Abstract
Internal ribosome entry sites (IRES) allow ribosomes to be recruited to mRNA in a cap-independent manner. Some viruses that impair cap-dependent translation initiation utilize IRES to ensure that the viral RNA will efficiently compete for the translation machinery. IRES are also employed for the translation of a subset of cellular messages during conditions that inhibit cap-dependent translation initiation. IRES from viruses like Hepatitis C and Classical Swine Fever virus share a similar structure/function without sharing primary sequence similarity. Of the cellular IRES structures derived so far, none were shown to share an overall structural similarity. Therefore, we undertook a genome-wide search of human 5′UTRs (untranslated regions) with an empirically derived structure of the IRES from the key inhibitor of apoptosis, X-linked inhibitor of apoptosis protein (XIAP), to identify novel IRES that share structure/function similarity. Three of the top matches identified by this search that exhibit IRES activity are the 5′UTRs of Aquaporin 4, ELG1 and NF-kappaB repressing factor (NRF). The structures of AQP4 and ELG1 IRES have limited similarity to the XIAP IRES; however, they share trans-acting factors that bind the XIAP IRES. We therefore propose that cellular IRES are not defined by overall structure, as viral IRES, but are instead dependent upon short motifs and trans-acting factors for their function.
INTRODUCTION
Viral infection often entails the hindering of the host cell's translation machinery in order that the viral genome is expressed more efficiently relative to the expression of the host's proteins. In the Picornaviridae family of single-stranded RNA viruses, a viral protease cleaves the scaffolding translation initiation factors of the eIF4G family, thereby reducing the efficiency of the host cell's cap-dependent translation initiation and favouring the alternate internal ribosome entry site (IRES) mechanism used by the virus (1). The importance of this mechanism for viruses has been best demonstrated in the attenuated Sabin poliovirus strains used for worldwide polio vaccination, which contains point mutations in the IRES (2–4) that allow efficient translation in the gut but not in neuronal cells.
Cap-dependent translation initiation is also rendered less efficient under several cellular conditions, but specific cellular mRNAs are still translated with relative efficiency using the IRES mechanism for translation initiation. Many fine reviews have been written on IRES in cellular messages (5), viruses (6,7), stress and apoptosis (8–10). Typical translation of cellular messages in eukaryotes begins with the association of translation initiation factors with the cap-binding protein factor, eIF4E, on the 5′end of the message where the ‘cap’, a methyl7GDP nucleotide resides. This complex, which includes eIF4E, eIF4G, eIF4A, eIF3, the 40S ribosomal small subunit and an activated start codon tRNAi, will scan along the untranslated region (UTR) of the mRNA until it finds a suitable start codon where the large ribosomal subunit will join and protein translation will begin. During mitosis, cellular perturbation or stress, and apoptosis, canonical initiation factors like eIF4E, 4E-BPs, eIF2α and the eIF4G family of proteins are either modified by changes in phosphorylation state or by protein cleavage (11) and are no longer available for efficient cap-dependent translation initiation. The ribosome may also undergo some modifications as well (12). At these times, other protein factors (13), many of which are part of the ribonucleoprotein complex, are required to enhance translation initiation through the IRES mechanism. An IRES is a stretch of sequence usually upstream of the AUG start codon in the 5′ untranslated region (UTR) of the messenger RNA that along with the IRES trans-acting protein factors (ITAFs), recruit the ribosome. It is not clear whether these factors need a sequence motif or a RNA secondary structure/sequence motif combination to bind to IRES-containing mRNAs. In viral IRESes, like HCV and EMCV, RNA secondary structure has been shown to be crucial for IRES function (14–19). These structures are also conserved in other viruses that do not share primary sequence similarity. For example, the HCV IRES structure is similar to CSFV and BVDV (20,21), whereas the EMCV IRES structure is shared with other cardio- picornaviruses (22).
The secondary structure of the cellular IRES of c-Myc (23), L-Myc (24), Apaf-1 (25), FGF-2 (26), FGF1 (27), Kv1.4 (28), Bag-1 (29), Igf2 (30), cat-1 (31), Mnt and MTG8a (32) have been empirically determined using enzymatic and chemical probing, but no similarities between the structures of these cellular IRES were identified. This could be due to a much wider regulatory range of translation initiation that is needed in distinct cellular contexts relative to a virus's need to translate its messages more efficiently than the host's transcripts. Therefore, the possibility exists that there are structural motifs that are shared in co-ordinately regulated as yet undiscovered IRES.
Determination of viral IRES structures can often benefit from the comparison of many sequences from the same virus, using the variation of sequence to determine which bases pair together. When a structure is preserved, a mutation in a base will be coupled with a second site mutation that preserves the base pairing, and therefore the IRES structure. This co-variation analysis is not always possible with a lower number of available mammalian sequences, and therefore secondary structure determination requires enzymatic or chemical probing to determine the secondary structure in lieu of tertiary structure analysis with NMR or X-ray crystallography. Computational de novo secondary structure prediction has still not achieved the accuracy needed to skip empirical structure determination, but it can be used for comparative purposes when at least one structure is known (33).
The X-linked inhibitor of apoptosis protein (XIAP) is the key inhibitor of apoptosis by virtue of binding to and inhibiting distinct caspases (34). It was shown that XIAP mRNA is translated by an IRES-dependent mechanism (35), and that this mode of XIAP translation is absolutely required for maintaining protective levels of XIAP protein in cells undergoing various forms of cellular stress (35–39). Thus, it is likely that functionally similar IRES exist that govern the expression of cellular genes involved in the control of cellular growth, proliferation and death. In this work, the secondary structure of the XIAP IRES was determined using enzymatic probing. This structure was then used to search a 5′UTR database using the RSEARCH program (40), which predicted that several mRNAs had some similar structure features. When tested in a bicistronic reporter construct, two of these UTRs from Aquaporin4 and the uncharacterized ELG1 exhibited IRES activity, while the 5′UTR of NRF was shown previously to contain an IRES element (41). Further structural and biochemical probing showed that XIAP, AQP4 and ELG1 share only limited RNA structure similarity; however, additional biochemical analyses demonstrated that they have several IRES trans-acting factors in common. These data prompt us to propose that, unlike viral IRES elements, the cellular IRES are primarily defined not by an overall common structure but rather by common short RNA motifs and shared trans-acting factors.
MATERIALS AND METHODS
Cell culture and transfection reagents
Human embryonic kidney (HEK) 293 and 293T cells were maintained in standard conditions in Dulbecco's modified Eagle's medium supplemented with heat-inactivated 10% fetal calf serum, 2 mM l-glutamine and 1% antibiotics (100 units/ml penicillin–streptomycin). Transient transfections were performed using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's protocol. Briefly, cells were seeded at a density of 3 × 105 cells/ml in 6-well plates and were transfected 24 h later in serum-free Opti-MEM medium (Invitrogen) with 2 µg of DNA and 4 µl of lipid per well. The transfection mixture was replaced 3 h later with fresh Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine and 1% antibiotics.
Bicistronic constructs
The pβgal/CAT bicistronic vector containing the minimal human XIAP IRES was described previously as pβgal/5′(−162)/CAT (42). The empty vector was modified to include an NheI site upstream of the original XhoI site to enable directional cloning of UTR sequences. The new UTRs were isolated by reverse transcribing total RNA from HEK 293T cells using the First-Strand cDNA Synthesis kit (Amersham Biosciences) with random hexamer primers and then PCR amplifying the UTRs with the following specific primers: AQP4-Nhe-F cgctagcAAGGACAGTTTGGATAAT, AQP4-Xho-R cctcgagCCACCATGATGTTCTCT, ELG1-Nhe-F cgctagcACTTTTGGTGGGCATTTA, ELG1-Xho-R cctcgagCTGGGCGGGGAATA, PCD10-Nhe-F cgctagCCTCAGTTGCTGGTAAG, PCD10-Xho-R cctcgagAAGCCAACTACAGTTGAA, VEGF-D-Nhe-F cgctagcACTTCTCTGCATTTTCT, VEGF-D-Xho-R cctcgagTTTCAATATCCACTGATT, ZINCF-Nhe-F cgctAGCCTGAGAAGATGATGC, ZINCF-Xho-R cctcgaGCTTTCCCACATTCACA. The bicistronic construct with the ATF4 5′UTR was provided as a gift from Dr Jamie Blais. The XIAP-PPT construct with the −47 to −1 sequence removed was previously described (35). The AQP4-PPT construct was created using the AQP4-Nhe-F primer and the AQP4-59-Xho-R primer CCTCGAGAGAAACAAATCAGCA and cloning into the pβgal/CAT bicistronic vector. The FLAG-tagged PTB-overexpressing construct was generated by inserting reverse transcription-PCR (RT-PCR) amplified PTB-1 cDNA into the pcDNA 3 vector (Invitrogen). The FLAG epitope was incorporated into the N-terminus of PTB using the following primers: 5′ –ggatccatggattacaaggac°gacgacgataaggacggcattgtcccagat (BamHI recognition site is underlined; FLAG-coding sequence is italicized) and 5′–ctcgagctagatggtggacttggagaag (XhoI recognition site is underlined). The monocistronic constructs were made by deleting out the β-galactosidase open reading frames from the bicistronic constructs using NotI. The hairpin sequence, which was previously described (43), was inserted in the NheI site, which is 5′ to the UTRs.
RNA secondary structure determination
RNA secondary structures were determined using enzymatic probing with RNase T1, RNase A, RNase T2 and RNase V1 according to the protocol supplied with the Ambion enzymes. RNA was in vitro transcribed using the MaxiShortScript kit (Ambion) following the manufacturer's protocol. All RNA structures were examined from both ends. Radioactive 5′ end labelling was performed on uncut RNA with Ambion's KinaseMax Kit. Instead of 3′ end-labelling, cold, RNase digested RNA was reverse transcribed using a fluorescent tagged primer from the 3′ end and the reaction was separated on a Licor IR2 DNA Sequencer alongside a control sequence reaction of the same sequence. RNase cut sites were used as constraints in either MFOLD (44) or RNASTRUCTURE (45) to predict secondary structure models. The XIAP mutant structures were probed with N-methylisatoic anhydride (NMIA) following the protocol of Wilkinson et al. (46) with the modification that reverse transcription took place using a fluorescent tagged primer from the 3′ end and the products were separated on a Licor IR2 DNA Sequencer alongside a control sequence reaction of the same sequence. RNA samples were reacted with 160, 65, 32 and 16 mM NMIA. Concentrations of 32 and 16 mM NMIA gave the best results for the amount of RNA used.
Structure database search
A human 5′UTR database was generated from the ENSEMBL genome annotation site for NCBI build 34. Exact duplicate UTRs were removed along with shorter UTRs that fully overlapped other entries. The database was supplemented with full-length UTRs of a published IRES dataset (33). CT structure files were converted to Stockholm format and used to search the UTR database using the default parameters of RSEARCH on a grid of 6 Sun workstations.
βgal, CAT and NeoPTII analysis
Transiently transfected cells were harvested 24 h post-transfection in CAT enzyme-linked immunosorbent assay (ELISA) kit lysis buffer (Roche), and cell extracts were prepared using the protocol provided by the manufacturer. β-Galactosidase enzymatic activity in cell extracts was determined by the spectrophotometric assay using ONPG (o-nitrophenyl-β-d-galactopyranoside) (47), and the CAT levels were determined by using the CAT ELISA kit (Roche) and the protocol provided by the manufacturer. Neomycin phosphotransferase II levels were detected using neomycin phosphotransferase II ELISA (Agdia) following the protocol of the manufacturer. Transient transfections were performed in HEK 293 cells where neomycin phosphotransferase II was being used because HEK 293T cells contain the NPTII gene. Relative IRES activity was determined as a ratio of CAT to β-galactosidase. Unless otherwise noted, the data represent mean ±S.E.M. of three independent experiments performed in triplicates.
Quantitative RT-PCR
Total RNA was isolated from cells that were previously transfected with the βgal/CAT bicistronic constructs as described above. For quantitative RT-PCR, reverse transcription was carried out using the First-Strand cDNA Synthesis kit (Amersham Biosciences) with NotI-d(T)18 primers. The quantitative PCR was performed using the QuantiTect SYBR green PCR kit (Qiagen) and analysed on an ABI Prism 7000 sequence detection system using the ABI Prism 7000 SDS Software. Quantitative PCRs were carried out to detect β-galactosidase (5′-ACTATCCCGACCGCCTTACT-3′ and 5′-CTGTAGCGGCTGATGTTGAA-3′) and CAT (5′-GCGTGTTACGGTGAAAACCT-3′ and 5′-GGGCGAAGAAGTTGTCCATA-3′) as described previously (48).
RNA-affinity chromatography
Isolation of IRES-binding proteins was performed using a modified RNA-affinity chromatography protocol (49). Briefly, XIAP IRES RNA, AQP4 5′UTR RNA, ELG1 5′UTR RNA, XIAPΔPPT RNA and AQP4ΔPPT RNA were transcribed in vitro with the MEGAShortscript transcription kit according to the manufacturer's protocol (Ambion), and were biotinylated at the 5′ end with the 5′ EndTag Nucleic Acid Labelling System according to the manufacturer's instructions (Vector Laboratories). The biotinylated RNAs (15 μg) were conjugated to Avidin-agarose beads (Sigma) in the presence of incubation buffer (10 mM Tris-Cl [pH 7.4], 150 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 0.05% [v/v] Nonidet P-40) at 4°C for 2 h with continuous rotation. Unbound RNAs were removed by washing beads twice with incubation buffer. Five hundred microgram of 293T protein extract (in incubation buffer) was added to the coated beads, along with 30 μg yeast tRNA (Sigma) and 200 units of Prime RNase inhibitor (Eppendorf). Reactions were incubated at room temperature with continuous rotation for 30 min, followed by incubation at 4°C with continuous rotation for 2 h. Beads were washed five times with incubation buffer, resuspended in 20 μl of 1× SDS-PAGE loading dye, and boiled for 5 min to elute bound proteins. Proteins were then separated by 10% SDS-PAGE and transferred to PVDF membrane for analysis by western blot using antibodies specific for La (50), hnRNP C1/C2 (AbCam), hnRNP A1 (Santa Cruz Biotechnology) and PTB (Zymed Laboratories).
RESULTS
Determination of the XIAP IRES secondary structure
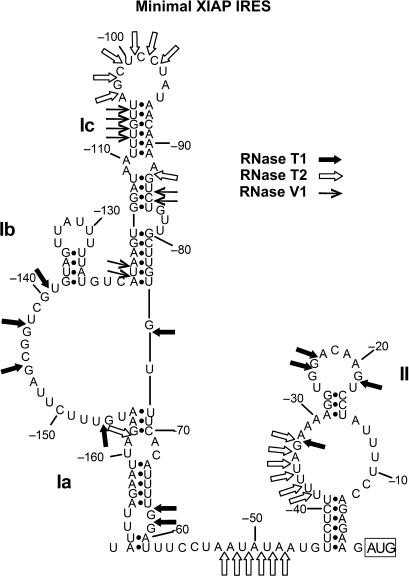
The 5′UTR of XIAP mRNA promotes cap-independent translation initiation through the activity of an IRES element located between nucleotides −162 to +3 (relative to the start codon) (42,51). Our recent investigations have addressed the role of trans-acting factors in XIAP IRES activity (42,51,52), yet little is known about the contribution of the RNA structure that is formed by this IRES sequence to the function of the XIAP IRES. Several studies have shown the importance of secondary structure for viral IRES activity (14–19); however, the importance of secondary structure for cellular IRES remains unclear. We therefore hypothesized that if secondary structure is an important determinant of cellular IRES activity, we should be able to identify new cellular IRES by searching for RNA sequences that form structures that are similar to a known IRES element (e.g. XIAP IRES). As a first approach to characterize the role of secondary structure in cellular IRES activity, we empirically determined the secondary structure of the XIAP IRES RNA. The sequence from −162 to +3 of the XIAP 5′UTR was in vitro transcribed and subjected to digestion with RNases T1, V1 and T2. The relative sensitivities of sites to these RNases are presented in Supplementary Table 1. We then used the information derived from the RNase T1, V1 and T2 digests as constraints for folding the XIAP IRES RNA sequence into a secondary structure using the MFOLD structure prediction webserver (Figure 1) (44). The model proposed has two domains with the first domain having two stems, Ia and Ib, flanking an unstructured region and the second domain, which is smaller and contains a polypyrimidine tract in its major stem that has previously been shown to be required for IRES activity (35).
Figure 1.
The proposed XIAP IRES RNA secondary structure model. The sites sensitive to RNase T1, which cuts after single-stranded guanine nucleotides, RNase T2, which cuts after single-stranded nucleotides, and RNase V1, which cuts within regions of double-stranded RNA, are shown on the derived structural model. These cut sites were used as constraints in the MFOLD (44) program to predict the secondary structure of the minimal XIAP IRES sequence. The full list of sensitive sites is shown in Supplementary Table 1.
We also examined the effect of flanking sequences on the structure of the XIAP IRES by including the first 50 nt of the CAT ORF, which are immediately downstream of the XIAP IRES in the bicistronic reporter plasmid pβgal/(−162)/CAT. This region of the CAT ORF was included so that the whole IRES region could be clearly seen on sequencing gels from the reverse transcription of RNA probed with RNases. As the CAT ORF would be common to other IRES cloned into our βgal/Cat bicistronic construct, our labelled primer could be used for all the structures analysed. We found that the sensitivity to the RNases remained the same when the first 50 nt of the CAT ORF were included in the XIAP IRES RNA (data not shown). Therefore, the flanking sequences do not influence the XIAP IRES structure.
A genome-wide search for structures similar to the XIAP IRES
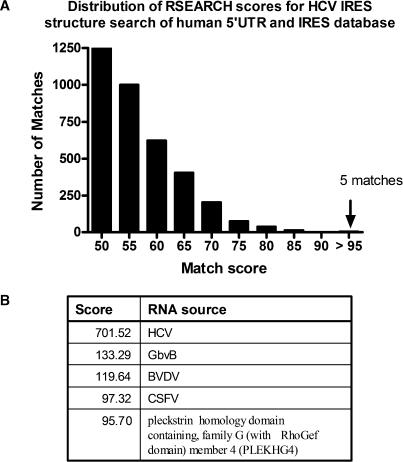
We were interested in identifying novel IRES that are structurally similar to the XIAP IRES, since the ability to do so would suggest an important role for secondary structure in cellular IRES function. Additionally, cellular IRES that share a similar secondary structure may also be regulated and/or function in a similar manner. We therefore developed a method for searching a database of 5′UTR sequences for structural features that are similar to a query sequence. A database of human 5′UTR sequences was compiled from the Ensembl database using Ensmart (now known as BioMart) (53). The sequences were filtered for redundancy by removing exact duplicate matches, and 5′UTR sequences that existed within longer 5′UTRs were discarded. The 5′UTRs from published cellular and viral IRES were added from an in-lab database (33) to the Ensembl-created database. To search this database we used the RSEARCH program, which is designed to search a database with a sequence or sequence alignment that has a known secondary structure (40). This program uses scoring matrices built with alignments of the small subunit rRNA, called RIBOSUM, to score for putative preserved base pairings in both the query structure and the database entry, as well as specific nucleotide matches in the alignment. RSEARCH outputs a sequence and structural alignment of the query structure and the RSEARCH-predicted structure of the database entry for each match that is above a user-defined cut-off score. We first tested efficacy of the RSEARCH program for our objectives by searching for 5′UTRs that are structurally similar to the well-studied HCV IRES structure, which was defined for this search by incorporating the latest NMR and crystallographic data (33). A distribution of the RSEARCH match scores is shown in Figure 2A. The top matches were the Hepatitis B isolate, GbvB, CSFV and the BVDV 5′UTRs (Figure 2B), all known to have similar structures to the HCV IRES (20,21,54). The primary sequences of either the CSFV 5′UTR or the BVDV 5′UTR lack sufficient similarity to the primary sequence of the HCV IRES to be found using BLAST (data not shown), and were therefore identified due to structural similarity and not sequence homology. The one high scoring match to the human PLEKHG4 5′UTR is perplexing. Although the structural alignment to domain III of HCV is quite strong, there are a couple of anomalies that suggest this match may not be significant. First, the two gaps of PLEKHG4 at the analogous IIId and IIIe loops of HCV are quite large, 86 and 225 nt, respectively, and the proposed structure may not form as RSEARCH has predicted. Second, the published IRESes so far have been fairly close to the AUG start codons and this structure is over 200 bases upstream of the known start codon for PLEKHG4. Although these differences suggest that PLEKHG4 would not have a structure that matches the HCV IRES, it must be noted that the match of PLEKHG4 to domain IIIb of the HCV IRES is quite good. Domain IIIb of HCV is known to bind directly to eIF3, the adapter protein that normally binds the 40S ribosomal subunit and eIF4G (14,55).
Figure 2.
The distribution of RSEARCH match scores from a search of a human 5′UTR and IRES sequence database with the HCV IRES structure. (A) A histogram of the number of matches to the HCV IRES structure for a given RSEARCH match score plotted in bins of 5 above the score of 50. The number of matches includes multiple matches to the same UTR. There were no matches with a score of 90 to 95. (B) The match score and identity of the matching 5′UTR sequence.
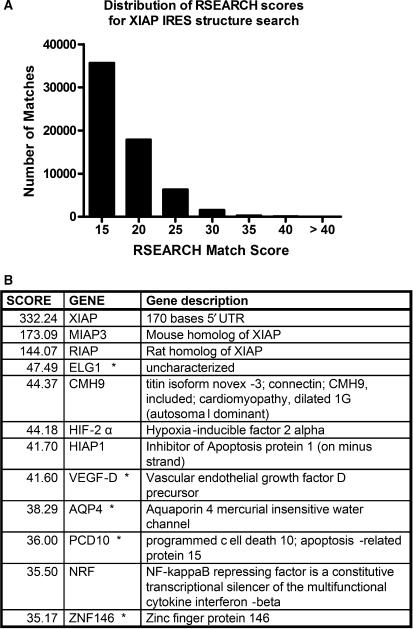
After confirming the utility of our search process using the HCV IRES structure, the XIAP IRES structure (Figure 1) was next used as a query to search the same database, and the distribution of the match scores for this search is displayed in Figure 3A. As the minimal XIAP IRES 170 nucleotide sequence we used is less than the 372 nt of the HCV IRES sequence, the likelihood of structure or sequence matches is much less and therefore the overall scores will be much lower. This is apparent, as the exact match to the XIAP IRES is 332 compared to the exact match to the HCV IRES with a score of 701. The top matches for the XIAP IRES structure were the mouse and rat XIAP IRES orthologs, which have similar primary sequence, with scores of 173 and 144, respectively. The highest match score for a 5′UTR of human sequence known to have an IRES was NRF (41), with a score of 35.5 and so we examined hits with scores over 35. There were ∼80 matches (in addition to the XIAP IRES orthologs) with scores higher than 35, which represent a range of 11–14% of an exact match score. In comparison with the HCV structure search, the score for CSFV is just under 14% of an exact match score of the HCV IRES structure.
Figure 3.
The distribution of RSEARCH match scores from a search of a human 5′UTR and IRES sequence database with the XIAP IRES structure. (A) A histogram of the number of matches to the XIAP IRES structure for a given RSEARCH match score plotted in bins of 5 above the score of 15. The number of matches includes multiple matches to the same UTR. (B) A selection of the top match scores above the score of 35 showing the score and identity of the matching 5′UTR sequence.
The NRF, AQP4 and ELG1 5′UTRs exhibit IRES activity
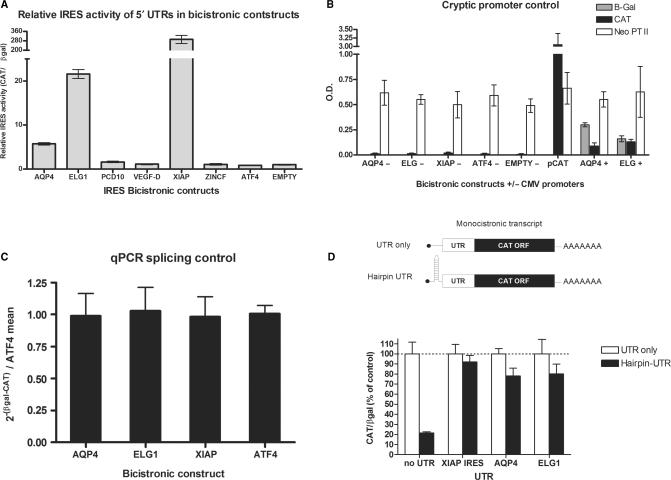
The 5′UTR of NRF was shown previously to contain IRES activity (41), confirming that our search could identify functional IRES. We therefore wished to test the identified 5′UTRs that had significant match scores (i.e. >35) for IRES activity. Several criteria were used to select five 5′UTRs (denoted by an asterisk in Figure 3B) from the list of potential structure matches for additional testing. As all characterized IRES elements are located just upstream of the AUG start codon, only matches that were present in the 3′ end of the 5′UTRs were chosen. Structure matches that had non-aligning inserts greater than 100 bases within the structural alignments were also not chosen for further testing, as these may represent structures unlikely to occur naturally. The sequences for each of the five chosen 5′UTRs were amplified by RT-PCR using RNA isolated from HEK 293T cells as a template, and the resulting products were subcloned into the pβgal/CAT bicistronic reporter vector (35). The 5′UTR from ATF4, which has been shown to not have IRES activity (Blais,J., personal communication), was also cloned into the pβgal/CAT bicistronic vector and serves as a negative control. These plasmids were subsequently transfected into HEK 293T cells, and relative IRES activity was determined by assessing the levels of β-galactosidase and CAT expression. Of the five UTRs tested, only the ELG1 and Aquaporin 4 (AQP4) 5′UTRs exhibit IRES activity, although this activity is much lower than that exhibited by the XIAP IRES (Figure 4A).
Figure 4.
Testing the IRES activity of the five selected 5′UTRs using bicistronic reporter constructs. (A) The IRES activity of each 5′UTR in βgal/UTR/CAT bicistronic construct measured as CAT protein levels relative to β-galactosidase activity. All values are relative to the empty vector, which is set to 1. The 5′UTR of ATF4 and the empty vector are negative controls. (B) Testing for cryptic promoter activity within the UTRs. Promoter activity was tested in bicistronic constructs in which the CMV promoter that drives expression of the bicistronic message was excised. These constructs were transfected into 293 cells and CAT and β-galactosidase expression was determined. A monocistronic plasmid in which CAT gene expression is driven by the CMV promoter was used as a positive control, as well as the bicistronic constructs of AQP4 and ELG1 that contain the CMV promoter. The neomycin phosphotransferase II (Neo PTII) gene, which also resides on the transfected plasmids, was used as a transfection control. ELISA measurements of Neo PTII protein levels are presented as O.D. values. (C) The AQP4 and ELG1 bicistronic messages were tested for spurious splicing events using quantitative RT-PCR of amplicons within the βgal and CAT ORFs. The fold differences of the CAT and βgal amplicons was calculated as 2−[Ct(βgal) – Ct(CAT)] and was plotted relative to the ATF4 negative splicing control, which has no IRES activity. Values represent the mean of nine biological samples and error bars represent ±S.E.M. of nine biological samples. (D) Monocistronic UTR-CAT constructs with and without a hairpin [ΔG = −60 kcal/mol as calculated by RNAStructure (45)] that was used to halt ribosome scanning were co-transfected with a βgal expressing plasmid. Each set of values is normalized to its non-hairpin constructs and β-galactosidase activity.
To ensure that the IRES activity exhibited by these two 5′UTRs is not due to the function of a cryptic promoter within the cloned UTR sequence, the CMV promoter was excised from the vector. In such a promoterless construct, any expression of CAT must be due to the presence of cryptic promoter activity within the cloned UTR sequence. The β-galactosidase and CAT protein levels in extracts from HEK 293 cells transfected with the promoterless bicistronic reporter constructs was measured, and is expressed relative to the levels of the neomycin phosphotransferase II (NPTII), which is also contained on the bicistronic vector. NPTII expression is driven by the SV40 promoter, and is therefore expressed independently from the βgal/CAT bicistronic RNA, allowing NPTII expression levels to serve as a control for transfection efficiency. None of the 5′UTRs tested displayed any cryptic promoter activity, as evidenced by the absence of CAT expression from the promoterless constructs (Figure 4B). The 5′UTRs exhibiting IRES activity were also tested for spurious splicing events by performing quantitative RT-PCR (qRT-PCR) analysis of mRNA isolated from transfected cells using oligonucleotide primers specific to each of the βgal and CAT open reading frames (48). We did not detect any splicing events as the ratio of CAT to βgal mRNA remained the same for all of the constructs tested, which also demonstrates the equivalent stability of all the messages (Figure 4C).
In order to confirm cap-independent activity of messages with the 5′UTRs of AQP4 and ELG1, we constructed monocistronic constructs with these UTRs and the CAT open reading frame with and without upstream hairpin structures. Cap-dependent initiation is inhibited in constructs where a thermodynamically stable hairpin is added upstream of the coding region, which hinders ribosome scanning initiated at the 5′ end of the message (43,56,57). The addition of an upstream hairpin in the control pCAT construct significantly inhibited translation of CAT (Figure 4D). In contrast, translation mediated by AQP4, ELG1 and XIAP 5′UTRs was resistant to the inhibition by the stable hairpin (Figure 4D). Therefore, these data indicate that the ELG1 and AQP4 5′UTRs exhibit bona fide IRES activity.
RNA secondary structure determination of the AQP4 and ELG1 5′UTRs, and comparison to the XIAP IRES structure
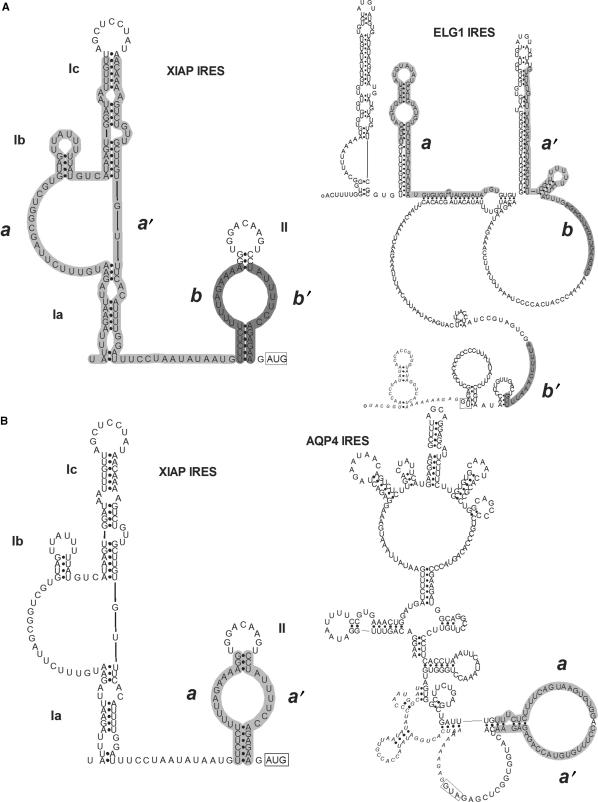
We have used XIAP IRES secondary structure to conduct a genome-wide search for novel cellular IRES elements that are structurally similar to the XIAP IRES. To determine if the IRES-containing 5′UTRs of AQP4 and ELG1 were indeed identified by the RSEARCH program due to actual structural similarities between these UTRs and the XIAP IRES, the secondary structures of the ELG1 and AQP4 5′UTR sequences were empirically determined by RNase probing (Supplementary Tables 3–6). The secondary structure models that were generated for the AQP4 and ELG1 5′UTRs, and the RSEARCH sequence/structure matches with the XIAP IRES structure, are shown in Figure 5. Surprisingly, the secondary structure of the ELG1 IRES does not exhibit any similarity to the structure of XIAP IRES (Figure 5A). The sequences of ELG1 that RSEARCH attributes to the base-paired stems of domain I or II of the XIAP IRES do not anneal in the empirically determined structure. Closer inspection of the ELG1 sequence shows that it contains 35 AUG repeats, which could theoretically allow it to form many structures, such as the one predicted by RSEARCH.
Figure 5.
Comparison of the RNA secondary structures of the ELG1, AQP4 and XIAP IRES. The RNA secondary structure models of AQP4 and ELG1 were determined with enzymatic probing using RNases T1, T2, V1 and A. The enzyme cut sites were used as constraints for an RNA secondary structure prediction program and are available in the Supplementary Data. (A) The XIAP IRES structure is compared to the ELG1 IRES structure. The RSEARCH-predicted matching sequence and structure between the XIAP and ELG1 IRES are outlined in grey and labelled as a/a′ and b/b′ on each structure. (B) The XIAP IRES structure is compared to the AQP4 IRES structure showing the RSEARCH-predicted matching sequence and structure in grey and labelled as a/a′ on each structure. The full list of sensitive sites is shown in Supplementary Tables 3 and 5.
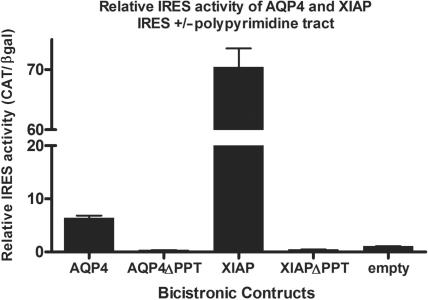
However, the AQP4 5′UTR structure contains a domain that displays significant similarity to the structure observed in the XIAP IRES domain II. Notably, a double-stranded sequence in the AQP4 5′UTR that contains a polypyrimidine tract is similar to the structure of a polypyrimidine tract in the XIAP IRES (Figure 5B). Both UTRs include the UUCUCUUUU motif. This polypyrimidine tract has been shown previously to be important for XIAP IRES activity, since upon deletion or mutation of these sequences IRES activity is lost (35). We therefore wished to determine if this polypyrimidine tract is also important for AQP4 IRES activity. To this end, the sequence between −59 and −3 within the AQP4 5′UTR (containing the polypyrimidine tract) was deleted in the bicistronic vector, and the resulting construct was then tested for IRES activity. We found that deletion of the polypyrimidine tract sequences in both the XIAP and AQP4 5′UTRs completely abrogates IRES activity (Figure 6). Therefore, the polypyrimidine tract is absolutely required for the IRES activity of both the AQP4 and XIAP 5′UTRs.
Figure 6.
Deletion of the polypyrimidine tracts in XIAP and AQP4 abrogates IRES activity. Bicistronic constructs, deleted of sequence from −47 to −1 in the XIAP IRES (XIAPΔPPT) and −50 to −3 in the AQP4 IRES (AQP4ΔPPT), which removes the matching polypyrimidine tracts, were tested for IRES activity. IRES activity is expressed as CAT protein levels relative to β-galactosidase activity. Values are given relative to the empty bicistronic vector, which was set to 1.
Changes in XIAP structure do not affect IRES activity
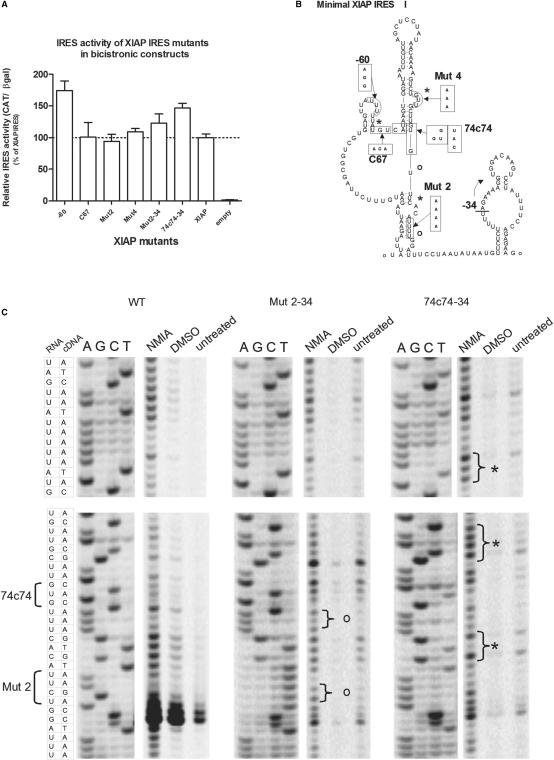
The comparison of the AQP4, ELG1 and XIAP IRES structures suggest that they do not share similar RNA structures. This could indicate that perhaps cellular IRES structures are not as important as viral IRES. Indeed, there exists previous evidence in support of this claim. The polypyrimidine tract upstream of position −34 in the XIAP IRES appears to form a stem (Figure 1). It is known that the deletion of position −1 to −34 relative to the start codon does not affect activity (35) but would most likely disrupt the stem formed with the polypyrimidine tract. In order to determine if more disruption in XIAP IRES's structure is possible without affecting IRES activity, we tested several XIAP IRES mutants previously made in our laboratory. It was apparent that several mutants had similar or slightly better activity than the wild-type XIAP IRES sequence (Figure 7A). The position of some of these mutations should not necessarily change the IRES structure as they are in loop regions; other mutants, however, are predicted to result in altered IRES structure (Figure 7B). Using N-methylisatoic anhydride (NMIA) to selectively probe for open regions of the RNA structure (46) we found that at least two of the mutants, Mut2-34 and 74c74-34, exhibited changes in their structure relative to the wild-type sequence. Importantly, these mutants did not exhibit any loss in IRES activity. Examples of these changes (denoted by either asterisks for mutant 74c74-34 or an ‘o’ for Mut2-34; Figure 7B and C) showed that the structural changes were not just at the site of sequence mutations but were elsewhere in the IRES as well. These data support our conclusion that overall structure is not of primary importance for the XIAP IRES's activity.
Figure 7.
Mutations in the XIAP IRES can alter the structure without affecting activity. (A) The relative IRES activity of each XIAP IRES mutant in the βgal/UTR/CAT bicistronic construct measured as CAT protein levels relative to β-galactosidase activity. All values are relative to the wild-type XIAP IRES sequence, which is set to 100%. (B) The location of the mutations for each of the IRES mutants on the proposed XIAP IRES structure is presented with the changed sequence. The –34 mutation refers to the mutants which have 34 nts deleted from the 3′ end of their UTR. (C) In vitro transcribed RNA was reacted with NMIA, which preferentially modifies bases that have an open conformation in the RNA structure. The RNA was reverse transcribed and the products separated on sequencing gels showing the relative sensitivity of bases to NMIA for the wild-type XIAP IRES and the mutants Mut2-34 and 74c74-34. The first four lanes show the sequence reactions from a DNA template followed by the reverse transcriptase reactions of RNA treated with either NMIA in DMSO, DMSO or untreated samples. Areas in the gel showing differences in NMIA sensitivity relative to the other XIAP IRES variants and controls are bracketed and labelled with an asterisks or an ‘o’ on the gel and the structure diagram in (B).
The XIAP and AQP4 IRES elements interact with the PTB ITAF via their polypyrimidine tracts
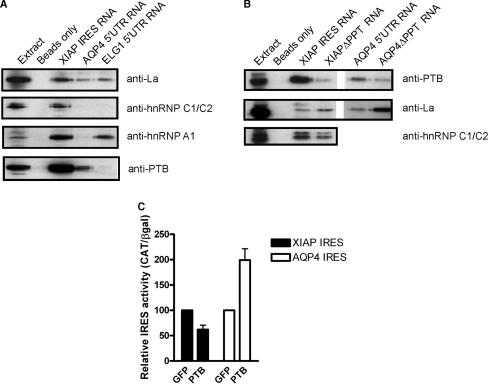
Although the structural similarity among the IRES elements of XIAP, AQP4 and ELG1 was weak, we cannot rule out that these sequences share IRES trans-acting factors (ITAFs), which allowed the ELG1 and AQP4 5′UTRs to be identified by our search protocol because the ITAFs may have weakly similar binding motifs (structural or sequence similarity) that would contribute to the RSEARCH match score. ITAFs are mRNA-binding proteins that are involved in a variety of processes dealing with mRNA processing, and are therefore quite promiscuous in the mRNA to which they bind. As the identity of ITAF-binding sites may be difficult to assess from the primary sequences, we have used RNA-affinity chromatography to isolate proteins that bind to the IRES RNA sequences and have probed for the presence of known ITAFs by Western blot analysis. It has already been shown that the La autoantigen (51) and hnRNP C1/C2 (42) bind to the XIAP IRES and enhance its activity, whereas hnRNP A1 binds to the XIAP IRES and reduces IRES activity (52). Western blot analysis showed that the La autoantigen associates with the XIAP, AQP4 and ELG1 IRES, whereas hnRNP A1 was found to be associated with the XIAP and ELG1 IRES elements, and hnRNP C1/C2 were only found to be associated with the XIAP IRES (Figure 8A). Since we have found that a polypyrimidine tract is required for both XIAP and AQP4 IRES activity, we assessed whether the polypyrimidine tract binding protein (PTB), a known ITAF for several IRES elements (25,32,58–61), could associate with these IRES sequences. Indeed, we found that PTB can associate with both the XIAP and AQP4 IRES, but not the ELG1 IRES (Figure 8A). To confirm that the polypyrimidine tracts of the XIAP and AQP4 IRES elements are required for PTB binding we performed RNA-affinity chromatography using XIAP and AQP4 IRES RNAs in which the polypyrimidine tracts have been deleted. We find that deletion of the polypyrimidine tract in either the XIAP or AQP4 IRES RNA abrogates PTB binding, but does not disrupt the binding of other ITAFs (Figure 8B). Therefore, the polypyrimidine tracts of the XIAP and AQP4 IRES elements are important for ITAF binding. Moreover, the 5′UTRs identified by our search share the ability to bind to a similar cohort of trans-acting factors.
Figure 8.
RNA-affinity chromatography isolation of XIAP, AQP4 and ELG1 IRES-binding proteins. (A) Pre-cleared protein extracts from HEK293T cells were incubated with either agarose beads coated with XIAP IRES RNA, agarose beads coated with AQP4 5′UTR RNA, agarose beads coated with ELG1 5′UTR RNA, or agarose beads alone. After protein binding, beads were washed extensively, pelleted, and proteins were eluted by boiling and resolved by SDS-PAGE. Following separation by SDS-PAGE, proteins were transferred to PVDF membrane and probed with anti-La, anti-hnRNP C1/C2, anti-hnRNP A1 and anti-PTB antibodies. (B) As in (A) only using agarose beads coated with RNA of the XIAP IRES, the XIAP IRES with the polypyrimidine tract deleted (XIAPΔPPT), the AQP4 IRES, or the AQP4 IRES with the polypyrimidine tract removed (AQP4ΔPPT). (C) HEK293T cells were co-transfected with βgal/CAT bicistronic constructs containing the XIAP or AQP4 IRES and GFP or PTB expressing plasmids. Each bicistronic construct is normalized to its relative IRES activity co-expressed with GFP.
To elucidate the role of PTB in XIAP and AQP4 IRES function, we tested the effect of PTB overexpression on the activity of these IRES elements. We find that PTB is a bona fide ITAF for both the XIAP and AQP4 IRES elements, as it affects their activities (Figure 8C). Surprisingly however, we find that overexpression of PTB represses XIAP IRES activity, whereas PTB overexpression enhances AQP4 IRES activity ∼2-fold (Figure 8C).
Together, our data suggest that motifs for ITAF binding and the proteins that bind to these motifs may be important determinants of IRES activity, rather than the overall structure of the IRES itself.
DISCUSSION
In this work, we empirically determined the secondary structure of the XIAP IRES and set out to identify novel cellular IRES by searching for human 5′UTR sequences that display a structural similarity to the XIAP IRES. We first tested the efficacy of our search protocol by using the well-characterized structure of the HCV IRES to search a database of human 5′UTRs and UTRs known to contain IRES. This search effectively identified IRES from GbvB, CSFV and BVDV, which have been previously shown to have structural similarity to the HCV IRES (20,21,54). This exercise established that structurally similar UTRs could be found using the RSEARCH program, thus validating our approach. Our search of the human 5′UTR database using the structure of the XIAP IRES resulted in the identification of five 5′UTRs in which the region with structural similarity was in the correct position (at the 3′ end of the UTR, proximal to the AUG codon) and orientation to exhibit IRES activity. Characterization of the IRES activity of these identified sequences showed that the 5′UTRs of AQP4, ELG1 and NRF promote IRES-dependent translation. However, empirical determination of the structure of the AQP4 and ELG1 5′UTRs and subsequent comparison to the structure of the XIAP IRES revealed that the structures of these sequences share little similarity. We then found that a polypyrimidine tract in the XIAP IRES and AQP4 IRES is absolutely necessary for IRES activity, and that similar trans-acting factors (ITAFs) can bind to these IRES sequences. Our results lead us to propose that, unlike the viral IRES, the overall structure of cellular IRES is not necessarily an important factor in determining IRES activity, but rather small motifs and the cohort of proteins that bind them define cellular IRES activity.
It has been recognized that both viral and cellular IRES do not share primary sequence homology, and therefore it is not possible to identify mRNAs that harbour IRES elements by comparison of sequence data using search programs such as BLAST. This realization has presented a barrier to the genome-wide identification of IRES, resulting in the identification of mRNAs that contain IRES in a piecemeal fashion—a message is suspected to be translated under conditions that repress cap-dependent translation and the 5′UTR is subsequently tested for IRES activity. However, several studies have highlighted the importance of secondary structure for the function of viral IRES (14,15,17) and some viral IRES share structural homology, suggesting that an underlying determinant for function of viral IRES is the presence of specific structural elements (62). We therefore hypothesized that cellular IRES may also share structural homology, and that comparison of the secondary structure of 5′UTRs may be a means to identify novel IRES on a genomic scale.
Our searches of a human 5′UTR genome, supplemented with 5′UTRs known to exhibit IRES activity from all species and viruses, with the structure of the XIAP IRES did indeed result in the identification of RNA sequences that exhibit IRES activity. Of the top matches, two were the 5′UTRs of the mouse XIAP ortholog (MIAP) and the rat XIAP ortholog (RIAP) that both display IRES activity (35) (Holcik,M., unpublished data), one that has previously been shown to have IRES activity [NRF;(41)], and we have demonstrated that two others (ELG1 and AQP4) also display IRES function. The IRES activity of AQP4 and ELG1 are much less than XIAP. However, we have only tested IRES activity in one cell line and under conditions of normal cell growth. AQP4, a water channel-forming protein expressed in the brain, has recently been shown to have a role in edema during eclampsia, as its protein levels are elevated during pregnancy (63). Interestingly, mRNA levels of AQP4 have not been shown to change (63), suggesting a possible IRES function. ELG1 is an uncharacterized transcript that exists in the database as a fully sequenced cDNA clone, accession # AK125048 and a portion of the UTR is represented by EST DB217710.1. While the presence of numerous AUGs in its 5′UTRs is unusual, it is nevertheless not completely uncommon as around 1% of all 5′UTRs have 30 to 100 upstream AUGs (33). Interestingly, we did not identify the 5′UTRs of p27(Kip1) (64) or Bcl-xL, both of which exhibit IRES activity (39), in our search (matching scores were between 16 and 17). Yoon et al. (39) recently showed that the IRES activity of XIAP, p27(Kip1) and Bcl-xL is specifically and severely impaired in cells harbouring mutations in the pseudouridinase gene Dkc1. This mutation was shown to prevent the proper pseudouridylation of rRNA and is thought to disrupt ribosome structure (39). The IRES-dependent translation of the XIAP, p27(Kip1) and Bcl-xL messages is specifically sensitive to these changes, as global translation is not affected. It was therefore hypothesized that these IRES may share some common feature, such as secondary structure, that is sensitive to changes in ribosome architecture (39). Based on our results, the secondary structures of these IRES elements may not be similar, and other factors, such as changes in the association of trans-acting factors with the modified ribosome or direct association of the IRES with the ribosome, may account for the coordinated reduction of XIAP, p27(Kip1) and Bcl-xL IRES activity in Dkc1 mutant cells.
To our surprise, comparison of the empirically determined secondary structures of the ELG1 and AQP4 5′UTRs to the secondary structure of the XIAP IRES showed only limited homology, with only small regions of AQP4 displaying some similarity within the overall structure. From the search of HCV IRES structure we learned that the score of CSFV, which is just over 13% of a perfect match, was significant. Our search for a similar IRES structure with a cut-off score of 35 had us examining matches with scores in the range of 11–14% of a perfect match. In retrospect, these values may have been too low to be significant, but represent the top matches available. As the search with the HCV structure validated this search protocol, the lack of results with the XIAP IRES structure search strongly supports the notion that there are no similar structures. This suggests that cellular IRESes do not share globally similar structure as do some viral IRESes. This conclusion is plausible when considering published evidence to date. First, no overall structural similarity has been found among the 11 known cellular IRES structures (23–32). Second, examination of the c-myc IRES has shown that its activity is not dependent on the overall structure but on distinct sequence modules (65). Third, mutations which opened up the Bag-1 IRES structure negated the need for the previously required ITAF, PCBP1, while still retaining full IRES activity (29). Fourth, non-overlapping segments of IRES sequence that would not preserve the overall structure still retain partial IRES activity (28,66), suggesting different modules may synergistically act to provide full IRES activity in vivo. In this light, it is not surprising that the XIAP IRES mutants we examined exhibited full IRES activity despite structural changes. Conversely, the point mutation of the IRES which attenuates the Sabin strain 3 of poliovirus does not change the structure (67) but it does effect the binding of PTB (68). It would be wrong to over generalize and suggest that structure will never be required for the function of cellular IRESes. Such an exception is the CAT-1 IRES, which is induced during amino acid starvation, where a specific stem is required for functional activity regardless of its sequence makeup (31).
Whereas a virus needs to compete efficiently to get its proteins expressed within an infected cell, cellular transcripts with an IRES are more likely translationally regulated, with several trans-acting factors contributing differently depending upon the cellular context. Studies of the HCV IRES have shown it to directly interact with the ribosome and induce conformational change of the ribosome (69). Cricket paralysis virus IRES has an RNA structure which allows it to interact with the ribosome and to initiate translation without an eIF2—initiator Met-tRNAi complex in the P-site (70). This mechanism may have evolved to overcome the antiviral response of the cell to shut off protein synthesis though PKR phosphorylation of eIF2α (71). The importance of structures seen in viruses has yet to be shown for any cellular IRES.
Importantly, we noted the presence of a polypyrimidine tract that was folded into a stem-loop structure in the both XIAP and AQP4 IRES. As we have previously found that the polypyrimidine tract is absolutely necessary for XIAP IRES activity (51), we tested whether deletion of this sequence in the AQP4 5′UTR would also abrogate IRES function. Indeed, deletion of the AQP4 5′UTR polypyrimidine tract causes a loss of PTB binding and a complete loss of IRES activity. Because this small region is required for IRES activity, we believe that short motifs may be critical determinants of IRES function. In agreement with this hypothesis, it has been found that a reiterated PTB-binding motif can induce internal ribosome entry (32). Moreover, a search by Mitchell et al. (32) for 5′UTRs that harbour this PTB-binding motif has resulted in the identification of novel IRES elements. This search, however, would not have found XIAP or AQP4 because the (CCU)n motif pattern considered only a small subset of sequence sites that PTB can bind and is not found in XIAP or AQP4 IRES. Even though the polypyrimidine tract region (UUCUCUUUU) is the same in XIAP and AQP4, PTB overexpression has an opposite effect on their IRES activity. This may possibly be due to competing ITAFs for sequences in this region, which can have either a positive or negative effect on translation initiation. Our observations, together with previously published findings, suggest that the recruitment of a particular cohort of trans-acting proteins is the critical factor in cellular IRES-mediated translation. Therefore, a cataloguing of the trans-acting factors required by each IRES may allow functional grouping of these elements and aid in the identification of common features required for cellular IRES-dependent translation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to E. Chan for the gift of anti-La A1 antibody and Jaime Blais for the pβgal/ATF/CAT plasmid construct. We thank the members of our laboratory for helpful discussions. This work was supported by grants from the Canadian Institutes of Health Research (CIHR MOP # 43984), Premier's Research Excellence Award, Canada Foundation for Innovation and Ontario Research and Development Challenge Fund. M.H. is a CIHR New Investigator. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawamura N, Kohara M, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. Determinants in the 5' noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 1989;63:1302–1309. doi: 10.1128/jvi.63.3.1302-1309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss EG, O’Neil RE, Racaniello VR. Mapping of attenuating sequences of an avirulent poliovirus type 2 strain. J. Virol. 1989;63:1884–1890. doi: 10.1128/jvi.63.5.1884-1890.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westrop GD, Wareham KA, Evans DM, Dunn G, Minor PD, Magrath DI, Taffs F, Marsden S, Skinner MA, et al. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J. Virol. 1989;63:1338–1344. doi: 10.1128/jvi.63.3.1338-1344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 6.Belsham GJ, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Salas E, Quinto SL, Ramos R, Fernandez-Miragal O. IRES elements: features of the RNA structure contributing to their activity. Biochimie. 2002;84:755–763. doi: 10.1016/s0300-9084(02)01408-6. [DOI] [PubMed] [Google Scholar]
- 8.Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- 9.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 10.Lewis SM, Holcik M. IRES in distress: translational regulation of the inhibitor of apoptosis proteins XIAP and HIAP2 during cell stress. Cell Death Differ. 2005;12:547–553. doi: 10.1038/sj.cdd.4401602. [DOI] [PubMed] [Google Scholar]
- 11.Morley SJ, Coldwel MJ, Clemens MJ. Initiation factor modifications in the preapoptotic phase. Cell Death Differ. 2005;12:571–584. doi: 10.1038/sj.cdd.4401591. [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Ji H, Doudna JA, Leary JA. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spriggs KA, Bushel M, Mitchel SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 14.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 15.Odreman-Macchioli F, Baralle FE, Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J. Biol. Chem. 2001;276:41648–41655. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- 16.Odreman-Macchioli FE, Tisminetzky SG, Zotti M, Baralle FE, Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875–885. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown EA, Day SP, Jansen RW, Lemon SM. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J. Virol. 1991;65:5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolupaeva VG, Hellen CU, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman MA, Palmenberg AC. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J. Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemon SM, Honda M. Internal ribosome entry sites within the RNA genomes of Hepatitis C virus and other flavivirues. Semin. Virol. 1997;8:274–288. [Google Scholar]
- 22.Pilipenko EV, Blinov VM, Chernov BK, Dmitrieva TM, Agol VI. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Quesne JP, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 24.Jopling CL, Spriggs KA, Mitchel SA, Stoneley M, Willis AE. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10:287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchel SA, Spriggs KA, Coldwel MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 26.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martineau Y, Le Bec C, Monbrun L, Allo V, Chiu IM, Danos O, Moine H, Prats H, Prats AC. Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol. Cell. Biol. 2004;24:7622–7635. doi: 10.1128/MCB.24.17.7622-7635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang GM, Leong LE, Hoang LT, Wang PH, Gutman GA, Semler BL. Structurally distinct elements mediate internal ribosome entry within the 5′-noncoding region of a voltage-gated potassium channel mRNA. J. Biol. Chem. 2004;279:47419–47430. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- 29.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen SK, Christiansen J, Hansen TO, Larsen MR, Nielsen FC. Human insulin-like growth factor II leader 2 mediates internal initiation of translation. Biochem. J. 2002;363:37–44. doi: 10.1042/0264-6021:3630037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, et al. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, Spriggs RV, Willis AE. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holcik M, Korneluk RG. XIAP, the guardian angel. Nat. Rev. Mol. Cell Biol. 2001;2:550–556. doi: 10.1038/35080103. [DOI] [PubMed] [Google Scholar]
- 35.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 36.Holcik M, Yeh C, Korneluk RG, Chow T. Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene. 2000;19:4174–4177. doi: 10.1038/sj.onc.1203765. [DOI] [PubMed] [Google Scholar]
- 37.Nevins TA, Harder ZM, Korneluk RG, Holcik M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 2003;278:3572–3579. doi: 10.1074/jbc.M206781200. [DOI] [PubMed] [Google Scholar]
- 38.Ungureanu NH, Cloutier M, Lewis SM, de Silva N, Blais JD, Bell JC, Holcik M. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. J. Biol. Chem. 2006;281:15155–15163. doi: 10.1074/jbc.M511319200. [DOI] [PubMed] [Google Scholar]
- 39.Yoon A, Peng G, Brandenburg Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 40.Klein RJ, Eddy SR. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oumard A, Hennecke M, Hauser H, Nourbakhsh M. Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol. Cell. Biol. 2000;20:2755–2759. doi: 10.1128/mcb.20.8.2755-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcik M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 2004;279:17148–17157. doi: 10.1074/jbc.M308737200. [DOI] [PubMed] [Google Scholar]
- 44.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 47.MacGregor GR, Nolan GP, Fiering S, Roederer M, Herzenberg LA. In: Methods in Molecular Biology. Murray EJ, Walker JM, editors. 1991. Vol. 7. Humana Press Inc., Clifton, N.J., pp. 217–235. [DOI] [PubMed] [Google Scholar]
- 48.Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 2004;24:7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan EK, Tan EM. Human autoantibody-reactive epitopes of SS-B/La are highly conserved in comparison with epitopes recognized by murine monoclonal antibodies. J. Exp. Med. 1987;166:1627–1640. doi: 10.1084/jem.166.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell. 2007;18:1302–1311. doi: 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clamp M, Andrews D, Barker D, Bevan P, Cameron G, Chen Y, Clark L, Cox T, Cuff J, et al. Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res. 2003;31:38–42. doi: 10.1093/nar/gkg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grace K, Gartland M, Karayiannis P, McGarvey MJ, Clarke B. The 5' untranslated region of GB virus B shows functional similarity to the internal ribosome entry site of hepatitis C virus. J. Gen. Virol. 1999;80(Pt 9):2337–2341. doi: 10.1099/0022-1317-80-9-2337. [DOI] [PubMed] [Google Scholar]
- 55.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 56.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K, Koumenis C, Harding HP, Ron D, et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol. Cell. Biol. 2006;26:9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giraud S, Greco A, Brink M, Diaz JJ, Delafontaine P. Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 2001;276:5668–5675. doi: 10.1074/jbc.M005928200. [DOI] [PubMed] [Google Scholar]
- 60.Pickering BM, Mitchell SA, Evans JR, Willis AE. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le SY, Liu WM, Maizel JV., Jr Phylogenetic evidence for the improved RNA higher-order structure in internal ribosome entry sequences of HCV and pestiviruses. Virus Genes. 1998;17:279–295. doi: 10.1023/a:1008073905920. [DOI] [PubMed] [Google Scholar]
- 63.Quick AM, Cipolla MJ. Pregnancy-induced up-regulation of aquaporin-4 protein in brain and its role in eclampsia. FASEB J. 2005;19:170–175. doi: 10.1096/fj.04-1901hyp. [DOI] [PubMed] [Google Scholar]
- 64.Miskimins WK, Wang G, Hawkinson M, Miskimins R. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 2001;21:4960–4967. doi: 10.1128/MCB.21.15.4960-4967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cencig S, Nanbru C, Le SY, Gueydan C, Huez G, Kruys V. Mapping and characterization of the minimal internal ribosome entry segment in the human c-myc mRNA 5′ untranslated region. Oncogene. 2004;23:267–277. doi: 10.1038/sj.onc.1207017. [DOI] [PubMed] [Google Scholar]
- 66.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pilipenko EV, Blinov VM, Romanova LI, Sinyakov AN, Maslova SV, Agol VI. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- 68.Guest S, Pilipenko E, Sharma K, Chumakov K, Roos RP. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol. 2004;78:11097–11107. doi: 10.1128/JVI.78.20.11097-11107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 70.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 71.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]