Abstract
There are some functional compatibilities between upstream and core promoter sequences for transcriptional activation in yeast, Drosophila and mammalian cells. Here we examined whether similar compatibilities exist in sea urchin embryos, and if so, whether they are dynamically regulated during early development. Two reporter plasmids, each containing a test promoter conjugated to either CFP or YFP, were concurrently introduced into embryos, and their expression patterns were studied by fluorescence microscopy. The upstream sequence of the Hemicentrotus pulcherrimus (Hp) OtxE promoter drives the expression of its own core promoter and that of Strongylocentrotus purpuratus (Sp) Spec2a in different embryonic regions, especially at the late gastrula stage. Interestingly, when the four putative transcription factor binding sites of this upstream sequence were individually mutated, the resulting sequences directed different spatiotemporal expression from the same set of two core promoters, indicating that combinations of upstream factors may determine core promoter usage in sea urchin embryos. In addition, the insertion or deletion of consensus or nonconsensus TATA sequences changed the expression profile significantly, irrespective of whether the upstream sequence was intact or mutated. Thus, the TATA sequence may serve as a primary determinant for core promoter selection in these cells.
INTRODUCTION
Eukaryotic promoters driven by RNA polymerase II are composed of two different functional units, the upstream and core promoter sequences. Cell type-specific transcription factors bind to the upstream sequence(s) and activate transcription via interactions with the basal transcriptional machinery that forms at the core promoter sequence (1). This machinery consists of a set of general transcription factors, i.e. TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH and RNA polymerase II (1). Among these, only TFIIB and TFIID can recognize core promoter sequence(s) in a sequence-specific manner (1). In metazoans, there are several distinct core promoter sequences, including the TATA element, TFIIB recognition element (BREu and BREd), downstream core element (DCE), initiator (Inr), motif ten element (MTE), downstream promoter element (DPE) and X gene core promoter element 1 (XCPE1) (2–5). These elements, with the exception of BRE and XCPE1, are thought to be recognized by TFIID.
Interestingly, previous studies showed that there are some functional compatibilities between the upstream and the core promoter sequences (2,6–10). In yeast, for instance, while the upstream sequence of the RPS5 promoter activates the ADH1 and CUP1 core promoter at the same levels as its own RPS5 core promoter, the upstream sequences of the latter two genes are largely unable to activate the RPS5 core promoter (7). Another example is the two distinct types of upstream sequences that act specifically with DPE- or TATA-dependent core promoters in Drosophila embryos (11). These functional compatibilities indicate that the core promoter architecture plays an important role in the transcriptional regulation of class II genes.
In this context, it is noteworthy that the utilization of the TATA element is altered during development. The shift from the utilization of a TATA-less promoter to a TATA-dependent promoter during early development (12–14) could be explained by the presence of DPE- or TATA-specific upstream sequences (11). If this were the case, functional compatibilities between the upstream and the core promoter sequences would provide a potential mechanism for the developmental regulation of differential core promoter usage (12–18).
Previously, we showed that an interplay between the upstream CACGTG element and the TATA element may determine the temporally specific expression profiles of the Hemicentrotus pulcherrimus (Hp) OtxE and OtxL promoters during early development of the sea urchin (18). Furthermore, the CACGTG element activates the HpOtxE core promoter specifically from a distal position, whereas it activates the Strongylocentrotus purpuratus (Sp) Spec2a core promoter from a proximal position in undifferentiated P19 embryonal carcinoma cells, which were derived from a mouse embryo (17). Intriguingly, these stimulatory effects of the CACGTG element disappear upon treatment with retinoic acid, which triggers the neuronal differentiation of P19 cells (17). These observations led us to believe that the HpOtxE and SpSpec2a promoters could provide an adequate model system to further analyze functional interactions between the upstream and core promoter sequences and their spatiotemporal regulation during early development of sea urchin embryos.
In this study, we examined the expression of two different test promoters containing upstream and core promoter sequences derived from HpOtxE or SpSpec2a in living sea urchin embryos. The promoters drove the expression of CFP and YFP reporter genes, which enabled us to monitor the activity of the two promoters concurrently and semi-continuously in single embryos, to verify the functional compatibility between the upstream and core promoter sequences, and to determine whether the interactions between the upstream and core promoter sequences are dynamically regulated during the early development of sea urchin embryos. The results reinforce the recent view that the combination of the upstream and core promoter sequences may determine the specific spatiotemporal expression profiles of a range of genes that are developmentally regulated.
MATERIALS AND METHODS
Embryo culture
Gametes of the sea urchin Hemicentrotus pulcherrimus were collected and inseminated as described previously (18). Fertilized eggs in which reporter constructs had been introduced (as described below) were cultured at 16°C in a petri dish (6 cm in diameter) at a cell concentration of 1.3% [v/v; 0.2 ml of eggs suspended in 15 ml of artificial sea water (ASW)].
Plasmid construction
YFP/CFP reporter genes
The NcoI-XbaI DNA fragment encoding GFP was amplified by PCR using the primer pair TK7264 and TK7265 and the pGreen Lantern-2 (Invitrogen) plasmid as a template. The PCR product was inserted into the EcoRV site of the pBluescript II SK(-) plasmid (Stratagene) to generate pM4642. The oligonucleotides used in this study are summarized in Supplementary Table 1. The NcoI and BamHI sites within the open reading frame (ORF) of GFP encoded by pM4642 were disrupted by site-specific mutagenesis (19) using TK7262 and TK7289 oligonucleotides, respectively, to generate pM4645.
Site-specific mutagenesis was conducted on pM4645 using the oligonucleotides TK7488 (T65G, V68L, S72A) and TK7489 (T203Y) to generate pM4682, encoding YFP. Similarly, site-specific mutagenesis was conducted on pM4645 using the oligonucleotides TK7490 (F64L, Y66W), TK7491 (N146I, M153T) and TK7492 (V163A) to generate pM4684, encoding CFP.
YFP/CFP reporter plasmids driven by the HpOtxE promoter
Site-specific mutagenesis was conducted on pM1249 (18) using oligonucleotide TK867 to generate pM1319, a construct with a truncated HpOtxE promoter fragment (base pairs −461 to +180, numbered with respect to the transcriptional initiation site as +1). The NcoI site was created at the initiating codon of the luciferase ORF of pM1319, and the XbaI site located upstream of the HpOtxE promoter was disrupted by site-specific mutagenesis using the TK6391 and TK6448 oligonucleotides, respectively, to generate pM4497. The SpeI-SmaI short DNA fragment containing the BamHI site of pM4497 was removed to generate pM7448 by enzyme digestion, followed by blunt end ligation.
The NcoI-XbaI DNA fragment of pM7448 encoding luciferase was replaced with the NcoI-XbaI DNA fragments of pM4682 and pM4684 encoding YFP or CFP to generate pM4692 and pM4705, respectively.
The TATTCA sequence at base pairs −28 to −23 in the HpOtxE promoter of pM4692/pM4705 was changed to TATAAA or TATAAAA by site-specific mutagenesis using the TK1150 or TK8923 oligonucleotides to generate two sets of YFP/CFP plasmids, pM4706/pM4694 and pM7071/pM7070, respectively.
YFP/CFP reporter plasmids driven by the chimeric promoter containing the HpOtxE upstream sequence and the SpSpec2a core promoter
pM4731 (YFP) and pM4725 (CFP) were generated from pM4692 (YFP) and pM4705 (CFP), respectively, by site-specific mutagenesis using the oligonucleotides AK312, AK313 and TK7598.
The AATAATA sequence at base pairs −30 to −24 in the SpSpec2a promoter of pM4731 and pM4725 was deleted by site-specific mutagenesis using the TK1488 oligonucleotide to generate pM4734 and pM4728, respectively. Similarly, the GAATAATAC sequence at base pairs −31 to −23 in the SpSpec2a promoter of pM4731 was changed to CTATAAAAG by site-specific mutagenesis using the TK1681 oligonucleotide to generate pM7073.
YFP/CFP reporter plasmids driven by the core promoter sequence alone
The SacI-PmacI DNA fragment containing the HpOtxE upstream sequence of pM4705, pM4692, pM4725 and pM4731 were removed to generate pM4914, pM4915, pM4916 and pM4917, respectively, by enzyme digestion and subsequent blunt end ligation. Note that PmacI recognizes the CACGTG sequence, a putative USF-binding site (E-box).
YFP/CFP reporter plasmids driven by the promoter carrying a mutation within the HpOtxE upstream sequence
The putative GATA-binding site (#1) at base pairs −360 to −355 in the HpOtxE promoter of pM4692/pM4705, pM4706/pM4694 and pM4731/pM4725 was changed to a HindIII site (AAGCTT) by site-specific mutagenesis using the oligonucleotide TK7989 to generate pM4938/pM4937, pM7065/pM7062 and pM4944/pM4943, respectively.
The putative Otx-binding site at base pairs −294 to −289 in the HpOtxE promoter of pM4692/pM4705, pM4706/pM4694 and pM4731/pM4725 was changed to a HindIII site (AAGCTT) by site-specific mutagenesis using the oligonucleotide TK7836 to generate pM4940/pM4939, pM7066/pM7063 and pM4946/pM4945, respectively.
The putative GATA-binding site (#2) at base pairs −250 to −245 in the HpOtxE promoter of pM4692/pM4705, pM4706/pM4694 and pM4731/pM4725 was changed to a HindIII site (AAGCTT) by site-specific mutagenesis using the oligonucleotide TK7837 to generate pM4942/pM4941, pM7067/pM7064 and pM4948/pM4947, respectively.
The E-box at base pairs −66 to −61 in the HpOtxE promoter of pM4692/pM4705, pM4706/pM4694 and pM4731/pM4725 was changed to a HindIII site (AAGCTT) by site-specific mutagenesis using the oligonucleotide TK1812 to generate pM4783/pM4782, pM4781/pM4780 and pM4789/pM4788, respectively.
Insulator plasmid
The BamHI-BamHI DNA fragment containing an insulator derived from the upstream region of the Ars gene (20,21) was amplified by PCR using the primer pair TK8181 and TK8182. The PCR product was inserted into the BamHI site of the pBluescript II KS(+) plasmid (Stratagene) to generate pM4932.
Introduction of DNA
Introduction of reporter plasmids into fertilized eggs was conducted as previously described (22) with slight modifications. The YFP/CFP reporter plasmids (2.5 μg each) were linearized with BamHI and mixed with 10 μg of Sau3A-digested calf thymus carrier DNA and 0.6 μg of insulator DNA (578 bp) excised from pM4932 by BamHI. Five milligrams of gold particles (1.5–3.0 μm in diameter, Sigma-Aldrich, USA) was coated with the mixture of DNA described above (total 15.6 μg) by co-precipitation in 7.5% polyethylene glycol 6000 and 0.94 M NaCl. After washes with ethanol, an ethanol suspension of 3.3 mg DNA-coated gold particles (∼20 μl) was placed on a projectile holder. Bombardment using a particle gun gene delivery system (GIE-III IDERA, Tanaka Co. Ltd) was carried out on ∼2 × 105 fertilized eggs. Bombarded embryos were cultured at 16°C in a petri dish (6 cm in diameter) of ASW containing penicillin (100 mg/l) and streptomycin (50 mg/l).
Quantitative RT-PCR analysis
At the indicated times after bombardment, ∼20 000 embryos were taken from the dishes, collected by centrifugation, and stored at −80°C until use.
Total RNA was prepared from collected embryos using the Isogen RNA extraction system (Nippon Gene, Japan). Samples were subsequently treated with 0.1 μg/μl DNase I for 18 h at 37°C to degrade contaminating genomic DNA. Specific messenger RNA (mRNA) transcripts were measured by quantitative RT-PCR using SYBR® ExScript® RT-PCR kit (Takara, Japan) and the iCycler® PCR system with a fluorescent detector lid (Bio-Rad Laboratories, USA). First-strand cDNA was synthesized from 50 ng total RNA with random hexamer primers. The specific primer pairs used to amplify YFP cDNA, +190 to +507 bp, (numbered relative to the A of the initiating methionine codon), and CFP cDNA, +190 to +456 bp, were TK9037/TK9043, and TK9036/TK9038, respectively. Amplification reactions were performed in triplicate, and the thermal cycling conditions were as follows: 94°C for 120 s, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 60 s.
Immobilization of embryos with solidified agar
Noble agar (0.45%; Difco, USA) in ASW was kept warm at 70°C and then cooled to 37°C just before use. Embryos were gently mixed with this agar solution and then swiftly transferred onto a chilled slide glass to solidify the agar (final concentration of ∼0.4%). The slide glasses with immobilized embryos were immersed in ASW containing penicillin (100 mg/l) and streptomycin (50 mg/l) and then cultured at 16°C. This method enabled us to monitor gene expression semicontinuously in the same embryos by taking photographs every few hours.
Fluorescence microscopy
To observe the expression of YFP/CFP reporter genes, the movement of embryos was stopped by adding formaldehyde to a final concentration of 0.1% when the embryos were not immobilized with agar. An aliquot of the embryo suspension containing 200–300 embryos was placed onto a slide glass, and the YFP- and/or CFP-expressing embryos were visualized using a Nikon Eclipse 80i microscope with an epifluorescence attachment. Embryos immobilized with agar were inspected using the same system.
RESULTS
Experimental design to visualize functional interactions between upstream and core promoter sequences in living embryos
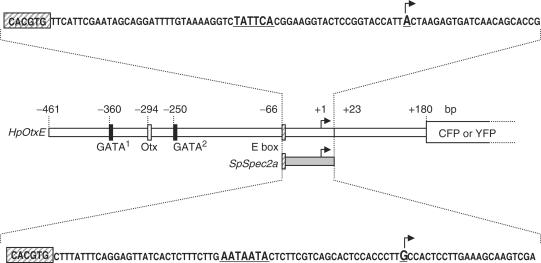
We chose the HpOtxE promoter as a model for studying functional interactions between upstream and core promoter sequences during early stages of development. The zygotic expression of HpOtxE begins at the unhatched blastula stage and gradually decreases by the gastrula stage (23). In contrast, the expression of SpSpec2a, a well-characterized target gene of SpOtx, begins at the late cleavage stage, exclusively in the aboral ectoderm territory, and is then largely extinguished after the late blastula stage (24). Fortuitously, the HpOtxE and SpSpec2a promoters contain the CACGTG element, a putative binding site for USF, at the same position (−66 to −61 bp) from the transcriptional start site (+1) (18,25). Thus, to investigate core promoter specificity, the region surrounding the transcriptional start site (−60 to +23 bp) of the HpOtxE promoter could be readily replaced with the corresponding region (−60 to +23 bp) of the SpSpec2a promoter without affecting the distance between the upstream and core promoter sequences (Figure 1).
Figure 1.
Schematic representation of the HpOtxE promoter (−461 to +180 bp, open rectangle) and the SpSpec2a core promoter (−66 to +23 bp, gray rectangle) used in this study to express the CFP or YFP reporter gene. The HpOtxE core promoter (−66 to +23 bp, open rectangle) was replaced with the SpSpec2a core promoter to test the functional interactions between the HpOtxE upstream sequence and the SpSpec2a core promoter sequence. The vertical rectangles on the promoters are putative binding sites for the GATA factor (−360 to −355 bp and −250 to −245 bp, black rectangles), the Otx factor (−294 to −289 bp, open rectangle) and USF (E-box, ∼−66 to −61 bp, hatched rectangle). The nucleotide sequences of the HpOtxE and SpSpec2a core promoters are shown at the top and the bottom, respectively. The transcriptional initiation site is highlighted with large bold letters and an arrow, the E-box with a hatched rectangle and the sequences that are mutated in this study with large bold letters.
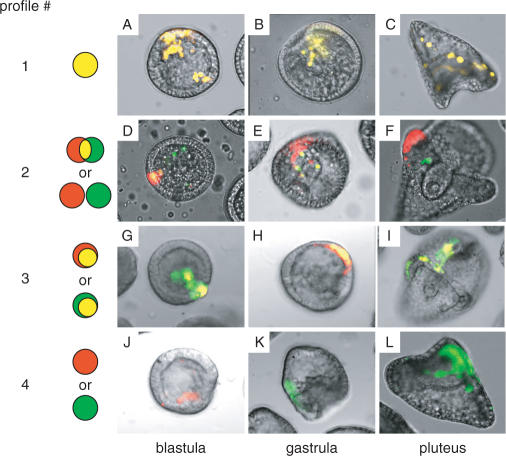
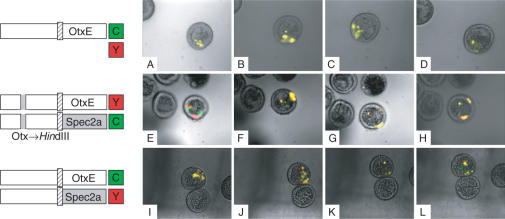
To compare the spatiotemporal expression patterns of two distinct test promoters in one embryo, two plasmids were introduced concurrently into fertilized eggs by particle gun bombardment. One plasmid carried a promoter connected to a CFP reporter gene, and the second plasmid carried a second promoter connected to YFP. Transfected embryos were then examined at different developmental stages for the expression of these two reporters by fluorescence microscopy and/or quantitative RT-PCR. The expression patterns were classified into four major groups (profiles #1–4, Figure 2): the signals of CFP (green) and YFP (red) completely overlapped (yellow; profile #1), overlapped partially or not at all (profile #2, note that no overlap was much more rare than partial overlap), one signal was included within the boundary of the other (profile #3), or only one signal (green or red) was observed (profile #4).
Figure 2.
Classification of the expression patterns of CFP/YFP reporter genes in transfected embryos. The expression of these two reporters were examined simultaneously in the same embryos using a fluorescence microscope (×200 magnification) at the blastula, gastrula and pluteus stages. Arbitrarily selected representative examples are shown as overlaid fluorescent and bright-field images. The CFP/YFP reporter plasmids transfected in these experiments are as follows: pM4694/pM4706 (A, C), pM4705/pM4692 (B), pM4947/pM4942 (D), pM4725/pM4692 (E), pM4705/pM4706 (F), pM7070/pM4706 (G, I), pM4694/pM7071 (H), pM4725/pM7073 (J), pM4694/pM7067 (K) and pM7070/pM7073 (L). Cells that express CFP or YFP are shown in green and red, respectively, whereas those that express both are shown in yellow. The expression patterns were classified into four groups as indicated: Profile #1, green and red signals completely overlapped; Profile #2, the signals partially overlapped or did not overlap; Profile #3, one signal was included within the boundary of the other and Profile #4, only one signal was observed.
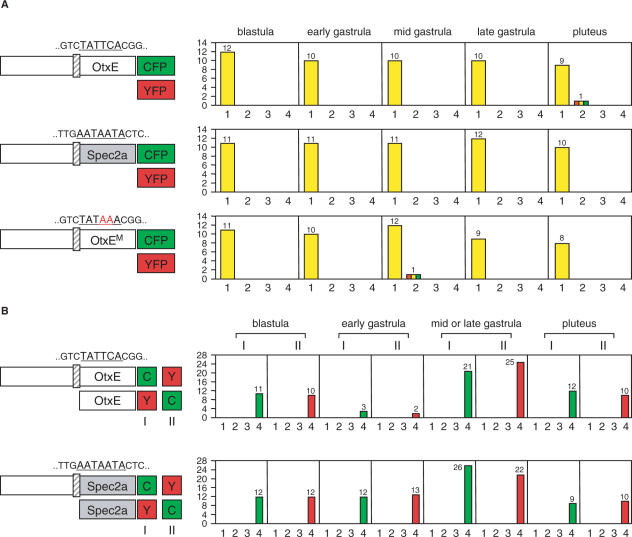
As expected, when we introduced the CFP and YFP reporters connected to the same promoter into embryos, they showed the same expression pattern (profile #1) with very few exceptions (e.g. profile #2) at any stage from blastula to pluteus (Figure 3A). Importantly, similar results were obtained irrespective of whether the core promoter region was derived from HpOtxE or SpSpec2a (Figure 3A). The fluorescent signals depended on functional interactions between the upstream and core promoter sequences, because no signals were observed when reporters were connected to either core promoter alone (Figure 3B). This was also confirmed by quantitative RT-PCR assays (Supplementary Figures 1 and 2). Therefore, we concluded that this system is adequate for analyzing the functional relationships between upstream and core promoter sequences in living embryos.
Figure 3.
Verification of the system for monitoring the functional interactions between the upstream and the core promoter sequences. In each panel, the schematic on the left indicates the reporter plasmids introduced into fertilized eggs. (A) Expression of CFP/YFP reporter genes driven by the same promoters. Three sets of CFP/YFP reporter plasmids were used: pM4705/pM4692, driven by the wild-type HpOtxE promoter; pM4725/pM4731, driven by the wild-type SpSpec2a promoter and pM4694/pM4706, driven by a mutated HpOtxE promoter bearing the TATAAA sequence instead of the TATTCA sequence (OtxEM; changed nucleotides are shown in red). The expression of these reporter genes were examined with fluorescence microscopy at the blastula, early gastrula, mid gastrula, late gastrula and pluteus stages, as indicated at the top. The number of embryos showing the expression pattern corresponding to each profile (defined in Figure 2) is summarized in the right panel. The vertical and horizontal axes indicate the number of embryos and expression pattern profiles, respectively. (B) Expression of CFP/YFP reporter genes driven by promoters containing or not containing the upstream sequence. (Upper panel) The reporter plasmids used were pM4705/pM4915 (I) or pM4914/pM4692 (II), which are driven by the wild-type HpOtxE promoter and its core promoter connected to CFP and YFP, respectively (I), or vice versa (II). The expression of the reporter genes was examined with fluorescence microscopy at different developmental stages, as indicated at the top. The number of embryos showing the expression patterns corresponding to each profile is summarized in the right panel as described in A. The color of the bars representing profile #4 reflect the signal that was observed (CFP, green; YFP, red). (Lower panel) Two CFP/YFP reporter plasmids were used, pM4725/pM4917 (I) and pM4916/pM4731 (II), which are driven by the wild-type SpSpec2a core promoter fused or not fused with the HpOtxE upstream sequence and connected to the complementary patterns of CFP and YFP. The number of embryos showing the expression patterns corresponding to each profile is summarized in the right panel as described above.
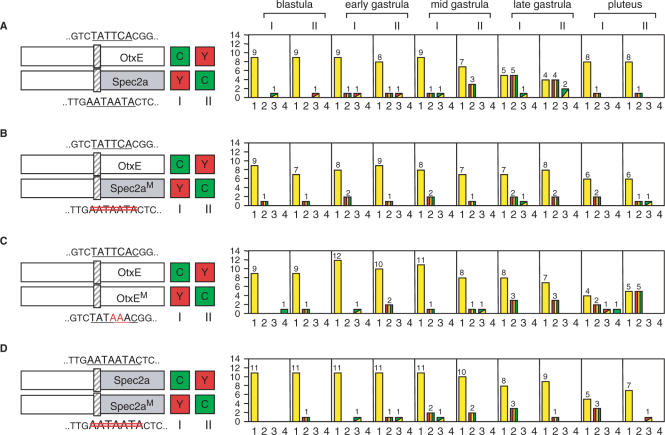
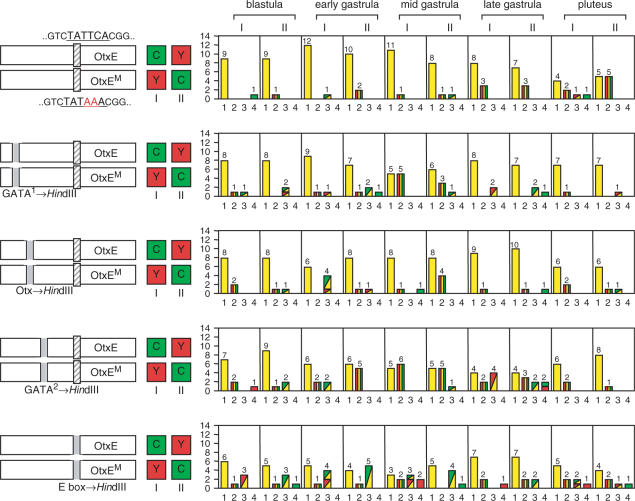
The TATA element alters core promoter usage of upstream sequences
When the expression of the HpOtxE promoter was compared with that of the chimeric promoter containing the SpSpec2a core promoter (−60 to +23 bp), a substantial number of embryos showed promoter-specific expression patterns (profile #2 or #3), especially at the late gastrula stage (Figure 4A). The reason for nonuniform gene expression in transfected embryos could be due to mosaic incorporation of reporter genes (26). In any case, this result indicates that cell type- and stage-specific transcription factors that bind to the upstream sequences of the HpOtxE promoter activate transcription in a core promoter-selective manner. Interestingly, this gastrula-specific enhancement of differential expression was suppressed when the nonconsensus TATA element (i.e. AATAATA) (27) was removed from the SpSpec2a core promoter (Figure 4B). Thus, the core promoter selectivity may be conferred, at least in part, by the TATA element.
Figure 4.
The effect of the TATA element on core promoter usage by the upstream sequence. In each panel, the schematic on the left indicates the reporter plasmids introduced into fertilized eggs. I and II refer to which reporter (CFP or YFP) was connected to each promoter, as indicated by the boxes marked C and Y. The number of embryos showing the expression pattern corresponding to each profile is summarized in the panel to the right of each schematic, as described in Figure 3. (A) The CFP/YFP reporter plasmids used were pM4705/pM4731 (I) and pM4725/pM4692 (II), which are driven by the wild-type HpOtxE promoter connected to CFP and the wild-type SpSpec2a core promoter fused with the HpOtxE upstream sequence. The bars representing profile #3 are shown as rectangles composed of two triangles; the left upper triangle corresponds to the signal that was observed in the broader region of the embryo (CFP; green, YFP; red). (B) The reporter plasmids used were pM4705/pM4734 (I) and pM4728/pM4692 (II), which are driven by the wild-type HpOtxE promoter and the mutated SpSpec2a core promoter (Spec2aM; the AATAATA sequence was removed as indicated with double red lines) fused with the HpOtxE upstream sequence. (C) The reporter plasmids were pM4705/pM4706 (I) and pM4694/pM4692 (II), which are driven by the wild-type HpOtxE promoter and the mutated HpOtxE promoter (OtxEM as described in Figure 3A). (D). Each set of the two CFP/YFP reporter plasmids, i.e. pM4725/pM4734 (I) or pM4728/pM4731 (II), which are driven by the wild-type and the mutated SpSpec2a core promoter (Spec2aM as described in B) fused with the HpOtxE upstream sequence.
To further confirm this assumption, we also tested the effects of creating the consensus TATA element (i.e. TATAAA) in the TATA-less HpOtxE core promoter (Figure 4C) or removing the nonconsensus TATA element (i.e. AATAATA) from the SpSpec2a core promoter (Figure 4D), in comparison with the expression of the native promoters. Here, other core promoter structures were left unaltered, in contrast to the experiments described above (Figure 4A and B). In both cases, increasing number of embryos showed differential expression patterns (profiles #2, #3 or #4; compare Figure 4C and D with Figure 3A). Substantial changes were observed again at late gastrula stage, although they were also evident at the pluteus stage, especially for the HpOtxE core promoter (Figure 4C and D). RT-PCR assays indicated that these differential expression patterns were largely qualitative rather than quantitative (Supplementary Figure 3). Collectively, these results indicate that at least some of the cell type- and stage-specific transcription factors can discriminate the presence or absence of the TATA element in the core promoter for transcriptional activation.
Multiple transcription factor binding sites are involved in spatiotemporal expression of the HpOtxE promoter
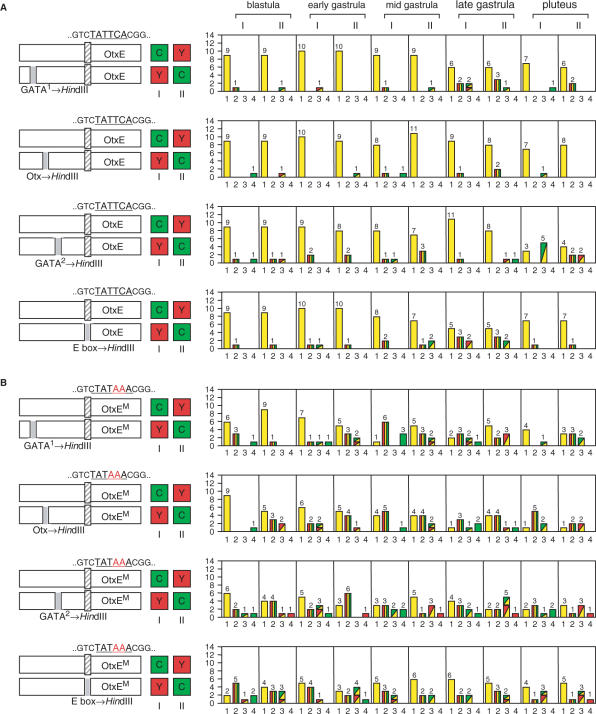
The upstream region of the HpOtxE promoter (−1819 to −3 bp) can activate both the HpOtxE and HpOtxL core promoters (28). Mutation analysis has also shown that the region of −397 to −64 bp, which contains the four putative transcription factor binding sites (GATA site 1, Otx site and GATA sites 2 and 3), functions as a strong enhancer for the HpOtxL core promoter at the blastula stage, when the Otx site plays the most important role in transcriptional activation (28).
To test whether these transcription factor binding sites or the CACGTG element (the E-box, a putative USF-binding site, Figure 1) are involved in spatiotemporal expression of the HpOtxE promoter, we introduced a CFP or YFP reporter connected to a mutated promoter in which one of these sites was changed to a Hind III recognition site, together with fluorescence reporter of the complementary color (i.e. YFP or CFP) connected to the intact promoter (Figure 5A). In each case, a small but significant number of embryos showed differential expression patterns (profiles #2, #3 or #4 in Figure 5A), suggesting that GATA, Otx and USF (or other closely related transcription factors) might be involved to varying extents in regulating the spatiotemporal expression from the intact HpOtxE promoter. Interestingly, the disruption of the GATA site 1 and E-box and of the GATA site 2 affected expression most strongly at the late gastrula and pluteus stages, respectively. Hence, they may correspond to binding sites for the cell type- and stage-specific transcription factors that we propose activate transcription at these developmental stages in a core promoter-selective manner (Figure 4).
Figure 5.
The effect of the four putative transcription factor binding sites on spatiotemporal expression of the TATA-less and TATA-containing HpOtxE promoters. In each panel, the schematic on the left indicates the reporter plasmids introduced into fertilized eggs. I and II refer to the particular combinations of reporter (CFP or YFP) and promoter, as indicated by the boxes marked C and Y. The number of embryos with each expression pattern profile is summarized in the panel to the right of each schematic. (A) (First panel) The plasmids used were pM4705/pM4938 (I) and pM4937/pM4692 (II), driven by the HpOtxE promoter fused with the intact or mutated HpOtxE upstream sequence within the putative GATA factor binding site #1 (−360 to −355 bp). (Second panel) Similar analysis using pM4940 (YFP) and pM4939 (CFP), in which the HpOtxE upstream sequence was mutated within the putative Otx factor binding site (−294 to −289 bp), instead of pM4938 (YFP) and pM4937 (CFP) in the first panel. (Third panel) Analysis using pM4942 (YFP) and pM4941 (CFP), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #2 (−250 to −245 bp). (Fourth panel) Analysis using pM4783 (YFP) and pM4782 (CFP), in which the HpOtxE upstream sequence was mutated within the E-box (−66 to −61 bp). (B) (First panel) pM4694/pM7065 (I) or pM7062/pM4706 (II), which are driven by the mutated HpOtxE core promoter (OtxEM as described in Figure 3A) fused with the intact or mutated HpOtxE upstream sequence within the putative GATA factor binding site #1. (Second panel) pM7066 (YFP) and pM7063 (CFP), in which the HpOtxE upstream sequence was mutated within the putative Otx factor binding site. (Third panel) pM7067 (YFP) and pM7064 (CFP), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #2. (Fourth panel) pM4781 (YFP) and pM4780 (CFP), the HpOtxE upstream sequence was mutated within the E-box.
Next, we tested the effects of the creation of the TATA element in the same sets of intact and mutated HpOtxE promoters (Figure 5B). Surprisingly, the transfected embryos showed differential expression to a much greater extent at all stages than those carrying the original TATA-less HpOtxE core promoter (compare Figure 5B with A). Such enhancement of differential expression might be partly explained (e.g. profiles #3 and #4) by the quantitative differences in expression levels that were detected in RT-PCR assays (Supplementaary Figure 4), but this cannot fully explain the occurrence of profile #2. Therefore, we conclude that upstream sequences lacking any one of the four transcription factor binding sites can display their own particular expression patterns more strongly when the TATA element is created in the HpOtxE core promoter.
The core promoter is selected in different ways by different combinations of upstream sequences
As described above, embryos displaying differential expression between the intact HpOtxE promoter and the mutated HpOtxE promoter lacking any one of the four transcription factor binding sites (Figure 5A) occur at similar frequencies as those displaying differential expression between the TATA-less and TATA-containing HpOtxE promoter (Figure 4C), but the frequency is much higher when the TATA element is created in the core promoter (Figure 5B). Therefore, we hypothesized that the mutated upstream sequences can discriminate the TATA-less and TATA-containing HpOtxE core promoters more strongly than the intact sequence can. To test this possibility, we compared the effects of mutations of each binding site (GATA site 1, Otx site, GATA site 2 and E-box) on the expression of the TATA-less and TATA-containing HpOtxE core promoters (Figure 6). As expected, the emergent frequencies of embryos displaying differential expression were increased to varying degrees by all mutations, especially at earlier developmental stages (e.g. blastula, early and mid gastrula; compare the top panel with the other four in Figure 6). Notably, mutations of the GATA site 2 and E-box affected expression at a broader range of stages than mutations of the other two sites (Figure 6), implying that GATA site 2 and the E-box might be recognized by more ubiquitously expressed transcription factors than GATA site 1 and Otx site.
Figure 6.
Comparison of the functional interactions between the different upstream sequences and the TATA element within the HpOtxE core promoter. In each panel, the schematic on the left indicates the reporter plasmids introduced into fertilized eggs. I and II refer to which reporter (CFP or YFP) was connected to each promoter, as indicated by the boxes marked C and Y. The number of embryos showing the expression pattern corresponding to each profile is summarized in the panel to the right of each schematic. (First panel) pM4705/pM4706 (I) or pM4694/pM4692 (II), which are driven by the wild-type and mutated HpOtxE core promoter (OtxEM as described in Figure 3A) fused with the intact HpOtxE upstream sequence. (Second panel) pM4937/pM7065 (I) or pM7062/pM4938 (II), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #1. (Third panel) pM4939/pM7066 (I) or pM7063/pM4940 (II), in which the HpOtxE upstream sequence was mutated within the putative Otx factor binding site. (Fourth panel) pM4941/pM7067 (I) or pM7064/pM4942 (II), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #2. (Fifth panel) pM4782/pM4781 (I) or pM4780/pM4783 (II), in which the HpOtxE upstream sequence was mutated within the E-box.
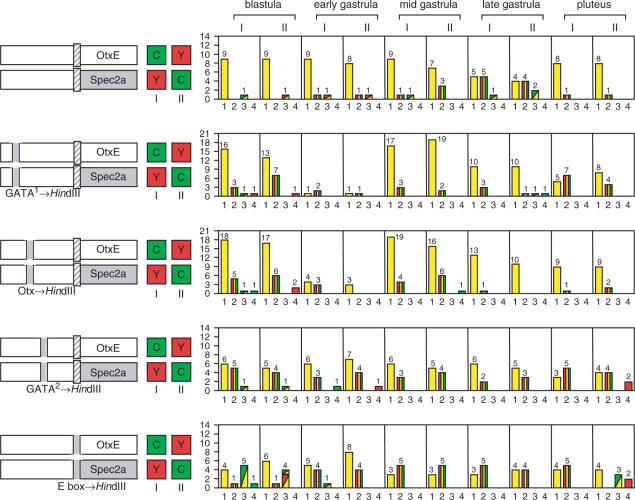
The same sets of mutations of upstream sequences were also tested against the TATA-less HpOtxE and the nonconsensus TATA-containing SpSpec2a core promoters (Figure 7). Although the details are different from the results shown in Figure 6, similar effects were observed; i.e. all mutations affected expression at earlier stages, and mutations of the GATA site 2 and E-box affected all stages. Collectively, these results indicate that different combinations of upstream sequences can select the core promoter in different ways, including through functional interactions with the TATA element.
Figure 7.
Comparison of functional interactions between the different upstream sequences and the HpOtxE/SpSpec2a core promoters. In each panel, the schematic on the left indicates the reporter plasmids introduced into fertilized eggs. I and II refer to which reporter (CFP or YFP) was connected to each promoter, as indicated by the boxes marked C and Y. The number of embryos showing the expression pattern corresponding to each profile is summarized in the panel to the right of each schematic. (First panel) pM4705/pM4731 (I) or pM4725/pM4692 (II), which are driven by the wild-type HpOtxE promoter and the wild-type SpSpec2a core promoter fused with the intact HpOtxE upstream sequence. (Second panel) pM4937/pM4944 (I) or pM4943/pM4938 (II), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #1. (Third panel) pM4939/pM4946 (I) or pM4945/pM4940 (II), in which the HpOtxE upstream sequence was mutated within the putative Otx factor binding site. (Fourth panel) pM4941/pM4948 (I) or pM4947/pM4942 (II), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #2. (Fifth panel) pM4782/pM4789 (I) or pM4788/pM4783 (II), in which the HpOtxE upstream sequence was mutated within the E-box.
Core promoter usage is dynamically regulated in sea urchin embryos during development
Core promoter usage appears to be altered during early development, because the frequencies of embryos displaying differential expression become greater in a stage-specific manner (e.g. see Figure 4A). To verify this developmental transition of core promoter usage, we entrapped transfected embryos in solidified agar and then observed the expression of the two fluorescent reporters in the same embryos semicontinuously (Figure 8). As expected, when the two reporters were driven by the same promoter, only overlapping signals (yellow) were observed, even though the region of expression changed during development (A–D). In contrast, when the two reporters were driven by a set of promoters bearing different core promoters, e.g. HpOtxE and SpSpec2a (E–H, I–L), promoter-specific fluorescent signals were observed in a stage-specific manner, e.g. at blastula (E) or late gastrula stages (K, L). In general, these data appear to be consistent with the results obtained from the analyses examining different embryos at each stage (Figures 4 and 7). Thus, we conclude that core promoter usage may be dynamically regulated in sea urchin embryos during early stages of development.
Figure 8.
Developmental changes in the expression of CFP/YFP reporter genes driven by the same or different pairs of promoters within single embryos immobilized in solidified agar. (A–D). The same set of the CFP/YFP reporter plasmids as used in Figure 3A (bottom panel), pM4705/pM4692, was introduced into fertilized eggs. The expression of these two reporters was monitored semicontinuously within the same embryos using fluorescence microscopy (×200 magnification). Photographs were taken 21 h (A), 25 h (B), 27 h (C) and 31 h (D) after fertilization, and are shown here as overlaid fluorescent and bright-field images as described in Figure 2. (E–H). The same set of CFP/YFP reporter plasmids used in Figure 7 (Third panel, II), pM4945/pM4940, was introduced into fertilized eggs. The expression of these two reporters was monitored as described above. Photographs were taken at 21 h (E), 24 h (F), 26 h (G) and 29 h (H) after fertilization. (I–L). The same set of CFP/YFP reporter plasmids used in Figure 4A (I), pM4705/pM4731, was introduced into fertilized eggs. The expression of these two reporters were monitored as described above. Photographs were taken at 21 h (I), 25 h (J), 27 h (K) and 29 h (L) after fertilization.
DISCUSSION
Here we developed a novel experimental system to simultaneously visualize two distinct functional interactions between upstream and core promoter sequences in real time in sea urchin embryos during early development. Similar but more finely tuned reporter systems based on YFP and CFP have been successfully developed for quantifying cellular variations in gene expression in bacteria (29–31) and yeast cells (32,33). Our system is thus far not as quantitative, but is sufficient to detect the spatiotemporal expression of the reporter genes in sea urchin embryos. Importantly, this system can be applied not only to freely moving embryos, which for technical reasons are not amenable to multiple inspections (Figures 4–7), but also to a single embryo immobilized in solidified agar that can be monitored semicontinuously (Figure 8).
By using this system, we showed that the upstream sequence of the HpOtxE promoter can activate its own promoter and the SpSpec2a core promoter in different embryonic regions, especially at the late gastrula stage (Figure 4A). Intriguingly, the TATA element appears to be involved in this selective expression from the core promoter, because the removal of a nonconsensus TATA element from the SpSpec2a core promoter reduced the level of differential expression at this stage (Figure 4B). The simplest explanation for this phenomenon would be that two or more transcription factors specifically expressed in different embryonic regions at the late gastrula stage bind to the upstream sequence of the HpOtxE promoter and activate TATA-less and TATA-containing core promoters in different ways. In fact, consistent with this explanation, it has been shown that there are two types of transcriptional enhancers in Drosophila, DPE specific and TATA specific (11). Furthermore, the cAMP responsive factor CREB and the oncoprotein c-Fos display substantial preferences for the TATA-containing core promoter, whereas the Ets family protein Elf-1 prefers the TATA-less core promoter for transcriptional activation in mammalian cells (34,35). Therefore, it is likely that at least some of the gastrula-specific transcription factors expressed in sea urchin embryos may also be selective for core promoter elements. In this regard, the GATA site 1 and/or Otx site may constitute a TATA-specific enhancer, as disruption of these sites reduced the level of differential expression, similar to the removal of the TATA element (Figure 7). It is also notable that the differential expression profiles of the two different sets of the TATA-less and TATA-containing promoters were not identical (e.g. compare expression patterns at the pluteus stage in Figure 4A and C). This might be due to the difference of core promoter structures other than the TATA element and/or that of the TATA sequence itself, as the TATA element appears to be functionally heterogeneous (36–38).
We showed that at least four transcription factor binding sites (GATA site 1, Otx site, GATA site 2 and E-box) are involved in the spatiotemporal expression of the HpOtxE promoter (Figure 5). Moreover, when any three of these four sites were combined, they activated transcription in a core promoter-selective manner (Figures 6 and 7). Intriguingly, the differential expression profiles of the TATA-less and TATA-containing core promoters became similar when the GATA site 1 and the Otx site were removed (Figures 6 and 7). Analogously, the expression profiles became similar when the GATA site 2 and the E-box were removed (Figures 6 and 7). However, the resulting two distinct sets of expression profiles were not similar to each other (e.g. compare expression profiles of the second and third panels with the fourth and fifth panels in Figures 6 and 7). These results indicate that the four sites can be divided into two separable functional units that work cooperatively to communicate with the core promoter. Consistent with this, previous studies done by E.H. Davidson's group on the SpEndo16 promoter demonstrated that multiple ‘cis-regulatory modules’, each of which contains ten or more binding sites for at least four transcription factors and confers a specific spatiotemporal expression pattern, communicate with each other to accomplish more complex and developmentally regulated networks of gene expression (39).
One of the most interesting observations from the current study is that core promoter usage determined by the upstream sequence of the HpOtxE promoter is not fixed, but rather is changed dynamically at different developmental stages (Figure 8). To our knowledge, this is the first evidence that the core promoter is differently utilized by the same upstream sequence in one embryo during development. We should also emphasize that these changes are not simply due to the differences of the ‘ON’ (e.g. profiles #2–4) and ‘OFF’ (e.g. profile #1) states of reporter genes but rather represent genuine alterations of the functional relationships between upstream and core promoter sequences, because the generation of detectable fluorescent signals in our assays totally depends on the presence of both sequences (Figure 3B). Our results can be interpreted as indicating that the core promoter is equipped with a filtering function that can sort out the signals coming from a wide range of transcription factors. Why do core promoters have such function? Consider that nearly 80% of all transcription factors (except zinc-finger proteins) encoded by the sea urchin genome are expressed by the late gastrula stage, and transcription factors are generally utilized multiple times and in different cells during development (40,41). Therefore, the filtering function may be indispensable for the core promoter to resolve such complexity and accomplish an elaborate regulatory network of gene expression. Further studies must be undertaken to elucidate the core promoter structures other than the TATA element that are involved in this filtering function for signals from transcription factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank the members of our laboratory for advice and comments on this work. We also thank Dr Masato Kiyomoto (Tateyama Marine Laboratory, Marine and Coastal Research Center, Ochanomizu University, Japan) and Dr Hideki Katow (Research Center for Marine Biology, Tohoku University, Japan) for providing us live sea urchins, and Emi Watanabe and Mamiko Yajima for providing us some of the plasmids used in this study. This study was supported by grants from the Japan Society for the Promotion of Science; the Ministry of Education, Culture, Sports, Science and Technology of Japan; CREST of Japan Science and Technology Corporation; and the Hayashi Memorial Foundation for Female Natural Scientists. Funding to pay the Open Access publication charges for this article was provided by the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Gross P, Oelgeschlager T. Core promoter-selective RNA polymerase II transcription. Biochem. Soc. Symp. 2006:225–236. doi: 10.1042/bss0730225. [DOI] [PubMed] [Google Scholar]
- 3.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat. Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 4.Tokusumi Y, Ma Y, Song X, Jacobson RH, Takada S. The new core promoter element XCPE1 (X Core Promoter Element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol. Cell. Biol. 2007;27:1844–1858. doi: 10.1128/MCB.01363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng W, Roberts SG. Core promoter elements recognized by transcription factor IIB. Biochem. Soc. Trans. 2006;34:1051–1053. doi: 10.1042/BST0341051. [DOI] [PubMed] [Google Scholar]
- 6.Cheng JX, Floer M, Ononaji P, Bryant G, Ptashne M. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 2002;12:1828–1832. doi: 10.1016/s0960-9822(02)01257-5. [DOI] [PubMed] [Google Scholar]
- 7.Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit BL, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 8.Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 9.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 10.Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 11.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis W, Jr, Schultz RM. Developmental change in TATA-box utilization during preimplantation mouse development. Dev. Biol. 2000;218:275–283. doi: 10.1006/dbio.1999.9486. [DOI] [PubMed] [Google Scholar]
- 13.Majumder S, DePamphilis ML. TATA-dependent enhancer stimulation of promoter activity in mice is developmentally acquired. Mol. Cell. Biol. 1994;14:4258–4268. doi: 10.1128/mcb.14.6.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan ZJ, Fang X, Rohde A, Han H, Stamatoyannopoulos G, Li Q. Developmental specificity of recruitment of TBP to the TATA box of the human gamma-globin gene. Proc. Natl Acad. Sci. USA. 2002;99:5509–5514. doi: 10.1073/pnas.072084499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen SK, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 16.Howcroft TK, Raval A, Weissman JD, Gegonne A, Singer DS. Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol. Cell. Biol. 2003;23:3377–3391. doi: 10.1128/MCB.23.10.3377-3391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi A, Kokubo T, Ota Y, Yokoyama S. Promoter-specific function of the TATA element in undifferentiated P19 cells. Biochem. Biophys. Res. Commun. 2003;310:458–463. doi: 10.1016/j.bbrc.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi A, Akasaka K, Kawaichi M, Kokubo T. Functional interaction between TATA and upstream CACGTG elements regulates the temporally specific expression of Otx mRNAs during early embryogenesis of the sea urchin,Hemicentrotus pulcherrimus. Nucleic Acids Res. 2002;30:3034–3044. doi: 10.1093/nar/gkf439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 20.Akasaka K, Nishimura A, Takata K, Mitsunaga K, Mibuka F, Ueda H, Hirose S, Tsutsui K, Shimada H. Upstream element of the sea urchin arylsulfatase gene serves as an insulator. Cell. Mol. Biol. (Noisy-le-grand) 1999;45:555–565. [PubMed] [Google Scholar]
- 21.Yajima M, Kiyomoto M, Akasaka K. Ars insulator protects transgenes from long-term silencing in sea urchin larva. Dev. Genes. Evol. 2007;217:331–336. doi: 10.1007/s00427-007-0140-9. [DOI] [PubMed] [Google Scholar]
- 22.Kurita M, Kondoh H, Mitsunaga-Nakatsubo K, Shimotori T, Sakamoto N, Yamamoto T, Shimada H, Takata K, Akasaka K. Utilization of a particle gun DNA introduction system for the analysis of cis-regulatory elements controlling the spatial expression pattern of the arylsulfatase gene (HpArs) in sea urchin embryos. Dev. Genes. Evol. 2003;213:44–49. doi: 10.1007/s00427-002-0281-9. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto N, Akasaka K, Mitsunaga-Nakatsubo K, Takata K, Nishitani T, Shimada H. Two isoforms of orthodenticle-related proteins (HpOtx) bind to the enhancer element of sea urchin arylsulfatase gene. Dev. Biol. 1997;181:284–295. doi: 10.1006/dbio.1996.8455. [DOI] [PubMed] [Google Scholar]
- 24.Brandhorst BP, Klein WH. Molecular patterning along the sea urchin animal-vegetal axis. Int. Rev. Cytol. 2002;213:183–232. doi: 10.1016/s0074-7696(02)13015-4. [DOI] [PubMed] [Google Scholar]
- 25.Kozlowski MT, Gan L, Venuti JM, Sawadogo M, Klein WH. Sea urchin USF: a helix-loop-helix protein active in embryonic ectoderm cells. Dev. Biol. 1991;148:625–630. doi: 10.1016/0012-1606(91)90280-g. [DOI] [PubMed] [Google Scholar]
- 26.Hough-Evans BR, Britten RJ, Davidson EH. Mosaic incorporation and regulated expression of an exogenous gene in the sea urchin embryo. Dev. Biol. 1988;129:198–208. doi: 10.1016/0012-1606(88)90174-1. [DOI] [PubMed] [Google Scholar]
- 27.Hardin PE, Angerer LM, Hardin SH, Angerer RC, Klein WH. Spec2 genes of Strongylocentrotus purpuratus. Structure and differential expression in embryonic aboral ectoderm cells. J. Mol. Biol. 1988;202:417–431. doi: 10.1016/0022-2836(88)90275-6. [DOI] [PubMed] [Google Scholar]
- 28.Hayashibara Y, Mitsunaga-Nakatsubo K, Sakamoto N, Shimotori T, Akasaka K, Yamamoto T. The Otx binding site is required for the activation of HpOtxL mRNA expression in the sea urchin,Hemicentrotus pulcherrimus. Dev. Growth. Differ. 2004;46:61–67. doi: 10.1111/j.1440-169x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 29.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 30.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 31.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 34.Conkright MD, Guzman E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 35.Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 36.Simon MC, Fisch TM, Benecke BJ, Nevins JR, Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 37.Wefald FC, Devlin BH, Williams RS. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature. 1990;344:260–262. doi: 10.1038/344260a0. [DOI] [PubMed] [Google Scholar]
- 38.Perera LP. The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62. J. Biol. Chem. 2000;275:487–496. doi: 10.1074/jbc.275.1.487. [DOI] [PubMed] [Google Scholar]
- 39.Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc. Natl Acad. Sci. USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard-Ashby M, Materna SC, Brown CT, Tu Q, Oliveri P, Cameron RA, Davidson EH. High regulatory gene use in sea urchin embryogenesis: implications for bilaterian development and evolution. Dev. Biol. 2006;300:27–34. doi: 10.1016/j.ydbio.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.