Abstract
The ankyrin repeats cofactor-1 (ANCO-1) was recently identified as a p160 coactivator-interacting protein that may inhibit transcriptional activity of nuclear receptors. Here we havev characterized the transcriptional regulatory domains of ANCO-1. Two intrinsic repression domains (RD) were identified: an N-terminal RD1 at residues 318-611 and a C-terminal RD2 at 2369-2663. ANCO-1 also contains an activation domain (AD) capable of stimulating transcription in both mammalian and yeast cells. The minimal AD was delimited to a 70-amino acid region at residues 2076-2145. Overall, full-length ANCO-1 exhibited transcriptional repressor activity, suggesting that RD domains may suppress the AD activity. We further demonstrated that ANCO-1 silencing by siRNA enhanced progesterone receptor-mediated transcription. Together, these results indicate that the transcriptional potential of ANCO-1 may be modulated by a combination of repression and activation signals.
Keywords: ANCO-1; Ankyrin Repeat Cofactors, p160 Coactivator; Transcriptional Activation Domains; Transcriptional Repression Domains; Histone Deacetylases
Introduction
Nuclear receptors (NRs) are DNA-binding transcription factors that control hormone-dependent gene expression in several important biological processes, including development, reproduction and hormonal responses. NRs share a common domain structure, including an N-terminal activation domain (AF-1), a central DNA binding domain (DBD), and a C-terminal ligand-binding domain (LBD). Several transcriptional coactivators and corepressors have been identified to regulate NR activity. For examples, the silencing mediator for retinoid and thyroid hormone receptor (SMRT) and the nuclear receptor corepressor (N-CoR) bind to unliganded NRs to promote repression [1-4] by recruiting histone deacetylases (HDACs) [5-7]. In contrast, the p160 coactivators, including SRC-1 [8], GRIP1/TIF2 [9-11], and RAC3/ACTR/AIB1/TRAM-1/SRC-3 [12-16] bind to liganded NRs to stimulate activation through recruitment of histone acetyltransferases [17]. The p160 coactivators utilize the LXXLL motif to interact with a hydrophobic cleft on the receptor's LBD [18, 19]. These coactivators cause modifications of local chromatin structure [20, 21], and are known to be involved in cancer [14, 22-24]. The p160 coactivators share a common domain structure, including a highly conserved basic-helix-loop-helix (bHLH)-Per-Arnt-Sim (PAS) domain [17]. The PAS domain is highly conserved among many PAS family proteins [25] and is implicated in mediating protein-protein interactions (34-36). We sought to explore mechanisms that regulate p160 activity through identification of RAC3-interacting proteins. Recently, we identified a novel family of ankyrin repeat-containing cofactors (ANCOs) as p160 coactivator-interacting proteins [26]. Interestingly, ANCO-1 interacts with both p160 coactivators and HDAC corepressors. Overexpression of ANCO-1 inhibits ligand-dependent transcriptional activation by several nuclear receptors. Here we have further characterized the transcriptional regulatory domains of ANCO-1.
Materials and Methods
Plasmids
The G4-ANCO-1 fragment fusions were created by PCR amplification of ANCO-1 fragments and subcloned into pCMXGal vector. The G4-ANCO-1F (full-length) fusion was created by subcloning ANCO-1 coding region (aa 3-2663) into pCMXGal and included an HA epitope (MDYPYDVPDYARA). The pCMXHA-ANCO-1 ΔRD2 was created by digesting pCMXHA-ANCO-1F with BamHI and NheI, followed by klenow “fill-in” reaction and self-ligation. The ANCO-1 ΔRD2 (aa 3-2234) insert was subcloned into pCMXGal vector at the Asp718 and BamHI sites to create G4-ANCO-1 (3-2235) construct. The ANCO-1AD (aa 1851-2145), ANCO-1Ank (aa 126-295), and ANCO-1C (aa 2369-2663) were subcloned into the pGBT9 vector. The ANCO-1 shRNA expression vector was created by ligating a double-stranded oligonucleotide into the pSUPER vector [27] at BglII and HindIII sites. The oligonucleotide sequences for the ANCO-1 shRNA are: 5′- gat ccc cgc gga agc tgc cct tca cct tca aga gag gtg aag ggc agc ttc cgc ttt ttg gaa a -3′ (sense) and 5′- agc ttt tcc aaa aag cgg aag ctg ccc ttc acc tct ctt gaa ggt gaa ggg cag ctt ccg cgg g -3′ (antisense).
Cell Culture and Transient Reporter Assays
HeLa and COS-7 cells were maintained in DME medium containing 10% fetal bovine serum (FBS) and 5-μg/ml gentamycin (Gibco BRL, Carlsbad, CA). For Gal4 reporter assay, cells (2×105) were seeded in 12-well plates and transfected with 2.5 μg DNA/well by the standard calcium phosphate precipitation method. Transfections proceeded overnight, and precipitates were removed by washing the cells with phosphate buffered saline (PBS) supplemented with calcium and magnesium. Cells were cultured for an additional 48 hrs, after which the cells were harvested and analyzed for luciferase and β-galactosidase activities as previously described [26].
Yeast One- and Two-Hybrid Assays
Yeast reporter assays were conducted in Y190 strain. Yeast competent cells were prepared in LiSORB buffer (100 mM lithium acetate, 1 M D-Sorbital in TE). Transformed yeast were spread on selection medium plates and incubated at 30°C for 2 days. Colonies were picked from each plate and grown in selection media for 48 hrs at 30°C. β-galactosidase activity was determined by filter X-Gal or liquid ONPG assay and filter assay. The reactions were terminated by adding 200 μl of 1M Na2CO3. OD420 was then measured by a plate reader (Dynetech, Inc.).
Results
Mapping of Transcriptional Regulatory Domains of ANCO-1
To characterize the transcriptional regulatory domains of ANCO-1, we generated a series of Gal4 DBD (G4) fusions with various ANCO-1 fragments (Fig. 1A), and then analyzed their transcriptional potentials on a Gal4-depedent luciferase reporter MH100-tk-luc in mammalian cells (Fig. 1B). Most of these G4-ANCO-1 fusions had little effect on the reporter, except fragments C (aa 318-611 or RD1) and J (aa 2369-2663 or RD2) exhibited strong repression activity while fragment H (aa 1851-2145 or AD) strong activation function. These results suggest that ANCO-1 contains both repression and activation domains. We demonstrated that the Gal4-RD2 displayed repression activity in a dose-dependent manner and this correlated with increased expression of the fusion protein (Fig. 1C). The presence of both repression and activation domains within ANCO-1 prompted us to test the overall transcriptional potential of the full-length protein. Thus, we generated a Gal4-ANCO-1 full-length fusion (construct K in Fig. 1A), and tested its ability to regulate expression of the Gal4 reporter. We found that full-length ANCO-1 repressed basal transcription of the reporter stronger that the RD2 domain (Fig. 1D, J fragment), suggesting that full-length ANCO-1 may repress transcription. We also determined the role of RD2 domain in transcriptional repression by full-length ANCO-1 by creating a RD2 deletion mutant (aa 3-2235, construct L). This construct exhibited virtually no repression activity, suggesting that RD2 is most critical for the repression by full-length ANCO-1. As expected, a combination of the AD and RD2 (aa 1802-2663, construct M) show neither repression nor activation function. These results suggest that the transcriptional potential of the full-length ANCO-1 may be determined by a combination of activation and repression domains.
Fig. 1. Delineation of Transcriptional Regulatory Domains of ANCO-1.
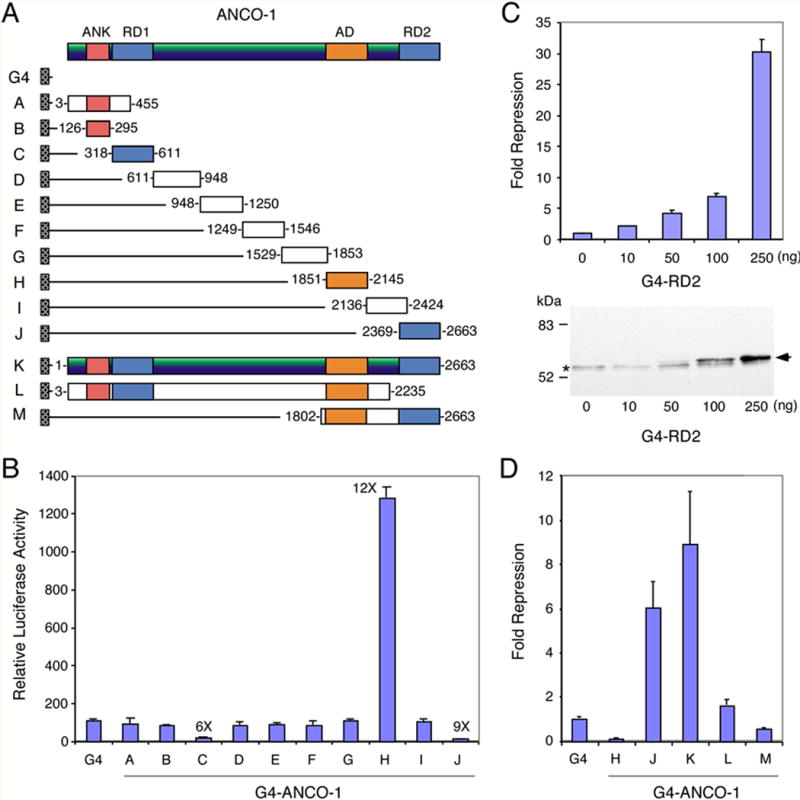
(A) Schematic representation of G4-ANCO-1 constructs. G4 represents the DNA binding domain (aa 1-147) of the yeast (S. cerevisiae) GAL4 protein. The starting and ending amino acids of individual ANCO-1 fragments are shown next to each fragment. (B) Transcriptional activities of G4-ANCO-1 constructs on a G4-dependent luciferase reporter MH100-tk-Luc. COS-7 cells were transfected with indicated plasmids and relative luciferase/β-galactosidase activities were determined. Fragments C (RD1 or repression domain-1) and J (RD2 or repression domain-2) show 6- and 9-fold repression activities relative to G4 alone, respectively. Fragment H (AD or activation domain) shows 12-fold activation relative to G4 alone. (C) Transcriptional activities of G4-ANCO-1 K (G4-ANCO-1F or full-length), L (ΔRD2), and M (AD + RD2) constructs, in comparison to G4 and ANCO-1 H and J fragments. Fold-repression of G4 reporter activity in comparison to G4 alone is represented. (D) Dose-dependent repression of the G4 reporter by G4-ANCO-1 J (RD2) fusion. Fold-repression relative to G4 alone is shown. Expression of G4-ANCO-1 J (RD2) fusion (indicated by an arrow at right) was detected by Western blot with anti-G4 antibody (Santa Cruz Biotechnology, Inc.). Asterisk indicates a nonspecific signal in all samples detected by the G4 antibody. The amounts of plasmid DNA used in each transfection are shown at bottom.
ANCO-1 Activation Domain Functions In Yeast
The finding of a potential transcriptional activation domain with ANCO-1 intrigued us to further characterize this domain. First, we verified that this activation domain was functional in yeast cells. The ANCO-1 AD domain (aa 1851-2145) was subcloned into the pGBT9 vector and expressed in Y190 cells. We found that this AD domain activated β-galactosidase expression about 4 times stronger than the RAC3 AD domain [12] (Fig. 2A, B). By contrast, the pGBT9 vector, the ANCO-1 ankyrin domain (ANK), and the ANCO-1 RD2 (C-terminal) domain did not activate the reporter. These results indicate that ANCO-1 AD is functional in yeast, suggesting that the mechanism of transcriptional activation by the AD is conserved between human and yeast. To pinpoint the region within ANCO-1 AD domain that is responsible for the observed transcriptional activation, we divided aa 1851-2145 region into five smaller fragments (Fig. 2C), and measured their transcriptional activities in Y190 (Fig. 2D) and mammalian COS-7 cells (Fig. 2E). We found that transcriptional activation was restricted to the C-terminal E fragment (aa 2076-2145). Thus, we have identified a minimal activation domain of ANCO-1 within a 70-aa fragment.
Fig. 2. ANCO-1 AD Domain Activates Transcription in Yeast.
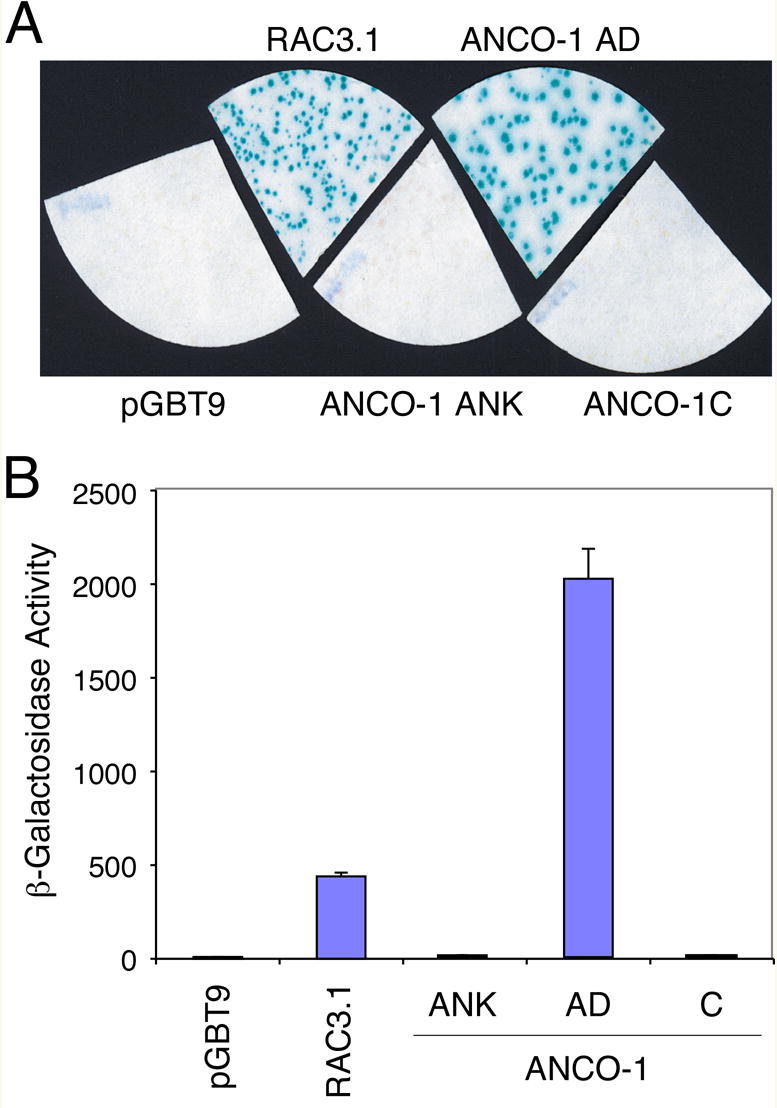
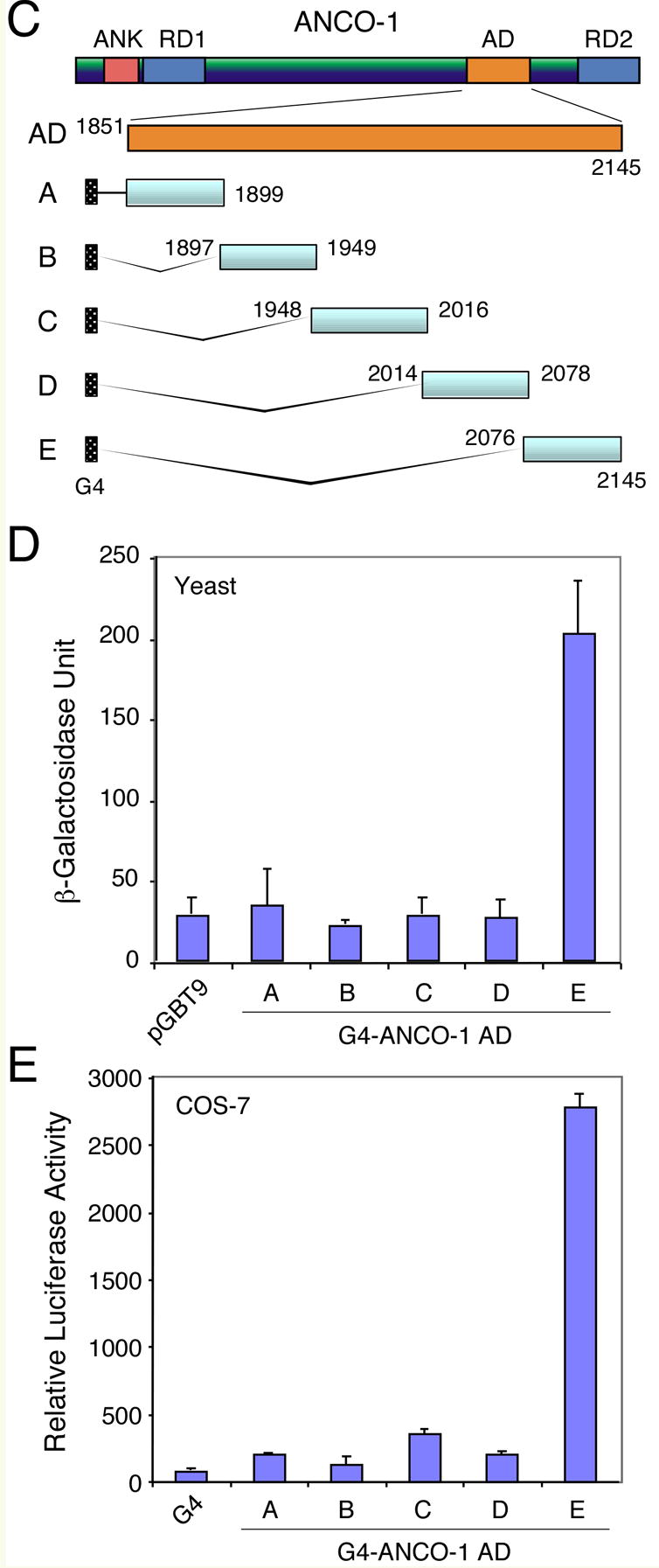
Yeast (S. cerevisiae) Y190 cells were transformed with plasmids encoding the indicated G4 fusion proteins or the empty vector pGBT9. Colony X-Gal filter assay (A) and liquid ONPG assay (B) were conducted to measure β-galactosidase activity as described in Materials and Methods. The control pGBT9 vector, ANCO-1 Ank (aa 126-295) and C (aa 2369-2663) domains had no detectable activity, while the RAC3.1 [12] and ANCO-1 AD (aa 1851-2145) activated lacZ gene expression strongly. (C) Schematic representation of G4-ANCO-1 AD sub-fragments fusions. Five ANCO-1 AD sub-fragments (A through E) are shown with indicated starting and ending amino acids beside each fragment. G4, Gal4 DNA binding domain. (D) Transcriptional activities of G4-ANCO-1 AD sub-fragments in yeast cells. Y190 cells were transformed with indicated expression vectors and average β-galactosidase activities were determined from three colonies. The E fragment 9aa 2076-2145) possesses the most transcriptional activation function. (E) Transcriptional activation by the G4-ANCO-1 AD sub-fragments in mammalian cells. COS-7 cells were transiently transfected with the G4 empty vector or G4-ANCO-1 AD sub-fragments and relative luciferase activities were determined from three independent transfections.
ANCO-1 RD2 Domain Contains Multiple Repressive Regions
The ANCO-1 RD2 domain had strongest repression activity and was essential for repression by the full-length protein. To map the minimal region required for RD2-mediated repression, we subdivided the RD2 region into several overlapping fragments (Fig. 3A), and tested their abilities to repress basal transcription in COS-7 cells (Fig. 3B). Consistently, G4-ANCO-1 RD2 fusion exhibited an approximately 8-fold repression activity. We found that aa 2368-2455 (A) and 2597-2663 (D) displayed better repression activity than the center fragments (B and C). The combination of C and D fragments showed the strongest repression activity. In contrast, combinations of BCD, ABC, AB, and BC fragments showed little or no repression activity. These results suggest that multiple regions may be involved in repression activity of ANCO-1 RD2 domain.
Fig. 3. Multiple Regions Are Involved in RD2-meidated Repression.
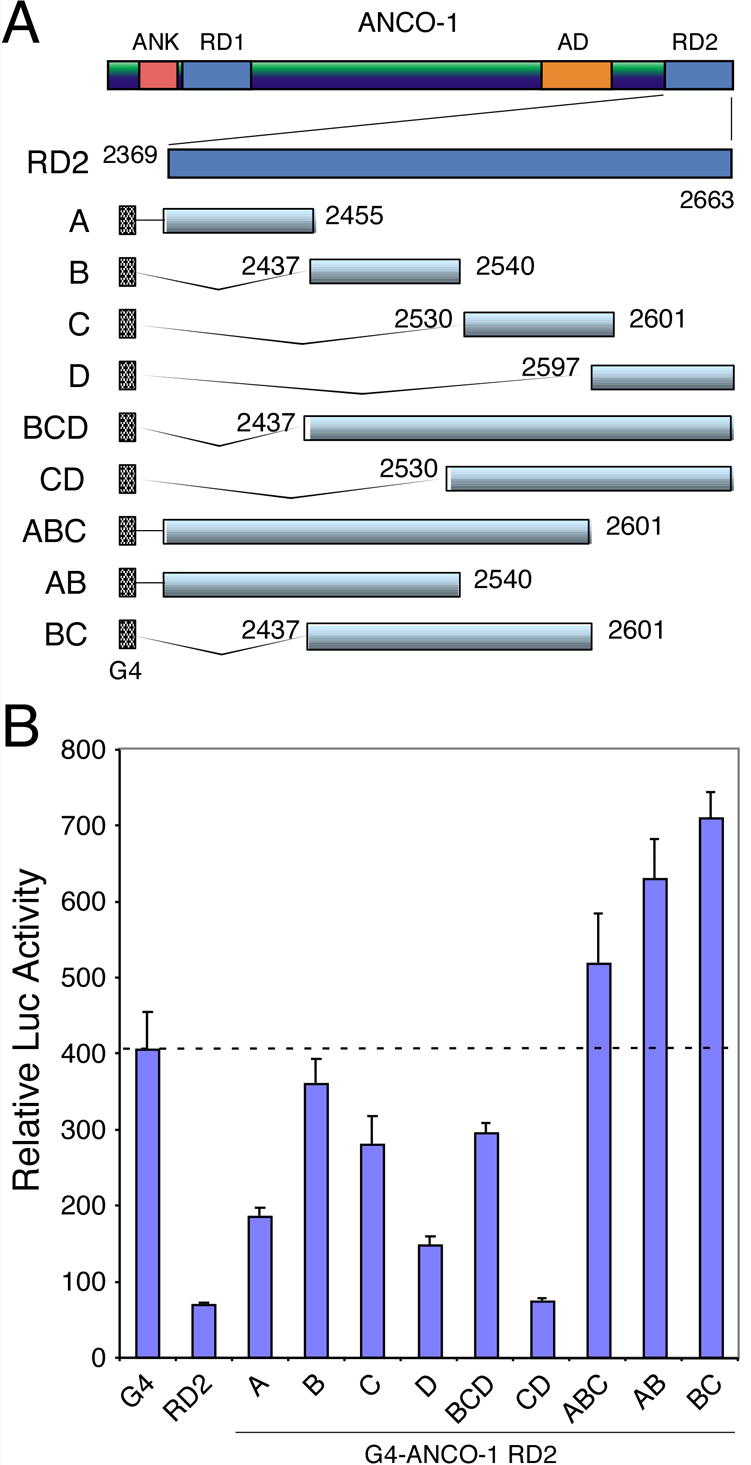
(A) Schematic representation of G4-ANCO-1 RD2 sub-fragments. The RD2 domain (aa 2369-2663) is subdivided into four smaller fragments (A through D). Each fragment is fused individually or in combination with Gal4 DNA binding domain (G4). The starting and ending amino acids are indicated beside each fragment. (B) Luciferase reporter assay showing repression activity of each RD2 sub-fragment. COS-7 cells were transfected with indicated G4-ANCO-1 RD2 sub-fragments. Normalized luciferase activities were determined from three independent transfections.
Silencing of ANCO-1 Enhances PR Transcriptional Activity
To determine if endogenous ANCO-1 plays a role in regulating nuclear receptor activity, we knocked down the expression of endogenous ANCO-1 by small hairpin (sh)RNA against ANCO-1 (Fig. 4A). The ANCO-1 shRNA specifically inhibited the expression of ANCO-1 and had no effect on the co-expressed Daxx protein (Fig. 4B). We determined the effects of ANCO-1 knockdown on PR-mediated transcriptional activation on MMTV-luciferase reporter (Fig. 4C), and found that progesterone stimulated expression of the reporter in a time-dependent manner. A maximal induction occurred at around 6 hours, which was followed by a 4-fold decline at 12 hours. Intriguingly, ANCO-1 shRNA was capable of enhancing PR-mediated transcriptional activation by 3 to 5-fold. Furthermore, we showed that the enhancement of PR transcriptional activity occurred in an ANCO-1 shRNA concentration-dependent manner (Fig. 4D). We have tested a second ANCO-1 shRNA and found similar results (data not shown). These results suggest that endogenous ANCO-1 indeed plays a role in regulating PR transcriptional activity.
Fig. 4. Silencing of ANCO-1 by siRNA Enhances PR Transcriptional Activity.
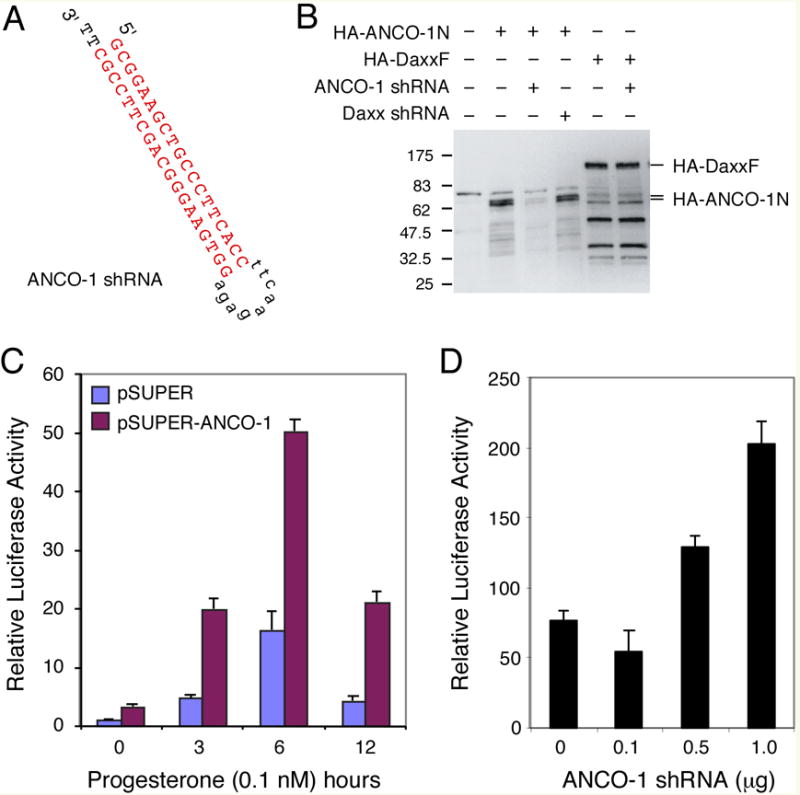
(A) ANCO-1 shRNA sequence and its predicted hairpin structure. The ANCO-1 shRNA targets nucleotide 458 to 478 of the ANCO-1 cDNA (corresponding to aa 57 to 63). The siRNA sequences that targets ANCO-1 mRNA are shown in red. (B) ANCO-1 shRNA inhibits expression of ANCO-1 in transfected cells. HeLa cells were transfected with indicated plasmids and analyzed by Western blot with anti-HA antibody. The ANCO-1 shRNA specifically inhibited the expression of HA-ANCO-1N but not HA-DaxxF. (C) Effects of ANCO-1 knock down on PR-mediated transcription. HeLa cells were transiently transfected with hPR-B, MMTV-luciferase reporter, and the ANCO-1 shRNA expressing vector or pSUPER vector as control. Progesterone (0.1 nM) was added at indicated hours before harvesting at 48 hours after transfection. (D) ANCO-1 shRNA enhances PR-mediated transcriptional activation in a concentration-dependent manner. Transfected cells were treated with progesterone (0.1 nM) for 6 hours before harvesting for luciferase assay.
Discussion
In this study, we have characterized the transcriptional regulatory domains of the ankyrin repeat cofactor ANCO-1. We demonstrated that ANCO-1 contains both repression and activation domains. The AD domain was mapped to residues 1851-2145 with a core AD identified at residues 2076 –2145. In addition, two ANCO-1 RD domains were identified at residues 318-611 (RD1) and 2369-2663 (RD2). Unlike the AD domain, the repression activity of the RD2 domain is determined by more than one region. We also showed that silencing of endogenous ANCO-1 enhanced PR-mediated transcriptional activation.
ANCO-1 was first identified as a p160 coactivator RAC3-interacting protein in a yeast two-hybrid screen [26]. Interestingly, ANCO-1 interacts with both the coactivators and HDAC corepressors. As a result, overexpression of ANCO-1 inhibited ligand-dependent transcriptional activation by several nuclear receptors. It is possible that the recruitment of HDACs by ANCO-1 may antagonize histone acetyltransferase activity, leading to transcriptional attenuation. Therefore, ANCO-1 represents a novel corepressor that could block ligand-dependent transcription by antagonizing coactivator' function through HDACs. Our effort to delineate the transcriptional regulatory domains of ANCO-1 resulted in the identification of two RDs and one AD domains. The ability of ANCO-1 AD domain to activate transcription was confirmed in multiple mammalian cell lines, as well as in yeast cells, suggesting that this AD domain may utilize a conserved mechanism for activating transcription. We delineated the minimal AD sequence to within a 70-aa region. Sequence analysis revealed no significant homology with known ADs, thus it is possible that the ANCO-1 AD might function through a novel mechanism. Unlike the AD domain, the RD2 domain contains multiple sequences that were capable of repressing transcription to various degrees. Both the RD2-A (aa 2368-2455) and D regions (aa 2597-2663) showed strongest repression activities. Inclusion of other fragments could also modulate the repression activity, suggesting multiple mechanisms may be involved in RD2-mediated repression. It is not without precedents that a transcriptional cofactor contains both autonomous repression and activation domains. The NR-binding, SET-domain-containing protein (NSD1) also contains both activation and repression domains and the characteristics of a corepressor and a coactivator [28, 29].
To confirm the role of ANCO-1 in NR-mediated transcriptional regulation, we conducted ANCO-1 knockdown experiments using shRNA against ANCO-1 and determined the effect on PR-mediated transcriptional activation. Two independent shRNA sequences targeting at different regions of ANCO-1 showed similar effects (Fig. 4 and data not shown). As expected, shRNA-mediated knockdown of ANCO-1 resulted in enhancement of PR transcriptional activity. Our results indicate that endogenous ANCO-1 may negatively regulate the transcriptional activity of nuclear receptors.
Based on the current study and our previous findings [26], we propose that ANCO-1 interacts with both p160 coactivators and HDAC corepressors to antagonize transcriptional activation by the receptors. It is known that nuclear receptor dimers activate gene expression in response to ligand binding by recruiting p160, p300/CBP, CARM1 and other coactivators to target promoters [17, 30]. These coactivators acetylate local chromatin to activate the promoter. ANCO-1 may localize to the NR/promoter complex by interaction with p160 through its RD2 region. The HDAC complex may then be recruited to the activated NR-promoter complex by interaction with another RD2 surface. The simultaneous interaction of ANCO-1 with p160 coactivators and HDAC corepressors may result in attenuation of ligand-activated transcription by nuclear receptors. Future study will be necessary to further test this hypothesis.
Acknowledgments
We thank R. Agami at Netherlands Cancer Institute for the pSUPER vector. We are grateful to Jianzhong Zhang, and Amy Chen for technical help. This work was supported by NIH (DK52888) and the Leukemia and Lymphoma Society to JDC.
Abbreviation
- ANCO-1
ankyrin repeats cofactor-1
- RAC3
nuclear receptor-associated cofactor-3
- PR
progesterone receptor
- HDAC
histone deacetylases
- DBD
DNA binding domain
- LBD
ligand binding domain
- RD
repression domain
- AD
activation domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [see comments] [DOI] [PubMed] [Google Scholar]
- 2.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 3.Park EJ, Schroen DJ, Yang M, Li H, Li L, Chen JD. SMRTe, a silencing mediator for retinoid and thyroid hormone receptors- extended isoform that is more related to the nuclear receptor corepressor. Proc Natl Acad Sci U S A. 1999;96:3519–3524. doi: 10.1073/pnas.96.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci U S A. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 6.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40- repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [In Process Citation] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oñate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 9.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a Transcriptional Coactivator for the AF-2 Transactivation Domain of Steroid, Thyroid, Retinoid, and Vitamin D Receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voegel JJ, Heine MJS, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci U S A. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear- receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [see comments] [DOI] [PubMed] [Google Scholar]
- 14.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 16.Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 17.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 18.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 19.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 20.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 21.Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tikkanen MK, Carter DJ, Harris AM, Le HM, Azorsa DO, Meltzer PS, Murdoch FE. Endogenously expressed estrogen receptor and coactivator AIB1 interact in MCF-7 human breast cancer cells. Proc Natl Acad Sci U S A. 2000;97:12536–12540. doi: 10.1073/pnas.220427297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- 25.Huang W, Sun GL, Li XS, Cao Q, Lu Y, Jang GS, Zahang FQ, Chai JR, Wang ZY, Waxman S, Chen Z, Chen SJ. Acute Promyelocytic Leukemia: Clinical Relevance of Two Major PML-RARα Isoforms and Detection of Minimal Residual Disease by Retrotranscriptase/Polymerase Chain Reaction to Predict Relapse. Blood. 1993;82:1264–1269. [PubMed] [Google Scholar]
- 26.Zhang A, Yeung PL, Li CW, Tsai SC, Dinh GK, Wu X, Li H, Chen JD. Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J Biol Chem. 2004;279:33799–33805. doi: 10.1074/jbc.M403997200. [DOI] [PubMed] [Google Scholar]
- 27.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 28.Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. Embo J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, Brown PH, Fuqua SA, Osborne CK. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem. 2001;276:27907–27912. doi: 10.1074/jbc.M104278200. [DOI] [PubMed] [Google Scholar]
- 30.McKenna NJ, O'Malley BW. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]


