Abstract
Shigella flexneri is an enteropathogen responsible for severe dysentery in humans. VirF is a key transcriptional regulator that activates the expression of the downstream virulence factors required for cellular invasion and cell-to-cell spread of this pathogen. There are several environmental factors that induce the translation of VirF including temperature, pH, osmolarity and post-transcriptional RNA modification. Durand and colleagues (vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase of Escherichia coli K-12. J. Bacteriol., 176, 4627–4634) have demonstrated a correlation between VirF and tRNA-guanine transglycosylase (TGT), which catalyzes the exchange of the hypermodified base queuine for the guanine in the wobble position of certain tRNAs. They characterized tgt- mutant S. flexneri strains in which the translation of VirF is markedly reduced and the bacteria are unable to invade host cells. Although the function of TGT is to modify tRNA, we report that the virF mRNA is recognized by the Escherichia coli TGT (99% identity to the S. flexneri TGT) in vitro. Further, we show that this recognition results in the site-specific modification of a single base in the virF mRNA. In the context of previous reports that small molecule binding motifs (‘riboswitches’) in mRNAs modulate mRNA conformation and translation, our observations suggest that TGT may modulate the translation of VirF by base modification of the VirF encoding mRNA.
INTRODUCTION
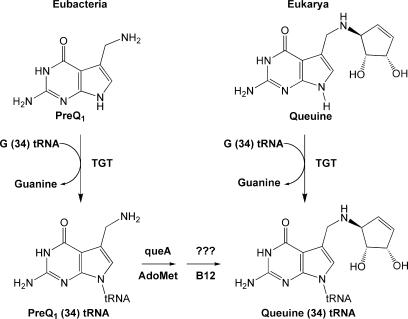
The occurrence of non-canonical nucleosides in RNA has been well-characterized (1–4), with ∼100 modified bases having been found in transfer RNA (tRNA) alone (5). Modifications occur at the post-transcriptional level, where some modifications are more simple chemical transformations (e.g. methylation) and still others are more complex (e.g. transglycosylation). One enzyme that performs a complex RNA modification (hypermodification) is tRNA-guanine transglycosylase (TGT, EC 2.4.2.29), catalyzing the exchange of the modified base queuine for the anticodon loop wobble position guanine in eukaryl and eubacterial tRNAAsp, Asn, His, Tyr (6). The proposed biochemical pathway for queuine incorporation in eubacterial tRNA is shown in Figure 1.
Figure 1.
Proposed biochemical pathway for queuine incorporation in eubacterial and eukaryl tRNA.
Although not yet fully understood, it is known that four genes are involved in the eubacterial biosynthesis of the queuine precursor, preQ1 (7). PreQ1 is incorporated into the tRNA by TGT (8). Two subsequent enzymes convert preQ1 to queuine in the tRNA (8). In contrast, eukaryl organisms acquire queuine from external routes such as diet, and this heterocyclic base is incorporated directly by the eukaryl TGT (9,10).
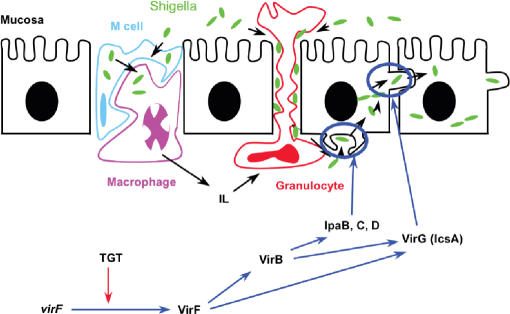
TGT plays a vital role in the pathogenesis of shigellosis (11), a disease that causes severe dysentery in humans. The pathogenic strain, Shigella flexneri, infects the cells of the human gastrointestinal tract following evasion of the host immune system defense mechanisms, such as the engulfment of foreign substances by macrophages. Shigella is able to escape from the macrophage endosome via the expression of certain virulence factors (Figure 2). There are several bacterial genes (including IcsA, IpaB, IpaC and IpaD) that mediate this escape as well as cell-to-cell spread of the organism (12). VirF, encoded on the primary pathogenicity island of Shigella, is a potent transcriptional regulator of the AraC family that regulates the expression of these virulence factors (Figure 2) (13,14).
Figure 2.
Schematic representation of Shigella pathogenesis. The invasion of the intestinal mucosal cells by S. flexneri is regulated by a number of virulence factors, including the transcriptional regulator, VirF. The VirF protein then activates the transcription of downstream virulence factors under the proper environmental conditions for invasion. Durand and colleagues (35) have shown that TGT is linked to the efficient translation of the VirF protein. Figure adapted from one provided by Prof. Ruth Brenk, University of Dundee.
There are several environmental factors that promote the expression of VirF, including oxygen and iron limitation, temperature, pH, osmolarity and post-transcriptional RNA modification (15). Durand and Björk (11) have demonstrated a positive correlation between VirF and TGT by characterizing a mutant Shigella flexneri strain with an inactivated tgt gene (termed vacC). In this mutant, the translation of VirF was markedly reduced whereas the levels of virF mRNA remained unchanged, and as a result, the bacteria were unable to invade host cells. In addition, when transformed with a plasmid containing a functional Shigella tgt gene, restoring queuine modification, the Shigella mutants exhibited both restored VirF expression and virulence, thus demonstrating a positive connection between virF mRNA translation and the presence of active TGT (and presumably queuine modification).
The role of modified nucleosides in RNA structure and stability has been well-studied (16–18). Agris and Brown (19) have identified key interactions between modified nucleosides and magnesium ions essential to the secondary structure of tRNA, in addition to facilitating key RNA–protein interactions. Mandal et al. (20,21) have characterized binding of small molecules and metabolites to RNA motifs termed ‘riboswitches’. Riboswitches are structural motifs in mRNAs [sometimes in the 5′ untranslated region (UTR) and extending into the start of the open reading frame] that can exist in at least two stable conformations. One of these conformations is stabilized by binding to a small molecule, thus altering the equilibrium between the conformations. One conformation supports translation of the protein while with the other conformation, translation is blocked. In this way, binding of the small molecule changes the conformation of the RNA and modulates its translation. In addition to the 5′ UTR of prokaryotic mRNAs, riboswitches have also been found in the 3′ UTR and introns in several eukaryotic species (22). Interestingly, the Breaker lab has just reported the discovery of a riboswitch that responds to the queuine precursor, preQ1 and appears to regulate the expression of the four genes that are involved in preQ1 biosynthesis (23). It certainly seems possible that base modification of an mRNA could modulate a similar conformational change. It therefore is feasible that such a control mechanism for gene expression might be involved in regulating the expression of virulence factors in pathogenic organisms, as is apparently seen with VirF.
The incorporation of modified nucleosides has been characterized more fully for tRNA (and some other RNAs, e.g. rRNA and snRNA), than for mRNA (5,24,25). The most common example of post-transcriptional processing in mRNA is the eukaryotic 7-methylguanosine 5′ cap structure, which aids in the binding to the small ribosomal subunit and is essential for the efficient synthesis of eukaryotic proteins (26–28). To date, the only known function of TGT is to catalyze the modification of tRNA with queuine. Previous work has shown that the eubacterial TGT will recognize a U-G-U sequence in the loop of an RNA hairpin structure that corresponds to the anticodon stem–loop of its cognate tRNAs (29,30). The eubacterial TGT will also recognize a U-G-U containing hairpin in the context of a dimeric form of a cognate tRNA (31).
It is conceivable that an mRNA may also be modified directly by TGT, provided the mRNA contained the appropriate recognition elements. An examination of the sequence of virF mRNA for the presence of U-G-U sequences revealed six unique U-G-U sites (Figure 3). Mfold analysis of the regions surrounding each of these U-G-U sequences reveal that nucleotides 410–433 could possibly fold into a hairpin structure with the U-G-U sequence in a position in a loop that is analogous to the anticodon loop of TGT-cognate tRNAs.
Figure 3.
Nucleotide sequence of virF mRNA. Six UGU sequences are found in the sequence, and are highlighted in red. The sequence of the virF MH is underlined.
As a first step towards probing the possibility that TGT may modulate the translation of VirF via modification of the virF mRNA, Michaelis–Menten kinetic analyses were conducted to probe this modification by TGT in vitro. We report that the Escherichia coli TGT, which has 99% sequence identity to the S. flexneri TGT, does indeed recognize the virF mRNA as a substrate in vitro. Further, we show that this recognition results in the site-specific modification of a single base in the virF mRNA.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, all reagents were ordered from Sigma or Aldrich. DNA oligonucleotides, agarose, dithiothreitol (DTT), T4 DNA ligase and DNA ladders were ordered from Invitrogen. All restriction enzymes and Vent® DNA polymerase were ordered from New England Biolabs. The ribonucleic acid triphosphates (NTPs) and pyrophosphatase were ordered from Roche Applied Sciences. The deoxyribonucleic acid triphosphates (dNTPs) were ordered from Promega. Low-melting Seaplaque agarose was ordered from Cambrex. Gelase™ Enzyme Prep, MasterAmp™ High Fidelity RT–PCR Kit, and Scriptguard™ RNase Inhibitor were ordered from Epicentre. Epicurian coli® XL2-Blue ultracompetent cells were ordered from Stratagene. Amicon Ultra Centrifugal Filter Devices were ordered from Millipore. Whatman GF/C Glass Microfibre Filters and all bacterial media components were ordered from Fisher. The QIAPrep® Spin Miniprep Kit was ordered from Qiagen. Tris–HCl Buffer was ordered from Acros Organics. [3H] PreQ1 was ordered from American Radiolabeled Chemicals, Inc. T7 RNA polymerase was isolated from E. coli BL21 cells containing the plasmid pBH161 according to the procedure of Prof. William McCallister, State University of New York, Brooklyn. The E. coli TGT was isolated with an amino terminal his-tag as previously described (32).
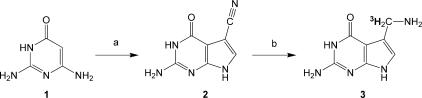
Synthesis of [3H] preQ1 (Figure 4)
Figure 4.
Synthesis of radiolabeled [3H] preQ1. (A) chloro(formyl)acetonitrile, NaOAc, H2O, 100°C; (B) 3H2, Pd/C, MeOH, 50 psi.
The cyano precursor (preQ0, 2) was synthesized according to the method of Migawa et al. (33) by the condensation of chloro(formyl)acetonitrile and pyrimidine 1 (33). Reduction of the cyano precursor with tritium gas gave the desired radiolabeled substrate preQ1 (3) with a specific activity of 500 mCi/mmol (34). The tritium reduction was performed commercially by American Radiolabeled Compounds Co.
Construction of pTZvirF
The plasmid pBDG302, containing the virf gene, was received from Prof. Glenn Björk (Umeå University, Sweden). The virf gene was amplified from the plasmid by polymerase chain reaction (PCR) under the following conditions: primers (20 pmol each), pBDG302 (500 ng), Mg2+ (2 mM), dNTPs (1 mM each), Vent DNA polymerase (4 U), brought to a final volume of 50 μl with deionized water. The sample was treated with 30 PCR cycles of the following sequence: 94°C (1 min), 50°C (1 min), and 72°C (2 min), followed by a final extension at 72°C (5 min). Following a double restriction enzyme digest with PstI and EcoRI (40 U each, 20-μl reaction) for 1 h at 37°C, the PCR product and vector were gel-purified from Seaplaque agarose with Gelase™ according to the vendor protocol. The purified virf gene was then ligated into digested pTZ19RAmp (5:1 volume ratio, 20 μl reaction) following overnight incubation with T4 DNA ligase (2 U) at 17°C. The ligated sample (10 μl) was transformed into 100 μl of Epicurian coli® XL2-Blue ultracompetent cells according to the Stratagene protocol. Cells were grown overnight at 37°C on L-Amp plates (50 μg/ml ampicillin). Individual colonies were isolated, and 3 mL 2xTY (16 g Bactotryptone, 10 g yeast extract, 5 g NaCl/liter of water with 50 μg/ml ampicillin) liquid cultures were inoculated at 37°C with shaking. Plasmid was isolated via miniprep, and the virf gene sequence was confirmed with DNA sequencing (University of Michigan DNA Sequencing Core Facilities).
In vitro transcription
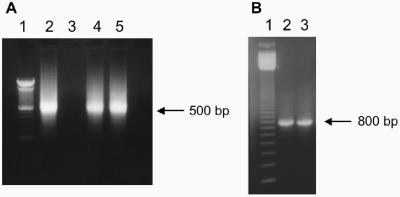
In vitro transcription reactions with pTZvirF were conducted by first linearizing the plasmid at the end of the virf sequence with the restriction enzyme EcoRI (40 U/100 μl DNA, 500 μl reaction). The sample was ethanol precipitated at −20°C, and the pellet was re-suspended in 250 μl of deionized water. In vitro 1 ml transcription conditions were as follows: pTZvirF template (100 μl), transcription buffer (4 mM Tris–HCl, pH 8.0; 2 mM MgCl2, 0.5 mM DTT, 0.1 mM spermidine), NTPs (4 mM each), T7 RNA polymerase (2500 U), inorganic pyrophosphatase (2 U) and RNase inhibitor (200 U). The reaction was incubated at 37°C for ∼4 h. The reaction was stored at −20°C following transcription. Best results were obtained when the 1 ml reaction was prepared and redistributed into 100 μl volumes prior to incubation at 37°C. The MasterAmp ™ High Fidelity RT–PCR Kit was used according to vendor protocol to generate virf DNA, which was confirmed with sequence analysis of the DNA product (Figure 5).
Figure 5.
Transcription of virF mRNA. (A) In vitro transcription of virF mRNA visualized with ethidium bromide on a 1% TAE agarose gel. The single-stranded mRNA ran at ∼500 bp in comparison to the 100-bp DNA ladder (lane 1). Lane 2: virF mRNA from a previous transcription reaction. Lanes 4 and 5: virF mRNA from replicate transcription reactions. (B) RT–PCR samples visualized with ethidium bromide on a 1% TAE agarose gel. RT–PCR was conducted using extracted mRNA from replicate samples (lanes 2 and 3), electrophoresed in comparison to a 100 bp DNA ladder (lane 1). Each RNA sample produced the corresponding 800 bp DNA species indicative of the virf ORF.
Mfold analysis and synthesis of virF MH
Analysis of the energetically favorable secondary structures within the virF mRNA sequence was performed using the biophysical web tool Mfold (M. Zuker, http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Sequences of ∼10 nucleotides surrounding either side of the six possible recognition motifs were analyzed by the web tool, and the hairpin structure determined to be most favorable was found between nucleotides 410–433 in the virF mRNA, which contains the potential recognition sequence U420G421U422. The RNAture Oligonucleotide Analyzer web tool was used to predict a Tm of 71°C for the virF minihelix hairpin. (Note: This web tool appears to be no longer available.) An Expedite™ Nucleic Acid Synthesis System was used to synthesize this 24-nucleotide sequence (5′-GGAGUAGUCUUUGUCGACUAUUUU-3′) using the vendor's protocols for the synthesis of RNA at the 1 μmol scale. The reagents were from Perkin–Elmer and the RNA amidites were from Glen Research. The extinction coefficient calculated for this RNA minihelix was ε260 = 265.3 OD/mM.
Generation of virF mRNA(G421A)
The single nucleotide mutation of guanine 421 to adenine in the virF mRNA sequence was generated via QuikChange site-directed mutagenesis (Stratagene), producing the new vector pTZvirF(G421A). The reactions conditions were as follows: complimentary oligonucleotides with desired mutation (175 ng), pTZvirF(wt) template (800 ng), dNTPs (0.25 mM) and Vent DNA polymerase (2 U), brought to a final volume of 30 μl with deionized water. The sample was treated with 25 PCR cycles of the following sequence: 94°C (30 s), 50°C (1 min), and 72°C (6.5 min). The PCR product was then incubated for 2 h at 37°C with Dpn I (40 U), and addition of NE Buffer 4 was required for proper digestion of wild-type plasmid. The digested sample (10 μl) was transformed into 100 μl of Epicurian coli® XL2-Blue ultracompetent cells according to the Stratagene protocol. Cells were grown overnight at 37°C on L-Amp plates (50 μg/ml ampicillin). Individual colonies were isolated, and 3 ml 2xTY (16 g bactotryptone, 10 g yeast extract, 5 g NaCl/liter of water with 50 μg/ml ampicillin) liquid cultures were inoculated at 37°C with shaking. Plasmid was isolated via miniprep, and the virF mRNA(G421A) mutation was confirmed with DNA sequencing (University of Michigan DNA Sequencing Core Facilities).
Kinetic assays
Assays were conducted by monitoring the incorporation of radiolabeled substrate, [3H] preQ1, into E. coli tRNATyr, dG34ECYMH (5′-GGGAGCAGACUdGUAAAUCUGCUCCC-3′) and various virF mRNA substrates. Samples from in vitro transcriptions were concentrated with Amicon Ultra Centrifugal Filters (10,000 MWCO). The concentration of virF mRNA was determined with a Cary UV-Visible Spectrophotometer, where the approximate concentration of a single-stranded RNA sample A260 = 1 OD is 40 μg/ml and the molecular weight of virF mRNA is 252 g/mmol virF mRNA. In brief, kinetic assays were set up under the following conditions: RNA substrate (various concentrations), [3H] preQ1 (10 μM, 296mCi/mmol stock), TGT (100 or 200 nM, as specified), and HEPES reaction buffer (100 mM HEPES, pH 7.3; 20 mM MgCl2; 5 mM DTT) to a final volume of 400 μl. All samples were incubated at 37°C for purposes of equilibration before initiating the reaction with the addition of TGT. Aliquots (70 μl) were removed at various points throughout the reaction and quenched in 3 ml of 5% TCA for 1 h before filtering on glass-fiber filters. Each filter was washed with three volumes of 5% TCA and a final wash of ethanol to dry the filter. The samples were analyzed in a scintillation counter (Beckman) for radioactive decay, where counts were reported in DPM and later converted to picomoles [3H] preQ1 by the following conversion (pmol = DPM × 0.00152, for the [3H] preQ1 stock with a specific activity of 296 mCi/mmol). Initial velocities were determined by converting the slopes of these plots (pmol/min) to units of second−1, taking into account the concentration of the enzyme and aliquot size. The individual data points from each trial were averaged, and the standard deviation was determined for each concentration of RNA substrate. The average data points (with error bars representing their standard deviations) were plotted. However, all of the individual data points were fit via non-linear regression to the Michaelis–Menten equation and the line for that fit is displayed (Figure 6). All non-linear regression fits with the Michaelis–Menten equation were determined using Kaleidagraph (Abelbeck Software).
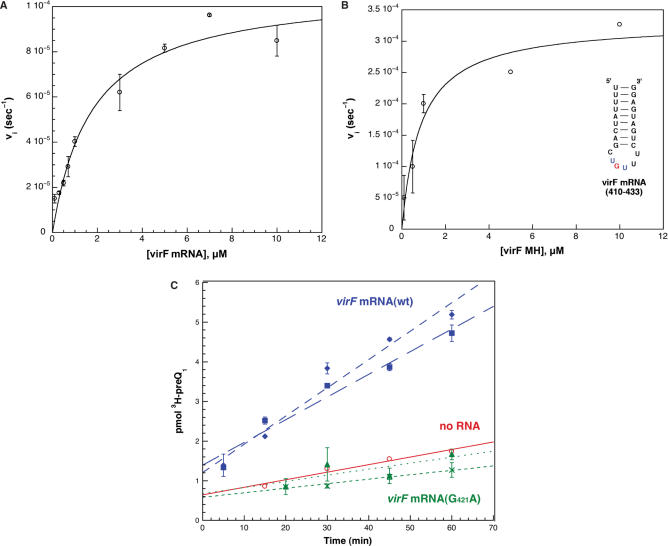
Figure 6.
Kinetic characterization of virF mRNA substrates with E. coli TGT and [3H] preQ1. (A) and (B) The individual data points were fit to the Michaelis–Menten equation by non-linear regression, and the average data points (with error bars representing the standard deviations of the averages) for each concentration of RNA substrate are depicted. The study was performed with duplicate kinetic trials at saturating concentrations of [3H] preQ1. The inset in B shows the structure of the RNA minihelix corresponding to bases 410–433 of the virF mRNA. (C) The comparison of radioactive substrate incorporated over time in virF mRNA(wt) (blue, squares and diamonds), virF mRNA(G421A) (green, crosses and triangles), and a no RNA control (red, circles), fit by linear regression. Each virF mRNA substrate was analyzed at two concentrations: 2 μM (squares and crosses) and 10 μM (diamonds and triangles). The average of duplicate trials is shown.
RESULTS
Construction and in vitro transcription of pTZvirF
To provide micromolar quantities of virF mRNA for our studies, we generated an in vitro transcription clone for the virF mRNA. The virf gene was subcloned from the plasmid pBDG302 containing the virf gene (a gift from Professor Glenn Björk, Umeå University, Sweden) into a plasmid suitable for in vitro transcription, generating pTZvirF. VirF mRNA was synthesized via in vitro run-off transcription following digestion with EcoRI, linearizing pTZvirF at the end of the virf gene sequence. The virF mRNA was physically characterized on an ethidium bromide stained, 1.2% formaldehyde agarose gel.RT–PCR was utilized to generate dsDNA from the in vitro transcription product using the same oligonucleotide primers initially designed for subcloning of the virf gene. Examples of formaldehyde and TAE agarose gels of the virF mRNA and the dsDNA from the RT–PCR are shown in Figure 5.
The single-stranded virF mRNA appears to run on the gel at ∼500 bp in comparison to the dsDNA ladder. The size of the virf gene is 789 bp, and the corresponding mRNA is 789 nucleotides in length. We hypothesize that the mRNA is running at a lower ‘apparent’ molecular weight due to the propensity of mRNA to adopt a variety of conformations, even in an agarose gel. This would explain why the observed molecular weight is a little larger than one half the size of the double-stranded virf DNA. The RT–PCR product (dsDNA) migrated to the anticipated molecular weight for a DNA sample of ∼800 bp. DNA sequencing of this product matched the virF mRNA sequence (data not shown).
Kinetic analysis of E. coli tRNATyr, ECYMH minihelix with preQ1
For comparison, Michaelis–Menten kinetic analyses were conducted with the natural RNA substrate E. coli tRNATyr (ECY) and the modified minihelix substrate dG34ECYMH (the anticodon stem–loop of ECY where the guanosine at position 34 contains a 2′-deoxyribose) with [3H] preQ1. It has been shown previously that a minihelix RNA consisting of the anticodon arm and loop of a queuine-cognate tRNA is a sufficient substrate for TGT (29). Aliquots were taken at various time points over a 15 min incubation of 100 nM E. coli TGT, various concentrations of ECY (0.05–1.5 μM) or dG34ECYMH (0.05–5 μM) and saturating concentrations of [3H] preQ1. The kinetic constants determined for the incorporation of [3H] preQ1 with ECY and dG34ECYMH are shown in comparison with the kinetic data for the virF substrates in Table 1.
Table 1.
RNA kinetic parameters with E. coli htTGT(wt) and [3H] preQ1 at pH 7.3
| KM (μM) | kcat (10−4 s−1) | kcat/KM (102 M−1 s−1) | |
|---|---|---|---|
| ECY | 0.16 (0.03)a | 40 (2) | 250 (40) |
| dG34ECYMH | 0.26 (0.07) | 70 (5) | 270 (60) |
| virF mRNA | 1.8 (0.4) | 1.1 (0.1) | 0.60 (0.1) |
| virF MH | 0.87 (0.3) | 3.3 (0.3) | 3.8 (1) |
aStandard errors are in parentheses.
Kinetic analysis of virF mRNA, virF MH minihelix with preQ1
Using the same approach described above, Michaelis–Menten kinetic analyses were conducted with virF mRNA. Aliquots were taken at various time points over a 1 h incubation of 200 nM TGT, various concentrations of virF mRNA (0.1–10 μM), and saturating concentrations of [3H] preQ1 (Figure 6A). Higher concentrations of virF mRNA were tested to obtain an accurate kinetic profile by characterizing the reaction over a large range of concentrations. In addition to characterizing the wild-type virF mRNA, a virF minihelix RNA (virF MH) corresponding to the 410–433 hairpin sequence (underlined in Figure 3) as well as a full-length virF mRNA mutant (G421A) were studied. The kinetic analyses were performed with 100 nM E. coli TGT, various concentrations of virF MH (0.1–10 μM) and saturating concentrations of [3H] preQ1 (Figure 6B). The full-length virF mRNA(G421A) was incubated under the same conditions as the wild-type mRNA, but only at concentrations corresponding to KM and 5xKM, as determined from the kinetic constants of virF mRNA(wt) (Table 1). A ‘no RNA’ control was also included to determine the background level of radioactivity present in the samples (Figure 6C). The data were fit by non-linear regression.
Both the full-length virF mRNA(wt) and the virF minihelix exhibited RNA concentration-dependent incorporation of [3H] preQ1 over time, and the Michaelis–Menten equation provided a good fit for the data. The full-length virF mRNA(G421A), which is full-length mRNA with a single nucleotide mutation at guanine 421, was analyzed at both 2 μM (∼KM) and 10 μM (∼5 × KM) mRNA. The virF mRNA(G421A) mutant showed no detectable activity greater than the ‘no RNA’ control (Figure 6C). The kinetic constants determined for the virF mRNA substrates are shown in Table 1.
Both the virF mRNA and virF MH have KM values in the low micromolar range, even though kcat and kcat/KM for both is lower than the corresponding values for the ECY substrates.
DISCUSSION
VirF is a critical transcriptional regulator responsible for activating virulence genes in Shigella flexneri. Durand and colleagues (35) demonstrated the involvement of TGT in modulating the translation of VirF via the observation that a mutant strain of Shigella with an inactivated tgt gene (termed vacC) showed decreased virulence. VirF protein levels were dramatically lower in the mutant as compared to wild-type, but the virF mRNA levels showed no detectable difference from wild-type Shigella. The lack of VirF protein resulted in a reduction of all downstream virulence gene expression, and thus exhibited a less virulent phenotype than that of the wild-type bacterium. When transformed with a plasmid encoding the Shigella tgt gene, both VirF translation and virulence were restored. It had previously been shown that the presence of modified nucleosides enhances translation (36,37). However, other studies have shown that growth rate and protein translation as a whole are not directly affected by a lack of queuine-modified tRNA (38). While this interesting correlation between VirF translation and TGT activity has been known for some time, yet the exact role TGT plays in the translation of this primary virulence factor remains unclear.
Our laboratory has previously demonstrated that TGT can modify substrates with more unusual structures than a canonical tRNA fold. We reported that a dimeric form of the ECY serves as a substrate for TGT, with a slightly higher KM and identical kcat, relative to the normal tRNA (31). It had previously been shown from NMR studies that the anticodon arms of the dimer subunits were intact and pointing away from the center of the dimer (39).
Those studies demonstrate that TGT can recognize a minihelix containing the requisite U-G-U sequence even in the context of a larger RNA structure. TGT is not the first tRNA modification enzyme to demonstrate recognition of alternative RNA structures. Gu and coworkers (24) have shown that, in vitro, the modification enzyme tRNA (m5U54)-methyltransferase will methylate 16S ribosomal RNA from E. coli in addition to its physiological tRNA substrate, although they found no evidence for this occurring in vivo. The enzyme pseudouridine synthase catalyzes the isomerization of specific uridines to the modified nucleoside ψ. A single pseudouridine synthase has been shown to have ‘dual specificity’, recognizing and modifying both tRNA and snRNA (40–42). Ofengand and colleagues (43,44) have also characterized a critical pseudouridine synthase responsible for site-specific modification with pseudouridine in vivo for both tRNA and 23S rRNA in E. coli. Such precedence for tRNA modification enzymes to recognize and modify other RNA species in vitro and in vivo, suggests that virF mRNA modification mediated by TGT may also occur. As a first step to probe for this possibility, we examined the virF mRNA sequence for the presence of a U-G-U sequence in a TGT recognition motif. Of the six U-G-U sequences in the virF mRNA, the one involving bases 410–433 (Figure 3) was predicted by Mfold analysis to be able to fold into a hairpin structure possibly suitable for recognition by TGT.
Incubation of virF mRNA with E. coli TGT and radiolabeled preQ1 revealed that the virF mRNA is indeed a substrate for TGT in vitro. The kinetic parameters (KM, kcat; Table 1) for virF mRNA were determined from a Michaelis–Menten analysis (Figure 6A). The virF mRNA substrates exhibit the same trend in kinetic parameters as the ECY substrates, where the minihelix substrates (dG34ECYMH and virF MH) have slightly higher values for both KM and kcat/KM with respect to the corresponding full-length RNA substrates (Table 1). The values of KM for both ECY and virF mRNA are very similar, both in the low micromolar range. This is encouraging considering the size and structural difference between the two substrates, where ECY is ∼80 nucleotides in length with a very well-defined tertiary structure common to tRNA and the virF mRNA is ca. 800 nucleotides in length and presumably does not have a compact tertiary structure. The Michaelis–Menten plot for the virF mRNA (Figure 6A) fits to a single KM, consistent with a single site of modification within the virF mRNA. The kcat for the virF mRNA is ∼ 40-fold lower than that for tRNA. It has previously been shown that altering the position of the U-G-U sequence within the minihelix loop of cognate tRNAs is correlated with a reduction in activity (45). All of the biochemical and structural data previously reported is consistent with a covalent intermediate, via Asp264, in the TGT reaction (32,46–48). The hairpin loop of the ECY substrate contains seven nucleotides, whereas the hairpin in the virF mRNA substrate contains only six nucleotides in the loop. This difference in loop length may result in a suboptimal orientation of the guanosine ribose in the U420-G421-U422 loop in the virF mRNA, making nucleophilic attack by Asp264 less likely. This difference in loop length and orientation could account for the reduced kcat that we have observed for the virF mRNA substrates.
Our analysis of the virF mRNA sequence predicts that there should be a single site of modification, guanine 421. We have taken two approaches to investigate this. In our first approach, a virF MH corresponding to the predicted hairpin structure within the native virF mRNA sequence (bases 410–433), was chemically synthesized (Figure 6B). The stem consists of nine base pairs, where the first four nucleotides in the stem are uridine residues forming wobble pair interactions with three guanines and one Watson–Crick pair with an adenosine. At first glance, the stability of a minihelix with three G-U wobble pairs might be questionable; however, in the context of the virF mRNA, the ends of the helix may be held in close proximity by other intramolecular interactions. Additionally, the virF MH by itself has a predicted melting temperature of 71°C, indicating the structure should be stable at physiological and assay temperatures. The virF MH is a substrate for TGT in vitro. The KM values for both the full-length virF mRNA and virF MH (1.8 and 0.87 μM, respectively; Table 1) are very similar, suggesting that the minihelix structure is likely a predominant conformation in the virF mRNA.
Although the recognition of the virF minihelix by TGT is consistent with the virF mRNA serving as a substrate for TGT, it does not provide conclusive evidence that guanine 421 is the site of modification in the virF mRNA. There are six UGU sequences within the virF mRNA that are possible recognition sites for TGT. Therefore, our second approach was to construct the point mutation, G421A, in the full-length virF mRNA to demonstrate the importance of guanine 421. The virF mRNA(G421A) mutant resulted in a complete loss of activity at two different concentrations of RNA (Figure 6C), indicating that G421 is indeed essential for recognition by TGT. Had a second exchangeable guanine existed in the sequence, we would have expected to see a decreased or possibly even unchanged activity of the mRNA. The relationship between the ‘no RNA’ negative control and the virF mRNA(G421A) indicates that guanine 421 is the only exchangeable nucleotide in the virF mRNA sequence (at least within the concentration ranges tested), and that the kinetic parameters observed for virF mRNA(wt) are due to specific recognition by TGT and could not be attributed to non-specific interactions with this large nucleic acid molecule. It should be noted that, under the conditions of the assay (Figure 6C), it appears that the enzyme is undergoing a limited number of turnovers. Two factors may be contributing to this. The first is that our calculations of kinetic parameters assume 100% active enzyme, which is almost certainly an over-estimate. The active enzyme concentration may be as much as 2-fold lower as recent studies suggest that the eubacterial TGT may exist as a homodimer with ‘half-of-sites’ reactivity (49). This would effectively double the turnovers per active site. Second, the off-rate for the modified mRNA may be sufficiently slower (relative to that for tRNA) such that the turnover rate may indeed be significantly slowed under these conditions. It remains to be seen if these observations hold under in vivo conditions.
From the results presented herein, it is clear that the virF mRNA does act as a substrate for the eubacterial TGT in vitro. Although there are six possible UGU recognition motifs, both the mutagenesis and virF mRNA minihelix studies are consistent with G421 serving as the sole site of modification within the mRNA. With a KM value in the low micromolar range, it is very possible that the modification of virF mRNA may be biologically relevant (e.g. may occur in vivo). These results provide the first ‘proof of principle’ evidence that post-transcriptional RNA modification may regulate mRNA function, as it has long been recognized to do for tRNA. The recent work characterizing the preQ1 riboswitch revealed that it is a fairly simple hairpin structure (the simplest riboswitch structure characterized to date) (23). Such a simple structure could feasibly occur in the coding region of an mRNA species. In fact, the TGT modification site that we have discovered in the virF mRNA is predicted to occur in a simple hairpin structural motif. Base modification in a hairpin structure in the coding sequence of virF mRNA could induce a similar structural switch as seen in the preQ1 riboswitch and thereby influence translation of VirF. Studies to determine the physiological significance of the virF mRNA modification by TGT that we have observed in vitro are currently in progress.
ACKNOWLEDGEMENTS
We gratefully acknowledge Professor Glenn Björk for the plasmid pBDG302 containing the virf gene. We thank Dr Ruth Brenk for use of her figure illustrating Shigella pathogenesis (Figure 2) and members of the Garcia laboratory for critical review of this manuscript. This research was supported by National Institutes of Health (GM065489 to G.A.G. and GM07767 J.K.H. trainee) and the University of Michigan, College of Pharmacy.
Funding to pay the Open Access publication charges for this article was provided by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia GA, Goodenough-Lashua DM. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: ASM Press; 1998. pp. 135–168. [Google Scholar]
- 3.Adamiak RW, Gornicki D. Hypermodified nucleosides of tRNA: synthesis, chemistry, and structural features of biological interest. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:27. doi: 10.1016/s0079-6603(08)60345-1. [DOI] [PubMed] [Google Scholar]
- 4.Grosjean H, Benne R. Modification and Editing of RNA: The Alteration of RNA Structure and Function. Washington, D.C: ASM Press; 1998. [Google Scholar]
- 5.Grosjean H, Sprinzl M, Steinberg S. Posttranscriptionally modified nucleosides in transfer RNA: Their locations and frequencies. Biochimie. 1995;77:139–141. doi: 10.1016/0300-9084(96)88117-x. [DOI] [PubMed] [Google Scholar]
- 6.Harada F, Nishimura S. Possible anticodon sequences of tRNAHis, tRNAAsn, and tRNAAsp from Escherichia coli B. Universal presence of nucleoside Q in the first position of the anticodons of these transfer ribonucleic acids. Biochemistry. 1972;11:301–308. doi: 10.1021/bi00752a024. [DOI] [PubMed] [Google Scholar]
- 7.Reader JS, Metzgar D, Schimmel P, de Crecy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2004;279:6280. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 8.Slany RK, Kersten H. Genes, enzymes and coenzymes of queuosine biosynthesis in procaryotes. Biochimie. 1994;76:1178–1182. doi: 10.1016/0300-9084(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 9.Reyniers JP, Pleasants JR, Wostmann BS, Katze JR, Farkas WR. Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing trnasfer RNAs in the mouse. J. Biol. Chem. 1981;206:11591–11594. [PubMed] [Google Scholar]
- 10.Gunduz U, Katze JR. Queuine salvage in mammalian cells. J. Biol. Chem. 1984;259:1110–1113. [PubMed] [Google Scholar]
- 11.Durand JMB, Bjork GR. Putrescine or a combination of methionine and arginine restores virulence gene expression in a tRNA modification-deficient mutant of Shigelia flexneri: a possible role in adaptation of virulence. Mol. Microbiol. 2003;47:519–527. doi: 10.1046/j.1365-2958.2003.03314.x. [DOI] [PubMed] [Google Scholar]
- 12.Dorman CJ, Porter ME. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 1998;29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 13.Jost BH, Adler B. Site of transcriptional activation of virB on the large plasmid of Shigella flexneri 2a by VirF, a member of the AraC family of transcriptional activators. Microb. Pathog. 1993;14:481–488. doi: 10.1006/mpat.1993.1047. [DOI] [PubMed] [Google Scholar]
- 14.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durand JM, Dagberg B, Uhlin BE, Bjork GR. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 2000;35:924–935. doi: 10.1046/j.1365-2958.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- 16.Agris PF. In: Progress in Nucleic Acid Research and Molecular Biology. Cohn WE, Moldave K, editors. Vol. 53. San Diego, CA 92101-4495: Academic Press Inc., 525 B Street, Suite 1900; 1996. pp. 79–129. [DOI] [PubMed] [Google Scholar]
- 17.Varani G, Tinoco I. Rna Structure And Nmr-Spectroscopy. Q Rev Biophys. 1991;24:479. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 18.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing Gnra loops. Science. 1991;253:191. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 19.Agris PF, Brown SC. In: Nuclear Magnetic Resonance and Nucleic Acids. James TL, editor. Vol. 261. San Diego, CA 92101-4495: Academic Press Inc., 525 B Street, Suite 1900; 1995. pp. 270–299. [Google Scholar]
- 20.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA. 2004;101:6421. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandal M, Breaker RR. Gene regulation by riboswitches. Nature Rev. Mol. Cell Biol. 2004;5:451. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 22.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth A, Winkler W, Regulski E, Lee B, Lim J, Jona I, Barrick J, Ritwik A, Kim J, et al. A riboswitch selective for the queuosine precursor preQ(1) contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 24.Gu XG, Ofengand J, Santi DV. In Vitro Methylation of Escherichia coli 16S rRNA by tRNA (m5U54)-Methyltransferase. Biochemistry. 1994;33:2255–2261. doi: 10.1021/bi00174a036. [DOI] [PubMed] [Google Scholar]
- 25.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 1996;236:747. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 27.Macejak DG, Sarnow P. Internal Initiation Of Translation Mediated By The 5′ Leader Of A Cellular Messenger-Rna. Nature. 1991;353:90. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 28.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 29.Curnow AW, Kung FL, Koch KA, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: gross tRNA structural requirements for recognition. Biochemistry. 1993;32:5239–5246. doi: 10.1021/bi00070a036. [DOI] [PubMed] [Google Scholar]
- 30.Curnow AW, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli - minimal tRNA structure and sequence requirements for recognition. J. Biol. Chem. 1995;270:17264–17267. doi: 10.1074/jbc.270.29.17264. [DOI] [PubMed] [Google Scholar]
- 31.Curnow AW, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: recognition of dimeric, unmodified tRNATyr. Biochimie. 1994;76:1183–1191. doi: 10.1016/0300-9084(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 32.Kittendorf JD, Barcomb LM, Nonekowski ST, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: Molecular mechanism and role of aspartate 89. Biochemistry. 2001;40:14123–14133. doi: 10.1021/bi0110589. [DOI] [PubMed] [Google Scholar]
- 33.Migawa MT, Hinkley JM, Hoops GC, Townsend LB. A two step synthesis of the nucleoside Q precursor 2-amino-5-cyanopyrrolo[2,3-d]pyrimidin-4-one (PreQ0) Synth. Commun. 1996;26:3317–3322. [Google Scholar]
- 34.Cheng CS, Hoops GC, Earl RA, Townsend LB. Synthesis of pyrrolo[2,3-d]pyrimidines that are structurally related to methylated guanosines from tRNA and the nucleoside Q analogs, PreQ(0) and PreQ(1) Nucleos Nucleot. 1997;16:347–364. [Google Scholar]
- 35.Durand JM, Okada N, Tobe T, Watarai M, Fukuda I, Suzuki T, Nakata N, Komatsu K, Yoshikawa M, et al. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (TGT) of Escherichia coli K-12. J. Bacteriol. 1994;176:4627–4634. doi: 10.1128/jb.176.15.4627-4634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Björk GR. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog. Nucleic Acids Res. Mol. Biol. 1995;50:263–338. doi: 10.1016/s0079-6603(08)60817-x. [DOI] [PubMed] [Google Scholar]
- 37.Qian QA, Curran JF, Bjork GR. The methyl group of the N-6-methyl-N-6-threonylcarbamoyladenosine in tRNA of Escherichia coli modestly improves the efficiency of the tRNA. J. Bacteriol. 1998;180:1808–1813. doi: 10.1128/jb.180.7.1808-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. J. Biol. Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- 39.Rordorff BF, Kearns DR. Nuclear Magnetic Resonance Investigation of the Base-Pairing Structure of Escherichia coli tRNATyr Monomer and Dimer Conformations. Biochemistry. 1976;15:3320–3330. doi: 10.1021/bi00660a024. [DOI] [PubMed] [Google Scholar]
- 40.Massenet S, Motorin Y, Lafontaine DLJ, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maden BEH. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 1990;39:241–300. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 42.Gu XR, Yu M, Ivanetich KM, Santi DV. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry. 1998;37:339–343. doi: 10.1021/bi971590p. [DOI] [PubMed] [Google Scholar]
- 43.Horne DA, Rood K, Levine E, Ofengand J. RNA substrate recognition by E-coli pseudouridine synthase (rluA) with dual substrate specificity. Abstr. Pap. Am. Chem. Soc. 1997;213:308. [Google Scholar]
- 44.Raychaudhuri S, Niu LH, Conrad J, Lane BG, Ofengand J. Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J. Biol. Chem. 1999;274:18880. doi: 10.1074/jbc.274.27.18880. [DOI] [PubMed] [Google Scholar]
- 45.Nonekowski ST, Garcia GA. tRNA Recognition by the E. coli TGT: the Role of U33 in U-G-U Sequence Recognition. RNA. 2001;7:1432–1441. [PMC free article] [PubMed] [Google Scholar]
- 46.Goodenough-Lashua DM, Garcia GA. tRNA-Guanine Transglycosylase from Escherichia coli: a Ping-Pong Kinetic Mechanism is Consistent with Nucleophilic Catalysis. Bioorg. Chem. 2003;31:331–344. doi: 10.1016/S0045-2068(03)00069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kittendorf JD, Sgraja T, Reuter K, Klebe G, Garcia GA. An essential role for aspartate 264 in catalysis by tRNA-guanine transglycosylase from Escherichia coli. J. Biol. Chem. 2003;278:42369–42376. doi: 10.1074/jbc.M304323200. [DOI] [PubMed] [Google Scholar]
- 48.Xie W, Liu XJ, Huang RH. Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate. Nat. Struct. Biol. 2003;10:781–788. doi: 10.1038/nsb976. [DOI] [PubMed] [Google Scholar]
- 49.Stengl B, Meyer EA, Heine A, Brenk R, Diederich F, Klebe G. Crystal Structures of tRNA-guanine Transglycosylase (TGT) in Complex with Novel and Potent Inhibitors Unravel Pronounced Induced-fit Adaptations and Suggest Dimer Formation Upon Substrate Binding. J. Mol. Biol. 2007 doi: 10.1016/j.jmb.2007.04.008. DOI:10.1016/j.jmb.2007.04.008. [DOI] [PubMed] [Google Scholar]