Abstract
Target of rapamycin (TOR) is a central regulator of cellular and organismal growth in response to nutrient conditions. In a genetic screen for novel TOR interactors in Drosophila melanogaster, we have identified the clathrin-uncoating ATPase Hsc70-4, which is a key regulator of endocytosis. We present genetic evidence that TOR signaling stimulates bulk endocytic uptake and inhibits the targeted endocytic degradation of the amino acid importer Slimfast. Thus, TOR simultaneously down-regulates aspects of endocytosis that inhibit growth and up-regulates potential growth-promoting functions of endocytosis. In addition, we find that disruption of endocytosis leads to changes in TOR and phosphatidylinositol-3 kinase activity, affecting cell growth, autophagy, and rapamycin sensitivity. Our data indicate that endocytosis acts both as an effector function downstream of TOR and as a physiologically relevant regulator of TOR signaling.
Introduction
Multicellular organisms must modulate cellular growth and proliferation in response to available nutrients, energy, and growth factor signaling. The regulatory kinase target of rapamycin (TOR) has emerged as a convergence point for the transduction of these signals into appropriate changes in cell metabolism and growth. TOR activity is stimulated by insulin-responsive phosphatidylinositol-3 kinase (PI3K)/Akt signaling, by nutrients such as amino acids, and by high cellular energy levels (for reviews see Martin and Hall, 2005; Sarbassov et al., 2005a). In response to these signals, TOR effects changes in a diverse number of downstream processes, including transcription, translation, nutrient import, and autophagy, to achieve the alignment of cellular energy utilization with available resources. Unsurprisingly, improper activation of TOR signaling has been implicated in the development of cancers and hamartoma syndromes and in metabolic diseases such as diabetes and obesity (Manning, 2004). For example, mutations in either component of the TSC1–TSC2 complex, which is an upstream inhibitor of TOR signaling, result in the formation of benign tumors in multiple tissues.
Although TOR is known to affect a wide range of cellular processes, the relative contribution of these processes and how they interact to result in a directed growth response remain poorly understood. Components of the translational machinery are well established downstream effectors of TOR signaling (Martin and Hall, 2005). TOR directly phosphorylates eukaryotic initiation factor-4E–binding protein (4E-BP) and ribosomal protein S6 kinase (S6K), thereby facilitating cap-dependent translation and ribosome biogenesis. Although these effects on protein synthesis are likely to contribute substantially to cellular growth capacity, they are unlikely to fully account for the growth effects of TOR. For example, whereas inactivation of TOR results in a nearly complete block of protein synthesis in yeast, this effect is more modest in mammalian cells, with an ∼15–50% decrease in translation rate (Jefferies et al., 1994; Terada et al., 1994). In Drosophila melanogaster, null mutations in 4E-BP are without effect on cell growth (Junger et al., 2003), and the growth phenotype of S6k-null mutants is significantly less severe than that of Tor mutants (Montagne et al., 1999; Oldham et al., 2000; Zhang et al., 2000). Activation of S6K only partially overcomes the growth arrest of Tor mutants in this system. Recent genetic studies in mouse have also shown that the ribosomal substrate of S6K, rpS6, does not appear to be a relevant mediator of the growth effects of this pathway (Pende et al., 2004).
These observations have motivated the search for other effector pathways and cellular processes downstream of TOR that might contribute to its effects on cell growth. A growing number of studies of TOR structure, function, and localization point to an important role for TOR signaling in controlling vesicular trafficking. Biochemical studies in yeast have found that TOR localizes to intracellular vesicles and cofractionates with endosomal markers (Kunz et al., 2000; Chen and Kaiser, 2003), which is consistent with a role in the endocytic compartment. In addition, TOR has a highly conserved function as a regulator of autophagy, which is a process of cytoplasmic degradation that involves the reorganization of intracellular membranes into autophagic vesicles (Noda and Ohsumi, 1998; Scott et al., 2004). Finally, TOR is structurally related to the class III PI3K/Vps34 family of lipid kinases, with well characterized roles in endocytosis. Interestingly, recent studies have identified a novel role for hVps34 in relaying intracellular nutrient status to TOR (Byfield et al., 2005; Nobukuni et al., 2005), indicating that this family of molecules may have common roles in nutrient sensing and membrane trafficking.
We report the identification of the clathrin-uncoating ATPase Hsc70-4 in a genetic enhancer/suppressor screen for novel TOR interactors in D. melanogaster. Hsc70-4 is a critical regulator of clathrin-mediated endocytosis, and we provide evidence that TOR signaling influences bulk endocytosis, as well as the targeted endocytic degradation of a specific amino acid transporter. Our results suggest that TOR controls growth, in part, by simultaneously down-regulating aspects of endocytosis that inhibit growth and up-regulating potential growth-promoting functions of endocytosis.
Results
Hsc70-4 mutants are dominant modifiers of TOR overexpression phenotypes
To better understand the regulation and downstream effects of TOR-mediated growth signaling, we took a genetic approach to reveal novel players in the TOR signaling pathway. We used a tissue-specific TOR overexpression phenotype as the sensitized background for a dominant-modifier ethyl methanesulfonate (EMS) mutagenesis screen. To create this background, P element–mediated transposition was used to introduce a copy of the wild-type Tor cDNA, which was preceded by multiple copies of the eyeless (ey) enhancer, into the D. melanogaster genome. ey-driven overexpression of TOR (eyTOR) led to an overall reduction in size of the adult eye (Fig. 1, A–B), demonstrating the paradoxical decrease in growth previously observed to result from wild-type TOR overexpression (Hennig and Neufeld, 2002). In addition to the reduction in eye size, ommatidial patterning was disorganized, with some ommatidia appearing fused or pitted. Immunohistochemistry of eyTOR imaginal discs revealed a delay in morphogenetic furrow progression and missing and disorganized photoreceptor cells, as well as an increased level of cell death (Fig. S1, available at http://www/jcb.org/cgi/content/full/jcb.200511140/DC1). The severity of the eyTOR phenotype was strongly enhanced by a heterozygous-null mutation of the negative TOR regulator Tsc1 (Fig. 1 C) and was suppressed by addition of rapamycin to the media (Fig. S1). These results suggest that the eyTOR phenotype results from inappropriately high levels of TOR signaling, and they demonstrate the potential utility of this overexpression phenotype as a dosage-sensitive genetic background.
Figure 1.
Mutations in endocytic regulators modify TOR overexpression phenotypes. (A–F) Disruption of Hsc70-4 or shibire dominantly enhances the small rough eye phenotype caused by ey TOR. Scanning electron microscopy images of D. melanogaster compound eyes: wild-type (A), eyTOR/+ (B), eyTOR/+; Tsc129/+ (C), eyTOR/+; Hsc70-4e19/+ (D), UAS-ShiK44A/+; eyGAL4/UAS-ShiK44A (E), and UAS-ShiK44A/+; eyTOR eyGAL4/UAS-ShiK44A (F). (G) Mutations in the e3/e19 complementation group disrupt the ATPase domain of the Hsc70-4 clathrin-uncoating ATPase. Shown is the domain structure of Hsc70-4, with the mutated residues indicated. (H and I) Mutation of Hsc70-4 does not lead to increased levels of TOR protein. (H) Loss of GFP expression marks a clone of cells homozygous for Hsc70-4e3 (arrow) from an eye imaginal disc expressing eyeless-driven FLAG-tagged TOR. Genotype: hsflp122/+; eyTOR/+; FRT82B Ubi-GFP/FRT82B Hsc70-4e3. (I) Extracts from wild-type and Hsc70-4e3/e19 third instar larvae were blotted with anti-TOR antibodies to detect endogenous TOR levels. (J–L) Expression of ShiK44A causes relocalization of TOR to endocytic vesicles. FLAG-tagged TOR expressed clonally in fat body cells (J) shows altered distribution upon coexpression of UAS-ShiK44A (K). TOR-containing vesicles near the cell surface can incorporate the endocytic tracer TR-avidin (L, arrow). Genotype: hsflp122/+; Act>CD2>Gal4, UAS-TOR/TM3-UAS-ShiK44A. Bars: (A–F) 100 μm; bar in (L) is 12.5 μm in H–K, and 6.25 μm in L.
We next sought to identify novel factors involved in TOR signaling by screening for the ability of EMS-generated mutations to dominantly modify the eyTOR phenotype. From ∼60,000 F1 progeny scored, we identified 23 eyTOR enhancers and 2 suppressors. Within this collection were two independent groups that failed to complement for lethality. We report the analysis of a third chromosome complementation group consisting of two alleles (e3 and e19) isolated as dominant enhancers of the eyTOR phenotype. Heterozygous mutation of either e3 or e19 caused a further reduction in eye size of eyTOR flies (Fig. 1 D and Fig. S1) and a similar enhancement of MS1096-Gal4-driven TOR misexpression phenotypes in the adult wing (Fig. S1). Through recombination and deficiency mapping we localized the e3 and e19 mutations to the 88E4 genomic region. Complementation tests with lethal mutations in this interval revealed that e3 and e19 disrupt the Hsc70-4 gene, which encodes a constitutively expressed member of the stress-induced Hsp70 family of ATPases. Independently isolated mutant alleles of Hsc70-4 also displayed a strong enhancement of the eyTOR phenotype (Fig. S1). In addition, both the dominant eyTOR enhancement and the recessive lethality of e3 and e19 could be rescued by a transgene carrying a wild-type copy of Hsc70-4 (Hing et al., 1999; Fig. S1). We found that the e3 and e19 mutations result in single amino acid substitutions within the ATPase domain of Hsc70-4 (Ser40 to Phe and Arg301 to Cys, respectively; Fig. 1 G). These residues are completely conserved in fungi, plant, and animal Hsc70-4 orthologues, indicating their likely importance for Hsc70-4 function. Therefore, from this point forth we refer to these mutations as Hsc70-4e3 and -4e19.
Hsc70-4 catalyzes the uncoating of clathrin-coated vesicles, which is an essential late step in clathrin-mediated endocytosis (Schlossman et al., 1984; Chang et al., 2002). In addition, Hsp70 family members can act as chaperones to regulate protein folding and stability (Mayer and Bukau, 2005). To determine whether either of these functions of Hsc70-4 contributes to its genetic interactions with TOR, we first assayed the ability of other endocytic factors to modify TOR misexpression phenotypes. Shibire (shi) encodes the D. melanogaster homologue of dynamin, which is a GTPase critical for the proper membrane closure and budding of endocytic vesicles from the plasma membrane (Chen et al., 1991). We found that expression of a dominant-negative form of shibire, ShiK44A (Moline et al., 1999), strongly enhanced the eyTOR phenotype (Fig. 1, E and F), suggesting that Hsc70-4 may influence TOR signaling through its role in endocytosis. In contrast, Hsc70-4 mutations did not lead to increased levels of TOR protein in eyTOR eye imaginal discs (Fig. 1 H), indicating that the enhancement of the eyTOR phenotype by Hsc70-4 mutants is not caused by increased stability or abundance of overexpressed TOR protein. We also observed no increase in total levels of endogenous TOR protein in Hsc70-4 mutants (Fig. 1 I). Interestingly, we found that expression of ShiK44A led to increased localization of TOR to vesicular structures, a subset of which were accessible to an endocytic tracer (Fig. 1, J–L; see below), further supporting a link between TOR signaling and endocytosis.
TOR signaling promotes bulk endocytosis
To further characterize the interplay between TOR and endocytosis, we tested the effect of TOR signaling on several endocytic markers in the D. melanogaster larval fat body. The fat body serves as a nutrient storage organ that is analogous to the vertebrate liver (Dean et al., 1985), and it has recently been shown to act as a nutrient sensor capable of affecting global growth through a TOR-dependent humoral mechanism (Colombani et al., 2003). We tested the effects of altered nutrient availability and TOR signaling on the intracellular localization of components of the endocytic machinery, using GFP-fusions to Rab5, Rab7, and clathrin. In fat body cells from fed control animals, we observed GFP-Rab5 localization both at the cell surface and throughout the cytoplasm (Fig. 2 A). In contrast, larvae subjected to a 5-h starvation displayed a variable redistribution of GFP-Rab5 toward the cell surface, with a less diffuse, punctate pattern often observed in these cells (Fig. 2 B). More direct alteration of TOR signaling, through overexpression of TSC1 and TSC2, or TOR itself, also affected GFP-Rab5 localization, resulting in the appearance of large aggregates near the cell surface (Fig. 2 C). The localization of GFP-Rab7 and clathrin-GFP was also altered in response to TOR inhibition (unpublished data). Overexpression of TOR in fat body cells also resulted in increased expression of Hsc70-4, as monitored by a GFP-Hsc70-4 fusion expressed from the endogenous Hsc70-4 locus (Clyne et al., 2003). Together, these data indicate that altered TOR signaling effects an endocytic response in the fat body.
Figure 2.
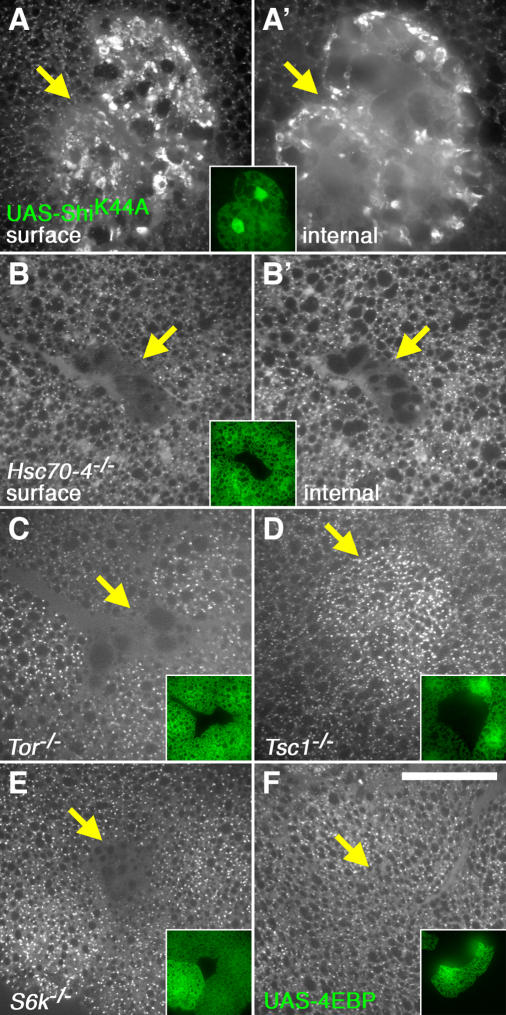
TOR signaling affects endocytosis. (A–C) Starvation and TOR overexpression affect GFP-Rab5 localization. Flp-dependent, spontaneously induced clones of GFP-Rab5–expressing cells in mid–third instar fat body tissue are shown. (A) Under fed conditions, GFP-Rab5 is uniformly distributed throughout both the cytoplasm and surface of these cells. (B) In larvae starved for 4.5 h, higher levels of GFP-Rab5 are observed near the cell surface. (C) TOR overexpression leads to the formation of large GFP-Rab5 aggregates at the cell surface. Images in A–C show the nuclear focal plane. Inset in C shows image taken at a focal plane just below the plasma membrane. Genotypes: hsflp122/+; Act>CD2>Gal4/UAS-GFP-Rab5 (A and B) and hsflp122/+; UAS-TOR, Act>CD2>Gal4/UAS-GFP-Rab5 (C). (D) TOR overexpression increases levels of GFP-Hsc70-4. The image shows a clone of cells expressing FLAG-tagged TOR (marked by FLAG staining in inset), resulting in increased levels of endogenously expressed GFP-Hsc70-4. Genotype: hsflp122/+; UAS-TOR, Act>CD2>Gal4/Hsc70-4Wee-P1. (E–J) Endocytic uptake in D. melanogaster fat body cells. Fat body tissue containing clones of overexpressing or mutant cells was incubated with fluorescently labeled avidin to monitor endocytic uptake. Images show surface (E and F) or cytoplasmic (E′, F′, and G–J) focal planes. Clones are demarcated by GFP expression (insets) and are indicated by yellow arrows. (E–F) Endocytosis is inhibited in response to disruption of shibire or Hsc70-4 function. A clone of ShiK44A-expressing cells (E) is marked by GFP expression, and a clone homozygous mutant for a null allele of Hsc70-4 (F) is marked by the absence of GFP. (G–J) TOR signaling positively affects bulk endocytic uptake. Shown are clones of cells homozygous for null mutations in Tor (G), Tsc1, (H), S6k (I), and a clone overexpressing 4E-BP (J). Bar, 25 μm. Genotypes: hsflp122/+; Act>CD2>Gal4, UAS-GFP/TM3-UAS-ShiK44A (E), hsflp122/+; Cg-Gal4/+; FRT82B UAS-GFP/FRT82B Hsc70-4 Δ16 (F), hsflp122/+; UAS-2XeGFP FRT40A fb-GAL4/Tor ΔP FRT40A (G), hsflp122/+; Cg-Gal4/+; FRT82B UAS-GFPnls/FRT82B Tsc129 (H), hsflp122/+; Cg-Gal4/+; UAS-2XeGFP FRT80B/S6Kl1 FRT80B (I), and hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-4E-BP (J). Bar, 25 μm.
To better understand the nature of these endocytic changes, we monitored the effects of TOR signaling on the ability of fat body cells to internalize a fluorescent endocytic tracer, Texas red–conjugated avidin (TR-avidin). In control experiments we found that genetic disruption of shi or Hsc70-4 prevented proper endocytic uptake in these cells, as was demonstrated previously in Garland cells (Kosaka and Ikeda, 1983; Chang et al., 2002). TR-avidin failed to be efficiently internalized in ShiK44A-expressing fat body cells, and instead accumulated near the cell surface, often in large aggregates (Fig. 2 E). TR-avidin uptake was also blocked in cells that were homozygous for a null allele of Hsc70-4 (Bronk et al., 2001), and, in this case, a decrease in both surface-bound and internalized tracer was observed (Fig. 2 F). A similar, but less extensive, decrease in TR-avidin uptake was observed in Hsc70-4e3 and -4e19 mutant cells (Fig. S2, available at http://www/jcb.org/cgi/content/full/jcb.200511140/DC1).
We next used this assay to monitor the effects of altered TOR signaling on bulk endocytic uptake. Mosaic clones mutant for a null allele of Tor showed a nearly complete block of TR-avidin uptake (Fig. 2 G). Similar effects were observed when TOR signaling was inhibited by overexpression of TSC1–TSC2 or TOR (Fig. S2). To see whether increased activation of TOR could further stimulate endocytic internalization, we generated mitotic clones homozygous for a Tsc1-null allele. Increased tracer uptake was observed in these cells (Fig. 2 H). Overexpression of Rheb, which is an upstream activator of TOR, caused a similar increase in TR-avidin uptake (Fig. S2). We also tested whether two targets of TOR, S6K and 4E-BP, affected endocytosis. Cells mutant for a null allele of S6k showed a strong decrease in TR-avidin uptake, suggesting that the effects of TOR on endocytosis are mediated in large part through S6K (Fig. 2 I). In contrast, overexpression of 4E-BP, whose activity is inhibited by TOR, had no effect in this assay (Fig. 2 J).
As an alternative approach to monitoring endocytosis in vivo, we tested the ability of fat body cells to internalize larval serum protein 2 (Lsp2). Lsp2 is present in the hemolymph during larval development, and is internalized through endocytosis by the fat body during the late third instar period (Levenbook, 1985). Anti-Lsp2 staining revealed a fine punctate appearance throughout the cytoplasm of wild-type cells (Fig. 3). In contrast, ShiK44A-expressing cells displayed large aggregates of Lsp2 protein at the cell surface and an absence of internal Lsp2 staining (Fig. 3 A). Hsc70-4 mutant cells were also defective in Lsp2 uptake, and, again, differed from ShiK44A-expressing cells in showing a decrease in Lsp2, both internally and at the surface (Fig. 3 B). The effects of TOR signaling on Lsp2 internalization were identical to the effects on TR-avidin; Lsp2 uptake was severely reduced in Tor or S6k mutant cells and in TSC1–TSC2–overexpressing cells, and was increased in cells mutant for Tsc1 or overexpressing Rheb (Fig. 3, C–E, and Fig. S2). Together, these results demonstrate that the Rheb–TOR–S6K pathway is required for proper endocytic uptake and indicate a novel role for TOR signaling in the positive stimulation of bulk endocytosis. Furthermore, they indicate that ShiK44A expression and loss of Hsc70-4 result in distinct endocytic phenotypes in the fat body.
Figure 3.
Endocytic regulators and TOR signaling control Lsp2 internalization. Images show surface (A and B) or cytoplasmic (A′, B′, and C–F) focal planes of third instar larval fat body stained with anti-Lsp2. (A and B) Endocytosis of Lsp2 from the larval hemolymph is inhibited in response to disruption of shibire (A) or Hsc70-4 (B). Note the accumulation of Lsp2 at the surface of ShiK44A-expressing cells, but not in Hsc70-4–null cells. (C–F) TOR signaling is required for Lsp2 uptake. Shown are clones of cells homozygous for null mutations in Tor (C), Tsc1 (D), S6k (E), and a clone overexpressing 4E-BP (F). Arrows indicate mutant or overexpressing cells. Bar, 25 μm. Genotypes are as in Fig. 2 E–J.
TOR signaling inhibits the endocytic degradation of Slimfast
In addition to the nonselective endocytic uptake described in the previous section, studies in yeast and cultured mammalian cells have shown that TOR can selectively influence the endocytic uptake and degradation of specific nutrient transporters (Beck et al., 1999; Edinger and Thompson, 2002). To investigate the effects of TOR signaling on targeted endocytosis, we generated an antibody to Slimfast, which is a cationic amino acid importer previously shown to positively affect organismal growth through effects on the TOR and PI3K signaling pathways (Colombani et al., 2003). We used this antibody to test whether TOR signaling may regulate the endocytic turnover of Slimfast in response to nutrient conditions. Antibody staining of fat body tissue confirmed that Slimfast is localized primarily to the cell surface, with peak levels just below the plasma membrane (Fig. 4 and Fig. S3, available at http://www/jcb.org/cgi/content/full/jcb.200511140/DC1). Disruption of endocytosis through ShiK44A expression resulted in a marked increase in Slimfast at the cell surface (Fig. 4 A), suggesting that endocytosis normally acts to antagonize the surface expression of Slimfast. In contrast, disruption of Hsc70-4 did not lead to increased surface levels of Slimfast (Fig. 4, B and C), which is consistent with the distinct effects of ShiK44A and Hsc70-4 mutants on bulk endocytic uptake.
Figure 4.
TOR signaling inhibits endocytic down-regulation of the amino acid transporter Slimfast. Fat body tissue from mid–third instar larvae stained with Slimfast antibody, which is shown in red. Nuclei are shown in blue. Insets show cellular genotypes; transgene-expressing clones (A, D–H, and J) are marked by GFP expression (green) and loss-of-function clones (B, C, and I) are marked by the absence of GFP. (A–C) High levels of Slimfast accumulate near the surface of cells expressing ShiK44A (A), but not in cells homozygous for two different alleles of Hsc70-4 (B and C). Mutant cells are indicated by arrows. (D and E) Slimfast levels are regulated by TOR signaling. (D) Overexpression of the positive TOR regulator Rheb leads to increased cell surface levels of Slimfast. (E) Overexpression of the negative TOR regulators Tsc1 and Tsc2 leads to decreased Slimfast levels. (F–H) Slimfast down-regulation resulting from TOR inactivation requires endocytosis. Coexpression of ShiK44A inhibits TSC1–TSC2-induced Slimfast down-regulation (F; compare to E). Expression of dominant-negative RabS43N does not affect Slimfast levels in fed animals (G), but prevents Slimfast down-regulation in response to 24-h starvation (H). (I and J) Slimfast turnover requires endocytic targeting/sorting factors. Loss of Hrs function (I) or expression of dominant-negative Nedd4 (J) leads to increased Slimfast at the plasma membrane. Slimfast is imaged just below the plasma membrane focal plane in these images; GFP expression in insets is imaged at the nuclear focal plane. Arrows mark mutant cells. Genotypes: hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-ShiK44A (A), hsflp122/+; Cg- Gal4/+; FRT82B UAS-GFP/FRT82B Hsc70-4e3 (B), hsflp122/+; Cg-Gal4/+; FRT82B UAS-GFP/FRT82B Hsc70-4Δ16 (C), hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-RhebEP50.084 (C and D), hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-Tsc1, UAS-Tsc2, (F) hsflp122/+; Tubulin>CD2>Gal4, UAS-GFP/+; TM3-UAS-ShiK44A/UAS-Tsc1, UAS-Tsc2 (E), hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-Rab5S43N (G and H), hsflp122/+; HrsD28 FRT40A/UAS-2XeGFP FRT40A fb-Gal4 (I), and hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-Nedd4C97411S (J). Bar, 25 μm.
We next asked whether changes in TOR signaling affect Slimfast levels. Activation of TOR through clonal overexpression of Rheb caused an increase in surface levels of Slimfast, which was similar to the effects of ShiK44A (Fig. 4 D). The relative effect of Rheb on Slimfast levels was especially pronounced in larvae subjected to a 24-h starvation (Fig. S3). Under these conditions, TOR activity is reduced in wild-type cells, but maintained in Rheb-overexpressing cells (Saucedo et al., 2003). To test whether Slimfast up-regulation is simply caused by increased protein synthesis in Rheb-overexpressing cells, we added the translation inhibitor cycloheximide during the 24-h starvation period. Under these conditions, Rheb overexpression still led to a modest relative increase in Slimfast levels (Fig. S3), which was consistent with a partial posttranslational effect. Reduction in TOR signaling also had a marked effect on Slimfast staining, as overexpression of TSC1–TSC2 led to a decrease in Slimfast levels (Fig. 4 E). Thus, the localization and levels of Slimfast are sensitive to TOR activity, with high levels of TOR signaling resulting in accumulation of Slimfast near the cell surface, and decreased TOR signaling leading to Slimfast down-regulation.
We next sought to determine whether TOR signaling might mediate these changes in Slimfast levels through effects on the targeted endocytosis of this importer. We tested whether the Slimfast down-regulation that results from TSC1–TSC2 overexpression requires endocytosis by cooverexpressing TSC1–TSC2 and ShiK44A. In the absence of functional endocytosis, TSC1–TSC2 overexpression no longer led to Slimfast down-regulation. Instead, Slimfast protein appeared to be trapped in large aggregates near the surface of these cells (Fig. 4 F). Similar results were observed when TOR activity was inhibited through starvation. In well fed larvae, partial disruption of endocytosis through expression of dominant-negative Rab5S43N had little effect on Slimfast levels (Fig. 4 G). In contrast, Rab5S43N expression led to a marked persistence of Slimfast at the plasma membrane in starved larvae (Fig. 4 H). Collectively, these results demonstrate that endocytosis is critical for Slimfast down-regulation in response to reduced TOR signaling, resulting either from poor nutrient conditions or inactivation by the TSC1–TSC2 complex.
To further investigate whether the endocytic down-regulation of Slimfast is a specific, targeted process, we examined the effects of altering components of the endocytic sorting/targeting machinery on Slimfast levels. Hrs (hepatocyte growth factor–regulated tyrosine kinase substrate) is an early endosome–associated, ubiquitin-binding protein that is critical for proper endocytic sorting. As shown in Fig. 4 I, homozygous mutation of Hrs led to a marked increase in Slimfast levels near the cell surface. Expression of a dominant-negative version of Nedd4, which is an E3 ubiquitin ligase, similarly caused persistence of Slimfast near the cell periphery (Fig. 4 J). Monoubiquitination by the Nedd4 family of enzymes is critical for the initial targeting of plasma membrane–localized proteins to clathrin-coated pits for internalization (Rotin et al., 2000). Together, these results demonstrate that components of the endocytic targeting/sorting machinery are critical in mediating Slimfast turnover.
Endocytosis affects cell growth
Our observations indicate that TOR signaling affects both bulk and targeted endocytosis. Interestingly, we note that TOR appears to exert opposing effects on these processes, stimulating bulk endocytosis (Figs. 2 and 3) and inhibiting the targeted endocytic degradation of Slimfast (Fig. 4). To test whether TOR's effects on endocytosis might contribute to its role in promoting cell growth, we assayed the growth properties of cells in which endocytosis was disrupted. Flow cytometric analysis of mosaic wing imaginal discs revealed an increased cell size and increased G2 population in Hsc70-4e3 and -4 Δ16 mutant cells, relative to wild-type cells (Fig. 5, B and C). These effects were acutely dosage-sensitive, as Hsc70-4 heterozygous cells were also larger than wild type (Fig S4). Expression of ShiK44A or of a dominant-negative Hsc70-4K71S transgene also resulted in increased cell size (Fig. 5, E and F). This cytometric profile of increased cell size and G2 content is similar to that of mutations in negative regulators of TOR such as Tsc1, Tsc2, and Pten (Gao et al., 2000; Gao and Pan, 2001), and to that of cells overexpressing Rheb (Saucedo et al., 2003).
Figure 5.
Inhibition of endocytosis leads to tissue-specific effects on cell growth. (A–F) Disruption of Hsc70-4 or shibire function in imaginal disc cells causes increased cell size and G2 population. Graphs show flow cytometric analysis of dissociated wing imaginal discs containing clones of Hsc70-4 mutant cells (B and C) or cells expressing dominant-negative Hsc70-4 or shibire (E and F). Histograms show DNA content (left) and forward light scatter (right; indicative of cell size). In (A-C) red traces represent GFP-negative Hsc70-4 mutant or control cell populations, and green traces represent GFP-positive wild-type cell populations. In D–F, green traces represent GFP-positive transgene-expressing cells and red traces represent GFP-negative wild-type cells. Genotypes: hsflp122/+; FRT82B Ubi-GFP/FRT82B (A), hsflp122/+; FRT82B Ubi-GFP/FRT82B Hsc70-4e19 (B), hsflp122/+; FRT82B Ubi-GFP/FRT82B Hsc70-4 Δ16 (C), hsflp122/+; Act>CD2>Gal4, UAS-GFP/+ (D), hsflp122/+; UAS-Hsc70-4K71S/+; Act>CD2>Gal4, UAS-GFP/+ (E), hsflp122/UAS-ShiK44A; UAS-ShiK44A/+; Act>CD2>Gal4, UAS-GFP/+ (F). (G–I) Hsc70-4 mutation and ShiK44A expression cause distinct effects on fat body cell size. Clones of Hsc70-4 mutant cells (G and H; GFP-negative cells marked by arrows) or ShiK44A expressing cells (I; GFP-positive cells) are shown. Cell outlines are marked by phalloidin staining (red). Genotypes: hsflp122/+; Cg-Gal4/+; FRT82B UAS-GFP/FRT82B Hsc70-4e3 (G), hsflp122/+; Cg-Gal4/+; FRT82B UAS-GFP/FRT82B Hsc70-4 Δ16 (H), hsflp122/+; Act>CD2>Gal4, UAS-GFP/TM3-UAS-ShiK44A (I). Bar, 25 μm. (J) Quantitation of fat body cell size data. Shown are area measurements of cells homozygous for Hsc70-4e3 or Hsc70-4 Δ16, or expressing ShiK44A, relative to surrounding wild-type cells. Error bars indicate the SEM. Number of cells scored are as follows: Hsc70-4e3, n = 116 (27 mutant and 89 wild type); Hsc70-4 Δ16, n = 109 (22 mutant and 87 wild type); ShiK44A, n = 175 (24 transgene-expressing and 151 wild type).
Disruption of endocytosis also led to dosage-dependent cell size changes in the fat body. Expression of ShiK44A caused a 1.5-fold increase in fat body cell size (Fig. 5, I and J), which is consistent with its growth effects in wing disc cells. In contrast, cells homozygous for mutant alleles of Hsc70-4 showed either a slight (e3) or a threefold decrease in cell size (Δ16 null; Fig. 5, G, H, and J). Given that ShiK44A expression and Hsc70-4 mutation result in a similar block of endocytic uptake, but show distinct effects on Slimfast expression, these results suggest that the relative contributions to growth of bulk and transporter-mediated nutrient uptake may differ between imaginal disc and fat body cells.
Together, the finding that endocytosis is both regulated by TOR and has effects on cell growth indicates that endocytosis functions as a downstream effector of TOR signaling. TOR simultaneously down-regulates aspects of endocytosis that inhibit growth and up-regulates potential growth-promoting functions of endocytosis.
Endocytosis affects TOR signaling
In addition to acting downstream of TOR, the potential role of endocytosis in controlling nutrient import suggested that it might also function upstream to regulate TOR activity, which is stimulated by nutrients in general and Slimfast in particular (Colombani et al., 2003). Endocytosis has also been shown to affect the levels and activity of the insulin receptor (Ceresa et al., 1998), which may function upstream of TOR by activating PI3K signaling (Inoki et al., 2002). Indeed, we observed an increase in insulin receptor levels similar to that of Slimfast in response to Rheb overexpression (Fig. S3). Therefore, we examined the effects of endocytosis on in vivo markers of PI3K and TOR activity. To test whether endocytosis influences PI3K signaling, we analyzed the effects of ShiK44A expression on the transcription factor FOXO, which is excluded from the nucleus in response to PI3K-dependent phosphorylation (Brunet et al., 1999). Clonal expression of ShiK44A in fat body cells resulted in relocalization of FOXO from the nucleus to the cytoplasm (Fig. 6 A), reflecting increased PI3K activity and, thus, indicating that endocytosis normally exerts a negative effect on PI3K signaling in these cells.
Figure 6.
Endocytosis regulates growth signaling. (A and B) Inhibition of shibire increases PI3K signaling and inhibits autophagy. (A) Antibody staining shows increased cytoplasmic localization of the transcription factor FOXO in ShiK44A-expressing clones. (B) Induction of autophagy by 5 h starvation is blocked in ShiK44A- expressing clones. Autophagic cells are indicated by the punctate staining of LysoTracker red. Genotype: hsflp122/+; Act>CD2>Gal4, UAS-GFP/UAS-ShiK44A. (C) Immunoblot showing levels and phosphorylation status of S6K and AKT from control (lane 1), Hsc70-4e3/e19 (lane 2), and Tsc129 (lane 3) larval extracts. (D) Hsc70-4 mutation increases the severity of Tsc1 cell size and cell cycle phenotypes. Flow cytometric histograms of Hsc70-4e3 and Tsc129 single- and double-mutant mitotic clones are shown. Genotypes: (Row 1) hsflp122/+; FRT82B Ubi-GFP/FRT82B Hsc70-4e3, (Row 2) hsflp122/+; FRT82B Ubi-GFP/FRT82B Tsc129, (Row 3) hsflp122/+; FRT82B Ubi-GFP/FRT82B Tsc129 Hsc70-4e3. (E) Hsc70-4 and Tsc1 mutants show synergistic effects on tissue growth. Shown are eyes containing eye-specific clones homozygous mutant for Hsc70-4 (top), Tsc1 (middle), or Hsc70-4 and Tsc1 (bottom). Mutant clones are marked by the lack of the white + marker, and thus appear white. Note the loss of anterior tissue in Hsc70-4 mutant eyes (top, arrow) and the overgrowth of anterior tissue in Hsc70-4 Tsc1 double mutants (bottom, arrow). Genotypes: ey-FLP/+; FRT82B Pw + l(3)clR3/FRT82B Hsc70-4e3 (top), ey-FLP/+; FRT82B Pw + l(3)clR3/FRT82B Tsc129 (middle), ey-FLP/+; FRT82B Pw + l(3)clR3/FRT82B Hsc70-4e3 Tsc129 (bottom). (F) Heterozygous mutation of Hsc70-4 dominantly confers rapamycin resistance. Wild-type and Hsc70-4e3/+ animals develop at a similar rate to adulthood (eclosion) when grown on normal media. Addition of rapamycin to the media results in a more severe delay in wild type than in Hsc70-4e3/+ animals. Bar in B is 25 μm in A, 50 μm in B, and 200 μm in E.
As a cellular readout of TOR signaling, we monitored the effects of endocytosis on autophagy, which is a process of cytoplasmic degradation that is inhibited by TOR (Noda and Ohsumi, 1998; Scott et al., 2004). Disruption of endocytosis through expression of ShiK44A prevented proper induction of autophagy after starvation (Fig. 6 B), which is indicative of high levels of TOR signaling in these cells. A similar inhibition of autophagy was observed in Hsc70-4-null cells and in cells expressing dominant-negative Hsc70-4K71S (Fig. S4, available at http://www/jcb.org/cgi/content/full/jcb.200511140/DC1).
To assess the effects of endocytosis on the kinase activity of TOR, we monitored the levels and the phosphorylation status of known TOR substrates. Inactivation of D. melanogaster TOR through mutation or starvation has previously been shown to cause a decrease in S6K phosphorylation at Thr398, as well as an increase in S6K protein levels, through an unknown mechanism (Oldham et al., 2000; Gao et al., 2002). More recently Akt (Ser505) has been identified as a substrate for TOR in association with its cofactor rictor (Sarbassov et al., 2005b). Accordingly, in control experiments we found that Tsc129 homozygous larvae showed a decrease in total S6K levels and a slight decrease in Thr398 phosphorylation relative to wild-type controls (Fig. 6 C); normalized for S6K levels, Thr398 phosphorylation was increased as expected. Tsc129 mutant larvae also showed a strong loss of Akt-Ser505 phosphorylation, which is consistent with recent studies showing that the TSC1–TSC2 complex promotes TOR–rictor signaling to Akt (Yang et al., 2006). We next examined these markers in extracts from Hsc70-4e3/e19 transheterozygous mutants, which are viable through the third instar larval stage. Similar to Tsc129 mutants, Hsc70-4e3/e19 larvae showed a decrease in overall S6K levels and a reduction in S6K and Akt phosphorylation; in this case, Thr398 phosphorylation relative to S6K abundance was reduced compared with wild type (Fig. 6 C).
The similarity between Hsc70-4 and Tsc1 mutants was further underscored through chemical and genetic interaction studies. Imaginal disc cells doubly mutant for Hsc70-4e3 and Tsc129 showed a synergistic increase in cell size as compared with their single mutant counterparts (Fig. 6 D). Similarly, using the ey-FLP system to generate homozygous mutant eye tissue, we observed an exacerbation of Tsc129-induced tissue overgrowth in the presence of Hsc70-4e3 mutation, in some cases resulting in a marked outgrowth of tissue in the anterior portion of the retina (Fig. 6 E). We also tested whether mutation of Hsc70-4 affects sensitivity to rapamycin, which inhibits TOR signaling through the formation of an inhibitory binding complex with the intracellular protein FKBP12 (Brown et al., 1994; Sabatini et al., 1994). Growth of wild-type D. melanogaster is delayed on media containing rapamycin, and this delay is sensitive to the dosage of TOR pathway genes (Zhang et al., 2000; Patel et al., 2003). We found that heterozygous Hsc70-4e3 mutation significantly alleviated the rapamycin-induced delay (Fig. 6 F), which is consistent with increased levels of TOR signaling. Collectively, the cellular, biochemical, and genetic effects of endocytosis are consistent with this process playing a significant role in cell growth and TOR signaling.
Discussion
Inactivation of TOR causes an inhibition of cellular growth, a reduction in cell size, and a suppression of cell cycle progression. In addition to well described changes in protein synthesis and ribosome biogenesis, recent studies have suggested that other cell processes are likely to contribute to these growth effects of TOR. The present study identifies endocytosis as one such process. Our results demonstrate that the clathrin-uncoating ATPase Hsc70-4 interacts genetically with TOR and Tsc1, and that bulk endocytosis is stimulated in cells with activated TOR signaling. Conversely, we find that TOR activity inhibits the endocytic degradation of nutrient transporters such as Slimfast. Together, these endocytic effects of TOR promote both the bulk and targeted uptake of nutrients and other biomolecules required for cell mass increase (Fig. 7). In addition to this direct role in cellular biosynthesis and growth, nutrients also act as potent regulators of TOR signaling. Indeed, Slimfast was previously identified as an upstream activator of TOR (Colombani et al., 2003). Our findings that disruption of endocytosis effects cell size, rapamycin sensitivity, and TOR kinase activity are consistent with an additional role for endocytosis upstream of TOR.
Figure 7.
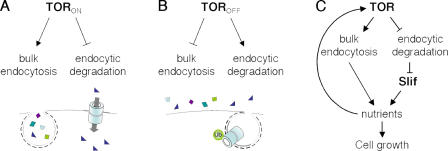
Inverse regulation of bulk endocytosis and targeted endocytic degradation. (A) Under conditions favorable for growth, TOR promotes bulk endocytic uptake and inhibits the endocytic turnover of specific nutrient importers such as Slimfast. (B) In growth-inhibitory conditions, inactivation of TOR leads to a decrease in bulk endocytosis and an increase in the targeted endocytic degradation of excess nutrient importers. (C) Together, the opposing actions of TOR on bulk and targeted endocytosis serve to facilitate nutrient import, providing energy and building blocks necessary for biosynthetic growth and leading to a further stimulation of TOR signaling.
Mutations that disrupt endocytosis are likely to have both positive and negative effects on nutrient uptake and cell growth because they inhibit bulk endocytic uptake, as well as degradation of nutrient transporters and other signaling molecules. Thus, the overall effects of endocytic disruption on nutrient uptake, cell growth, and TOR signaling are difficult to predict a priori. Our results suggest that both the cellular context and the specific step at which endocytosis is blocked influence the growth response. Thus, in fat body cells, expression of ShiK44A resulted in an increase in cell size, whereas loss of Hsc70-4 function caused reduced cell size. We note that these changes mirror the effects of these mutations on Slimfast levels; whereas both ShiK44A expression and Hsc70-4 mutation decreased bulk endocytic uptake, only ShiK44A resulted in increased levels of Slimfast. In contrast, both ShiK44A and Hsc70-4 mutants led to the increased size of wing imaginal disc cells, suggesting that in these cells the growth-inhibitory effects of endocytic degradation of membrane proteins such as Slimfast predominate over the potential positive effects of increased bulk uptake. Similarly, our results indicate a complex effect of endocytosis on TOR signaling. Partial reduction in Hsc70-4 levels lead to an increase in TOR signaling, as was evident in the eyTOR interaction and rapamycin resistance. In contrast, larvae that are homozygous mutant for Hsc70-4 show a decrease in TOR kinase activity. These results suggest that modest inhibition of endocytosis may increase TOR signaling, whereas a complete block of endocytosis may reduce it.
A striking parallel to the inverse regulation of bulk and targeted endocytic processes by TOR can be observed in its effects on autophagy in yeast. Through autophagy, random portions of cytoplasm are nonselectively engulfed within double membrane–bound vesicles for delivery to the lysosome. Activation of TOR causes this nonselective form of autophagy to be suppressed, and, instead, the autophagic machinery engages in a selective type of autophagy known as the cytoplasm–vacuole targeting (CVT) pathway, which is responsible for lysosomal delivery of specific hydrolases (Klionsky and Emr, 2000). Thus, TOR acts as a switch between selective and nonselective autophagy. TOR may also be involved in switching between clathrin-and caveolae/raft-mediated endocytosis in higher eukaryotes. A genome-wide survey of protein kinases found that RNAi-mediated inactivation of TOR in HeLa cells inhibited clathrin-dependent processes such as transferrin uptake and vesicular stomatitis virus infection, and stimulated cavelolae/raft-dependent events (Pelkmans et al., 2005). Together, these findings suggest that TOR may control the specificity of membrane trafficking components. In addition, our results show that S6K, which is an important TOR substrate, acts downstream of TOR in promoting bulk endocytosis, but is not involved in the suppression of starvation-induced autophagy.
The identification of endocytosis as a TOR-controlled function adds to the growing list of cell processes regulated by TOR, including protein synthesis, ribosome biogenesis, autophagy, metabolic gene expression, and cytoskeletal organization. How these distinct functions interact to achieve a coordinated growth response is only beginning to be understood. One likely mechanism involves the common use of molecular components and cellular substrates by different cell functions, as in the case of selective and nonselective autophagy, bulk endocytosis, and endocytic degradation. Two or more distinct branches of TOR signaling may also act cooperatively to control the same target, as in the case of Slimfast regulation by both translation and endocytosis, or may act in opposition, as previously observed for the role of S6K in limiting autophagy. Finally, distinct TOR complexes may converge on the same targets with opposing effects, as in the regulation of Akt by TOR-raptor versus TOR–rictor complexes (Shah et al., 2004; Sarbassov et al., 2005b). The finding that TOR signaling regulates the levels of Slimfast, which was previously shown to function upstream of TOR, adds another layer of complexity to the TOR signaling network.
Materials and methods
D. melanogaster genetics and culture
Strains used were as follows: dpp-lacZBS3.0, HrsD28, Hsc70-454.1, Hsc70-4 Δ16, Hsc70-4e195, Hsc70-4Wee-P1, P[w + ]Hsc70-4.14, S6kl1, slif1, Tor ΔP, Tsc129, UAS-EGFP-Clc, UAS-Hsc4K71S, UAS-Nedd4C974FS, UAS-GFP-Rab5, UAS-Rab5S43N, UAS-GFP-Rab7, UAS-RhebEP50.084, UAS-shiK44A, UAS-slif, UAS-Tor, UAS-Tsc1, UAS-Tsc2, UAS-4EBP, Act>CD2>GAL4 UAS-GFP, Act>CD2>GAL4 UAS-myrRFP, Cg-GAL4.A2, en-GAL4, fb-GAL4, MS1096-GAL4, and ey-FLP; FRT82B Pw + l(3)clR3. Flies were incubated at 25°C on standard cornmeal–yeast medium, unless otherwise indicated. Starvation experiments were performed essentially as previously described (Scott et al., 2004). In brief, larvae were transferred to fresh fly food supplemented with yeast paste, allowed to feed for 24 h, and then transferred to a 20% sucrose solution and starved for the indicated time before dissection. For rapamycin treatment, larvae were cultured in standard fly medium supplemented with 2 μM rapamycin.
Generation of eyTOR flies
A 1.5-kb KpnI (blunted)–BamHI fragment containing four copies of the eye-specific enhancer fragment from the ey gene and the noninducible hsp70 promoter was isolated from pBD1915 (Newsome et al., 2000), and ligated into the unique SalI (blunted) and XbaI sites located immediately upstream of a FLAG-tagged D. melanogaster Tor cDNA in pBluescript (Zhang et al. 2000). The ey-hsp70-Tor sequence was excised as a 9.3-kb XbaI–PspOMI fragment and cloned into the transformation vector pCasper4 digested with XbaI and NotI. P element–mediated transposition was used to introduce the eyTOR transgene into yw flies by standard methods. CyO and TM3 balancers carrying eyTOR insertion lines were created through transposase-mediated mobilization of an X chromosome eyTOR insert.
Mutagenesis, mapping, and sequencing
Mutagenesis.
yw males isogenized for the second and third chromosomes were starved for 2 h before being fed a solution of 25 mM EMS in 0.01 M Tris, pH 7.5, and 1% sucrose overnight. Batches of 50 mutagenized males were then mated to 50 CyO-eyTOR/Sp virgins. Crosses were reared in bottles at 25°C, being transferred once daily for 4 d before adults were discarded. F1 progeny were screened for the dominant enhancement or suppression of the eyTOR small eye phenotype. Flies carrying EMS-induced eyTOR-modifying mutations were back crossed to CyO-eyTOR and TM3-eyTOR stocks, allowing mutations to be recovered and mapped to chromosome through segregation of the eyTOR interaction phenotype against the second and third chromosome markers.
Mapping.
e3 and e19 were localized to map position 57 centimorgans on the third chromosome by genetic recombination, and this region was further refined to the 87E8-88E6 interval by deficiency mapping. In complementation tests against available lethal mutations in this region, e3 failed to complement multiple P element-, EMS-, and x-ray–induced mutations in Hsc70-4. e19 showed complete failure to complement e3 and Hsc70-454.1, which is an antimorphic allele of Hsc70-4, and partial complementation with other Hsc70-4 alleles. Based on this pattern of complementation and on the strength of eyTOR interactions, we classify e3 as an antimorph and e19 as a hypomorphic allele of Hsc70-4.
Sequencing e3 and e19 mutations. Genomic DNA was isolated from e3 and e19 homozygous mutant larvae, and the second exon of Hsc70-4 (containing the coding region) was PCR amplified using the primers CCATTTTCTCAGTATTACTTCTCCTCTGGC and GAGAACTGTTACTGTATGGTTGCATTGAGG and sequenced using the primers CGAGAAAAGGAAAATTAGAATTGTAAAACACACC, GGAGATCTCTTCGATGGTGCTTACC, CAAGCACAAGAAGGATCTGACCACC, CATTCTGCACGGCGACAAGTCG, and GTCGTCTCTCCAAGGAGGACATC.
Overexpression and loss-of-function clones in D. melanogaster fat body tissue
Gain-of-function clones.
Clonal transgene expression was achieved using the Gal4-UAS system. Act>CD2>GAL4, UAS-GFP or Act>CD2>GAL4, UAS-myrRFP virgins were crossed to males carrying hs-FLP and UAS-driven transgenes of interest. FLP-mediated excision of the CD2 insert is induced spontaneously in ∼10% of fat body cells at 25°C, leading to Act5c-driven constitutive Gal4 expression in GFP- or myrRFP-marked single cells or small clones (Britton et al., 2002). GFP-Rab5 expressing cells were generated as above but in the absence of UAS-GFP.
Loss-of-function fat body clones.
hs-FLP/FRT–mediated mitotic recombination was induced in 0–8 h embryos, before initiation of endoreplication in fat body tissue, through a 2 h, 37°C heat shock in an air incubator. For uniform GFP marker expression, the fat body drivers Cg-Gal4 or fb-Gal4 were used to activate UAS-GFP lines on FRT-bearing chromosomes. Loss-of-function clones were marked by the absence of GFP.
Histology
Fixation.
Third instar larvae were dissected in PBS, inverted, and transferred to 1.5-mL tubes containing 3.7% formaldehyde in PBST (PBS + 0.1% Tween 20) for 4-h (imaginal discs) or overnight (fat body) fixation at 4°C, with gentle agitation. Fix was removed through four 5-min washes in PBST.
Antibody staining.
Fixed carcasses were blocked for 2–4 h in PBSTG (PBST + 5% normal goat serum) before incubation in PBSTG + primary antibody at 4°C overnight. Carcasses were washed four times for 20 min in PBST and blocked in PBSTG for 1–2 h before incubation in PBST + secondary antibody + 1 μM Hoechst 33258 at 4°C overnight. Carcasses were rinsed in PBST four times for 20 min. Tissue was dissected in PBS and mounted in FluoroGuard reagent (Bio-Rad Laboratories).
Antibodies.
Anti-Slimfast (1:400 dilution of rabbit polyclonal sera raised against a bacterially produced fusion between GST and the COOH-terminal cytoplasmic tail of Slif [amino acids 554–604]), anti-FOXO #3015 (1:300; gift from O. Puig, University of California, Berkeley, Berkeley, CA), anti-FLAG M5 (1:500; Sigma-Aldrich), anti-Elav 9F8A9 (1:50; Developmental Studies Hybridoma Bank), anti B-gal (1:500; Calbiochem), anti InR 386 (1:2,000; gift from R. Fernandez, New York University Medical Center, New York, NY), anti-Lsp2 (1:500; gift from H. Benes, University of Arkansas, Little Rock, AR).
Phalloidin staining.
Carcasses were dissected, fixed, washed, and blocked for 2–4 h in PBSTG, before being incubated at 4°C overnight in 0.165 μM Alexa Fluor 568 phalloidin (Invitrogen) in PBSTG containing 1 μM Hoechst 33258. Carcasses were rinsed in PBST four times for 20 min before final dissection, mounting, and imaging.
Live tissue staining.
Lysotracker staining was performed as in Scott et al. (2004). For acridine orange staining, dissected eye discs were incubated in PBS containing 1 μM acridine orange for 5 min at room temperature. Tissue was rinsed in PBS, transferred to a drop of PBS on a microscope slide, and covered loosely with a coverslip for imaging.
Endocytic uptake assay
5–10 larvae per genotype were bisected and inverted in PBS, and then transferred to a 1.5-mL tube containing 80 μg/ml TR-avidin (Invitrogen) in M3 insect medium (Sigma-Aldrich) containing 5% fetal calf serum, 1× insect medium supplement (Sigma-Aldrich) and penicillin/streptomycin antibiotics (Invitrogen). Carcasses were incubated for 15 min at room temperature with gentle agitation, rinsed two times and washed three times for 5 min with ice-cold PBS + 0.5% BSA at 4°C, and then fixed, washed, and mounted in FluoroGuard Reagent.
Microscopy
Confocal images were acquired on a microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) equipped with a digital camera (ORCA-ER; Hamamatsu) and a spinning disc confocal system (CARV; BD Biosciences). Axiovision v3.1 acquisition software, Plan-Apochromat 63×, 1.40 NA, and Plan-Neofluar 40×, 0.75 NA, objectives were also used (all from Carl Zeiss MicroImaging, Inc.). All other images were taken with a digital camera (DXM1200; Nikon) attached to an epifluorescence microscope (Axioscope 2; Carl Zeiss MicroImaging, Inc; ACT-1 acquisition software [Nikon], Plan-Neofluar 40×, 0.75 NA, 20×, 0.50 NA, and 5×, 0.15 NA, objectives were also used) or a digital camera (Coolpix 990; Nikon) attached to a dissecting microscope (Stemi 200C; Carl Zeiss MicroImaging, Inc.). All images were processed in Photoshop v7.0 (Adobe).
Scanning electron microscopy.
Whole flies were passed through an ethanol series to achieve hydration in 100% EtOH. Flies were then dehydrated using a critical point dryer (model 780A; Tousimis), mounted on carbon tape, and gold sputter–coated using a high vacuum deposition system (DV-502A; Denton). Eyes were imaged using a Field Emission Gun Scanning Electron Microscope (S-4700; Hitachi; FE-PC SEM v3.3 control software, v2.10 operating software) and transferred to PCI v4.2 (Quartz) for processing.
Western blotting
Larvae were transferred to fresh medium supplemented with yeast paste and allowed to feed for 24 h before extraction. Extracts were prepared by homogenizing equal masses of wild-type or mutant larvae directly in 1× SDS sample buffer. Extracts were boiled, run on an 8% SDS-polyacrylamide gel, and transferred to PVDF membrane using the Mini PROTEAN 3 electrophoresis and wet transfer system (Bio-Rad Laboratories). Membranes were blocked in PBST containing 5% dry milk for 4 h before overnight incubation in primary antibody. Primary antibodies were diluted in PBST containing 5% BSA. The primary antibodies used were as follows: Akt, P-Ser505-dAkt, P-Thr398-dS6K (1:1,000; Cell Signaling Technology), dS6K (1:1,000, mouse monoclonal; gift from G. Thomas, University of Cincinnati, Cincinnati, OH), dTOR (1:1,000; gift from D. Pan, Johns Hopkins University, Baltimore, MD), tubulin DMIA (1:5,000; Calbiochem). Membranes were washed four times for 20 min in PBST and blocked for 1 h before 4 h incubation in HRP-conjugated secondary antibody. Membranes were washed four times for 20 min in PBST. SuperSignal ECL substrate solutions (Pierce) were applied to membranes to allow protein detection on BioMax light film (Kodak).
Flow cytometry
Mitotic loss-of-function clones were induced at 3.5 d after laying through a 2-h heat-shock in a 37°C air incubator, and were allowed to grow for 45 h postinduction at room temperature. Overexpression clones were induced at 3.5 d after laying through a 1 h 15-min heat-shock, and were allowed to grow for 48 h at 25°C. Approximately 25 wing imaginal discs per genotype were dissected and processed for FACS analysis, essentially as previously described (Neufeld et al., 1998). In brief, discs were added to 250 μL of PBS in 5 mL polystyrene round-bottom tubes, and kept on ice until 275 μL trypsin solution (9× Trypsin-EDTA [Sigma-Aldrich], 1× PBS, and 2 μM Hoechst 33342) was added to each tube to allow tissue dissociation into single cells. Samples were mixed on a Nutator (Clay Adams) at room temperature for a minimum of 2 h before flow cytometric analysis using a FACSAria (BD Biosciences). FlowJo v4.4 was used for data analysis.
Online supplemental material
Fig. S1 displays additional effects of TOR overexpression in eye and wing tissues, interactions with additional Hsc70-4 alleles, and rescue by a genomic Hsc70-4 construct. Fig. S2 displays additional TR-avidin and Lsp2 internalization experiments. Fig. S3 shows Slimfast antibody specificity controls and additional Slimfast and insulin receptor localization data. Fig. S4 displays effects of Hsc70-4 on autophagy induction and cell size. Online supplemental material is available at http://www/jcb.org/cgi/content/full/jcb.200511140/DC1.
Supplementary Material
Acknowledgments
We thank Henry Chang and Marcos Gonzalez-Gaitan for unpublished fly stocks, Rafael Fernandez, Helen Benes, Duojia Pan, Oscar Puig and George Thomas for antibodies, Jennifer Stafford for screening assistance, and Pierre Leopold and Do-Hyung Kim for helpful discussions.
This work was supported by National Institutes of Health grant RO1 GM062509 and Tuberculosis Sclerosis Alliance grant 04-04 to T.P. Neufeld and by National Research Service Award institutional training grant T32 HD007480 to K.M. Hennig.
Abbreviations used in this paper: EMS, ethyl methanesulfonate; Hrs, hepatocyte growth factor–regulated tyrosine kinase substrate; Lsp, larval serum protein; PI3K, phosphatidylinositol-3 kinase; TOR, target of rapamycin; TR-avidin, Texas red–conjugated avidin.
References
- Beck, T., A. Schmidt, and M.N. Hall. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146:1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, J.S., W.K. Lockwood, L. Li, S.M. Cohen, and B.A. Edgar. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell. 2:239–249. [DOI] [PubMed] [Google Scholar]
- Bronk, P., J.J. Wenniger, K. Dawson-Scully, X. Guo, S. Hong, H.L. Atwood, and K.E. Zinsmaier. 2001. Drosophila Hsc70-4 is critical for neurotransmitter exocytosis in vivo. Neuron. 30:475–488. [DOI] [PubMed] [Google Scholar]
- Brown, E.J., M.W. Albers, T.B. Shin, K. Ichikawa, C.T. Keith, W.S. Lane, and S.L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 369:756–758. [DOI] [PubMed] [Google Scholar]
- Brunet, A., A. Bonni, M.J. Zigmond, M.Z. Lin, P. Juo, L.S. Hu, M.J. Anderson, K.C. Arden, J. Blenis, and M.E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 96:857–868. [DOI] [PubMed] [Google Scholar]
- Byfield, M.P., J.T. Murray, and J.M. Backer. 2005. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 280:33076–33082. [DOI] [PubMed] [Google Scholar]
- Ceresa, B.P., A.W. Kao, S.R. Santeler, and J.E. Pessin. 1998. Inhibition of clathrin-mediated endocytosis selectively attenuates specific insulin receptor signal transduction pathways. Mol. Cell. Biol. 18:3862–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.C., S.L. Newmyer, M.J. Hull, M. Ebersold, S.L. Schmid, and I. Mellman. 2002. Hsc70 is required for endocytosis and clathrin function in Drosophila. J. Cell Biol. 159:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.J., and C.A. Kaiser. 2003. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161:333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.S., R.A. Obar, C.C. Schroeder, T.W. Austin, C.A. Poodry, S.C. Wadsworth, and R.B. Vallee. 1991. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 351:583–586. [DOI] [PubMed] [Google Scholar]
- Clyne, P.J., J.S. Brotman, S.T. Sweeney, and G. Davis. 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 165:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani, J., S. Raisin, S. Pantalacci, T. Radimerski, J. Montagne, and P. Leopold. 2003. A nutrient sensor mechanism controls Drosophila growth. Cell. 114:739–749. [DOI] [PubMed] [Google Scholar]
- Dean, R.L., M. Locke, and J.V. Collins. 1985. Structure of the fat body. In Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Vol. 4. G.A. Kerkut and L.I. Gilbert, editors. Pergamon Press, New York. 155–210.
- Edinger, A.L., and C.B. Thompson. 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell. 13:2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., and D. Pan. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15:1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., T.P. Neufeld, and D. Pan. 2000. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 221:404–418. [DOI] [PubMed] [Google Scholar]
- Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi, R.S. Yeung, B. Ru, and D. Pan. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4:699–704. [DOI] [PubMed] [Google Scholar]
- Hennig, K.M., and T.P. Neufeld. 2002. Inhibition of cellular growth and proliferation by dTOR overexpression in Drosophila. Genesis. 34:107–110. [DOI] [PubMed] [Google Scholar]
- Hing, H.K., L. Bangalore, X. Sun, and S. Artavanis-Tsakonas. 1999. Mutations in the heatshock cognate 70 protein (hsc4) modulate Notch signaling. Eur. J. Cell Biol. 78:690–697. [DOI] [PubMed] [Google Scholar]
- Inoki, K., Y. Li, T. Zhu, J. Wu, and K.L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648–657. [DOI] [PubMed] [Google Scholar]
- Jefferies, H.B., C. Reinhard, S.C. Kozma, and G. Thomas. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA. 91:4441–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger, M.A., F. Rintelen, H. Stocker, J.D. Wasserman, M. Vegh, T. Radimerski, M.E. Greenberg, and E. Hafen. 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J., and S.D. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka, T., and K. Ikeda. 1983. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J. Cell Biol. 97:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, J., U. Schneider, I. Howald, A. Schmidt, and M.N. Hall. 2000. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275:37011–37020. [DOI] [PubMed] [Google Scholar]
- Levenbook, L. 1985. Insect storage proteins. In Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Vol. 10. G.A. Kerkut and L.I. Gilbert, editors. Pergamon Press, Oxford. 307–346 pp.
- Manning, B.D. 2004. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.E., and M.N. Hall. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17:158–166. [DOI] [PubMed] [Google Scholar]
- Mayer, M.P., and B. Bukau. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62:670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moline, M.M., C. Southern, and A. Bejsovec. 1999. Directionality of wingless protein transport influences epidermal patterning in the Drosophila embryo. Development. 126:4375–4384. [DOI] [PubMed] [Google Scholar]
- Montagne, J., M.J. Stewart, H. Stocker, E. Hafen, S.C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science. 285:2126–2129. [DOI] [PubMed] [Google Scholar]
- Neufeld, T.P., A.F. de la Cruz, L.A. Johnston, and B.A. Edgar. 1998. Coordination of growth and cell division in the Drosophila wing. Cell. 93:1183–1193. [DOI] [PubMed] [Google Scholar]
- Newsome, T.P., B. Asling, and B.J. Dickinson. 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 127:851–860. [DOI] [PubMed] [Google Scholar]
- Nobukuni, T., M. Joaquin, M. Roccio, S.G. Dann, S.Y. Kim, P. Gulati, M.P. Byfield, J.M. Backer, F. Natt, J.L. Bos, et al. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 102:14238–14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963–3966. [DOI] [PubMed] [Google Scholar]
- Oldham, S., J. Montagne, T. Radimerski, G. Thomas, and E. Hafen. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14:2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, P.H., N. Thapar, L. Guo, M. Martinez, J. Maris, C.L. Gau, J.A. Lengyel, and F. Tamanoi. 2003. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 116:3601–3610. [DOI] [PubMed] [Google Scholar]
- Pelkmans, L., E. Fava, H. Grabner, M. Hannus, B. Habermann, E. Krausz, and M. Zerial. 2005. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 436:78–86. [DOI] [PubMed] [Google Scholar]
- Pende, M., S.H. Um, V. Mieulet, M. Sticker, V.L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S.C. Kozma, and G. Thomas. 2004. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24:3112–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1–17. [DOI] [PubMed] [Google Scholar]
- Sabatini, D.M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S.H. Snyder. 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin- dependent fashion and is homologous to yeast TORs. Cell. 78:35–43. [DOI] [PubMed] [Google Scholar]
- Sarbassov, dos D., S.M. Ali, and D.M. Sabatini. 2005. a. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596–603. [DOI] [PubMed] [Google Scholar]
- Sarbassov, D., D.A. Guertin, S.M. Ali, and D.M. Sabatini. 2005. b. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- Saucedo, L.J., X. Gao, D.A. Chiarelli, L. Li, D. Pan, and B.A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566–571. [DOI] [PubMed] [Google Scholar]
- Schlossman, D.M., S.L. Schmid, W.A. Braell, and J.E. Rothman. 1984. An enzyme that removes clathrin coats: purification of an uncoating ATPase. J. Cell Biol. 99:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.C., O. Schuldiner, and T.P. Neufeld. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7:167–178. [DOI] [PubMed] [Google Scholar]
- Shah, O.J., Z. Wang, and T. Hunter. 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14:1650–1656. [DOI] [PubMed] [Google Scholar]
- Terada, N., H.R. Patel, K. Takase, K. Kohno, A.C. Nairn, and E.W. Gelfand. 1994. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. USA. 91:11477–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., K. Inoki, E. Kim, and K.L. Guan. 2006. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. USA. 103:6811–6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., J.P. Stallock, J.C. Ng, C. Reinhard, and T.P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.