Abstract
Ribosomes must dissociate into subunits in order to begin protein biosynthesis. The enzymes that catalyze this fundamental process in eukaryotes remained unknown. Here, we demonstrate that eukaryotic translocase, eEF2, which catalyzes peptide elongation in the presence of GTP, dissociates yeast 80S ribosomes into subunits in the presence of ATP but not GTP or other nucleoside triphosphates. Dissociation was detected by light scattering or ultracentrifugation after the split subunits were stabilized. ATP was hydrolyzed during the eEF2-dependent dissociation, while a non-hydrolyzable analog of ATP was inactive in ribosome splitting by eEF2. GTP inhibited not only ATP hydrolysis but also dissociation. Sordarin, a fungal eEF2 inhibitor, averted the splitting but stimulated ATP hydrolysis. Another elongation inhibitor, cycloheximide, also prevented eEF2/ATP-dependent splitting, while the inhibitory effect of fusidic acid on the splitting was nominal. Upon dissociation of the 80S ribosome, eEF2 was found on the subunits. We propose that the dissociation activity of eEF2/ATP plays a role in mobilizing 80S ribosomes for protein synthesis during the shift up of physiological conditions.
INTRODUCTION
The dissociation of ribosomes into subunits is an indispensable event in translation. During the resting phase or under stress conditions, almost all of the ribosomes exist in the 80S form (1–3). However, when conditions favor growth, and the protein synthesis must ramp up again, the 80S ribosomes rapidly dissociate into subunits to enter the cycle of translation (4,5).
In bacteria, dissociation of free 70S ribosomes is catalyzed by elongation factor G (EF-G) and ribosome recycling factor (RRF), powered by GTP hydrolysis. This dissociation of 70S ribosomes by EF-G·GTP/RRF is transient and requires stabilization by initiation factor 3 (IF3) to keep the subunits apart (6). IF3 binds to the 30S subunit and prevents its re-association with the 50S subunit (7). IF3 by itself can split 70S ribosomes. However, this dissociation is extremely slow and less extensive in comparison with EF-G·GTP/RRF-induced ribosome splitting (8).
In eukaryotes, several translation initiation factors, such as eIF1A, eIF1 and eIF3, have been shown to possess ribosome anti-association/dissociation activity similar to that of bacterial IF3 (9–11). Furthermore, translation factor eIF6 has a strong ribosome anti-association activity (12,13). This factor participates in the biogenesis of the 60S subunit (14,15) and remains associated with it until the late event of the initiation step (16). However, the process of active dissociation of the 80S ribosome into its subunits, analogous to the EF-G·GTP/RRF-dependent splitting of 70S ribosomes, has not been described. Importantly, the homolog of RRF in eukaryotes appears to be localized in organelles, and is not found in the cytoplasm (17,18).
The eukaryotic elongation factor 2 (eEF2), like its bacterial homolog EF-G, catalyzes translocation of tRNA and mRNA in the presence of GTP (19). eEF2 is larger than EF-G (20) and, in contrast to EF-G, has a binding site specific for adenine nucleotides in addition to the binding site for GTP/GDP. Thus, it has been shown that eEF2 of yeast (21) and mammalian (22) cells can bind ATP or ADP in the absence of ribosomes and in the presence of GTP. The functional importance of the ATP/ADP-binding site of eEF2 remained unknown so far.
In this report, we describe a novel activity of eEF2, the dissociation of 80S ribosomes into subunits in the presence of ATP. This splitting is transient and dependent on the energy released by hydrolysis of ATP. The resulting subunits can then be stabilized either by adding the ribosome anti-association factor or by treating the ribosomes/eEF2/ATP reaction mixture with glutaraldehyde. GTP inhibits the eEF2/ATP-dependent dissociation of 80S ribosomes. We propose that eEF2/ATP-dependent ribosome splitting is involved in catalysis of 80S ribosome dissociation during the shift up at the onset of favorable physiological conditions.
MATERIALS AND METHODS
Buffers
Buffers were prepared from reagent grade chemicals: buffer 5/100 (20 mM HEPES-KOH, pH 7.6, 5 mM MgCl2, 100 mM KCl, 2 mM DTT); buffer 5/500 (buffer 5/100 with 500 mM KCl); buffer 2/150 (buffer 5/100 with 2 mM MgCl2 and 150 mM KCl); buffer 10/150 (buffer 5/100 with 10 mM MgCl2 and 150 mM KCl).
Purification of yeast ribosomes, eEF2 and recombinant eIF6
Yeast ribosomes, eEF2 and eIF6 were prepared as described in the Supplementary Data.
Sedimentation profiles of ribosomes
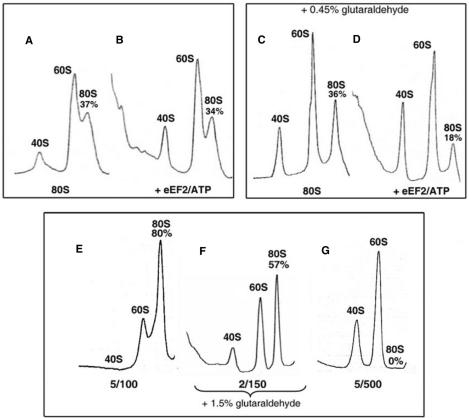
Prior to use, frozen ribosomes were incubated in buffer 2/150 for 5 min at 30°C, then sedimented at 4500 × g for 4 min at room temperature. Ribosomes were treated with or without eEF2, eIF6 and nucleotides in 60 µl of buffer 2/150, 5/100 or 5/500 as specified in the figure legends (or tables). Then the reaction mixtures were loaded on a 5–30% (w/v) sucrose density gradient prepared in the same buffers, and were sedimented for 2 h at 4°C (Beckman rotor SW50.1, 46500 r.p.m.). In Figure 2C and D, after incubation, the reaction mixtures were treated for 2 min on ice with a cold glutaraldehyde solution (final concentration 0.45%, v/v) prepared in 100 mM Tris-HCl (pH 7.6), 2 mM MgCl2 and 150 mM KCl. In Figure 2F, ribosomes in buffer 2/150 were treated with glutaraldehyde under identical conditions except that the final concentration of glutaraldehyde was 1.5% (v/v) and fixation was performed in the presence of 0.3 mg/ml bovine serum albumin. The sedimentation behavior of ribosomes was monitored using an ISCO UA-6 spectrophotometer at 254 nm. The percentage of 80S ribosomes was calculated from the areas corresponding to 40S, 60S and 80S ribosomes measured by ImageJ 1.31j software (Bethesda, USA).
Figure 2.
Splitting of 80S ribosomes by eEF2/ATP: sedimentation studies. (A and B) Ribosomes (0.05 μM) were incubated in buffer 2/150 alone (A) or with 1 µM eEF2 and 0.5 mM ATP (B) for 20 min at 30°C and subjected to SDGC. (C and D) Conditions in (C) and (D) were identical to (A) and (B), respectively, except that the reaction mixtures were treated with 0.45% glutaraldehyde (v/v) before SDGC. (E–G) The stock solution of ribosomes (0.5 μM in buffer 5/100) was diluted to a final concentration 0.05 μM in the buffers 5/100, 2/150 and 5/500. In buffer 2/150, ribosomes were treated with 1.5% glutaraldehyde (v/v). The ribosomes were then sedimented as in (A–D). The percentages of 80S ribosomes relative to the total ribosome content are indicated.
Light scattering analysis
The light scattering experiments were performed at room temperature with a spectrofluorometer (Photon Technology International, incoming slits: 1 mm × 0.1 mm, outgoing slits: 0.3 mm × 1 mm; wavelength: 436 nm, angle: 90°). ‘Factor mixture’ (180 μl) containing eEF2, eIF6, ATP and/or ADP was mixed manually with 20 μl of 0.5 μM 80S ribosomes or 0.25 μM of 40S or 60S subunits. In each case, the final concentrations of MgCl2 and KCl were adjusted to 2 and 150 mM, respectively. The resultant mixture (200 μl) was placed in a cuvette immediately and the intensity of the light scattering (counts per second, CPS) was continuously recorded without stirring, beginning at 20 s after the mixing. The light scattering was calculated as the percentage of the initial value (×10−2) at 20 s is expressed as %CPS. Apparent rate constants of ribosome splitting were obtained using Kyplot software (Tokyo, Japan) by fitting data to a double exponential equation.
Localization of eEF2 analyzed by western blotting
Ribosomes (0.05 µM) were incubated at 30°C in 80 µl of buffer 2/150 with 1 µM eEF2 alone or eEF2 with 0.5 mM ATP, GTP or a nonhydrolyzable analog of ATP, 5′-adenylyl-[β, γ-imido]diphosphate (ADPNP). After 15 min of incubation, the reaction mixtures were treated with 0.45% glutaraldehyde (see ‘Sedimentation profiles’ section). Gradients were fractionated from the bottom (10 drops per fraction) and the protein content in each fraction was precipitated by the addition of 100% TCA to a final concentration of 10%. Protein pellets were washed with a mixture of ether and ethanol (1:1) and resolved on 10% SDS-PAGE. eEF2 was detected by western blotting using rabbit antibodies against yeast eEF2 (dilution 1:20 000). The intensities of the bands corresponding to the antibody bound to eEF2 were determined using ImageJ 1.31j software and amounts of eEF2 were estimated using standard eEF2.
ATP hydrolysis assay
The release of inorganic phosphate from [γ-32P]ATP upon hydrolysis was analyzed as described (23). A standard reaction mixture (15 µl) contained 0.05 µM ribosomes, 2.5 µM eEF2 and 0.5 mM [γ-32P]ATP (specific activity 0.12 µCi/nmol). Incubation was at 30°C for 1, 2, 4, 6 and 10 min.
RESULTS
eEF2/ATP splits 80S ribosomes: detection by light scattering
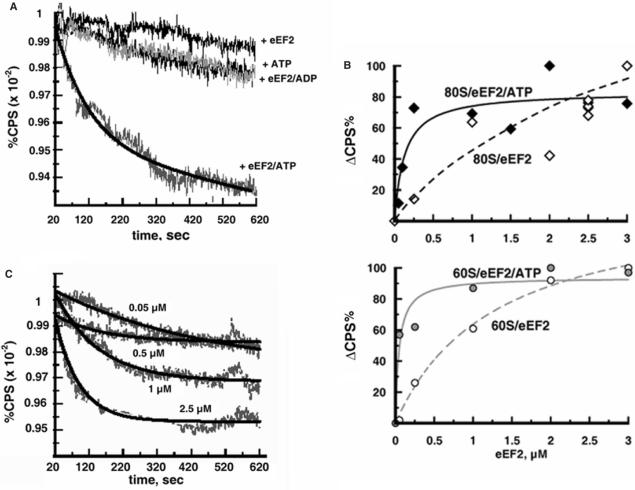
Light scattering of ribosomes decreases upon the dissociation into subunits because the 80S ribosome scatters more light than its subunits due to the larger size (8,24). As shown in Figure 1A, incubation of ribosomes with eEF2 and ATP caused a decrease in the scattered light (lower curve), suggesting that eEF2/ATP promoted splitting of the 80S ribosomes into subunits. In contrast, when ribosomes were incubated with eEF2 or ATP alone or eEF2 and ADP, the change in the light scattering was negligible during the incubation.
Figure 1.
eEF2 dissociates 80S ribosomes in the presence of ATP: light scattering analysis. (A) Ribosomes (0.05 μM) were incubated with 2.5 μM eEF2 and 0.5 mM ATP in buffer 2/150 containing 0.8% glycerol. In the control experiments, the ribosomes were incubated with ATP or eEF2 alone or with eEF2/ADP. The light scattering (CPS) is expressed as the percentage of the initial value at 20 s after mixing and is plotted against the time of incubation. (B) The instantaneous increase in light scattering (ΔCPS) due to the binding of eEF2 to ribosomes was measured at 20 s after mixing of the ribosomes and eEF2 (or eEF2/ATP). ΔCPS is expressed as the percentage of the maximum light scattering increase observed at the point of ribosome saturation with eEF2, and is plotted against the amount of eEF2 added. Upper panel: binding of eEF2 to 80S ribosomes with (closed diamonds) and without (open diamond) ATP. Lower panel: binding of eEF2 to 60S subunits with (gray circles) and without (open circles) ATP. Experimental conditions were the same as in (A). (C) As in (A) except that various amounts of eEF2 (in the absence of glycerol) were added as indicated.
Since the light scattering change of ribosomes is an indirect method of assessing ribosome splitting and could occur for a number of reasons, further experiments were required before a definite conclusion could be reached. As shown in Figure S1 (Supplementary Data), addition of eEF2 to ribosomes alone, or together with ATP, caused an instantaneous increase in the light scattering. A similar increase in light scattering of Artemia and mammalian ribosomes upon addition of eIF3 was reported (25) and used for studying the binding of the factor to ribosomes (26). Figure 1B shows the binding of eEF2 to the ribosomes measured by the light scattering increase. As can be seen, ATP stimulated the binding of eEF2 to 80S ribosomes or 60S subunits. The binding of eEF2 to 40S subunits with or without ATP was not detected (data not shown). The difference of the affinity of eEF2 to the ribosomes due to ATP, however, does not influence the light scattering results shown in Figure 1A, because 99% of ATP still remained at 10 min after the onset of the reaction (see Figure 4B). Therefore, it was concluded that the decrease in light scattering observed in Figure 1A indicates dissociation of 80S ribosomes into subunits.
Figure 4.
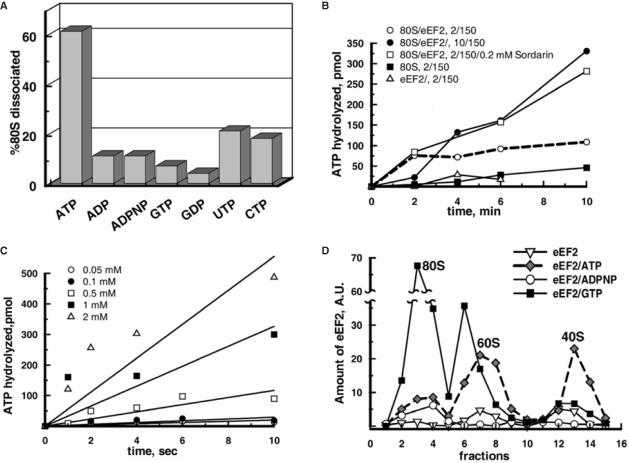
ATP but not other nucleotides are essential for the eEF2-dependent dissociation of 80S ribosomes and the binding of eEF2 to subunits: ATP hydrolysis during the splitting. (A) Ribosomes (0.05 μM) were pre-incubated in buffer 2/150 with 2.5 μM eEF2 and 0.5 mM of one of the specified nucleotides for 6 min at 30°C and then eIF6 was added to 2 μM and further incubated for 15 min. The reaction mixtures were analyzed by SDGC. The percentages of dissociation of 80S ribosomes were determined as in Figure 3A. Standard error was ±5%. (B) Reaction mixtures (15 µl) containing 0.05 μM 80S ribosomes and 2.5 μM eEF2 were incubated at 30°C with 0.5 mM [γ-32P]ATP in buffer as indicated. Sordarin was purchased from Sigma. (C) Ribosomes were incubated with eEF2 as in (B) with indicated amounts of [γ-32P]ATP in buffer 2/150. (D) Ribosomes (0.05 μM) were incubated with eEF2 (1 μM) and 0.5 mM nucleotide as indicated in buffer 2/150 followed by treatment with 0.45% glutaraldehyde (v/v) and SDGC. The presence of eEF2 in each fraction was estimated by western blotting. In (B) and (C), each curve represents the average of three independent experiments.
For determination of the extent of the ribosome splitting by eEF2/ATP, the light scattered by completely dissociated 80S ribosomes must be compared with that by ribosomes in the buffer containing 2 mM MgCl2 and 150 mM KCl (2/150), in which the splitting reaction took place (Figure 1A). Such comparison was performed as described in Figure S1 (Supplementary Data). These results indicated that 92% of the 80S ribosomes were converted into subunits by 2.5 µM eEF2 and ATP. It should be noted that the 80S preparation used had extra 60S subunits (see Figure 2E) because of the procedure of the 80S ribosomes purification. The presence of the excess of 60S subunits inhibits dissociation of 80S ribosomes (see Supplementary Data, Figure S3A). However, even under these conditions, 2.5 µM eEF2 almost completely converted the existing 80S ribosomes into subunits.
In the experiment described in Figure 1C, various amounts of eEF2 were added and the rate and extent of the ribosome splitting were determined. From these data, the Michaelis-Menten constant (KM) value was 0.24 ± 0.10 µM. This matches the estimated dissociation constant (Kd) value of eEF2 to 80S ribosomes in the presence of ATP (Figure 1B), which was 0.26 ± 0.04 µM.
Transient nature of eEF2/ATP-dependent splitting of 80S ribosomes: detection by sucrose density gradient ultracentrifugation (SDGC)
To confirm the eEF2/ATP-dependent splitting of 80S ribosomes deduced from the measurements of the light scattering change, we compared the sedimentation patterns of ribosomes incubated alone and with eEF2/ATP (Figure 2A and B). In the presence of eEF2/ATP, no significant dissociation of 80S ribosomes could be detected. These results are reminiscent of the finding that the transient splitting of bacterial 70S ribosomes by EF-G·GTP/RRF cannot be observed by the SDGC analysis (8). Therefore, these results suggest that the dissociation of 80S ribosomes by eEF2/ATP detected by the light scattering decreases (Figure 1A) is transient and that the separated subunits appear to re-associate during the SDGC analysis.
Even if the splitting of 80S ribosomes by eEF2/ATP is transient, it should still be detectable by SDGC upon stabilization of the dissociated subunits. In the experiment shown in Figure 2D, the reaction mixture containing 80S ribosomes, 1 µM eEF2 and ATP was treated with 0.45% glutaraldehyde. This treatment is known to fix eEF2 on the 80S ribosome (27) without influencing the ribosomal sedimentation pattern. Figure 2C is a control, where the ribosomes were treated with glutaraldehyde without eEF2/ATP. It is clear that upon incubation of ribosomes with eEF2/ATP, the apparent amount of 80S ribosomes decreased from 36 to 18% of the total ribosomes. This is equal to a 50% conversion of 80S ribosomes into subunits. The amounts of 80S ribosomes, 36 and 18% of the total ribosomes, actually correspond to 60 and 30% because of the induced dissociation of the 80S ribosomes during the ultracentrifugation analysis under the buffer conditions used (see the next section ‘Determination of …’). Therefore, the results shown in Figure 2D indicate that eEF2/ATP-dependent splitting of 80S ribosomes is indeed transient and can be observed by SDGC only following stabilization of eEF2 on the subunits.
It should be noted that the results obtained by the SDGC analysis correlate well with the light scattering data. Thus, when ribosomes were incubated with 1 µM eEF2 and ATP, the decrease of the light scattering was ∼50% of the signal recorded at 2.5 µM eEF2 (Figure 1C), which corresponds to almost complete dissociation of the 80S ribosomes (see Supplementary Data, Figure S1).
Determination of the quantity of 80S ribosomes converted into subunits
The experiments described in the preceding sections were carried out in buffer 2/150. These ionic conditions are close to the optimum for in vitro protein synthesis (28). To determine the amount of 80S ribosomes split under conditions 2/150 (Figure 1A), it was necessary to estimate the exact quantity of 80S ribosomes present in this buffer. As noted in Figure 2A, under these conditions only 37% of the total ribosomes were detected as the 80S form by SDGC. This value does not represent the actual amount of 80S ribosomes present in buffer 2/150 because of the well-known effect of ultracentrifugation—dissociating ribosomes into subunits (29).
In Figure 2E–G, the ribosomes were analyzed with SDGC under three ionic conditions 5/100, 2/150 and 5/500. In buffers 5/100 (tightly associated 80S ribosomes; Figure 2E) and 5/500 (complete dissociation into subunits; Figure 2G), analyses were performed without glutaraldehyde fixation. To avoid induced ribosome dissociation during centrifugation in buffer 2/150, the ribosomes in this buffer were fixed with 1.5% glutaraldehyde as described previously (30). The result showed that 57% of the ribosomes existed in the 80S form in buffer 2/150 (Figure 2F). It is noted that the fixation with this high concentration of glutaraldehyde cannot be used for detection of the eEF2/ATP activity because treatment of the ribosomes/eEF2 mixture with 1.5% glutaraldehyde produces artifacts presumably due to fixation of eEF2 to 80S ribosomes (data not shown).
The determination of the amount of 80S ribosomes in buffer 2/150 by two other methods, light scattering analysis and gel-filtration, revealed similar values (see Supplementary Data, Figure S2). It is worth mentioning here, that the obtained value of 60% of 80S ribosomes correlates well with the data of light scattering by Artemia ribosomes (31). In those experiments, it was shown that at 2 mM Mg2+, 65% of 80S ribosomes exist in equilibrium with ribosomal subunits. Therefore, under the experimental conditions used in Figure 1, the starting concentration of 80S ribosomes was estimated at 0.03 µM. Using this value and the calculated maximum velocity of the splitting reaction (Figure 1C), the turnover number (kcat) of the eEF2/ATP-dependent dissociation was estimated as 0.72 ± 0.10 min−1.
Ribosomes transiently split by eEF2/ATP can be stabilized by the ribosome anti-association factor
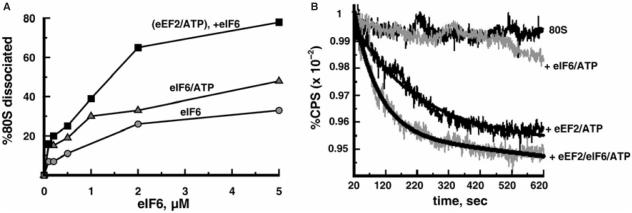
In the experiment described in Figure 3A, after the eEF2/ATP-dependent splitting of 80S ribosomes, increasing amounts of anti-association factor eIF6, which binds to 60S subunits (13,32), were added followed by the SDGC analysis. As can be seen, stable subunits were formed in an eIF6-dose dependent manner to the maximum of 80% conversion of the existing 80S ribosomes into subunits. This is somewhat lower than what we observed with the light scattering analysis, possibly, due to the partial release of the bound eIF6 during the ultracentrifugation. The control experiments where eEF2 or eEF2/ATP was omitted revealed noticeably less dissociation, indicating that eEF2 increases the extent of the 80S ribosome dissociation into subunits in the presence of anti-association factor eIF6.
Figure 3.
The 80S ribosomes transiently dissociated by eEF2/ATP are stabilized by eIF6. (A) Ribosomes (0.05 μM) were pre-incubated with 2.5 μM eEF2 and 0.5 mM ATP for 6 min at 30°C in buffer 2/150, then various amounts of eIF6 were added as indicated and incubated further for 15 min. The reaction mixtures were analyzed by SDGC. In the control experiments, ribosomes were exposed for 15 min to eIF6 or eIF6/ATP. Percentages of 80S ribosomes dissociated (Z) were calculated as follows: Z = 100 × (1 − (Y/W)), where (Y) is the amount of 80S ribosomes expressed as a percentage of total ribosomes remaining after the addition of factors and (W) is the amount of 80S ribosomes (percentage of total ribosomes) without the addition of factors. (B) Ribosomes (0.05 µM) were mixed with factors as indicated in buffer 2/150 containing 0.8% glycerol and the light scattering change was measured as in Figure 1A. Final concentrations of eEF2, eIF6 and ATP were 2.5 μM, 2.5 μM and 0.5 mM, respectively.
The effect of eIF6 on the eEF2/ATP-dependent dissociation of 80S ribosomes was confirmed by light scattering measurements of ribosomes as described in Figure 3B. This figure shows that when eIF6 was added, the initial rate and the final value of the eEF2/ATP-dependent dissociation of 80S ribosomes were increased. We should emphasize that the rate of dissociation by eIF6/ATP alone is much slower than that by eEF2/ATP. These results indicate that the dissociation is performed by eEF2/ATP, which plays the major role in facilitating the splitting of 80S ribosomes, but not by eIF6, which stabilizes dissociated subunits.
Energy dependence and nucleotide specificity of eEF2-dependent 80S ribosome dissociation
In Figure 4A, ATP was substituted by its non-hydrolyzable analog ADPNP or ADP and the reaction mixtures were analyzed by SDGC as described for Figure 3A. The results showed that incubation of ribosomes with eEF2/ADPNP or eEF2/ADP did not lead to the splitting of 80S ribosomes. In the presence of other nucleotides, the splitting did not occur either (Figure 4A, see results for GTP or GDP) or was much less than with ATP (Figure 4A, see results for UTP or CTP). This indicates that the dissociation of 80S ribosomes by eEF2 is an energy-dependent process specifically involving ATP.
The fact that the non-hydrolyzable ATP analog was inactive in the splitting of 80S ribosomes strongly suggested that ribosome dissociation is accompanied by the hydrolysis of ATP. This is shown in Figure 4B, where [γ-32P]ATP was converted to inorganic phosphate (32Pi) during the dissociation reaction (open circles). When incubated separately, eEF2 or ribosomes alone hydrolyzed much less ATP (open triangles and closed squares, respectively). As can be noted, the ribosome/eEF2-dependent hydrolysis of ATP in buffer 2/150 almost leveled off at 2 min (Figure 4B, dashed line). Meanwhile, the splitting reaction went on up to 10 min (see Figure 1A). The mechanism of such slow down of the ATPase reaction is not clear. Since the molar concentration of ATP is far higher than that of ribosomes in the ATPase reaction mixture, the background level of ATP hydrolysis must be sufficient for the ribosome splitting. It is possible that the major activity of ATP hydrolysis takes place upon binding of eEF2 to 80S ribosomes. However, after most ribosomes are split, the hydrolysis subsides.
When 80S ribosomes and eEF2 were incubated in the buffer with 10 mM MgCl2 (i.e. under the conditions where the splitting of 80S ribosomes by eEF2/ATP does not occur; data not shown and see Supplementary Data, Figure S3C), hydrolysis of ATP was considerably higher than in buffer 2/150 (Figure 4B, closed circles). This indicates that ATP hydrolysis is not always coupled with the splitting reaction. Importantly, sordarin, which is inhibitory to the dissociation of 80S ribosomes (Table 2), stimulated the ATPase reaction significantly (open squares) indicating again that the ATPase activity is not entirely coupled with the ribosome splitting.
Table 2.
Effect of elongation inhibitors and polyamines on the eEF2/ATP-dependent 80S ribosome dissociation
| Inhibitor added | mM | 80S ribosome dissociated, % | Inhibition, % |
|---|---|---|---|
| None | – | 76 | 0 |
| Cycloheximide | 0.2 | 24 | 68 |
| Sordarin | 0.2 | 14 | 82 |
| Fusidic acid | 0.2 | 68 | 11 |
| Paromomycin | 0.2 | 0* | 100 |
| Spermidine | 0.4 | 24 | 68 |
| Putrescine | 2 | 68 | 11 |
Ribosomes (0.05 µM) in buffer 2/150 were pre-incubated with eEF2 (2.5 µM), ATP (0.5 mM) and one of the indicated inhibitors for 6 min at 30°C, then eIF6 was added to 2.5 µM. Incubation was continued for another 15 min. The reaction mixtures were analyzed by SDGC and the percentage of dissociation of 80S ribosomes was estimated as described for Figure 4A. (*), in the presence of paromomycin, the amount of 80S ribosomes was 1.4 times more than that in the control without any inhibitor. Standard error was ±3%.
When various concentrations of ATP were added to ribosomes and eEF2 under the dissociation conditions, the rate of the 32Pi release increased at higher concentrations of ATP (Figure 4C). From these data, the turnover number of ribosome/eEF2 ATPase was estimated as 40.2 ± 4 min−1, which is significantly higher than the kcat value described above of the splitting reaction catalyzed by eEF2/ATP (0.72 ± 0.1 min−1).
Binding of eEF2 to the subunits during ribosome splitting in the presence of ATP
To better understand the mechanism of the 80S ribosome dissociation by eEF2/ATP, the binding of eEF2 to subunits during the splitting reaction was analyzed. The ribosomes were incubated under the same conditions as shown in Figure 2D (with 0.45% glutaraldehyde fixation), where 50% of the existing 80S ribosomes were converted to subunits (Figure 4D). This condition was specifically chosen to examine the presence of eEF2 on the remaining 80S ribosomes in the presence of ATP. In parallel, the ribosomes were incubated with eEF2 alone or with GTP or ADPNP. The results showed (Figure 4D) that in the presence of ATP, the binding of eEF2 to the 80S ribosomes is significantly less than that to the subunits. In contrast, GTP strongly stimulated the binding of eEF2 to the 80S ribosome. In the presence of ADPNP, a small amount of eEF2 was localized only on the 80S ribosomes suggesting that the binding of eEF2 to subunits observed in the presence of ATP is related to the splitting reaction. It should be noted that with ATP, eEF2 was found not only on 60S subunits but also on 40S subunits (Figure 4D, diamonds). On the other hand, we did not observe the binding of eEF2 to isolated 40S subunits by the light scattering technique (data not shown). Hence, further studies will be necessary to assess the significance of the binding of eEF2 to 40S subunits under the splitting conditions.
Inhibition of eEF2/ATP-dependent splitting of 80S ribosomes
As pointed out in the preceding section, GTP stimulated binding of eEF2 more to the 80S ribosomes than to the subunits (Figure 4D, squares). On the contrary, ATP stimulated the eEF2 binding mostly to the subunits. These results suggest that the effect of eEF2/GTP on the ribosomes is different from that of eEF2/ATP. Therefore, it was important to examine the effect of GTP on the splitting and the ATPase activity. The results are shown in Table 1. With increasing concentrations of GTP, the eEF2/ATP-dependent ribosome splitting as well as the ATPase activity was progressively inhibited. The inhibition of the ATPase activity was more efficient than that of the dissociation reaction.
Table 1.
Inhibitory effect of GTP on the eEF2/ATP-dependent splitting of 80S ribosomes and the ATPase activity
| eEF2/ATP-catalyzed reaction | GTP, mM | ||||
|---|---|---|---|---|---|
| 0 | 0.005 | 0.01 | 0.025 | 0.05 | |
| (A) 80S ribosome dissociated,% | 76 | 67 | 57 | n.d. | 44 |
| % inhibition | 0 | 12 | 25 | n.d. | 42 |
| (B) ATP hydrolyzed, pmol | 109 | 61 | 40 | 19 | 11 |
| % inhibition | 0 | 44 | 63 | 83 | 90 |
(A) Ribosomes (0.05 µM) in buffer 2/150 were pre-incubated with 2.5 µM eEF2, 0.5 mM ATP and indicated amounts of GTP for 6 min at 30°C, then eIF6 was added to the final concentration 2.5 µM and the incubation continued for 15 min. The reaction mixtures were analyzed by SDGC as described for Figure 4A. Standard error was ± 3%. (B) The indicated amounts of GTP were added and hydrolysis of [γ-32P]ATP by ribosomes/eEF2 was estimated after 10 min and expressed as total pmoles of ATP hydrolyzed. Each value represents the average of three independent experiments; n.d.—not determined.
Various agents known to inhibit the conventional eEF2 activity were tested on the dissociation reaction catalyzed by eEF2/ATP (Table 2). Cycloheximide and sordarin at 0.2 mM inhibited more than 60%, while fusidic acid at the same concentration inhibited only 11%. In the presence of the aminoglycoside paromomycin, which prevents EF-G·GTP/RRF-dependent splitting of 70S ribosomes (33), the dissociation of 80S ribosomes by eEF2/ATP was completely prevented and the amount of 80S ribosomes increased 40% over the initial amount of 80S ribosomes.
Since the 80S ribosome is in equilibrium with its subunits, conditions and factors that influence this equilibrium should have an effect on the eEF2/ATP-dependent splitting (see Supplementary Data). Polyamines, which stimulate association of the subunits, are expected to inhibit the dissociation of the 80S ribosomes by eEF2/ATP. However, as it is shown in Table 2, in the presence of spermidine at a concentration higher than the in vivo concentration of this polyamine (34), the eEF2/ATP-dependent splitting still occurs. Furthermore, putrescine at a concentration stimulatory for in vitro protein synthesis (35), had very little effect on dissociation.
Because magnesium ions have a similar effect as polyamines, shifting the equilibrium towards the 80S ribosomes, the increase in Mg2+ led to the gradual inhibition of ribosome splitting by eEF2/ATP (Supplementary Data, Figure S3C). Thus, at 4 mM Mg2+ only 10% of the existing 80S ribosomes were split. However, it should be pointed out that at this Mg2+ concentration, in vitro protein synthesis is very inefficient (28). As can be noted from Figure S3B, in the presence of higher concentrations of ribosomes, less dissociation was observed. At the same time, when the eEF2 concentration was increased, the splitting of ribosomes by eEF2/ATP was significantly restored even at a high ribosome concentration (Figure S3B).
DISCUSSION
Biological significance of the eEF2/ATP-dependent splitting of 80S ribosomes
There are physiological and genetic data consistent with the present finding that eEF2 may play an essential role in the dissociation of free 80S ribosomes in cells. Under stress conditions (3,4) or during resting phase (2), the ribosome pool is primarily preserved in the 80S form. Nutrient depletion stress dramatically increases free 80S ribosomes, and decreases the in vivo ATP concentration (4). In a separate experiment, the depletion of ATP elevated phosphorylation of eEF2 (36), which inactivates the factor (37). Similarly, the expression of a non-functional form of eEF2 (mutations H699K or V488A) led to an increase in 80S monomers (38). All of these data point to the fact that inactivation of eEF2 with simultaneous decrease of ATP results in the accumulation of the ribosomes in the 80S form. Conversely, when the conditions are favorable for cell growth and there is enough ATP, eEF2 is activated, resulting in the rapid conversion of the accumulated 80S ribosomes to polysomes (4,5).
The translation factors eIF3, eIF1A and eIF6 have been reported to dissociate vacant 80S ribosomes into their subunits (9,10,13). We propose that eEF2, but not the these factors, plays the active catalytic role in the splitting of 80S ribosomes for the following six reasons. First, eIF3 is an anti-association factor that exerts its activity from the solvent side of the 40S subunit, preventing subunits joining (39,40). In contrast, eEF2 translocates tRNA and mRNA through the inter-subunit space of the 80S ribosome (41). The binding site of eEF2 is located mostly in the inter-subunit space of the 80S ribosome with domain IV contacting the broadest inter-subunit bridge B2a (42–44). This makes eEF2 more suitable than eIF3 to get into the inter-subunits space to split the ribosome. Second, the cellular content of a core subunit of eIF3 (eIF3g) is less than the in vivo ribosome concentration (45). There are only 2400 molecules of eIF3i, which is also a core subunit of eIF3, per yeast cell, but 78 100 molecules of eEF2 per cell (http://www.yeastgenome.org/). Third, the rate of the binding of eIF3 to the 40S subunit is 12-times faster than that to the 80S ribosome (26) suggesting that the main target of eIF3 is the 40S subunit, not the 80S ribosome. Fourth, recent evidence indicates that eIF3 splits 80S ribosomes into subunits only in the presence of polyuridylic acid (46). Fifth, the dissociation activity of eIF1A is weaker than that of eIF3 (10). Finally, the speed of the 80S ribosome dissociation by eIF6 is incomparably slower than that by eEF2/ATP (Figure 3B).
The kinetic parameters of the eEF2/ATP-dependent dissociation are consistent with the proposed physiological function. ATP stimulates binding of eEF2 to 80S ribosomes (27). We estimated that KM of eEF2/ATP for the 80S ribosome is 0.2 µM, which is comparable with Kd of eEF2 for the 80S ribosome in the post-translocation state in the presence of GDP (0.4 µM) or a non-hydrolyzable analog of GTP (0.3 µM) (47). The turnover number (kcat) of eEF2/ATP for the 80S ribosome splitting is ∼0.7 min−1. This is comparable with the turnover number of the peptide bond formation in the yeast in vitro system (2 min−1) (48). Due to the re-association of the split subunits into 80S ribosomes under our dissociation condition, the calculated kcat value is much smaller than the actual turnover number of the splitting in vivo where the formed subunits are rapidly ‘removed’ by the initiation step. In addition, our kcat value of the ribosome/eEF2 ATPase activity (40 min−1) is 4 times faster than kcat of the GTPase activity of the ribosome/eEF2 complex (9.6 min−1) (49). Furthermore, in yeast, all ribosomes accumulate in the 80S form under conditions of glucose depletion. However, they are completely converted to polysomes within 10 min after a shift up of the culture conditions to normal glucose levels (4). This can be explained on the basis of the known cellular content of ribosomes (45) and eEF2, with the kcat value we estimated for the eEF2/ATP-dependent 80S ribosome splitting.
One may argue that the spontaneous dissociation of 80S ribosomes into subunits under the optimum ionic conditions can be sufficient for the splitting, especially under the conditions where the formed subunits are efficiently ‘utilized’ for the initiation step. However, the experiment shown in Figure 3B demonstrated that combining the spontaneously produced 60S subunits with eIF6, which prevents ribosome re-association, is not sufficient for rapid completion of 80S ribosome dissociation. The rate of the splitting by eIF6 is extremely slow compared with that of the splitting by eEF2/ATP.
The possibility that eEF2/ATP-dependent splitting produces inactive subunits is highly unlikely. First, eEF2 and ATP are present in vivo; it is difficult to imagine that these natural components might harm the ribosomes. Second, we have shown that the 60S subunits formed by eEF2/ATP are capable of binding eIF6 (Figure 3A). Third, the split subunits can re-associate to form 80S ribosomes (Figure 2A and B). However, despite these facts and the evidences cited above, the idea that the eEF2/ATP-dependent splitting of the 80S ribosomes is a part of the utilization process of the dormant ribosomes should be tested by further experiments indicating that the formed subunits can engage in the initiation and elongation phases. Until this is accomplished, our proposal will remain a hypothesis.
The dissociation of 70S ribosomes of prokaryotes is catalyzed by EF-G, GTP and RRF (8,50,51). It is therefore reasonable to expect that eEF2, the eukaryotic homolog of EF-G, can dissociate 80S ribosomes. In a similar manner to prokaryotic IF3, eukaryotic factors (eIF3, eIF1 (11), eIF1A or eIF6) can stabilize the ribosome subunits once they are separated by eEF2/ATP. In the experiment described in Figure 3, we used eIF6 (a strong ribosome anti-association factor (13)) as a convenient tool to keep apart the subunits formed by the eEF2/ATP-dependent reaction. Which factors play this role in vivo is a subject of a separate study.
Since the EF-G·GTP/RRF-dependent splitting of 70S ribosomes is a part of the recycling step of the protein synthesis in bacteria, it is quite possible that the eEF2/ATP-dependent dissociation of 80S ribosomes is a part of the eukaryotic ribosome-recycling step. Our preliminary data obtained on yeast model post-termination complexes [puromycin-treated polysomes (52)] strongly support this possibility. However, we should emphasize that in this article we do not intend to implicate the eEF2/ATP-dependent splitting of 80S ribosomes as a part of the eukaryotic recycling system until further solid evidence for this idea becomes available.
Energy requirement for the splitting of 80S ribosomes
In our earlier studies of the dissociation of bacterial ribosomes, we showed that the splitting of vacant 70S ribosomes is dependent on GTP hydrolysis (8). In analogy with this process, in the present article, it is demonstrated that the dissociation of the eukaryotic ribosome is dependent on the energy released upon hydrolysis of ATP. The specific involvement of ATP was shown by the inability of GTP, CTP and UTP to split 80S ribosomes (Figure 4A) and the ATPase activity of the ribosome/eEF2 complex. Sordarin stimulated the ATPase activity (Figure 4B), while it inhibited the dissociation (Table 2). The stimulatory effect of sordarin on the ATPase is of interest in view of the finding that this antibiotic apparently competes with ATP (or ADP) for binding to eEF2 in the solution (21). Our results indicate that sordarin does not inhibit ATP binding to the complex of the 80S ribosome and eEF2. Cryo-EM studies of the stalled complex of yeast 80S ribosome/eEF2/sordarin suggested that sordarin restricts conformational changes of eEF2 by blocking rotation of domains III, IV and V of the factor (44). It is possible that similar conformational changes are the key movements for the splitting activity of eEF2 in the presence of ATP, which would explain the inhibitory effect of sordarin on ribosome dissociation. The seemingly contradictory effects of sordarin (i.e. inhibition of the splitting and stimulation of the ATPase activity) are reminiscent of the similar effect of sordarin on the GTPase of the post-translocation form of 80S ribosome/eEF2 complex and the translocation (53).
eEF2 exerts different effects on the 80S ribosome depending on the bound ligand, ATP or GTP
From the data presented in this study, it appears that eEF2/GTP exerts a different effect on ribosomes from that of eEF2/ATP. How does normal peptide elongation take place in the presence of the eEF2/ATP-dependent dissociation of 80S ribosomes? During the elongation step, the splitting reaction cannot occur because of the stabilizing effect of mRNA and peptidyl-tRNA on the ribosome. In addition, the affinity of eEF2/GTP to the pre-translocation ribosome is far greater than that of eEF2/ATP to the ribosome.
We showed that GTP inhibited the ATP-dependent splitting and the ATPase activity (Table 1). How then can eEF2/ATP mobilize 80S ribosomes in the presence of GTP? We assume that most of the free ribosomes in vivo are in the post-translocation state. The affinity of eEF2/ATP and that of eEF2/GTP to these ribosomes are approximately the same [Figure 1B and (47)]. During shift up conditions, the concentration of ATP is ∼6-fold higher than that of GTP (54). The subunits produced by eEF2/ATP-dependent dissociation are rapidly utilized for the initiation step, driving the splitting reaction to the right by depleting the products. These considerations led us to suggest that the mobilization of the vacant 80S ribosomes by eEF2/ATP can occur in vivo.
There are a number of differences in the effect of ATP and GTP on eEF2 action on the ribosome. First, fusidic acid facilitates ribosome·eEF2/GTP complex formation and keeps eEF2 bound to the ribosome (55), thereby stimulating subunits association activity by eEF2 (unpublished data). In contrast, the effect of fusidic acid on the eEF2/ATP-dependent splitting is nominal (Table 2). Second, the binding site for ATP on eEF2 is different from that for GTP (21,22). Depending on the binding site, nucleoside triphosphates exert different structural conformational changes on eEF2. In support of this hypothesis, GTP inhibited eEF2/ATP-dependent dissociation, as well as ATPase activity (Table 1). Third, GTP hydrolysis by ribosome/eEF2 is assumed to take place at the binding site of GTP on eEF2 (56), but ribosomes may play a larger role in the hydrolysis of ATP during the dissociation by eEF2. It is known that the 80S ribosome possesses intrinsic ATPase activity (57,58), which is higher than the intrinsic GTPase activity. This ATPase is presumably associated with the 5S RNP complex and is stimulated by eEF2 (59). The central protuberance, which involves the 5S RNP complex and forms B1a and B1b/c inter-subunit bridges (43), undergoes substantial conformational changes upon binding of eEF2/sordarin (44). These data suggest that eEF2/ATP-dependent splitting of the 80S ribosome might be triggered by the conformational change of the central protuberance of the 60S subunit.
It is known that ATP is required for the initiation (60,61) and elongation (62–64) steps of polypeptide synthesis. In this study, we show that ATP is also consumed for splitting of 80S ribosomes into subunits, which may be important for utilization of 80S ribosomes for the initiation step. Thus, in contrast to prokaryotic protein synthesis, ATP plays critical roles in eukaryotic translation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr T.G. Kinzy (Robert Wood Johnson Medical School, USA) for generously providing us with yeast strain TKY675 for the eEF2 purification and antiserum against eEF2, Dr F. Fasiolo (Institut de Biologie Moleculaire et Cellulaire du CNRS, France) for the plasmid pGEX-4T-3 carrying the TIF6 gene. We would like to thank Dr Charles P. Scott (Thomas Jefferson University, USA) for the use of the spectrofluorometer. We are grateful to Dr C. Yanofsky (Stanford University, USA), Dr M. Kiel (Marywood University, USA), Dr E. Wickstrom and Dr N. Iwakura (Thomas Jefferson University, USA) for helpful comments on the manuscript. This work was supported by NIH grant GM60429 (A.K.) and by the Nippon Paint Research Fund (H.K.). Funding to pay the Open Access publication charges for this article was provided by A.K. and H.K.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eliasson E, Bauer GE, Hultin T. Reversible degradation of polyribosomes in Chang cells cultured in a glutamine-deficient medium. J. Cell. Biol. 1967;33:287–297. doi: 10.1083/jcb.33.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frerichs KU, Smith CB, Brenner M, DeGracia DJ, Krause GS, Marrone L, Dever TE, Hallenbeck JM. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc. Natl Acad. Sci. USA. 1998;95:14511–14516. doi: 10.1073/pnas.95.24.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uesono Y, Toh EA. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- 4.Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 6.Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A. The ribosome recycling step: consensus or controversy? Trends Biochem. Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Kaempfer R. Initiation factor IF-3: a specific inhibitor of ribosomal subunit association. J. Mol. Biol. 1972;71:583–598. doi: 10.1016/s0022-2836(72)80025-1. [DOI] [PubMed] [Google Scholar]
- 8.Hirokawa G, Nijman RM, Raj VS, Kaji H, Igarashi K, Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson HA, Sadnik I, Scheinbuks J, Moldave K. Studies on native ribosomal subunits from rat liver. Purification and characterization of a ribosome dissociation factor. Biochemistry. 1977;16:2221–2230. doi: 10.1021/bi00629a028. [DOI] [PubMed] [Google Scholar]
- 10.Thomas A, Goumans H, Voorma HO, Benne R. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur. J. Biochem. 1980;107:39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- 11.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell DW, Spremulli LL. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J. Biol. Chem. 1979;254:8796–8800. [PubMed] [Google Scholar]
- 13.Valenzuela DM, Chaudhuri A, Maitra U. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6) J. Biol. Chem. 1982;257:7712–7719. [PubMed] [Google Scholar]
- 14.Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, et al. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 15.Basu U, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. Release of eIF6(p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 17.Rolland N, Janosi L, Block MA, Shuda A, Teyssier E, Miege C, Cheniclet C, Carde J, Kaji A, et al. Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl Acad. Sci. USA. 1999;96:5464–5469. doi: 10.1073/pnas.96.10.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teyssier E, Hirokawa G, Tretiakova A, Jameson B, Kaji A, Kaji H. Temperature sensitive mutation in yeast mitochondrial ribosome recycling factor (RRF) Nucleic Acids Res. 2003;31:4218–4226. doi: 10.1093/nar/gkg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skogerson L, Moldave K. Evidence for aminoacyl-tRNA binding, peptide bond synthesis, and translocase activities in the aminoacyl transfer reaction. Arch. Biochem. Biophys. 1968;125:497–505. doi: 10.1016/0003-9861(68)90607-3. [DOI] [PubMed] [Google Scholar]
- 20.Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 21.Soe R, Mosley RT, Justice M, Nielsen-Kahn J, Shastry M, Merrill AR, Andersen GR. Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2. J. Biol. Chem. 2007;282:657–666. doi: 10.1074/jbc.M607830200. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalo P, Sontag B, Lavergne JP, Jault JM, Reboud JP. Evidence for a second nucleotide binding site in rat elongation factor eEF-2 specific for adenylic nucleotides. Biochemistry. 2000;39:13558–13564. doi: 10.1021/bi000896k. [DOI] [PubMed] [Google Scholar]
- 23.Raj VS, Kaji H, Kaji A. Interaction of RRF and EF-G from E. coli and T. thermophilus with ribosomes from both origins-insight into the mechanism of the ribosome recycling step. RNA. 2005;11:275–284. doi: 10.1261/rna.7201805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godefroy-Colburn T, Wolfe AD, Dondon J, Grunberg-Manago M. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol. 1975;94:461–478. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- 25.Goss DJ, Rounds D, Harrigan T, Woodley CL, Wahba AJ. Effects of eukaryotic initiation factor 3 on eukaryotic ribosomal subunit equilibrium and kinetics. Biochemistry. 1988;27:1489–1494. doi: 10.1021/bi00405a014. [DOI] [PubMed] [Google Scholar]
- 26.Goss DJ, Rounds DJ. A kinetic light-scattering study of the binding of wheat germ protein synthesis initiation factor 3 to 40S ribosomal subunits and 80S ribosomes. Biochemistry. 1988;27:3610–3613. doi: 10.1021/bi00410a012. [DOI] [PubMed] [Google Scholar]
- 27.Nygard O, Nilsson L. Nucleotide-mediated interactions of eukaryotic elongation factor EF-2 with ribosomes. Eur. J. Biochem. 1984;140:93–96. doi: 10.1111/j.1432-1033.1984.tb08070.x. [DOI] [PubMed] [Google Scholar]
- 28.Shenvi CL, Dong KC, Friedman EM, Hanson JA, Cate JH. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations – implications for the study of ribosome dynamics. RNA. 2005;11:1898–1908. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Infante A, Baierlein R. Pressure-induced dissociation of sedimenting ribosomes: effect on sedimentation patterns. Proc. Natl Acad. Sci. USA. 1971;68:1780–1785. doi: 10.1073/pnas.68.8.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian AR. Glutaraldehyde fixation of ribosomes. Its use in the analysis of ribosome dissociation. Biochemistry. 1972;11:2710–2714. doi: 10.1021/bi00764a025. [DOI] [PubMed] [Google Scholar]
- 31.Goss DJ, Harrigan T. Magnesium ion dependent equilibria, kinetics, and thermodynamic parameters of Artemia ribosome dissociation and subunit association. Biochemistry. 1986;25:3690–3695. doi: 10.1021/bi00360a032. [DOI] [PubMed] [Google Scholar]
- 32.Groft CM, Beckmann R, Sali A, Burley SK. Crystal structures of ribosome anti-association factor IF6. Nat. Struct. Biol. 2000;7:1156–1164. doi: 10.1038/82017. [DOI] [PubMed] [Google Scholar]
- 33.Hirokawa G, Kaji H, Kaji A. Inhibition of anti-association activity of translation initiation factor 3 by Paromomycin. Antimicrob. Agents Chemother. 2007;51:175–180. doi: 10.1128/AAC.01096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Comm. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 35.Tuite MF, Plesset J, Moldave K, McLaughlin CS. Faithful and efficient translation of homologous and heterologous mRNAs in an mRNA-dependent cell-free system from Saccharomyces cerevisiae. J. Biol. Chem. 1980;255:8761–8766. [PubMed] [Google Scholar]
- 36.McLeod LE, Proud CG. ATP depletion increases phosphorylation of elongation factor eEF2 in adult cardiomyocytes independently of inhibition of mTOR signalling. FEBS Lett. 2002;531:448–452. doi: 10.1016/s0014-5793(02)03582-2. [DOI] [PubMed] [Google Scholar]
- 37.Carlberg U, Nilsson A, Nygard O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- 38.Ortiz PA, Kinzy TG. Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res. 2005;33:5740–5748. doi: 10.1093/nar/gki882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol. 1992;226:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- 40.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 41.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 42.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 43.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae – tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 44.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von der Haar T, McCarthy JE. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 46.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nygard O, Nilsson L. Characterization of the ribosomal properties required for formation of a GTPase active complex with the eukaryotic elongation factor 2. Eur. J. Biochem. 1989;179:603–608. doi: 10.1111/j.1432-1033.1989.tb14589.x. [DOI] [PubMed] [Google Scholar]
- 48.Dresios J, Panopoulos P, Suzuki K, Synetos D. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 2003;278:3314–3322. doi: 10.1074/jbc.M207533200. [DOI] [PubMed] [Google Scholar]
- 49.Nygard O, Nilsson L. Kinetic determination of the effects of ADP-ribosylation on the interaction of eukaryotic elongation factor 2 with ribosomes. J. Biol. Chem. 1990;265:6030–6034. [PubMed] [Google Scholar]
- 50.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Hirashima A, Kaji A. Factor dependent breakdown of polysomes. Biochem. Biophys. Res. Commun. 1970;41:877–883. doi: 10.1016/0006-291x(70)90165-8. [DOI] [PubMed] [Google Scholar]
- 53.Dominguez JM, Gomez-Lorenzo MG, Martin JJ. Sordarin inhibits fungal protein synthesis by blocking translocation differently to fusidic acid. J. Biol. Chem. 1999;274:22423–22427. doi: 10.1074/jbc.274.32.22423. [DOI] [PubMed] [Google Scholar]
- 54.Ditzelmuller G, Wohrer W, Kubicek CP, Rohr M. Nucleotide pools of growing, synchronized and stressed cultures of Saccharomyces cerevisiae. Arch. Microbiol. 1983;135:63–67. doi: 10.1007/BF00419484. [DOI] [PubMed] [Google Scholar]
- 55.Justice MC, Hsu MJ, Tse B, Ku T, Balkovec J, Schmatz D, Nielsen J. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 1998;273:3148–3151. doi: 10.1074/jbc.273.6.3148. [DOI] [PubMed] [Google Scholar]
- 56.Nilsson L, Nygard O. Affinity labelling of the eukaryotic elongation factor EF-2 with the guanosine nucleotide analogue 5′-p-fluorosulfonylbenzoylguanosine. Biochim. Biophys. Acta. 1984;782:49–54. doi: 10.1016/0167-4781(84)90105-2. [DOI] [PubMed] [Google Scholar]
- 57.Rodnina MV, Serebryanik AI, Ovcharenko GV, El'Skaya AV. ATPase strongly bound to higher eukaryotic ribosomes. Eur. J. Biochem. 1994;225:305–310. doi: 10.1111/j.1432-1033.1994.00305.x. [DOI] [PubMed] [Google Scholar]
- 58.Kovalchuke O, Chakraburtty K. Comparative analysis of ribosome-associated adenosinetriphosphatase (ATPase) from pig liver and the ATPase of elongation factor 3 from Saccharomyces cerevisiae. Eur. J. Biochem. 1994;226:133–140. doi: 10.1111/j.1432-1033.1994.tb20034.x. [DOI] [PubMed] [Google Scholar]
- 59.Ogata K, Terao K, Uchiumi T. Stimulation by aminoacyl-tRNA of the GTPase and ATPase activities of rat liver 5S RNA protein particles in the presence of EF-2. J. Biochem. (Tokyo) 1980;87:517–524. doi: 10.1093/oxfordjournals.jbchem.a132773. [DOI] [PubMed] [Google Scholar]
- 60.Kozak M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- 61.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamath A, Chakraburtty K. Role of yeast elongation factor 3 in the elongation cycle. J. Biol. Chem. 1989;264:15423–15428. [PubMed] [Google Scholar]
- 63.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 64.El'Skaya AV, Ovcharenko GV, Palchevskii SS, Petrushenko ZM, Triana-Alonso FJ, Nierhaus KH. Three tRNA binding sites in rabbit liver ribosomes and role of the intrinsic ATPase in 80S ribosomes from higher eukaryotes. Biochemistry. 1997;36:10492–10497. doi: 10.1021/bi970631e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.