Abstract
Development and refinement of methods to analyse differential gene expression has been essential in the progress of molecular biology. A novel approach called iGentifier is presented for profiling known and unknown transcriptomes, thus bypassing a major limitation in microarray analysis. The iGentifier technology combines elements of fragment display (e.g. Differential Display or RMDD) and tag sequencing (e.g. SAGE, MPSS) and allows for analysis of samples in high throughput using current capillary electrophoresis equipment. Application to epidermal tissue of wild-type and mlo5 barley (Hordeum vulgare) plants, infected with powdery mildew [Blumeria graminis (DC.) E.O. Speer f.sp.hordei], led to the identification of several 100 genes induced or repressed upon infection with many well known for their response to fungal pathogens or other stressors. Ten of these genes are suggested to be classified as marker genes for durable resistance mediated by the mlo5 resistance gene.
INTRODUCTION
Transcriptome analysis can be performed in a knowledge-based manner [‘closed systems’ such as microarrays (1,2)] or independent of any assumptions [‘open systems’, (3–6)] as reviewed in (7). The advantage of the latter for less well-analysed organisms is obvious. But even when investigating the human transcriptome, analysis of a number of important processes such as alternative splicing, polyadenylation and the generation of antisense transcripts requires technologies that utilize only a few assumptions regarding the transcriptome's composition. Current ‘open systems’ comprise fragment display, tag sequencing and subtractive hybridization (7). Fragment display technologies (3,4,8,9) rely on the generation of cDNA fragments either by arbitrary priming or by the use of restriction enzymes. Fragments are labelled and subjected to size separation by gel electrophoresis, and corresponding fragment patterns (‘fingerprints’) from different biological sources are compared. Differentially expressed genes are represented and, thus, identified by fragments differing in abundance between samples. These technologies are particularly appealing given their technical simplicity, low costs for primary analysis involving the comparison of fingerprint patterns obtained from different samples and robustness. Nonetheless, secondary analysis for the assignment of corresponding genes to displayed signals indicating differential expression remains a major bottleneck. The traditional approach of physical isolation followed by reamplification and sequencing (3) is cumbersome and error-prone. Identification of fragments by their mobility and thus their predicted physical length (8,9) is of little use for unknown transcripts and has proved unreliable since electrophoretic mobility is strongly influenced by a fragment's base composition (10), rendering length predictions inaccurate. Very much like fragment display technologies, tag-sequencing approaches rely on the comparative quantitation of cDNA fragments. However, in this case the quantitation is achieved by determining the frequency of occurrence of a given sequence tag (sometimes called a ‘signature’) within a large population of sequenced tags. Sequencing of the tags can occur in a serial manner [SAGE, (5)] or in a massively parallel fashion [MPSS, (6)]. Tag sequences are then used to identify the corresponding transcripts by database searches.
iGentifier could be regarded as a hybrid technology which combines elements of both fragment display and tag sequencing. Our approach allows for the identification of displayed fragments by assigning to each an 18-bp sequence tag as a gene identifier. The parallel architecture of iGentifier avoids the massive oversampling of abundant transcripts typical for tag sequencing methods, thus overcoming the major costs of SAGE and its variants (11–13).
MATERIALS AND METHODS
Plant treatment
A pair of near-isogenic lines of barley, cv Ingrid Mlo and cv Ingrid BC7 mlo5, were grown in pots of compost soil (from the IPK nursery) in a greenhouse with automatic shading and supplementary light (sodium-halogen lamps) with a light period of 16 h. Temperature ranged from 18°C at night to 21°C during the day. Seven-day-old seedlings were used for all inoculation experiments. Blumeria graminis (DC.) E.O. Speer f.sp. hordei, strain 4.8 carrying AvrMla9, was cultivated by weekly inoculation of 7-day-old seedlings of barley cv Golden Promise. Seven days post-inoculation, conidia were used for inoculating test plants by shaking inoculated plants over test plants in a settling tower of ∼60 × 60 × 60 cm in dimension. A split-plot design was used for the simultaneous inoculation of both barley lines. Control and inoculated plants were incubated at a constant temperature of 20°C and exposed to natural daylight until RNA extraction.
RNA isolation
The abaxial epidermis of primary leaves was stripped 12 h post-inoculation and immediately frozen in liquid nitrogen. Care was taken to strip all four samples [two genotypes each with two treatments (control and inoculated)] concurrently to prevent artefacts from genes under circadian regulation. Ground material was suspended in a hot (80°C) 1:1 mixture of phenol and extraction buffer (100 mM LiCl, 10 mM EDTA, 1% SDS, 100 mM Tris pH 9.0). After addition of 0.5 vol. chloroform, samples were mixed for 30 min. at room temperature. Extraction was repeated and then samples were adjusted to 2 M LiCl and precipitated. Digesting RNA preparations with DNase I proved to be not required and thus was omitted from our protocol.
Display reactions
cDNA synthesis, fragmentation and amplification were performed according to (4) with the following exceptions: fragmentation was achieved with 20 U of AluI (New England Biolabs); and ligation of blunt-ended linker DAL4138 (upper strand: 5′-TGGATAGAGCAGTGGTAGCACACGTGAGCGATGACTATGAG-3′, lower strand: 5′-CTCATAGTCATCGCTCACGTGTCGGCACATCTCATATA-3′) was performed with 1 U of T4 DNA Quick ligase (New England Biolabs) in the reaction buffer. PCR was executed with 1 U of Taq polymerase (Invitrogen) pre-incubated with TaqStart antibody (Clontech) in a total volume of 20 μl using microtitre plates and PE 9700 thermocyclers (Perkin-Elmer). All four extension bases were positioned on the fragments’ 3′-end. For example, subpool ‘GCAT’ was generated using the primer CP28GCAT (5′-ACCTACGTGCAGATTTTTTTTTTTTTTTGCAT-3′). To visualize amplification products, linker primers were labelled at their 5′-end with a fluorescent FAM group. Reactions were mixed with GeneScan 500 length standard (Applied Biosystems) and analysed on an ABI 3100 capillary sequencing apparatus (Applied Biosystems). Separation of cDNA fragments was performed according to the manufacturer's recommendations. Trace file peaks were recognized by the DAX software package (van Mierlo Software, Eindhoven, NL) using a 4-fold local baseline as a general threshold. The Java-based inhouse iGentifierMATCH software imported the detected signals as unique addresses into a Sybase database. A native Java driver allowed for a higher performance than the usual JDBC/ODBC Bridge. After calibration of each fluorogram via a third-degree polynomial function, an automated QC algorithm checked data integrity. Passed fluorograms were then aligned following an algorithm which processed peaks according to the calibrated fragment lengths, peak patterns and normalized signal intensities calculated on the fly. Alignment was accomplished in three stages: (i) display alignment, (ii) indexing alignment and then (iii) merging of aforementioned. Each match (comprising all display and indexing peaks of the same size, from the same subpool, and with assignment to each other) was given a unique match ID and comprised expression data of one particular transcript across all analysed samples as well as the corresponding sequencing data, i.e. the respective signature. Data were then visualized in a signature list linked to a fluorogram view, comprising output data of a BLAST search against the HarvEST database (http://harvest.ucr.edu). Final data analysis and identification of gene regulation events was effected with GeneSpring software (Silicon Genetics). Copies of the iGentifierMATCH software are available upon request at no charge from A.F.
Indexing reactions
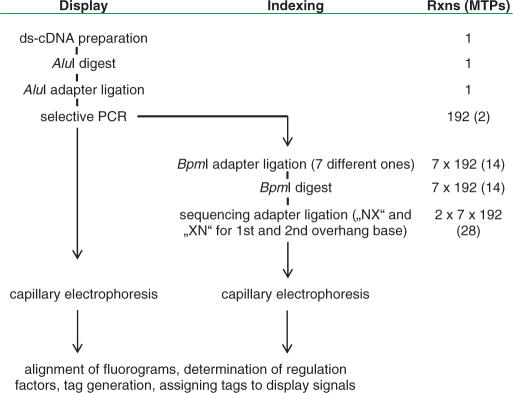
An overview of the workflow and of the interrelation of display reactions and sequencing reactions can be taken from Table 1 and Figure 1. Equal volumes of display reactions of all four conditions (MLO/mlo5 and infected/non-infected) each were pooled subpool-wise (i.e. pooling involved reactions from corresponding subpools out of the total 196 subpools) and cleaved with AluI to remove the adaptor sequence. Reactions were split into seven aliquots and subjected to IIs adaptor ligation with each of seven different adaptors per reaction. Each of the adaptors contained a recognition site for restriction endonuclease BpmI at a different position from the ligatable end Adapter ligation was conducted for 1 h at 20°C, using 1 U of T4 DNA ligase (Roche) in a total volume of 20 μl. IIs adaptors were released using 5 U BpmI (New England Biolabs) in a reaction volume of 50 μl for 1 h at 37°C. Reactions were heat-inactivated for 20 min at 65°C. Each digest was split into two aliquots and subjected to ligation of a sequencing adaptor with the first aliquot to be interrogated for each fragment's first (‘inner’) overhang base and the second aliquot interrogated for the overhang's second (‘outer’) base. Ligation of sequencing adaptors also took place for 1 h at 37°C using 1 U of T4 DNA ligase in a volume of 20 μl. Unligated adaptors were removed using Montage PCR96 plates (Millipore). A 2 μl amount of each sequencing reaction was mixed with 1-μl GeneScan 500 size standard and 14-μl HiDi (Applied Biosystems) and subjected to capillary electrophoresis on an ABI 3100 sequencer. Trace files were handled as described above, except that the detected signals were subjected to an automated base-calling step. Corresponding fluorograms (representing bases 5–18 of the displayed fragments) were aligned and, by adding the already known 4-bp sequence of restriction endonuclease AluI, used to assemble 18-bp signatures of the displayed fragments.
Table 1.
Flowchart of iGentifier
Flowchart of iGentifier. In the ‘Rxns (MTPs)’ column, the number of reactions performed in parallel at the respective stage of the iGentifier protocol are indicated. Numbers in brackets refer to the number of microtitre plates used for performing the enzymatic reactions, corresponding to the number of runs on a 96-capillary sequencing machine. Generating a gene expression fingerprint requires two runs per sample, a full indexing requires 28 runs per species and type of tissue.
Figure 1.
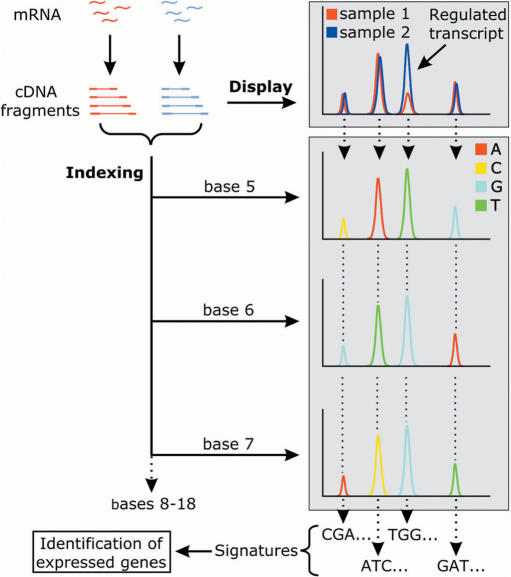
Architecture of iGentifier. In display reactions, RNA is used for synthesis of oligo(dT)-primed ds-cDNA, cDNA is fragmented with a frequently cutting restriction enzyme (AluI), and adaptors are ligated to provide a primer-binding site. With an adaptor primer and a primer derived from the cDNA primer sequence, specific amplification of the 3′-most fragment of each cDNA species is achieved. The enzymatic steps for generating signatures for fragment identification are described in the text. For each of the 14 of 18 signature bases to be determined, an individual indexing reaction is performed. Each indexing reaction provides one base-pair sequence information for any detected fragment.
Signature annotation
Gene identity was deduced from iGentifier's 3′ signatures with a method that relied on BLAST as a search engine. The procedure avoided BLAST's limitations in searching short fragments but still allowed handling of ambiguities in the signature or mismatches between signature and database sequences. The procedure first expanded all ambiguous nucleotides and thus created a set of non-ambiguous deduced signatures. A BLASTN search against the selected database was then performed with default parameters as well as ‘−F F’, ‘-q −1’ and ‘−W 7’. The first parameter guarantees that low complexity fragments within the signatures do not interfere with the search while the second allows the recognition of ‘close to signature end’ mismatches and the third ensures that sufficient consecutive nucleotides match despite a mismatch in the middle region of the signature. Since mismatches in the first or last bases are not reported by BLASTN, the results here were post-processed by extending the alignment over the length of the signature by adding matches corresponding to the BLASTN alignment. The matching database entries were then sorted according to hit- and database-entry-quality. While we did not allow for gaps to take place in the alignment, up to two mismatches per signature were allowed to compensate for sequence polymorphisms as well as for sequencing errors.
Reamplification of 3′-ends
For each fragment to be reamplified, a PCR primer was synthesized corresponding to the respective 18-bp signature sequence. Using 1 µl of a 1:30 dilution of the respective display reaction as a template, 50 µl reactions were assembled containing 10 µM each of fragment specific primer and subpooling primer, 1.5 mM MgCl2, and 2.5 U Taq polymerase in 1× PCR buffer (Invitrogen). Amplification was effected over 25 cycles with duration 30 s at 94°C, 30 s at 60°C and 1 min at 72°C for each cycle.
Real-time PCR
Quantitative RT–PCR was performed using the Roche LightCycler according to the manufacturer's recommendations.
Identification of regulated genes
All data were derived from three independent inoculation experiments. Pathogenesis-related genes were selected based on two criteria: (i) up- or down-regulation by at least 3-fold which was based on average signal intensities from control and inoculated samples; and (ii) a statistically significant difference between control and inoculated samples with P < 0.05 (student's t-test). Resistance- or susceptibility-related genes were respectively selected utilizing the two criteria described above, as well as a difference in the signal intensities (by a factor of at least five) between inoculated barley lines in the presence or absence of the mlo5 resistance gene.
RESULTS
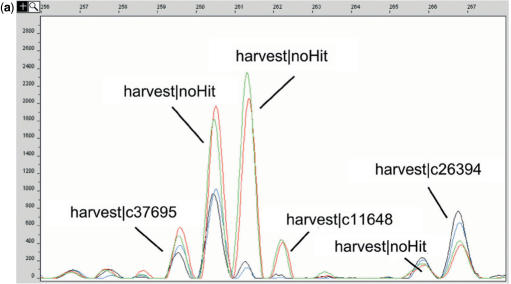
Comparison of biological samples in iGentifier employs a fragment display procedure similar to the already published RMDD protocol (4). RNA is first converted to double-stranded cDNA which is cut using a frequently cutting restriction endonuclease, AluI. So-called linkers (double-stranded oligonucleotides with one blunt end, which allows unambiguous orientation of the linkers upon ligation) are ligated to the fragment ends, and cDNA 3′-fragments are amplified by PCR. Amplification is achieved by use of PCR primers capable of binding to a sequence corresponding to either one of the linker strands or to the reverse complement of the oligo(dT) cDNA primer. Since linkers are not phosphorylated, only one of the two linker strands is covalently attached to the cDNA fragments. Thus, amplification of a fragment takes place only when, upon denturation, the potential primer binding site remains attached to the respective strand. This is the case only for the reverse complement of the oligo(dT) cDNA primer, thus allowing selective amplification of cDNA 3′-fragments, while internal fragments remain unamplified. In order to reduce complexity, PCR primers carrying additional ‘selective’ 3′-bases are employed. The rationale behind this step is to use primers which consist of (i) a ‘universal’ sequence which is capable of hybridizing to the cDNA primer sequence incorporated into any cDNA 3′ fragment plus (ii) one or more ‘selective’ bases at the primers’ 5′-ends which allow for primer extension only when the corresponding base or bases on the template strand is/are complementary to the selective base(s) of the primer. Thus, subdivision of all cDNA fragments to be amplified into 3 × 4 × 4 × 4 = 192 different reduced-complexity subpools is achieved, corresponding to two 96-well plates per sample. Use of fluorescently labelled primers allows reactions to be ‘displayed’ on automated sequencers (Figure 1). Thus, each RNA preparation is represented by a set of 192 fluorograms providing a fingerprint of the respective transcriptome. After normalization, comparison of the signal intensities (‘peak heights’) between corresponding reactions allows for straightforward detection of gene regulation events (Figure 2a). Peak calling, normalization, cross-sample alignment of corresponding signals and calculation of relative expression levels are performed automatically. With the use of current automatic sequencers, expression profiles can be generated with high throughput. A single 96 capillary sequencer allows for analysis of more than 300 biological samples per month. Spiking experiments, employing in vitro transcribed, oligo-adenylated RNAs as ‘pseudo-mRNAs’, demonstrated a sensitivity sufficient to detect transcripts at a relative abundance of 1:100 000, which is in line with published fragment display technologies (4,8).
Figure 2.
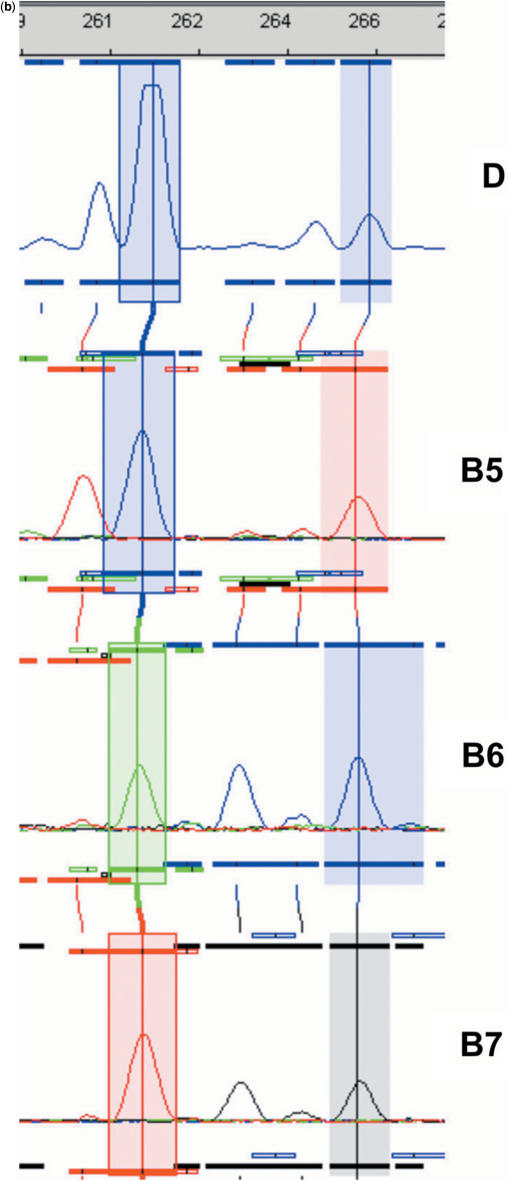
iGentifier fluorograms. (a) Display of expressed barley genes. The window width corresponds to 12 bp. Coloured lines indicate plant conditions: black, wild-type plants non-infected; blue, mlo5 plants non-infected; green, wild-type plants infected; and red, mlo5 plants infected. (b) Signature generation (only 3 of 14 bases shown). D, display reaction; B5–B7, indexing reaction of bases 5–7. After addition of four bases corresponding to the AluI site, signatures for the two highlighted fragments read as (AGCT)GTA …, and (AGCT)AGC ….
For gene identification, the basic idea was to implement a so-called ‘orthogonal sequencing approach’ for generating cDNA-specific sequence tags. While, in standard sequencing, fluorescence signals encoding for the identity of a template's consecutive bases are obtained from one capillary and independent templates are analysed by electrophoresis in independent capillaries, iGentifier's orthogonal sequencing provides sequence information for one particular cDNA fragment using one capillary per base, while many fragments are analysed simultaneously in the same set of capillaries. For example, determination of 14 bases each for a set of 100 fragments different in size requires no more than 14 capillaries, the first capillary providing the signals encoding for the identity of each of the fragments’ first base, the second capillary disclosing the identity of each of the 100 fragments’ second base, and so forth (Figures 1 and 2b). Thus, iGentifier takes advantage of the power and throughput of the highly sophisticated latest-generation automatic capillary sequencers, without the requirement of any specialized instruments. Employing this orthogonal sequencing approach, each displayed fragment (representing an expressed gene) is assigned an 18-bp long sequence tag or ‘signature’. The first four signature bases are defined by the restriction enzyme used for fragment generation, while the remaining 14 bases are determined by orthogonal sequencing of fragment mixtures.
In practice, each of the 192 display reactions is split into seven identical aliquots (Table 1). For each case, one of seven different adaptors carrying a recognition site for a type IIs enzyme is then attached. As IIs enzyme, BpmI was chosen which has the cutting characteristics CTGGAG(N)16/14 (i.e. cutting the ‘upper’ strand 16 bases away and the ‘lower’ strand 14 bases away of its recognition site, CTGGAG, and thereby generating 2-base overhangs). After cutting with BpmI, a defined portion of each fragment (the exact position of which depends on the position of the BpmI recognition site within the attached adaptor) is converted into a 2-base overhang amenable to base identification. Successive adaptor's BpmI recognition sites are displaced by two bases each: The position of the BpmI site within the set of seven adaptors is 13, 11, 9, 7, 5, 3 or 1 from the adaptors’ ligatable ends. Thus, digest of the fragments linked to the ‘first’ IIs-adaptor exposes all the fragments’ ‘first two bases’ as an overhang; digest of the same fragments linked to the ‘second’ IIs-adaptor exposes ‘bases three and four’ of each fragment, and so forth (Figure 3a, Supplementary Data). Identification of the overhanging bases is achieved by sequence-specific ligation of fluorescently labelled ‘sequencing adapters’. A sequencing adapter is characterized by a single-stranded overhang of defined length and sequence, and a fluorescence label specific for a particular overhang sequence. In this case, overhangs are two bases long, which corresponds to the length of the fragments’ overhangs having been generated by the BpmI digest. Sequencing adaptors have the structure Fluo-Core-NX or Fluo-Core-XN, wherein ‘Core’ refers to a double-stranded core sequence common to all adaptors; ‘N’ to equimolar mixture of A, C, G and T; ‘X’ to a sequencing nucleotide (either A, C, G or T); and ‘Fluo’ to fluorophore encoding the sequencing nucleotide. Upon ligation, each fragment is linked to one of four fluorophores, each in turn serving as a base identifier for a given base position within the overhang, which depends on the set of sequencing adaptors employed (Figure 3b, Supplementary Data). Capillary electrophoresis then allows for identification of one base, each at a predetermined fixed position, for all fragments simultaneously (Figure 2b). Performance of 14 sequencing reactions for a given display reaction (seven digests and two bases per overhang) followed by alignment of corresponding fluorograms and execution of a base-calling step provides a contiguous stretch of sequence information for any cDNA fragment present. With the addition of the known first four bases, an 18-bp signature for each expressed gene can be determined. Performing all enzymatic steps in 96-well microtitre plates allows for automation of the complete indexing protocol by use of liquid handling stations. The determination of all signatures detectable within a given transcriptome requires 28 96-well plates (Table 1).
Sixty percent of the barley iGentifier signatures identified in our study could be assigned to a corresponding HarvEST database entry. To allow for gene identification of the remaining signatures, the full sequence of the corresponding fragments can easily be obtained by using the signature for oligonucleotide primer design and selectively amplifying the corresponding fragment. This approach was tested with 23 randomly chosen fragments which yielded no hit with a signature BLAST search against the HarvEST database (http://harvest.ucr.edu). In 11 cases (48%), a corresponding database entry could be found. In the remaining 12 cases, a PCR product and a clearly readable sequence were obtained, but no BLAST hit found in HarvEST (data not shown).
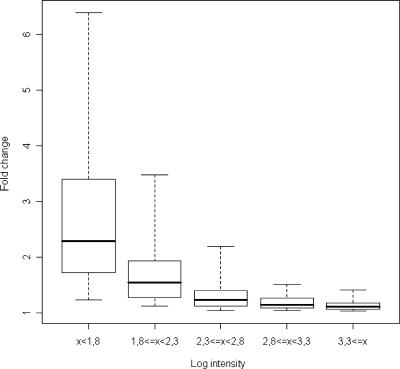
Five technical replicates were prepared to examine the reproducibility of iGentifier. From these reproducibility data the confidence of measured regulation factors, depending on the signal intensity and number of replicates, was estimated. For example, signals of average intensity yielded a lower limit for reliably detectable regulation events (α = 0.001) near 1.5-fold if no replicates were utilized (Figure 4).
Figure 4.
Reproducibility of iGentifier. To generate a performance figure for the reproducibility of iGentifier data it was assumed that the SD (and, thus, the confidence) is a function of signal intensity. Under this assumption, gene-specific effects on the SD are neglected. The total range of logarithmic signal intensities was subdivided into five sub-ranges of equal size (x-axis). The standard deviations of the replicate measurements of the genes in each of the sub-ranges were used to calculate confidence values, which are displayed in the box-whisker plots on non-logarithmic scale. In each case, the horizontal bar marks the median of the confidence values. The range marked by the box comprises 50% of all confidence values and 90% of all confidence values lie within the range marked by the dotted lines.
iGentifier was applied in the analysis of barley epidermal cells infected by barley powdery mildew (Blumeria graminis hordei, Bgh). The study included a comparison of two near-isogenic barley lines differing in the absence or presence of the mlo5 resistance gene, one of several recessive loss-of-function alleles of the Mlo gene which mediates durable and race-non-specific resistance to Bgh (15–17). Plant material from three independent experiments was included to assess biological variability. Since the primary events of pathogen attack and plant response take place in the epidermis of affected leaves, RNA was extracted from such stripped tissue of treated and untreated leaves.
A large number of genes were detected as either up- or down-regulated upon infection. Response of both lines to infection was similar. In mutant plants, 291 signatures (235 signatures in wild-type plants) indicating genes up- or down-regulated at least 3-fold upon infection were identified, 189 of which (145 in wild-type plants) were detected in the HarvEST database (Tables 2 and 3, Supplementary Data). The lower number of differentially expressed genes in the susceptible line mostly resulted from less robust expression patterns with subsequent removal of several genes by data filtration. The group of genes strongly regulated in resistant plants was further characterized by determining the percentage of genes that were similar to barley expressed sequence tags (ESTs) encoding for known pathogen–response (PR) proteins (Table 4, Supplementary Data). A major fraction of ESTs corresponded to novel defence-related genes not previously described (Table 5, Supplementary Data).
In order to verify results by an independent method, 14 transcripts indicated as up-regulated were tested by real-time PCR in all four experimental conditions (mutant and wild type, each coupled with infected and non-infected). Although for some transcripts regulation factors as determined by iGentifier deviated more than 2-fold from RT–PCR factors, up-regulation per se could be clearly confirmed in all cases (Table 6, Supplementary Data). This finding is not surprising since regulation factors determined with different methods (e.g. northern blot versus microarray versus RT-PCR data) frequently deviate from each other, which appears to reflect sequence-specific bias inherent to the particular method employed (18). A recent study shows that even within one given microarray platform, the choice of the data processing method has a non-neglectable influence on the gene regulation data obtained (19).
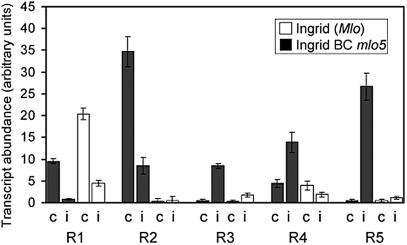
Comparing inoculated and resistant plants with those that were susceptible revealed six and 10 marker genes of susceptibility and mlo-mediated resistance that differed in transcript abundance by a factor of at least four (Table 7, Supplementary Data). The transcript abundance of some resistance-related and pathogen-regulated marker genes with the strongest genotype-dependent differences is shown in Figure 5.
Figure 5.
Genes associated with mlo-mediated resistance. Pathogen–response genes were selected that differed in their transcript abundance at least 4-fold amongst inoculated, susceptible and inoculated, resistant barley lines. Mean values and corresponding SDs for three independent inoculation experiments are shown where ‘c’ refers to the non-inoculated control and ‘i’ indicates inoculation with Bgh. BLASTX, which was applied to barley ESTs and sequence signatures of fragments R1 to R5, revealed sequence identity of R1 to a glutaredoxin-like protein (Acc. No. T48552). Fragments R2–R5 either did not match a barley EST sequence or could not be used for BLASTN analysis due to signature sequence ambiguities.
DISCUSSION
In contrast to SAGE and MPSS, iGentifier signature sequencing is required only once per species and tissue type, rendering this approach much less expensive than other tag-sequencing technologies. Even if hundreds of different biological samples are to be investigated, the identity of expressed genes can be determined by one single indexing experiment. Thus the deposition of iGentifier signature catalogues of a specific biological material in a central database would allow scientists to generate their own sets of display data and interpret them on the basis of publicly available indexing data.
One of the major technical hurdles during iGentifier development turned out to be the extremely high dynamic range of gene expression, i.e. the difference in copy numbers of high and low abundance transcripts. We estimate this dynamic range to be approximately several thousand to one, which clearly exceeds the dynamic range of commercially available sequencing machines. It turned out that a very simple way of solving this problem is to overload the capillaries in a way that the few fragments representing the most abundant transcripts fall out of the detector's linear range. The respective signals get clipped and are excluded from quantitative analysis. Under these conditions, signals from low abundance genes are strong enough to allow for reliable quantitation. It is interesting to note in this context that the sensitivity of iGentifier regarding low abundance transcripts appears to be higher than the sensitivity of MPSS, since MPSS signature collections are seriously dominated by a small number of very high abundance signatures. On the other hand, low abundance signatures with an abundance of <100 copies per million turned out to be very unreliable due to sequencing errors (our unpublished data). In the past, the only way to improve the sensitivity of MPSS was to collect data from many runs which caused prohibitive costs. It actually was the astronomically high reagent costs and poor data quality of MPSS that triggered development of iGentifier in our group.
While a high dynamic range still can be easily managed on the level of the display reactions, it is more difficult to cope with when it comes to signature identification of low abundance transcripts. For unknown reasons, a certain reproducible fluctuation of signal intensities is observed upon performing signature sequencing. In other words, there is no strict linearity between signal intensity of a display reaction and signal intensity of the corresponding sequencing reactions. While this does not impair correct signature identification of medium and high abundance transcripts, signature bases of very low abundance transcripts sometimes ‘drop out’, rendering correctly calling the affected signature base(s) impossible. Thus, future modifications of iGentifier will have to address signature sequencing of low abundance transcripts. One possibility to achieve an improved sequencing of low abundance transcripts might be to subject fragment mixtures to a normalization step before entering the sequencing branch of the protocol.
An important question is the usefulness of our signatures for correctly identifying the corresponding expressed gene. It has been shown that a signature length of 16–17 bp is sufficient for unambiguous gene identification in ∼92% of human genes (14), rendering the iGentifier approach suitable for analysis of any complex eukaryotic organism. However, this does not exclude ambiguity of a certain fraction of signatures, and this fraction may differ from organism to organism. While EST libraries proved valuable for identification of (differentially) expressed genes, other sorts of databases such as, e.g. genomic libraries might be useful as well. Even without any sequence information at all available of the species under investigation, iGentifier would allow identification of expressed genes by taking the approach described above of reamplifying and sequencing cDNA fragments of interest by use of gene-specific signature primers. The sequences obtained by this route could be subjected to a ‘heterologous BLAST’ search in sequence databases of related species. It might turn out, however, that the lesser degree of inter-species sequence conservation within cDNA 3′-ends, as compared to the degree of conservation within open reading frames, would necessitate the adaptation of iGentifier to ‘internal’ cDNA fragments with a higher likelihood of containing coding information. Also, due to the effort required for reamplification and sequencing of many individual fragments, such an analysis could not be called a high throughput approach any more.
Also, any expression profiling technology has to address the issue of comprehensiveness. Theoretically, the strength of open systems is their inherent comprehensiveness. In practice, several factors may compromise perfect comprehensiveness of a given technology. For example, SAGE experiments are subject to cost constraints, which in practice reduces the number of detectable genes. The fact that, by nature of SAGE's experimental design, abundant sequence tags have to be sequenced repeatedly when rare tags are to be identified as well, forces the experimentator to define a balance of cost and sensitivity. In a typical SAGE experiment, 10 000–30 000 tags are sequenced, which seriously limits the coverage of low abundance transcripts. While repeated sequencing of the more abundant tags is avoided by our orthogonal sequencing approach, which dramatically reduces the cost per tag, iGentifier shares a restriction step with all other signature sequencing technologies as well as with the more recent, restriction-based fragment display technologies. This means that those transcripts will escape detection which lack the corresponding restriction enzyme recognition site. We chose AluI as restriction enzyme for cDNA fragmentation since simulations unveiled that, when applied to barley cDNA, ∼85% of cDNAs result in a cDNA fragment in the ‘displayable’ size range between 60 and 750 bp. Coverage could be even increased to ∼97% by repeating the analysis on the basis of another frequent cutter, however, at the cost of doubling the effort for each sample. It should be pointed out that comprehensiveness of iGentifier is virtually not limited by the size of this technology's fragment space, i.e. the number of different fragments which can be independently displayed. In this context, it has to be kept in mind that, in capillary electrophoresis, fragments are not separated by size per se, but by mobility. While fragment size is quantized, mobility is a continuum, depending on factors such as a fragment's charge, size, A/T content, secondary structure and others. Our experiments indicated that effective resolution of state-of-the-art capillary sequencers is ∼0.3 bp. In other words, two fragments differing in apparent size (calculated, with the help of an internal size standard, from the measured mobility) by 0.3 bp can be reliably and reproducibly separated and distinguished from each other. Thus, analysing fragments in the apparent size range from 60 to 750 bp provides a fragment space of (750 − 60) × (1/0.3) × 192 = 441 600 ‘theoretical bins’. Even when fragment size distribution is not homogenous, but rather resembles a bell-shaped distribution, the number of theoretical bins greatly exceeds the number of different mRNA species expected to occur in a given cell type [according to (3) ∼15 000 species]. Accordingly, the overlap of signals representing two similarly sized fragments turned out to be an extremely rare event.
Employing iGentifier for the analysis of powdery mildew-infected barley plants, we could identify several hundred signatures indicating transcripts up- or down-regulated upon infection. Interestingly, the BLAST hit-rate of these signatures was higher among down-regulated genes (75%) than up-regulated genes (58%). This trend was confirmed when analysing strongly (>10-fold) regulated genes in mlo5 plants (Table 4, Supplementary Data). It is suggested that, in addition to a number of fungal genes covered by the presented indexing approach, many up-regulated genes escaped detection in EST collections due to a combination of epidermis-specific and pathogenesis-specific expression patterns. The proportion of epidermal RNA in a whole-leaf preparation is estimated at no more than 5%, causing a significant dilution of transcripts involved in the primary response to pathogen attack in preparations typically utilized for leaf library construction.
Remarkably, comparison of regulation patterns of wild-type and mlo5 mutant plants suggests that several of these transcripts might be qualitative markers for resistance, in contrast to a recent cDNA array-based study on mlo-mediated resistance where only quantitative differences were found (20). A possible explanation for this is that the array (complexity ∼3200 unigenes, which is only a small fraction of the barley transcriptome) represents a closed resource, thereby minimizing the chance to identify rare, qualitative resistance markers. Cloning the marker genes for resistance coupled with functional analysis by reverse genetics (21) should elucidate their function in the response of epidermal cells to Bgh. In addition, a group of genes could be identified that show different expression behaviours between barley lines independent of inoculation (Tables 2 and 3, Supplementary Data), allowing a general study of the effect of the Mlo gene on cell physiology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank A. Lobüscher, K. Herbster and T. Lorenz for excellent technical assistance and D. Newrzella for performing calculations involving data reproducibility. We are also highly indebted to A. Bach's clear visions, remarkable professionalism and never-ending support. Funding to pay the Open Access publication charges for this article was provided by the Institut für Kulturpflanzenforschung und Pflanzenzüchtung (IPK) in Gatersleben.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M, Shalon D, Davis RW, Brown P. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 3.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 4.Fischer A. Restriction-mediated differential display (RMDD) In: Ausubel FM, et al., editors. Current Protocols in Molecular Biology. New York, USA: 25B.4.1 (supplement 56), Wiley; 2001. [DOI] [PubMed] [Google Scholar]
- 5.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 7.Scheel J, Von Brevern MC, Hoerlein A, Fischer A, Schneider A, Bach A. Yellow pages to the transcriptome. Pharmacogenomics. 2002;3:791–807. doi: 10.1517/14622416.3.6.791. [DOI] [PubMed] [Google Scholar]
- 8.Shimkets RA, Lowe DG, Tai JT, Sehl P, Jin H, Yang R, Predki PF, Rothberg BE, Murtha MT, et al. Gene expression analysis by transcript profiling coupled to a gene database query. Nat. Biotechnol. 1999;17:798–803. doi: 10.1038/11743. [DOI] [PubMed] [Google Scholar]
- 9.Sutcliffe JG, Foye PE, Erlander MG, Hilbush BS, Bodzin LJ, Durham JT, Hasel KW. TOGA: an automated parsing technology for analyzing expression of nearly all genes. Proc. Natl Acad. Sci. USA. 2000;97:1976–1981. doi: 10.1073/pnas.040537997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitoh H, Ueda S, Kurosaki K, Kiuchi M. The different mobility of complementary strands depends on the proportion AC/GT. Forensic Sci. Int. 1998;94:155–156. doi: 10.1016/s0379-0738(97)00178-3. [DOI] [PubMed] [Google Scholar]
- 11.Spinella DG, Bernardino AK, Redding AC, Koutz P, Wei Y, Pratt EK, Myers KK, Chappell G, Gerken S, et al. Tandem arrayed ligation of expressed sequence tags (TALEST): a new method for generating global gene expression profiles. Nucleic Acids Res. 1999;27:e22. doi: 10.1093/nar/27.18.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;19:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura H, Reich S, Ito A, Saitoh H, Kamoun S, Winter P, Kahl G, Reuter M, Kruger DH, et al. Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc. Natl Acad. Sci. USA. 2003;100:15718–15723. doi: 10.1073/pnas.2536670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unneberg P, Wennborg A, Larsson M. Transcript identification by analysis of short sequence tags - influence of tag length, restriction site and transcript database. Nucleic Acids Res. 2003;31:2217–2226. doi: 10.1093/nar/gkg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorgensen JH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. [Google Scholar]
- 16.Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 17.Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 1993;239:122–128. doi: 10.1007/BF00281610. [DOI] [PubMed] [Google Scholar]
- 18.Maguire TL, Grimmond S, Forrest A, Iturbe-Ormaetxe I, Meksem K, Gresshoff P. Tissue-specific gene expression in soybean (Glycine max) detected by microarray analysis. J. Plant Physiol. 2002;159:1361–1374. [Google Scholar]
- 19.Qin L-X, Beyer RP, Hudson FN, Linford NJ, Morris DE, Kerr KF. Evaluation of methods for oligonucleotide array data via quantitative real-time PCR. BMC Bioinformatics. 2006;7:23. doi: 10.1186/1471-2105-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zierold U, Scholz U, Schweizer P. Transcriptome analysis of mlo-mediated resistance in the epidermis of barley. Mol. Plant Pathol. 2005;6:139–151. doi: 10.1111/j.1364-3703.2005.00271.x. [DOI] [PubMed] [Google Scholar]
- 21.Schweizer P, Pokorny J, Schulze-Lefert P, Dudler R. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 2000;24:895–903. doi: 10.1046/j.1365-313x.2000.00941.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.