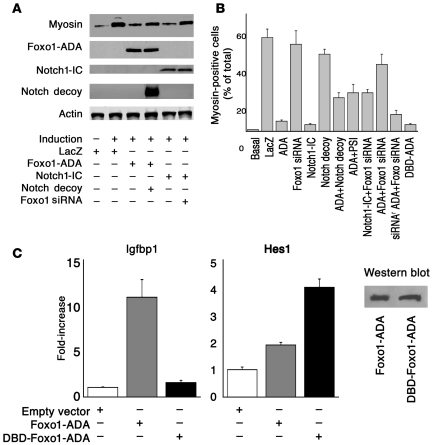
To examine the effect of Foxo1 on Notch signaling, we cotransfected Foxo1 siRNA and Notch1-IC. Foxo1 siRNA rescued inhibition of myoblast differentiation and myosin expression by Notch1-IC (Figure 1I) while control siRNA had no effect (data not shown). To rule out nonspecific effects of Foxo1 siRNA on myoblast differentiation, we generated an siRNA-resistant Foxo1-ADA (Supplemental Figure 5). Foxo1 siRNA reversed the effects of Foxo1-ADA (Figure 1J) but failed to rescue inhibition of C2C12 differentiation caused by siRNA-resistant Foxo1-ADA (Figure 1K). We present a quantitative analysis of these data in Figure 2A, showing that Foxo1 and Notch1-IC decreased myosin levels by more than 80% while Notch decoy and Foxo1 siRNA restored them to approximately 70% of fully differentiated cells. We obtained similar data by performing a morphometric analysis of myosin-positive cells (Figure 2B). These data indicate that Foxo1 is required for the effect of Notch on myoblast differentiation.
Figure 2. Quantitative analysis of C2C12 differentiation.
(A) Western blotting analysis of myosin expression in C2C12 cells. (B) Morphometric analysis of myosin-positive cells. Results from differentiation experiments were analyzed by scoring the number of myosin-immunostained cells as a percentage of all DAPI-positive cells. (C) DBD-Foxo1-ADA reporter gene assays. We carried out reporter gene assays using the canonical Foxo1-responsive Igfbp1 promoter (left panel) and the Hes1 promoter (right panel) in cells cotransfected with Foxo1-ADA or DBD-Foxo1-ADA. Western blot (inset) demonstrates that expression levels of the 2 proteins are similar. An asterisk indicates P < 0.01 by ANOVA.
We next determined whether Foxo1 affects differentiation via its transcriptional function. To this end, we generated a DNA-binding deficient (DBD) mutant in the backbone of the ADA mutant by replacement of N208A and H212R (DBD-Foxo1-ADA) (6, 35). We confirmed that this mutant is unable to bind DNA by measuring insulin-like growth factor–binding protein 1 (Igfbp1) promoter activity, a canonical Foxo1 target. Foxo1-ADA increased Igfbp1 promoter activity by 10-fold whereas DBD-Foxo1-ADA was unable to do so (Figure 2C). Surprisingly, this mutant was as effective as the DNA binding–competent Foxo1-ADA at inhibiting differentiation (Figure 1L). These data indicate that Foxo1 controls differentiation independently of its ability to bind DNA in a sequence-specific manner.
Foxo1 binds to Csl and is recruited to the Hes1 promoter.
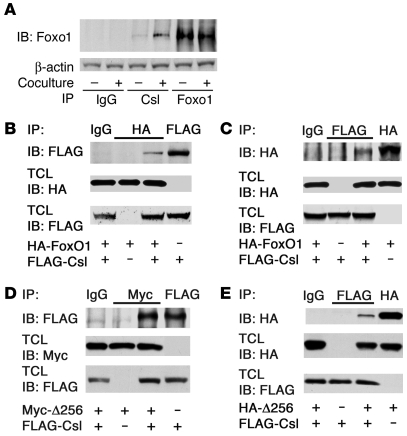
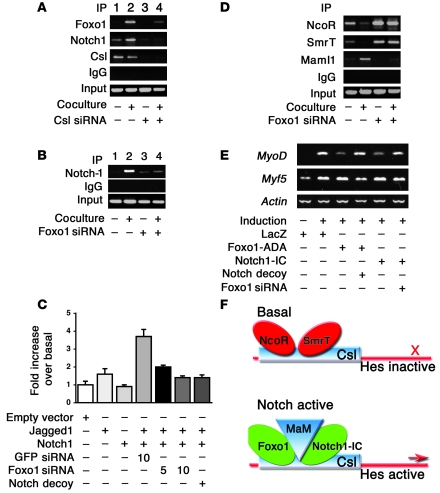
Notch1-IC binds to and coactivates Csl to promote Hes and Hey expression (11). Based on the results with the DBD-Foxo1-ADA mutant, we determined whether Foxo1 interacts with Csl in a Notch-dependent manner using coculture of C2C12 cells expressing Notch1 receptor with HEK293 cells expressing the Notch ligand Jagged1 or LacZ as a negative control. We provide several lines of evidence that Foxo1 and Csl interact in cultured cells. We detected endogenous Foxo1 in endogenous Csl immunoprecipitates, and the coimmunoprecipitation was significantly enhanced by activation of Notch signaling (Figure 3A). To confirm the specificity of the interaction, we expressed HA-tagged Foxo1 and FLAG-tagged Csl in C2C12 cells. Following immunoprecipitation with anti-HA (Foxo1) antiserum, we detected FLAG-Csl in immunoblots (Figure 3B). Conversely, following immunoprecipitation with anti-FLAG (Csl) antiserum, we detected HA-Foxo1 in immunoblots (Figure 3C). The ability to coimmunoprecipitate with Csl appears to be specific to Foxo1, as we failed to detect other Foxo isoforms in Csl immunoprecipitates (Supplemental Figure 6). A truncated Foxo1 mutant (Δ256, encoding aa 1–256) (36) retained the ability to interact with Csl. We detected FLAG-Csl in immunoprecipitates (Figure 3D) and HA-Δ256 in FLAG-Csl immunoprecipitates (Figure 3E), indicating that Csl interacts with the Foxo1 N terminal domain.
Figure 3. Foxo1 coimmunoprecipitates with Csl.
(A) Coimmunoprecipitation of endogenous Foxo1 and Csl in C2C12 cells cocultured with LacZ-expressing (denoted by the minus sign) or Jagged1-expressing HEK293 cells (denoted by the plus sign). (B and C) Coimmunoprecipitation experiments in C2C12 cells cotransfected with FLAG-Csl and HA-Foxo1. (D and E) Coimmunoprecipitation experiments in C2C12 cells cotransfected with FLAG-Csl and the truncated mutant Myc- or HA-tagged Δ256 Foxo1. TCL, total cellular lysate.
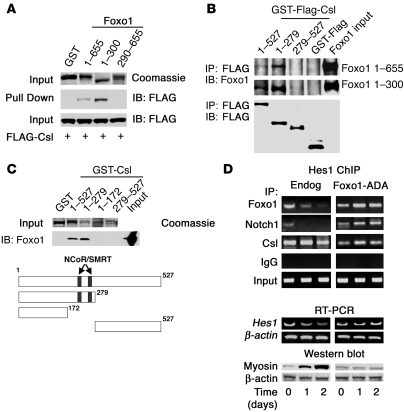
To determine whether this is a direct protein-protein interaction and map the interaction domain(s), we first carried out pull-down assays with affinity-purified glutathione-S-transferase–Foxo1 (GST-Foxo1) produced in bacteria and FLAG-Csl expressed in HEK293 cells. We detected Csl association with full-length and N terminal Foxo1 (aa 1–300) but not with C terminal Foxo1 (aa 290–655) or GST (Figure 4A). We next mapped the Csl domain that interacts with Foxo1 using a cell-free system with GST-Foxo1 and GST-Flag-Csl purified from bacterial cultures. Again, we recovered full-length (aa 1–655) and N terminal (aa 1–300) but not C terminal (aa 290–655) Foxo1 in Csl immunoprecipitates. Conversely, N terminal Foxo1 interacts with N terminal Csl (Figure 4B).
Figure 4. Foxo1 binds directly to Csl.
(A) GST pull-down assays of GST-Foxo1 fusion protein with Csl immunoprecipitated from HEK293 cells. (B and C) Binding of GST-Foxo1 and GST-FLAG-Csl in a cell-free system and mapping of the Csl interaction domain. Full-length and truncated fragments of GST-Foxo1 and GST-FLAG-Csl were purified from bacteria and coincubated. Thereafter, Csl was isolated using anti-FLAG antibody, and the immunoprecipitate was analyzed by immunoblotting with anti-Foxo1 or anti-FLAG antibodies. (D) Hes1 promoter ChIP assay spanning the Csl-binding site in C2C12 cells to detect endogenous Foxo1, Csl, and Notch1 (Endog) or following transduction with Foxo1-ADA during myoblast differentiation. Input represents DNA extracted from chromatin prior to immunoprecipitation. Hes1 (semiquantitative RT-PCR) and myosin (Western blot) expression corresponding to each time point are shown. Day 0 is defined as the time when cells were serum deprived to induce myoblast fusion.
We used Csl deletion mutants to map the Foxo1-binding domain in Csl. These studies indicate that Foxo1 binds to a domain encompassing aa 172–279 (Figure 4C), which is contained within the Csl NH2 terminal domain (NTD) domain (37) (Figure 4C). Interestingly, this domain is required for DNA and corepressor binding but does not contribute to Notch binding (38, 39).
Csl binds to a consensus sequence in the Hes1 promoter (40), which thus provides a useful readout assay of the Foxo/Csl interaction. If the latter were required to regulate C2C12 differentiation, 3 predicted conditions should be met: (a) Foxo1 should be detected in chromatin immunoprecipitation (ChIP) assays spanning the Csl element in the Hes1 promoter; (b) the interaction should be differentiation dependent; and (c) inhibition of differentiation by Foxo1-ADA should be accompanied by constitutive binding to the Csl element in the Hes1 promoter. Figure 4D demonstrates that all predictions are fulfilled. First, we performed ChIP assays using primers spanning the Csl-binding site of Hes1 in differentiating C2C12 cells. We detected endogenous Foxo1, Notch1, and Csl in immunoprecipitates from undifferentiated cells (Figure 4D). As the PCR-amplified sequence contains no forkhead binding sites, we concluded that Foxo1 binds to this DNA fragment via Csl. Moreover, binding of both Foxo1 and Notch1 decreased as cells became differentiated (days 1 and 2). When we transduced cells with constitutively nuclear Foxo1-ADA, differentiation was inhibited (Figure 1C) and the mutant Foxo1 was persistently bound to the Hes1 promoter, as were Csl and Notch1 (Figure 4D).
We next analyzed Hes1 expression. The prediction was that Hes1 levels should correlate with occupancy of the Hes1 promoter by Foxo1 and Notch1. Indeed, Hes1 mRNA expression declined as Foxo1 and Notch1 binding to Csl decreased while myosin protein levels increased (Figure 4D). To rule out a direct effect of Foxo1 on Csl transcription, we carried out reporter gene assays with the Csl promoter. Foxo1 failed to activate expression of a Csl reporter gene despite the presence of 10 repeats of a forkhead binding site in the Csl promoter (ref. 41 and data not shown). Moreover, Csl expression was unaffected in C2C12 cells expressing Foxo1-ADA (data not shown). These data indicate that Foxo1 regulates Notch-dependent differentiation via protein/protein interactions with Csl.
Foxo1 is required for Notch induction of Hes and Hey genes via Csl.
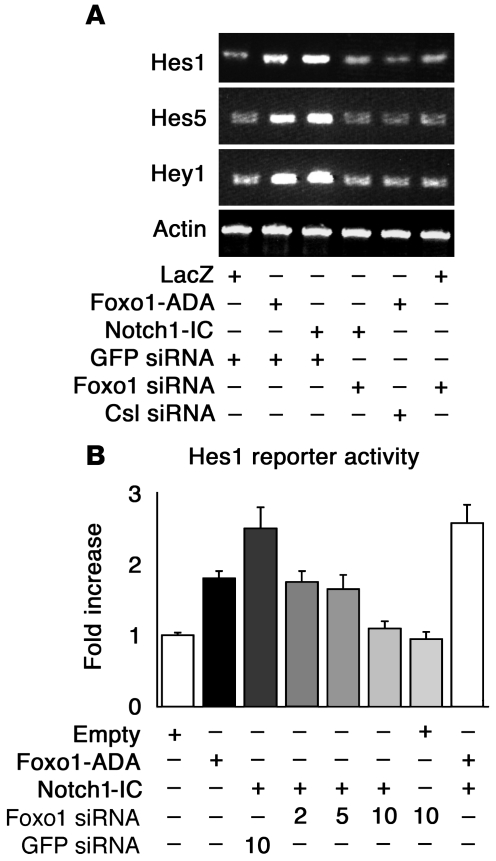
We examined the ability of Foxo1-ADA to promote expression of endogenous Hes1, Hes5, and Hey1 in C2C12 cells. Both Foxo1-ADA and Notch1-IC increased the expression of the 3 genes while Foxo1 siRNA inhibited Hes1, Hes5, and Hey1 expression induced by Notch1-IC (Figure 5A). Foxo1 siRNA had no effect on Hes1, Hes5, and Hey1 expression in growth factor–deprived cells (Figure 5A).
Figure 5. Foxo1 regulates Notch-induced Hes1, Hes5, and Hey1 expression.
(A) Hes1, Hes5, and Hey1 expression measured by semiquantitative RT-PCR in C2C12 cells transduced with Foxo1-ADA or Notch1-IC following transfection of GFP, Foxo1, or Csl siRNA as indicated. (B) Hes1 reporter gene assays in HEK293 cells transduced with Foxo1-ADA, Notch1-IC, Foxo1 siRNA, GFP siRNA, or control plasmid (empty). We measured luciferase activity and normalized it by β-galactosidase activity. The data represent arbitrary units relative to control empty vector.
We focused the next set of experiments on Hes1, as a prototypical Notch target gene. We tested Foxo1’s ability to regulate Hes1 transcription using reporter assays with the Hes1 promoter as well as measurements of Hes1 expression. Foxo1-ADA and Notch1-IC induced Hes1 promoter activity by 1.8- and 2.5-fold, respectively. Cotransfection of Foxo1-ADA with Notch1-IC caused a 2.5-fold increase (Figure 5B). Cotransfection of Foxo1 siRNA suppressed Notch-induced Hes1 activity in a dose-dependent manner while control siRNA had no effect (Figure 5B). We obtained similar results with a synthetic Hes1 reporter containing 4 tandem repeats of the Csl-binding motif (Supplemental Figure 7). Moreover, DBD-Foxo1-ADA was able to induce Hes1 reporter gene activity to an even greater extent than Foxo1-ADA, confirming that direct DNA binding is not required for Foxo1 activation of Hes1 (Figure 2C).
The failure of Notch1-IC to induce Hes1 expression in cells expressing Foxo1 siRNA suggests that Foxo1 is required for Csl/Notch interaction. Thus, we investigated the binding of Foxo1 and Notch1 to the Hes1 promoter in a coculture system. We cocultured C2C12 cells expressing Notch1 with HEK293 cells expressing the Notch ligand Jagged1 to induce activation of endogenous Notch signaling. Coculture in the presence of Jagged1-expressing cells increased endogenous Foxo1 (Figure 6A) and Notch1 binding to the Hes1 promoter in ChIP assays (Figure 6, A and B) (42). These data are consistent with the observation that Foxo1 coimmunoprecipitation with Csl increased upon coculture (Figure 3A). To determine whether Foxo1 binding to the Hes1 promoter is Csl dependent, we inhibited Csl expression with siRNA (Supplemental Figure 8). Transfection of Csl siRNA inhibited both Foxo1 and Notch1 binding to Hes1 promoter (Figure 6A), indicating that they are Csl dependent. Moreover, Foxo1-ADA failed to induce Hes1 expression in the presence of Csl siRNA (Figure 5A). The results of ChIP experiments were corroborated by Hes1 promoter assays. Expression of Jagged1 or Notch1 alone had no effect on Hes1 activity, but coculturing yielded a 3.7-fold increase in Hes1 reporter gene activity (Figure 6C). Foxo1 siRNA abolished Notch binding to the Hes1 promoter in ChIP assays (Figure 6B) and induction of Hes1 promoter activity (Figure 6C). These results suggest that Foxo1 is required for binding of Notch1 to the Hes1 promoter and provide a mechanism whereby inhibition of Foxo1 expression restores differentiation of myoblasts expressing Notch1-IC. The ability of Foxo1 siRNA to inhibit Notch induction of Hes1 in a coculture system rules out the possibility that the effects observed in differentiation experiments with Notch1-IC are due to nonphysiologic activation of Notch signaling by the truncated intracellular Notch1 mutant (15).
Figure 6. Foxo1 is required for Notch binding to the Hes1 promoter and activation of Hes1 target genes.
(A) ChIP assays of endogenous Foxo1 and Notch1 in C2C12 cells cocultured with LacZ-expressing (denoted by a minus sign) or Jagged1-expressing HEK293 cells (denoted by a plus sign) in the absence (lanes 1 and 2) and presence (lanes 3 and 4) of Csl siRNA. (B) ChIP assays of endogenous Notch1 in coculture system in the absence (lanes 1 and 2) and presence (lanes 3 and 4) of Foxo1 siRNA. (C) Hes1 promoter assays following coculture in the absence and presence of Foxo1 or GFP siRNA. (D) ChIP assays of NcoR and Smrt and Maml1 binding to Hes1 in the coculture system in the absence (lanes 1 and 2) and presence (lanes 3 and 4) of Foxo1 siRNA. (E) Expression of MyoD, Myf5, and β-actin in C2C12 cells by semiquantitative RT-PCR. (F) Model of Foxo1 and Notch regulation of Hes1 promoter.
Foxo1 promotes corepressor clearance and Maml1 binding to Csl.
To clarify the molecular mechanism of Foxo1-dependent activation of Hes1 expression, we investigated corepressor/coactivator exchange at the Hes1 promoter. Activation of Notch cleared the corepressors nuclear corepressor (NcoR) and silencing mediator for retinoid and thyroid hormone receptor (Smrt) (43) and recruited the coactivator mastermind-like 1 (Maml1) (42) to the Hes1 promoter. Foxo1 siRNA prevented Notch-induced corepressor exchange (Figure 6D). These data are consistent with the observation that Foxo1 binds to the region 172–279 of Csl (Figure 4C), which has been shown to contain the NcoR/Smrt binding sites (38, 39).
To demonstrate that the observed changes in the transcriptional complex result in changes in Hes1 activity, we investigated expression of Hes1 target genes involved in myogenesis. Hes1 has been proposed to suppress myoblast differentiation by inhibiting the basic helix-loop-helix transcription factor MyoD without affecting Myf5 (16, 17). Expression analyses revealed that Notch1-IC or Foxo1-ADA suppressed MyoD, while Myf5 was unaffected. Notch decoy or Foxo1 siRNA partly restored MyoD expression (Figure 6E).
Altered fiber type composition in skeletal muscle lacking Foxo1.
Based on the cellular data, we undertook to probe Foxo1 function in muscle differentiation in vivo using conditional gene inactivation. The predicted outcome of this experiment was accelerated differentiation of MyoD-containing but not Myf5-containing myoblasts. Because MyoD is the predominant myogenic factor in fast fibers while myogenin is the predominant factor in slow fibers (44), the removal of Foxo/Notch inhibition on MyoD expression should result in increased formation of fast fibers, potentially at the expense of slow fibers.
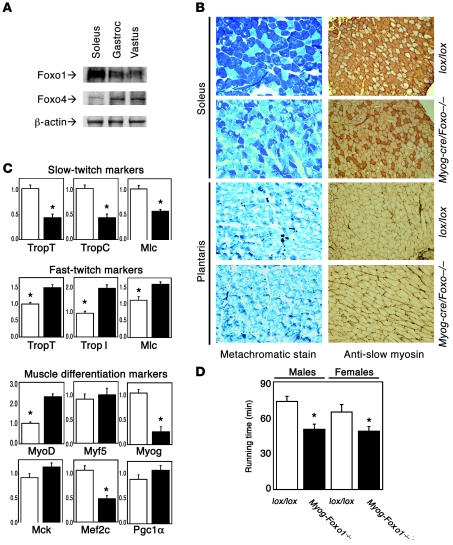
There are 3 Foxo isoforms in mice: Foxo1, Foxo3, and Foxo4 (8, 9). The latter is predominant in most muscle types (45) except soleus, where Foxo1 is the most abundant (Figure 7A). Coincidentally, soleus is also physiologically enriched in slow-twitch fibers and thus allowed us to readily test our hypothesis. We inactivated Foxo1 expression in skeletal muscle by crossing mice homozygous for a floxed Foxo1 allele with myogenin-cre transgenics. mRNA analysis indicated that the knockout occurred as planned (data not shown). Histological analyses revealed a reduction of type I (slow-twitch) fibers in soleus of myogenin-Foxo1 (Myog-Foxo) mice while type II fiber–enriched muscles were unaffected (Figure 7B). Consistent with the histological findings, expression of type I fiber markers decreased while type II fiber markers increased in Myog-Foxo1 mice (Figure 7C). We then analyzed expression of the myogenic transcription factors MyoD, Myf5, and myogenin. MyoD is the predominant factor in fast fibers and myogenin in slow fibers (44). Consistent with the histopathology, we found a 2-fold increase in MyoD expression and an approximately 80% decrease in myogenin while Myf5 expression was unchanged (Figure 7C). Moreover, expression of the Foxo1 coactivator Pgc1α, which regulates type I fiber determination (30) was unchanged, indicating that the phenotype of Myog-Foxo1 mice cannot be accounted for by decreased Foxo1-dependent Pgc1α transcription (Figure 7C) (46). As a functional correlate of the observed fiber type switch, we examined running performance on a treadmill. Indeed, Myog-Foxo1 mice displayed reduced running capacity, as predicted from the reduction in type I (endurance) fibers (Figure 7D).
Figure 7. Conditional ablation of Foxo1 in skeletal muscle.
(A) Western blot analysis of Foxo1 and Foxo4 expression levels in various muscle types. Gastroc, gastrocnemius muscle; Vastus, vastus lateralis muscle. (B) Metachromatic and immunohistochemical analysis of soleus and plantaris muscle from Myog-Foxo1 mice and control (lox/lox) littermates. Original magnification, ×10. (C) Gene expression analysis of Myog-Foxo1 (black bars) and control mice (white bars); TropC, troponin-C; TropT, troponin-T; Mlc, myosin light chain; Myog, myogenin; Mck, muscle-type creatine kinase. Data are means ± SEM of 3 independent measurements (n = 6 for each genotype). An asterisk indicates P < 0.05 by ANOVA. (D) Treadmill performance test in 8-week-old Myog-Foxo1 mice and lox/lox littermates (n = 6 for each genotype). An asterisk indicates P < 0.05 by ANOVA.
Finally, to determine whether these changes reflected developmental alterations in fiber-type specification as opposed to adaptive or cell-nonautonomous factors, we determined MyoD expression in Foxo1 (24) and Notch1 knockout (25) embryos at E9.5. In Foxo1–/– embryos, MyoD levels increased 3.1 ± 1.1–fold, and in Notch1–/– embryos 7.3 ± 2.9–fold compared with controls (P < 0.05 in both mutants versus wild type, n = 4). The increase in MyoD expression observed in vivo is consistent with the physical and functional interactions between Foxo1 and Notch at this key signaling nexus in myoblast differentiation. Thus, we propose that the fiber-type switch in Myog-Foxo1 mice is the result of accelerated differentiation of MyoD-containing myoblasts during embryonic development.
Discussion
This study provides biochemical, cellular, and genetic evidence that Foxo and Notch pathways cooperate in the regulation of muscle differentiation. The data reveal what we believe is a novel mode of Foxo1 action to promote corepressor exchange at the Hes1 promoter via direct binding to the Csl NTD region (Figure 6F). We propose that Foxo1 binding to this domain stabilizes the Notch/Csl complex and promotes corepressor clearance and Maml1 recruitment, consistent with the proposed role of NTD from structural studies (37). The findings also provide a mechanism by which 2 major biochemical pathways, the phosphoinositol 3-kinase/Akt pathway and the Notch/Hes pathway, converge in a synergistic manner to control cellular differentiation in vivo.
The proposed role for Foxo1 is independent of its transcriptional function and involves a direct interaction with Csl. While our studies have focused on Hes1 as a prototypical effector of Notch1 signaling, our data should not be construed to indicate that Hes1 is the sole mediator of the Notch/Foxo interaction. For example, we have observed a similar Foxo/Notch epistasis in the differentiation of preadipocytes, PC-12, and HUVECs, suggesting that Foxo interacts with Notch in multiple cell contexts (data not shown). We propose that Notch/Foxo cooperation integrates environmental cues through Notch with metabolic cues through Foxo1 to regulate progenitor cell maintenance and differentiation. This 2-tiered mechanism allows committed progenitor cells in various tissues to avoid differentiation in response to developmental cues (Notch) when Foxo1 is active, i.e., in the absence of growth factors. These cells would then persist in a dormant state in adult tissues, where they can terminally differentiate in response to a combination of Notch ligand and hormonal/nutritional cues leading to Foxo1 inhibition. This interpretation is consistent with the fiber-type switch observed in Foxo1-deficient muscle, an observation that appears to position Foxo1 as a fate decider within the myogenic lineage, as opposed to an inducer of the myogenic program. It remains to be seen whether other Foxo and Notch isoforms also interact and how they contribute to this process.
The demonstration that Foxo1 is a coregulator of gene expression provides a potential explanation for the protean functions of this transcription factor. Interesting questions emerging from our studies involve how the switch from one function to the other is effected and how the complex posttranslational modifications of Foxo1 in response to growth factors, hormones, and nutrients impinge on this process. The findings have broad implications for the pathophysiology of disease processes that involve Foxo1 signaling. A potential implication of our observation is the ability to explore the use of agents that inhibit Notch signaling (47) as a treatment of metabolic disorders characterized by excessive Foxo function (48).
Methods
Animal generation and analysis.
Myogenin-cre (49) and Foxo1flox mice have been described (9). The wild-type, null, and Foxo1flox alleles were detected using PCR with primers 5′-GCTTAGAGCAGAGATGTTCTCACATT-3′, 5′-CCAGAGTCTTTGTATCAGGCAAATAA-3′, and 5′-CAAGTCCATTAATTCAGCACATTGA-3′. Prior to the treadmill performance test, mice were trained for 2 days (Columbus Instruments). The test was performed at 15 m/min for the first 30 minutes, followed by 1 m/min increases at 10 minute intervals until exhaustion. Skeletal muscle samples were quickly frozen in OCT matrix, and 7-μm serial sections were obtained. Muscle fibers were typed using metachromatic ATPase (50) or immunostaining with anti-skeletal slow myosin (Sigma-Aldrich). For embryonic studies, we set up timed matings of heterozygous Foxo1 (24) or Notch1 (25) mice and recovered embryos at E9.5. mRNA was isolated from whole embryos, and real-time RT-PCR was performed as described below. All animal experiments were approved by the Columbia University Animal Care and Utilization Committee.
Viral expression studies.
C2C12 cells were differentiated as described (3, 4). Foxo1-ADA, Notch1-IC, Jagged1, Csl, and Notch decoy adenoviral and mammalian expression vectors have been described (36, 51). We generated retroviruses expressing Foxo1-ADA and Notch1-IC using the pQCXIH vector (Clontech). To generate Notch decoy (pAdlox Notch1ECD-Fc), the extracellular domain of Notch1 (bp 241-4229, GenBank accession number X57405) was fused in frame with human IgG Fc tag and cloned into pAdlox. Retroviral supernatant was produced from cells transiently cotransfected with pVSV-G vector (Clontech) and designated pQCXIH vector into GP2-293 cells (BD Biosciences). To generate the DNA binding–deficient Foxo1, we replaced N208 and H212 with alanine and arginine, respectively, using QuikChange Mutagenesis Kit (Stratagene). The mutations were then cloned in the backbone of the Foxo1-ADA mutant.
Luciferase assay and coculture assay.
We transfected HEK293 cells with Hes1-luciferase (–194 to 160 from transcription start site) (Hes1/pGL2 basic [Stratagene]), synthetic Hes1-luciferase (containing a 4× Csl binding site, 4× Csl/pGL2 basic) or Csl-luciferase (–1536 to 22, Csl/pGL2 basic) reporter genes along with pCMV5, pCMV5-Foxo1-ADA, pQNC–Notch1-IC, pHyTc (51), Notch decoy, or Foxo1 siRNA. We used plasmid pRSV–β-galactosidase as a control of transfection efficiency (51). For coculture assay, we expressed Notch1 in C2C12 cells and Jagged1 or LacZ in HEK293 cells by transfection. We then harvested HEK293 cells and seeded them on C2C12 cells. After 1 hour incubation, we used the cocultured cells for experiments.
Western blotting and immunoprecipitation.
We performed these assays according to standard techniques using anti-myosin (MF-20), anti-HA (12CA5; Boehringer Mannheim), anti-FLAG (M2; Sigma-Aldrich), anti-Foxo1 (H128 and N20; Santa Cruz Biotechnology Inc.), anti-Notch1 (C-20; Santa Cruz Biotechnology Inc.), anti-Csl (Millipore and Santa Cruz Biotechnology Inc.), anti-NcoR (Santa Cruz Biotechnology Inc.), anti-Smrt (Santa Cruz Biotechnology Inc.), or anti-MAML1 (Millipore) antibodies. For Foxo/Csl coimmunoprecipitation, we used purified nuclear fractions (52). Because Csl migrates close to IgG heavy chain on SDS-PAGE, we used dimethylpyrimilidate (DMP; Pierce) to cross-link antibodies to protein A beads and avoid IgG contamination of eluted protein complexes (52).
ChIP assays.
We performed ChIP assays in C2C12 cells as described previously (4) and in cocultured cells as described by Fryer (42). The primer pairs employed to amplify the Csl-binding site of the Hes1 promoter are as follows: 5′-GCAAAGCCCAGAGGAAAGAGTTAG-3′ and 5′-AGGAGAGAGGTAGACAGGGGATTC-3′.
siRNA transfection and siRNA-resistant Foxo1.
The Foxo1-specific siRNA sequence is 5′-ACGGAGGATTGAACCAGTATA-3′. The Csl-specific siRNA sequence is 5′-TAGGGAAGCTATGCGAAATTA-3′. siRNA was transfected using lipofectamine-plus reagent (Invitrogen). We generated siRNA-resistant Foxo1 by replacing 3 residues (underlined) in the sequence 5′-ACGGCGGTCTGAACCAGTATA-3′. Primer sequences employed for real-time RT-PCR studies are in the Supplemental Methods.
Recombinant proteins and interaction assays.
We generated GST-FLAG-Csl encompassing aa 1–527, 1–279, 1–172, and 279–527 fragments by cloning into pGEX6P-1. GST-Foxo1 constructs have been described (53). Following bacterial culture and iso-propyl-thio-galactose induction, we purified GST fusion proteins and incubated them together. Thereafter, we isolated GST-FLAG/Csl by immunoprecipitation with anti-FLAG antibody, washed the immune pellets extensively, and performed immunoblot with anti-Foxo1 antiserum.
Statistics.
All results are presented as ± SEM. P values were calculated by 1-factor ANOVA.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK57539, DK64819, 5 RO1 HL062454 (JK), the Columbia Diabetes and Endocrinology Research Center (DK63608), and the Berrie Program in Cell-Based Therapies at Columbia University. We thank E. Olson (University of Texas Southwestern) for Myog-cre mice and members of the Accili and Kitajewski laboratories for stimulating discussions.
Footnotes
Nonstandard abbreviations used: ChIP, chromatin immunoprecipitation (assay); DBD, DNA binding deficient; Foxo, forkhead box O; GST, glutathione-S-transferase; Hes, Hairy and Enhancer of split; Hey, Hes-related; Maml1, mastermind-like 1; Myf5, myogenic factor 5; MyoD, myogenic determination gene; Myog, myogenin; NcoR, nuclear corepressor; Notch1-IC, constitutively active Notch1; NTD, NH2 terminal domain; Pgc1α, Pparγ coactivator 1α; Smrt, silencing mediator for retinoid and thyroid hormone receptor.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2477–2485 (2007). doi:10.1172/JCI32054
T. Kitamura and Y.I. Kitamura contributed equally to this work.
References
- 1.Singec I., Jandial R., Crain A., Nikkhah G., Snyder E.Y. The leading edge of stem cell therapeutics. Annu. Rev. Med. 2007;58:313–328. doi: 10.1146/annurev.med.58.070605.115252. [DOI] [PubMed] [Google Scholar]
- 2.Accili D., Arden K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/S0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 3.Hribal M.L., Nakae J., Kitamura T., Shutter J.R., Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J. Cell Biol. 2003;162:535–541. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakae J., et al. The forkhead transcription factor foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/S1534-5807(02)00401-X. [DOI] [PubMed] [Google Scholar]
- 5.Potente M., et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z.P., Wang Z., Yanagisawa H., Olson E.N. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Castrillon D.H., Miao L., Kollipara R., Horner J.W., DePinho R.A. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 8.Tothova Z., et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Paik J.H., et al. Foxos are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shawber C.J., Kitajewski J. Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays. 2004;26:225–234. doi: 10.1002/bies.20004. [DOI] [PubMed] [Google Scholar]
- 11.Lai E.C. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen J., et al. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 13.Ross D.A., Rao P.K., Kadesch T. Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol. Cell. Biol. 2004;24:3505–3513. doi: 10.1128/MCB.24.8.3505-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtsuka T., et al. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shawber C., et al. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 16.Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 17.Kuroda K., et al. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 18.Nofziger D., Miyamoto A., Lyons K.M., Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 19.Wilson-Rawls J., Molkentin J.D., Black B.L., Olson E.N. Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol. Cell. Biol. 1999;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsinger E., et al. Notch signalling acts in postmitotic avian myogenic cells to control MyoD activation. Development. 2001;128:107–116. doi: 10.1242/dev.128.1.107. [DOI] [PubMed] [Google Scholar]
- 21.Conboy I.M., Rando T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002;3:397–409. doi: 10.1016/S1534-5807(02)00254-X. [DOI] [PubMed] [Google Scholar]
- 22.Shawber C.J., Das I., Francisco E., Kitajewski J. Notch signaling in primary endothelial cells. Ann. N. Y. Acad. Sci. 2003;995:162–170. doi: 10.1111/j.1749-6632.2003.tb03219.x. [DOI] [PubMed] [Google Scholar]
- 23.Limbourg F.P., et al. Essential role of endothelial Notch1 in angiogenesis. Circulation. 2005;111:1826–1832. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosaka T., et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krebs L.T., et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 26.McKinsey T.A., Zhang C.L., Olson E.N. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 2001;11:497–504. doi: 10.1016/S0959-437X(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 27.Rudnicki M.A., Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 28.Bassel-Duby R., Olson E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 29.Schiaffino S., Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 30.Lin J., et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 31.Lagouge M., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Nickoloff B.J., Osborne B.A., Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 33.Nickoloff B.J., et al. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 2002;9:842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- 34.Pan Y., et al. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Dowell P., Otto T.C., Adi S., Lane M.D. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J. Biol. Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- 36.Nakae J., Kitamura T., Silver D.L., Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 2001;108:1359–1367. doi: 10.1172/JCI200112876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovall R.A., Hendrickson W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh J.J., Hayward S.D. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 39.Kao H.Y., et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tun T., et al. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawaichi M., et al. Genomic organization of mouse J kappa recombination signal binding protein (RBP-J kappa) gene. J. Biol. Chem. 1992;267:4016–4022. [PubMed] [Google Scholar]
- 42.Fryer C.J., White J.B., Jones K.A. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Liang Y., Chang J., Lynch S.J., Lukac D.M., Ganem D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 2002;16:1977–1989. doi: 10.1101/gad.996502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes S.M., et al. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 45.Kitamura T., et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic β cell growth. . J. Clin. Invest. 2002;110:1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daitoku H., Yamagata K., Matsuzaki H., Hatta M., Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 47.Miele L., Miao H., Nickoloff B.J. NOTCH signaling as a novel cancer therapeutic target. Curr. Cancer Drug Targets. 2006;6:313–323. doi: 10.2174/156800906777441771. [DOI] [PubMed] [Google Scholar]
- 48.Accili D. Lilly lecture 2003: the struggle for mastery in insulin action: from triumvirate to republic. Diabetes. 2004;53:1633–1642. doi: 10.2337/diabetes.53.7.1633. [DOI] [PubMed] [Google Scholar]
- 49.Knapp J.R., et al. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006;133:601–610. doi: 10.1242/dev.02249. [DOI] [PubMed] [Google Scholar]
- 50.Ogilvie R.W., Feeback D.L. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 1990;65:231–241. doi: 10.3109/10520299009105613. [DOI] [PubMed] [Google Scholar]
- 51.Das I., et al. Notch oncoproteins depend on gamma-secretase/presenilin activity for processing and function. J. Biol. Chem. 2004;279:30771–30780. doi: 10.1074/jbc.M309252200. [DOI] [PubMed] [Google Scholar]
- 52.Chi T., Yan Z., Xue Y., Wang W. Purification and functional analysis of the mammalian SWI/SNF-family of chromatin-remodeling complexes. Methods Enzymol. 2004;377:299–316. doi: 10.1016/S0076-6879(03)77018-9. [DOI] [PubMed] [Google Scholar]
- 53.Puigserver P., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.