Abstract
The nuclear receptors LXRα (NR1H3) and LXRβ (NR1H2) are attractive drug targets for the treatment of diabetes and cardiovascular disease due to their established role as regulators of cholesterol and lipid metabolism. A large body of literature has recently indicated their important roles in glucose metabolism and particularly LXRβ is important for proper insulin production in pancreas. In this study, we report that glucose induces transcription via the LXRB gene promoter. The transcription start site of the human LXRB gene was determined and we identified two highly conserved, and functional, ETS and Elk1 binding sites, respectively, in the LXRB gene promoter. The Elk1 binding site also bound the serum responsive factor (SRF). Mutation of these sites abolished binding. Furthermore, mutation of the binding sites or siRNA knockdown of SRF and Elk1 significantly reduced the promoter activity and impaired the glucose response. Our results indicate that the human LXRB gene is controlled by glucose, thereby providing a novel mechanism by which glucose regulates cellular functions via LXRβ.
INTRODUCTION
The occurrence of hyperlipidemia, hyperglycemia, insulin resistance and its metabolic complications such as type-2 diabetes mellitus (T2DM) increases dramatically in the western world. A deeper understanding of the pathogenesis causing these diseases and development of drugs targeting metabolic disorders currently has high priority. Nuclear receptors (NRs), including liver X receptors (LXRs), have been suggested as potential drug targets for the treatment or prevention of T2DM (1). LXRα and LXRβ are established regulators of cholesterol and lipid metabolism and activation of LXRs promotes conversion of cholesterol to bile acids, lipid/triglyceride biosynthesis and reverse cholesterol transport from peripheral cells to the liver and subsequent elimination of cholesterol via the gall bladder [reviewed in (2)].
A large body of literature establishes an important physiological role of LXR in carbohydrate metabolism. The carbohydrate-response element-binding protein (ChREBP) mediates glucose activated lipogenesis via the xylulose 5-phosphate pathway (3) and has been identified as an LXR target gene (4). Recently, glucose itself was shown to be an LXR agonist activating LXRs at physiological concentrations (5). Activation of LXR promoted glucose uptake and glucose oxidation in muscle (6). As skeletal muscle constitutes 40% of the human body weight and is the major site for glucose utilization, this observation suggests that LXR might have a considerable impact on overall glucose oxidation in the body. Expression of the insulin responsive glucose transporter GLUT4 in adipocytes was induced by LXR while the basal expression of GLUT4 was lower in LXRα−/− mice compared to wild type mice (7,8). Increased glucose uptake in adipocytes and muscle cells as well as reduced hepatic gluconeogenesis due to suppressed expression of gluconeogenic genes including PEPCK, G6P and PGC1α were observed in response to treatment with an LXR agonist (6,8,9). Moreover, activation of LXR increased glucose dependent insulin secretion in vitro from pancreatic β-cell line cultures (10) and lead to increased plasma insulin concentrations in mice (11). It was also shown that LXRβ−/− mice have less basal insulin levels and, on a normal diet, are glucose intolerant due to impaired glucose-induced insulin secretion (12).
LXR signaling seems more prominent in disease where, for instance, impaired lipid oxidation was seen in isolated muscle cells from T2DM patients compared to control cells when the muscle cells were treated with an LXR agonist (6). Further, improved glucose tolerance was observed in obese C57Bl/6 mice in response to treatment with an LXR agonist, but not in lean C57Bl/6 mice (8) and similar results were observed in db/db mice, Zucker diabetic and obese rats and ob/ob mice (9,13,14). Improved whole body insulin sensitivity was observed in ob/ob mice upon activation of LXRs, but not in lean mice (13). Together, these observations suggest an anti-diabetic role of LXRs.
Elk1 is a well-studied member of the ETS family of transcription factors. Elk1 activity is tightly regulated by phosphorylation and dephosphorylation which have been extensively studied in the context of cellular signaling. Elk1 has been shown to be positively regulated by activation of the MAPK pathway including Erk1/2, p38 and JNK, which has been shown to be dysfunctional in T2DM (15,16). Here we identify a 5′-ETS site and a 3′-Elk1 binding site in the human LXRB gene promoter and show that Elk1 can bind both sites while SRF only binds to the 3′-Elk1 site. We show that binding of SRF and Elk1 to the identified binding sites is important for LXRB transcription. Furthermore, we report that glucose significantly induces transcription via the LXRB gene promoter and that the identified binding sites are important for proper glucose responsiveness.
MATERIALS AND METHODS
Rapid amplification of cDNA ends (RACE)
The LXRB gene specific primers 5′-CGGCCTCTCGCGGAGTGAACTACTCCTGTT-3′ and nested 5′-AGGCTGAGCTGGCCTCATCAGTGCCTGGGA -3′ were used to amplify 5′-transcript from full-length cDNA from human testis, ovary and thymus using Marathon ready cDNA kits (Clontech, Mountain View, CA, USA) with the Expand Long Template PCR System (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer's instructions. The PCR products were cloned into the pGEM-T easy vector (Invitrogen, Carlsbad, CA, USA), and the identity of cloned products determined by DNA sequencing.
Plasmid constructs
The pcDNA-Elk1 plasmid was generously provided by Dr Robert Hipskind (Institut deGénétique Moléculaire de Montpellier, FRANCE). The SRF plasmids were a gift from Dr Eric Olson (UT Southwestern Medical Center at Dallas, USA). PCR fragments of the human LXRB gene promoter were cloned into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI, USA) using the KpnI and MluI sites with forward primers (−3839) 5′-ATCAGGTACCCTTTTACCTCATTTAGTCATAAGAGTAAGGCAACAAGGTCA-3′, (−1673) 5′ATCAGGTACCAAAACAGCATATGCAGTAAAGAAGTCAGCCAGATCCCAGCA-3′ and (−245) 5′-ATCAGGTACCGGCCGCAGGCTCAGAGAAGCGCATGAATGAGCTAA-3′ and reverse (+1163) 5′-ATCACTCGAGGGTGGGGTCACGGAGCAGCCTGTAGAATACAGGGGATTGAGAG-3′ with the restriction enzyme sites underlined. All mutations were introduced using the QuickChange™ XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The -245/+1163 construct was further mutated to destroy the putative Ets binding site using primers 5′- GATCTACCCGGTAAACTTTTGGTGAGTTTCCAACTTCCG-3′ and the corresponding reverse compliment. The Elk1 binding site was mutated using 5′-GGCAGCAGCTTCGGCTGGTCCTAAGCGGTTTTTTTGTTCGTCAAGTTTCACGCTCCGCCCCTCTTCCGG-3′ and the reverse compliment primers. DNA sequencing confirmed the identity of all clones.
Transient transfections
The mouse MIN6 insulinoma cell line was maintained in Dulbecco's modified Eagle's medium (DMEM, 4.5 g/l glucose), (GIBCO-BRL cat no. 41965-039), and the rat INS1E insulinoma cell line was maintained in RPMI 1640, including l-glutamine and 11.1 mM glucose, (GIBCO-BRL, cat no. 21875-034). Media were supplemented with fetal bovine serum (INS1E: 10%, MIN6: 15%), 50 μM β-mercaptoethanol and penicillin/streptomycin at a final concentration of 100 U/ml and 100 μg/ml, respectively. MIN6 medium also contained 2 mM L-glutamine while 10 mM HEPES and 1 mM Sodium Puryvate were added to INS1E medium. For serum and glucose starvation, INS1E cells were grown in plain RPMI 1640 (11879-020) containing no serum or glucose. Cells were grown under 5% CO2 at 37°C. Total 4 × 104 MIN6 and 25 × 104 INS1E cells were seeded in 24-well plates and transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Each well received 125 ng of reporter vector and 500 ng of expression vector. Empty vehicle vector was added to ensure equal amounts of DNA in each transfection. Cells were transfected for 24 h and thereafter lysed in 25 mM TAE, 1 mM EDTA, 10% glycerol, 1% Triton X-100 and 2 mM DTT. Luciferase activities were measured using a Luciferase Assay Kit (BioThema, Umeå, Sweden) in a luminometer (Luminoscan Ascent, Thermo electron Corporation, Waltham, MA, USA).
Whole cell extracts (WCE) and in vitro translation/transcription
Cells were grown in 24-well plates, washed with PBS and incubated in TEN buffer (40 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl) for 4 min. Cells were mechanistically removed with a cell scraper and pelleted by centrifugation at 3500 r.p.m. for 2 min at 4°C. Cell pellets were freeze dried on dry ice and resuspended in 50 μl ice-cold buffer C (10 mM HEPES-KOH pH 7.9, 0.4 M NaCl, 0.1 mM EDTA, 5% glycerol, 1 mM DTT, 0.5 mM PMSF). After another round of freeze drying, cell debris was removed by centrifugation for 5 min at 13 000 r.p.m. at 4°C. The supernatant corresponds to whole cell extracts.
Electro mobility shift assay (EMSA)
WT and mutated (‘Mut’) oligos (mutated nucleotides underlined) were; ETS1:5′-GATCTACCCGGTAAACTTCCGGTGAGTTT-3′, Elk1:5′-GGTCCTAAGCGGACCGGAAGTTCGTCAAGTTTCA -3′, Mut ETS1:5′- GATCTACCCGGTAAACTTTTGGTGAGTTT -3′ and Mut Elk1:5′-GGTCCTAAGCGGTTTTTTTGTTCGTCAAGTTTCA-3′. Five microgram of the respective forward and reverse oligos were annealed in 20 mM Tris-HCl pH 7.8, 2 mM MgCl2, 50 mM NaCl by heating to 95°C for 5 min and slow cooling by 1.5°C/min for 47 cycles. Oligonucleotide probes were labeled by mixing 0.2 μg annealed oligo with 250 μM non-radioactive dATP, dGTP, dTTP, respectively, 1× Klenow buffer, 20 μCi 32P labeled dCTP (GE Lifesciences, Piscataway, NJ, USA) and 1 Unit Klenow polymerase. Samples were incubated for 20 min at room temperature (RT) and the reactions terminated by adding 0.5 M EDTA. Probes were purified using G-25 Nick Columns (GE Lifesciences) and the efficiency of labeling determined using the 1214 Rackbeta liquid scintillation counter (LKB Wallac, Markham, Ontario, Canada). For binding reactions, 2 μg of whole cell extracts were incubated with 4 × 104 c.p.m. of radiolabeled oligonucleotide in binding buffer pH 8.0 (10 mM Tris-HCl, 1 mM DTT, 1 mM EDTA, 50 mM KCl, 0.3% BSA, 5% glycerol) including 1 μg poly(dI/dC) and 1× Proteinase Inhibitor Cocktail (PIC). One microgram DNA template was in vitro translated in a 50 μl reaction using the TNT® Coupled Reticulocyte Lysate Systems (Promega). From this, 5 μl was used in EMSA binding reactions. Binding reactions were incubated for 20 min at RT and protein–DNA interactions separated by electrophoresis at 240 V for 4 h at 4°C using 8% polyacrylamide gels. The gels were dried and analyzed by autoradiography. In supershift assays, 1 μg of the respective antibodies were added prior to the addition of WCE or IVT protein.
Chromatin immunoprecipitation (ChIP) assay
INS1E cells were transfected with the LXRB gene promoter containing reporter vectors and expression vectors for 24 h and protein–DNA were crosslinked using 1% formaldehyde for 20 min at RT. Cells were washed and harvested in cold 1× PBS and pelleted. The pellet was resuspended and incubated in cold RIPA buffer (50 mM Tris pH 8.0, 1 mM EDTA, 0.5 mM EGTA pH 8.0, 1% Triton X100, 0.1% Na deoxycholate, 140 mM NaCl and 1 × PIC for 10 min). DNA was sheared by sonication, centrifuged for 10 min at 13 000 r.p.m. at 4°C and the supernatant incubated with 20 μl protein A/G sepharose/agarose (50% slurry in RIPA buffer) on a rotating wheel for 2 h at 4°C. Fifty microliter of the supernatant was immunoprecipitated with 25 μg salmon sperm DNA, 100 μg BSA and 10 μg of Elk1 antibody in RIPA buffer at 4°C on a rotating wheel over night. Twenty-five microliter protein A/G slurry was added and incubated for an additional 1 h. The samples were centrifuged at 5000 r.p.m. for 2 min, the precipitates washed twice with 1 ml TSE I (1% Triton X100, 2 mM EDTA, 20 mM Tris pH 8, 150 mM NaCl), once with LiCl buffer (20 mM Tris-HCl pH 8, 1 mM EDTA, 250 mM LiCl, 1% NP40, 1% Na deoxycholate) and twice with TE buffer pH 7.4 and the protein–DNA complexes were eluted with 100 μl freshly prepared 1% SDS/TE by incubation for 30 min on a rotating wheel at RT following 65°C over night. For input control, 10% of saved samples were treated similarly to the immunoprecipitated samples. Supernatants were purified using QIAQUICK columns (QIAGEN, Hilden, Gemany). Five microliter of the elution was used in each real-time qPCR reaction using primers covering conserved sites in the rat promoter (Forward:5′-AGGCATCTCATTCGGTGGC-3′ and Reverse:5′-GGAAAGGTGACAGACTTCCGG) or the human promoter (Forward:5′-CCGGAAGTTCGTCAAGTTTCA and Reverse:5′-TTGCGTCACGTCCGGAA).
Quantitative PCR (qPCR)
Total RNA was prepared from cells using the RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. Here, 0.5 μg total RNA was reverse transcribed into cDNA using SuperscriptII and random hexamer primers (Invitrogen). The concentration and quality of the purified total RNA were determined spectrophotometrically at OD260 nm and by the OD260/280 ratio, respectively. mRNA expression levels were quantified using the ABI 7500 instrument and the SYBR green technology (Applied Biosystems, Foster City, CA, USA). All primers were designed with the Primer Express® Software version 2.0, a program specifically provided for primer design using ABI qPCR instruments. Hundred nanomolar of SYBR green assay primers were used and for each primer pair a dissociation curve analysis was carried out to ensure the specificity of the qPCR amplification. All primer pairs were designed over exon–exon boundaries. All real time qPCR reactions were performed in triplicates. We calculated relative changes employing the comparative CT method using 18S as the internal reference gene.
siRNA
INS1E cells were transfected for 4 days with mouse siElk1 and siSRF (both SMRT pool) oligos (Dharmacon, Lafayette, CO, USA) using DharmaFECT™ buffer 4 (Dharmacon) according to the manufacturer's instructions. Importantly, R&D at Dharmacon confirmed that the oligo sequences used in the mouse SMRT pools for Elk1 and SRF matched the rat sequence as well. Non-targeting control (D-001210-01), siLuciferase and siGAPDH were used as controls at corresponding concentrations. After incubation, cells were either used for WCE extraction for western blot analysis or used for RNA preparation and subsequent real-time qPCR analysis of knockdown.
RESULTS
Identification of transcription start sites in the human LXRB gene promoter
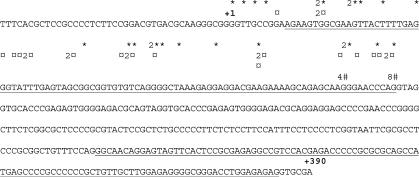
Rapid amplification of 5′-cDNA ends (5′-RACE) was performed using different tissue libraries to identify transcription start sites and, consequently, the proximal promoter region of the human LXRB gene promoter. No exact transcriptional start site was observed, rather transcription was initiated within a confined region of the promoter, in keeping with observations from other TATA-less promoters and previous observations for the mouse Lxrb gene promoter (17). We designated the most 5′-transcription start site observed as +1 (Figure 1).
Figure 1.
Characterization of the transcriptional start site of the human LXRB gene using 5′ RACE. 5′ RACE was performed using ovary (*), testis (¤) and thymus (#) cDNA libraries as described in Materials and Methods Section. Numbers indicate how many transcripts (if more than one) with a specific start site that were identified by sequencing of RACE products. Published exon sequences found in the NCBI database are underlined. The translational start site (ATG) is at +1339 but not shown in this figure.
The human LXRB gene promoter contains conserved and functional Elk1 and ETS binding sites
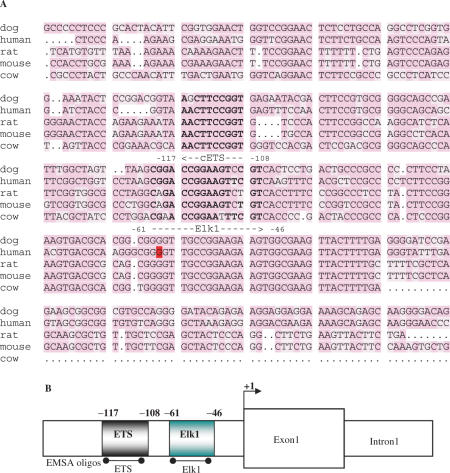
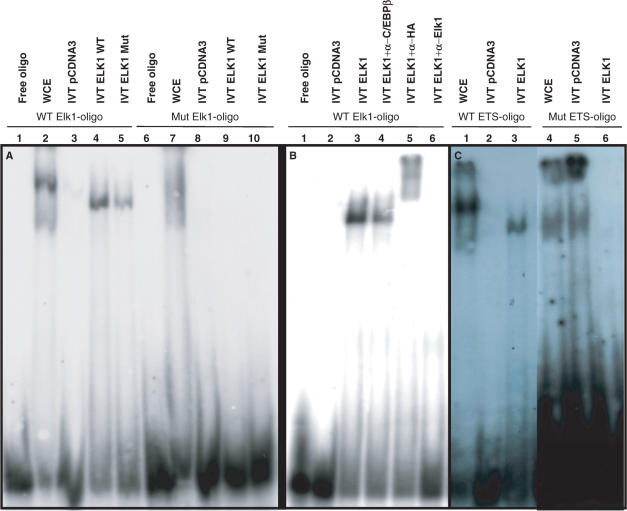
The genomic sequences from mouse, rat, dog and cow were aligned with the corresponding identified proximal promoter region of human LXRB. Using a theoretical transcription factor binding site search [Transcription Element Search System (TESS); http://www.cbil.upenn.edu/cgi-bin/tess/tess] two highly conserved binding sites were identified, Elk1 and ETS (Figure 2A). The ETS site is located 5′ of the Elk1 site in the LXRB gene promoter (Figure 2B). Next, we used EMSA to analyze protein–DNA interactions at the identified binding sites using independent DNA oligos covering these sites depicted in Figure 2B. Bands representing protein–DNA interactions at both the wild type Elk1 and ETS binding sites were observed using whole cell extract (WCE) and in vitro translated (IVT) Elk1 protein (Figure 3A, lanes 1–5 and 3C, lanes 1–3) and the interactions were abolished when these binding sites were mutated (Figure 3A, lanes 6–10 and 3C, lanes 4–6). The IVT Elk1 interactions were supershifted using a specific Elk1 antibody or an HA antibody (Elk1 cDNA was HA-tagged), but no supershift was observed using an antibody directed against the transcription factor C/EBPβ indicating a specific binding of Elk1 to this site (Figure 3B, lanes 1–6).
Figure 2.
Two conserved Elk1 and ETS sites are found in the LXRB gene promoter. (A) The LXRB promoter sequences from human, dog, rat, mouse and cow are aligned. Highly conserved Elk1 and an ETS transcription factor binding sites were identified. The binding sites are shown in bold where the ETS and Elk1 sites are in the 3′-5′ and 5′-3′ orientation, respectively as indicated by the arrows. The transcriptional start site at G (+1) is marked in red. (B) The identified 5′-ETS site and 3′-Elk1 site in the human LXRB gene promoter are schematically depicted. The location of the DNA oligos used in EMSA experiments is indicated.
Figure 3.
There are functional Elk1 and ETS binding sites in the LXRB gene promoter. (A) An oligo covering the wild type LXRB gene promoter Elk1 binding site (WT Elk1-oligo) and a mutated Elk1 site (Mut Elk1-oligo) were incubated with whole cell extracts (WCE) from the rat insulinoma INS1 cell line (lanes 2 and 7) or in vitro translated (IVT) Elk1 (lanes 4, 5, 9 and 10) or the empty control plasmid (lanes 3 and 8). IVT Elk1 Mut has three mutated regulatory phosphorylation sites. (B) WT Elk1-oligo was incubated with empty control plasmid (lane 2) or IVT Elk1 protein (lanes 3–6) in the presence of antibodies directed against the transcription factor C/EBPβ (lane 4), the HA-tag (lane 5) or Elk1 (lane 6). (C) An oligo covering the wild type LXRB gene promoter ETS binding site (WT ETS-oligo) and a mutated ETS site (Mut ETS-oligo) was incubated with whole cell extracts (WCE) from the rat insulinoma INS1 cell (lanes 1 and 4) or IVT ETS (lanes 3 and 6) or the empty control plasmid (lanes 2 and 5).
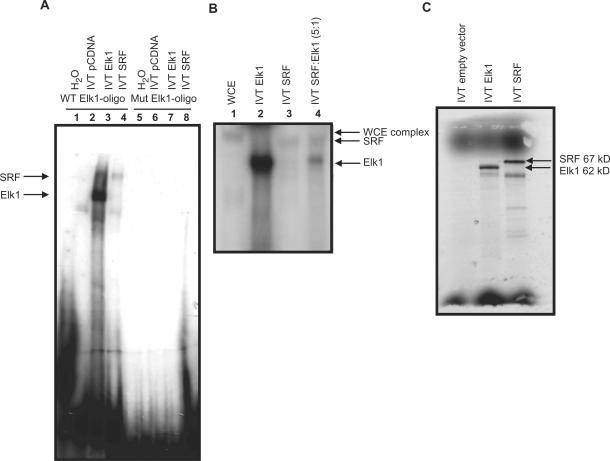
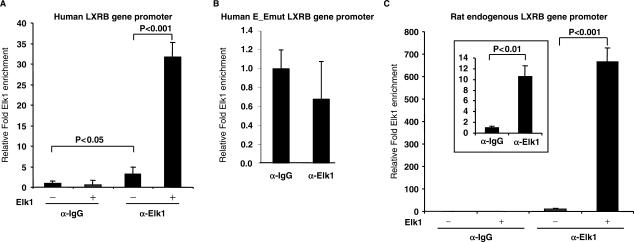
WCE yielded a complex which migrated more slowly compared to the pure IVT Elk1 protein indicating that additional proteins forming larger complexes were responsible for the interaction observed using WCE. Figure 4A, lanes 1–4 show that IVT Elk1 and SRF bind to the wild type Elk1 binding site, although the Elk1 interaction seems to be stronger. Both proteins were equally expressed in our in vitro transcription/translation system (Figure 4C) suggesting that this is not due to a molar difference for the two proteins, rather, this might simply be due to the composition of the EMSA binding buffer used, favoring Elk1 binding. Both SRF and Elk1 binding was abolished when the Elk1 binding site was mutated (lanes 5–6). Combining both IVT Elk1 and SRF yielded two bands of smaller size than observed using WCE but of the same size as when IVT Elk1 and SRF were used separately (Figure 4B, lanes 1–4) suggesting that in vitro translated Elk1 and SRF do not by themselves form the same complex as seen in WCE. SRF did not interact with the ETS binding site (data not shown). Furthermore, we performed ChIP assays in the INS1 cell line to analyze the interaction of Elk1 and SRF on the transfected human LXRB proximal promoter and the native rat promoter in the INS1 cell line using a non-specific IgG antibody as control. The LXRB gene promoter was transiently transfected into INS1 cells before crosslinking of DNA and proteins. Elk1 was enriched at the identified binding sites and the enrichment was strongly increased upon overexpression of Elk1 before crosslinking (Figure 5A). Similar results were seen on the endogenous rat Lxrb gene promoter where endogenous Elk1 was found to be enriched (Figure 5C and scaled up in the inserted frame) and this enrichment was strongly enhanced upon overexpression of Elk1. No enrichment of Elk1 was seen when the LXRB gene promoter with mutated binding sites for Elk1 and ETS was transiently transfected (Figure 5B). This indicates that Elk1 is associated with its binding site at the endogenous promoter. No enrichment was seen using primers amplifying the luciferase gene (used as control for the overexpressed reporter gene experiment) or primers amplifying an exon in the Lxrb gene (used as control for the rat native Lxrb promoter experiments) (data not shown) indicating that the enrichment is specific for the identified binding sites SRE. Unfortunately, we could not get any of the antibodies directed against SRF to work in the ChIP assay.
Figure 4.
There is a functional SRF binding site in the LXRB gene promoter. (A) An oligo covering the wild type LXRB gene promoter Elk1 binding site (WT Elk1-oligo) and a mutated Elk1 site (Mut Elk1-oligo) was incubated with IVT Elk1 (lanes 3 and 7) or IVT SRF (lanes 4 and 8) or the empty control plasmid (lanes 2 and 6). (B) The WT Elk1-oligo was incubated with WCE from the INS1 cell line (lane 1) IVT Elk1 (lane 2), IVT SRF (lane 3) or both (lane 4). (C) Empty vector or a vector containing Elk1 or SRF were in vitro transcribed/translated and separated on a 8% gel to monitor the levels of expression.
Figure 5.
The endogenous LXRB gene promoter recruits Elk1. The INS1 cell line was transfected with (A) the LXRB gene promoter reporter vector and expression vectors for Elk1 (+) or the empty pcDNA vector (−), (B) the mutated LXRB gene promoter reporter vector [Elk1mut and ETSmut (E_Emut) binding site mutated] and (C) only the Elk1 expression vector and control vector and ChIP performed using a non-specific igG according to Materials and Methods Section using a non-specific IgG antibody or an Elk1 specific antibody. Sample values were divided by their respective input value to normalize each sample for differences in the amount of DNA between samples used in each qPCR reaction. The relative fold enrichment of Elk1 at the binding site is compared to the value of α-IgG control without Elk1 overexpression which is set to 1.0 ±SEM.
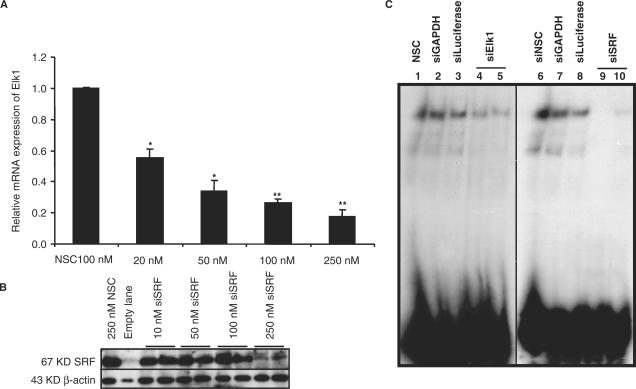
Next we investigated the effect of knocking down Elk1 and SRF in the INS1 cell line. A significant knockdown of either Elk1 or SRF was observed with siRNA targeting Elk1 or SRF but not with unrelated siRNA used as controls. The efficacy of siRNA knockdown was anlyzed at the RNA level using qPCR for Elk1 (Figure 6A) and at the protein level using western analysis for SRF (Figure 6B); in the latter case β-actin was used as a control. No cytotoxicity was observed even at 500 nM siRNA (data not shown). Using WCE from the INS1 cell line after transfection with siRNA targeting either Elk1 or SRF almost completely abolished binding to the Elk1 site in the LXRB gene promoter (Figure 6C, lanes 4, 5, 9 and 10) while control siRNAs did not affect the protein–DNA interaction at the Elk1 site (lanes 1, 2, 3, 6, 7 and 8). These results suggest that both Elk1 and SRF must be present for adequate transcription factor complex formation at the binding sites in the LXRB gene promoter.
Figure 6.
Knockdown of the Elk1 and SRF proteins abolishes binding. The INS1 cell line was transfected with increasing concentrations of siRNA targeting Elk1, SRF or a non-silencing control (NSC). The level of knockdown was analyzed at the mRNA levels using qPCR for Elk1 (A) and at the protein level using western blot analysis for SRF (B). (C) An oligo covering the wild type Elk1 binding site in the LXRB gene promoter was incubated with WCE from the INS1 cell line previously transfected with 250 nM siRNA of an NSC (lanes 1 and 6), siRNA controls targeting GAPDH and luciferase (lanes 2, 3, 7 and 8) or siRNA targeting Elk1 (lanes 4 and 5) and SRF (lanes 9 and 10). * Indicates P < 0.05 and ** Indicates P < 0.01 using the student t-test ±SEM.
The identified Elk1 and ETS binding sites induce transcription
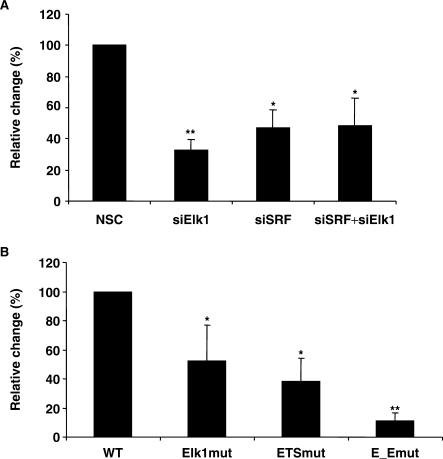
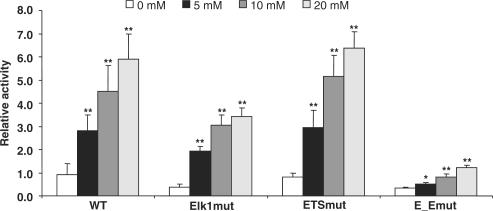
In order to characterize the importance of the identified transcription factor binding sites for transcription of the LXRB gene we cloned the −245 to +1163 LXRB gene regulatory region in front of the luciferase reporter gene. We knocked down expression of Elk1 and SRF in the INS1 cell line by siRNA targeting Elk1, SRF or both and then transiently transfected the −245/+1163 LXRB gene promoter construct. A significant reduction in promoter activity was observed when the levels of Elk1 and SRF were reduced (Figure 7A). Second, the Elk1 site, the ETS site or both sites (the same mutations were used here as the ones which showed abolished binding in Figures 3 and 4) were mutated in the −245/+1163 LXRB promoter constructs and transiently transfected into the INS1 cell line. The individually mutated Elk1 or ETS sites reduced promoter activity by 50–60% while the activity was reduced by 90% in the double mutation (Figure 7B), suggesting that both binding sites are necessary for full activity and that both Elk1 and SRF induce transcription via these binding sites in the LXRB gene promoter.
Figure 7.
Reduction of endogenously expressed Elk1 and SRF or disruption of their binding decreases LXRB gene promoter activity. (A) The INS1 cell line was transfected using 200 nM siRNA targeting Elk1, SRF, both or a non-silencing control (NSC) siRNA for 72 h and then transfected with −245/+1163 LXRB gene promoter construct for 24 h. (B) Wild type −245/+1163 LXRB gene promoter construct or mutated construct where the Elk1 site (Elk1mut), the ETS site (ETSmut) or both (E_Emut) were mutated by in vitro mutagenesis were transiently transfected. The promoter activities were analyzed by a luciferase assay. *Indicates P < 0.05 and **Indicates P < 0.01 using the student t-test ±SEM.
Glucose induces transcription via the LXRB gene promoter
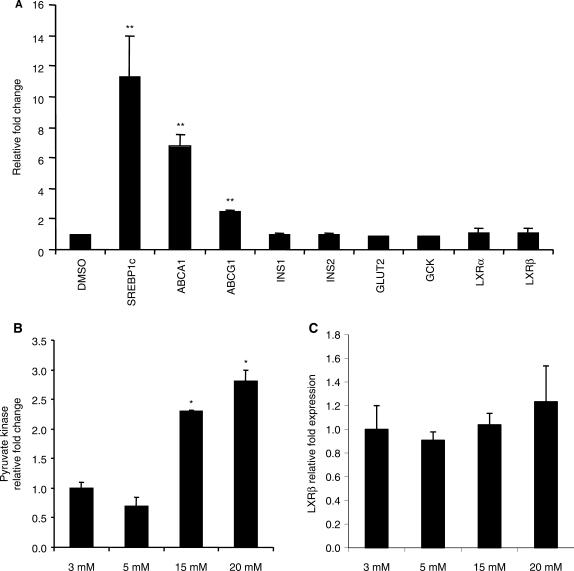
A strong induction in expression of known LXR target genes including the ATP-binding cassette (ABC) transmembrane cholesterol and lipid transporters (ABCA1 and ABCG1) and the lipogenic sterol regulatory element-binding protein 1c (SREBP1c) transcription factor was observed when INS1 cells were treated with an LXR agonist (Figure 8A) indicating that LXR signaling in these cells is working properly. Furthermore, the INS1 cells showed the expected induction in expression of pyruvate kinase upon treatment with increasing concentrations of glucose (18) (Figure 8B) while the endogenous expression of LXRβ was not affected by glucose treatment (Figure 8C). Neither was the endogenous expression of LXRβ using primary pancreatic β-cells from rat affected with glucose treatment (data not shown). LXRB gene promoter constructs with wild type or mutated Elk1, ETS or mutations of both sites were transiently transfected into INS1 cells and the cells were treated with increasing concentrations of glucose (Figure 9). As expected and in keeping with Figure 7B, the basal activities of the mutated constructs were reduced. Surprisingly, a significant concentration dependent induction of the wild type promoter activity was observed with glucose treatment whereas the double mutated Elk1 and ETS construct showed reduced response to glucose. This shows that glucose significantly induces transcription via the wild type LXRB gene promoter. Furthermore, both the ETS site and the Elk1 site are involved in proper glucose response as the activity of the double mutant construct at 20 mM glucose only showed an activity similar to that of the WT promoter under starved conditions.
Figure 8.
The INS1 cell line responds to both an LXR agonist and glucose. (A) INS1 cells were treated with 2 μM of the GW3965 LXR agonist for 24 h and the effect on the endogenous expression of known LXR target genes as well as genes involved in insulin signaling analyzed by qPCR. The endogenous expression of the pyruvate kinase (B) and LXRβ (C) in INS1 cell treated with various concentrations of glucose was measured by qPCR. The relative fold change of the GW3965 or the glucose treatments was compared to the DMSO treatment (A) or 3 (mM) glucose (B and C), respectively, the values of which were set to 1.0 ±SEM. *Indicates P < 0.05 and **Indicates P < 0.001 using the student t-test.
Figure 9.
The human LXRB gene promoter is responsive to glucose. Wild type −245/+1163 LXRB gene promoter construct (WT) or a mutated construct where the Elk1 site (Elk1mut), the ETS site (ETSmut) or both (E_Emut) were mutated by in vitro mutagenesis were transfected into INS1 cells for 24 h. Cells were serum and glucose starved for 10 h, treated with increasing concentrations of glucose for 16 h and lysed for luciferase assays. The activities of the mutant constructs as well as their response to glucose were compared to the activity of the WT construct with 0 mM glucose which was set to 1.0 ±SEM. *Indicates P < 0.05 and **Indicates P < 0.01 using the student t-test.
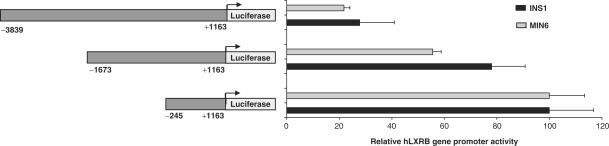
The effect of glucose on the promoter was surprising since endogenous expression of LXRβ in the INS1 cell line was not affected by increasing concentrations of glucose (Figure 8C). Therefore, we cloned larger 5′-regions of the human LXRB gene promoter as indicated in Figure 10 to look for regions which might cause repression of transcription. Transient transfections of equimolar amounts of the promoter–reporter vectors showed that inclusion of upstream regions significantly reduced the activity of the promoter. The −3839/+1163 construct still mediated a glucose response, but this response was markedly reduced compared to that seen with the −245/+1163 construct (data not shown). We speculate that these repressive elements and others located outside the fragments we have cloned suppress endogenous activation by glucose. Thus, additional cell signaling pathways could be responsible for targeting these repressive functions which apparently overrun the stimulating effects of glucose on the LXRB promoter.
Figure 10.
The 5′-regions in the human LXRB gene promoter reduce its activity. INS1 and MIN6 cells were transfected with reporter vectors representing three different lengths of the LXRB gene promoter for 24 h. The promoter activities were analyzed by a luciferase assay and the activity of the various lengths of the promoter related to that containing the short fragment which was set to 100% ±SEM.
DISCUSSION
Together with the SRF, the ETS family of transcription factors is known to form a transcription complex which can bind the serum response element (SRE). An SRE normally consists of a 5′-binding site which can recruit members of Elk1 and ETS transcription factor family and a 3′-binding site which recruits SRF. SRF binds to the 3′-binding site and associates with a member of the Elk1-ETS-family of transcription factors at the 5′-binding site and consequently the transcription complex occupies both binding sites (15). The identified 3′- binding site in the LXRB gene promoter was not a consensus SRF binding site as reported in the literature (16). Thus, our identified binding sites do not represent a classical SRE. Nevertheless, in this study we identify two binding sites in the human LXRB gene promoter which are highly conserved between species. Using mammalian cellular systems our results indicate that the ETS site and the Elk1 site are involved in increasing transcription via the LXRB gene promoter. Interestingly, we also show that the promoter is strongly responsive to glucose, partially through mediation by these binding sites in the promoter.
Mutating both the Elk1 and ETS binding sites in the LXRB, gene promoter strongly reduced its activity and suppressed its response to glucose indicating that these sites are important for both basal promoter activity and promoter responses to glucose. However, the mutations did not completely abolish the glucose response, indicating involvement of additional transcriptional regulatory factors. Both the INS1 and MIN6 cell lines were analyzed for changes in endogenous levels of LXRβ after treatment of glucose. Both overexpression of Elk1 and/or SRF as well as siRNA targeting both factors was performed in presence or absence of glucose. Surprisingly, none of these treatments had any effect on the endogenous expression of LXRβ. Neither did glucose treatment lead to any changes in recruitment of Elk1 or SRF to the endogenous rat LXRβ promoter in the INS1 cells or the transfected human LXRβ promoter as analyzed by ChIP (data not shown). Immortal cell lines do not always reflect responses in normal cells from where the immortal cells originate. Interestingly, however, neither was any effect of glucose treatment seen on LXRβ expression in primary pancreatic β-cells from rat (data not shown). Therefore, it cannot be excluded that a glucose responsive human LXRB promoter is confined to human cells since these cells have the necessary transcriptional network for the response in question. Unfortunately, human primary pancreatic β-cells are very difficult to obtain so we could not test this notion in a human primary cell system. However, the identified binding sites are highly conserved between species from mouse, rat, cow, dog and human (Figure 2) and, therefore, we do not expect to see any species specific effect using primary islets from human. This is also supported by our observation that Elk1 is enriched at the endogenous Lxrb gene promoter in rat (Figure 5B). Rather, we speculate that more complex cell signaling mechanisms including additional transcription factors, which modify the chromatin structure on the native promoter, are necessary for endogenous glucose response. The biological effects of glucose, directly affecting transcriptional regulation of target genes (for instance via ChREBP) or via insulin-mediated signaling, are pivotal for overall energy homeostasis. Therefore, it is conceivable that the glucose-activated transcriptional regulatory pathways are under strict and complex control.
Treatment of the MIN6 insulinoma cell line with glucose activates Elk1 by phosphorylation of the Ser368 residue which could lead to induced expression of Elk1 target genes (19). We show that Elk1 with the phosphorylation sites Ser324, Ser383 and Ser389 mutated to alanine interacts with its binding site in the LXRB gene promoter (Figure 3A), but has weaker effects on transcriptional regulation of the LXRB gene promoter (data not shown). Thus, phosphorylation of Elk1 could alter the effect of Elk1 on transcription. Furthermore, insulin has been shown to phosphorylate Elk1, thereby inducing its transcriptional activity (20). Accordingly, several important signaling events in response to metabolic processes may influence Elk1 activity and potentially alter the expression of LXRβ. Thus, the regulation of Elk1 activity is important for its effect on the targeted promoter.
Multiple signaling cascades involved in cell growth and proliferation including the mitogen-activated protein kinases (MAPKs) have been identified as activators of the SRF/TCF complex and factors that form these complexes (15). These signals can work via SRF/TCF to regulate gene expression. For instance, a dominant negative Elk1 was shown to inhibit cell proliferation and induce apoptotic cell death (21) and the TCF is a docking site for the pro-proliferative Wnt signaling cascade via β-catenin, which is known to interact with several members of the NR family and regulate cell proliferation events (22). LXRs were recently shown to induce growth arrest and promote apoptosis in the INS1 insulinoma cell line (23) and several additional studies report that LXRs mediate anti-proliferative effects (24–26). Hence, cell signaling cascades targeting TCF might also elicit anti-proliferative effects by enhancing expression of LXRβ.
The role(s) of LXRs in various aspects of glucose metabolism and as mediators of biological effects of glucose render LXRs highly interesting to study in the attempts to define molecular mechanisms behind insulin resistance and diabetes.
ACKNOWLEDGEMENTS
This work was supported by The Swedish Science Council, KaroBio AB, The Royal Physiographic Society in Lund (to M.N.), Golje foundation (to M.N.) and Lars Hierta foundation (to M.N.). K.R.S. holds a Norwegian Research Council fellowship. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest statement. Jan-Åke Gustafsson is shareholder and consultant of KaroBio AB.
REFERENCES
- 1.Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. N. Engl. J. Med. 2005;353:604–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- 2.Steffensen KR, Gustafsson J-Å. Liver X receptors: new drug targets to treat Type 2 diabetes? Future Lipidol. 2006;1:181–189. [Google Scholar]
- 3.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Cha JY, Repa JJ. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 2007;282:743–751. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- 5.Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V, Kreusch A, Saez E. The nuclear receptor LXR is a glucose sensor. Nature. 2007;445:219–223. doi: 10.1038/nature05449. [DOI] [PubMed] [Google Scholar]
- 6.Kase ET, Wensaas AJ, Aas V, Hojlund K, Levin K, Thoresen GH, Beck-Nielsen H, Rustan AC, Gaster M. Skeletal muscle lipid accumulation in type 2 diabetes may involve the liver X receptor pathway. Diabetes. 2005;54:1108–1115. doi: 10.2337/diabetes.54.4.1108. [DOI] [PubMed] [Google Scholar]
- 7.Dalen KT, Ulven SM, Bamberg K, Gustafsson J-Å, Nebb HI. Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor alpha. J. Biol. Chem. 2003;278:48283–48291. doi: 10.1074/jbc.M302287200. [DOI] [PubMed] [Google Scholar]
- 8.Laffitte BA, Chao LC, Li J, Walczak R, Hummasti S, Joseph SB, Castrillo A, Wilpitz DC, Mangelsdorf DJ, et al. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl Acad. Sci. USA. 2003;100:5419–5424. doi: 10.1073/pnas.0830671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao G, Liang Y, Broderick CL, Oldham BA, Beyer TP, Schmidt RJ, Zhang Y, Stayrook KR, Suen C, et al. Antidiabetic action of a liver x receptor agonist mediated by inhibition of hepatic gluconeogenesis. J. Biol. Chem. 2003;278:1131–1136. doi: 10.1074/jbc.M210208200. [DOI] [PubMed] [Google Scholar]
- 10.Efanov AM, Sewing S, Bokvist K, Gromada J. Liver X receptor activation stimulates insulin secretion via modulation of glucose and lipid metabolism in pancreatic beta-cells. Diabetes. 2004;53(Suppl. 3):S75–S78. doi: 10.2337/diabetes.53.suppl_3.s75. [DOI] [PubMed] [Google Scholar]
- 11.Loffler M, Bilban M, Reimers M, Waldhausl W, Stulnig T. Blood glucose lowering nuclear receptor agonists only partially normalize hepatic gene expression in db/db mice. J. Pharmacol. Exp. Ther. 2005;316:797–804. doi: 10.1124/jpet.105.093831. [DOI] [PubMed] [Google Scholar]
- 12.Gerin I, Dolinsky VW, Shackman JG, Kennedy RT, Chiang SH, Burant CF, Steffensen KR, Gustafsson J-Å, Macdougald OA. LXR{beta} Is Required for adipocyte growth, glucose homeostasis, and {beta} cell function. J. Biol. Chem. 2005;280:23024–23031. doi: 10.1074/jbc.M412564200. [DOI] [PubMed] [Google Scholar]
- 13.Grefhorst A, van Dijk TH, Hammer A, van der Sluijs FH, Havinga R, Havekes LM, Romijn JA, Groot PH, Reijngoud DJ, et al. Differential effects of pharmacological liver X receptor activation on hepatic and peripheral insulin sensitivity in lean and ob/ob mice. Am. J. Physiol. Endocrinol. Metab. 2005;289:E829–E838. doi: 10.1152/ajpendo.00165.2005. [DOI] [PubMed] [Google Scholar]
- 14.Chisholm JW, Hong J, Mills SA, Lawn RM. The LXR ligand T0901317 induces severe lipogenesis in the db/db diabetic mouse. J. Lipid. Res. 2003;44:2039–2048. doi: 10.1194/jlr.M300135-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- 16.Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Alberti S, Steffensen KR, Gustafsson J-Å. Structural characterisation of the mouse nuclear oxysterol receptor genes LXRalpha and LXRbeta. Gene. 2000;243:93–103. doi: 10.1016/s0378-1119(99)00555-7. [DOI] [PubMed] [Google Scholar]
- 18.Sandberg MB, Fridriksson J, Madsen L, Rishi V, Vinson C, Holmsen H, Berge RK, Mandrup S. Glucose-induced lipogenesis in pancreatic beta-cells is dependent on SREBP-1. Mol. Cell Endocrinol. 2005;240:94–106. doi: 10.1016/j.mce.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Bernal-Mizrachi E, Wen W, Srinivasan S, Klenk A, Cohen D, Permutt MA. Activation of Elk-1, an Ets transcription factor, by glucose and EGF treatment of insulinoma cells. Am. J. Physiol. Endocrinol. Metab. 2001;281:E1286–E1299. doi: 10.1152/ajpendo.2001.281.6.E1286. [DOI] [PubMed] [Google Scholar]
- 20.Xi XP, Graf K, Goetze S, Hsueh WA, Law RE. Inhibition of MAP kinase blocks insulin-mediated DNA synthesis and transcriptional activation of c-fos by Elk-1 in vascular smooth muscle cells. FEBS Lett. 1997;417:283–286. doi: 10.1016/s0014-5793(97)01303-3. [DOI] [PubMed] [Google Scholar]
- 21.Vickers ER, Kasza A, Kurnaz IA, Seifert A, Zeef LA, O'Donnell A, Hayes A, Sharrocks AD. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol. Cell Biol. 2004;24:10340–10351. doi: 10.1128/MCB.24.23.10340-10351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr. Rev. 2005;26:898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 23.Wente W, Brenner MB, Zitzer H, Gromada J, Efanov AM. Activation of liver X receptors and retinoid X receptors induces growth arrest and apoptosis in insulin secreting cells. Endocrinology. 2006;148:1843–1849. doi: 10.1210/en.2006-1247. [DOI] [PubMed] [Google Scholar]
- 24.Blaschke F, Leppanen O, Takata Y, Caglayan E, Liu J, Fishbein MC, Kappert K, Nakayama KI, Collins AR, et al. Liver X receptor agonists suppress vascular smooth muscle cell proliferation and inhibit neointima formation in balloon-injured rat carotid arteries. Circ. Res. 2004;95:e110–e123. doi: 10.1161/01.RES.0000150368.56660.4f. [DOI] [PubMed] [Google Scholar]
- 25.Fukuchi J, Kokontis JM, Hiipakka RA, Chuu CP, Liao S. Antiproliferative effect of liver X receptor agonists on LNCaP human prostate cancer cells. Cancer Res. 2004;64:7686–7689. doi: 10.1158/0008-5472.CAN-04-2332. [DOI] [PubMed] [Google Scholar]
- 26.Vigushin DM, Dong Y, Inman L, Peyvandi N, Alao JP, Sun C, Ali S, Niesor EJ, Bentzen CL, et al. The nuclear oxysterol receptor LXRalpha is expressed in the normal human breast and in breast cancer. Med. Oncol. 2004;21:123–131. doi: 10.1385/MO:21:2:123. [DOI] [PubMed] [Google Scholar]