Abstract
The binding of transcription factor (TF) IIIA to the internal control region of the 5 S RNA gene is the first step in the assembly of a DNA–TFIIIA–TFIIIC– TFIIIB transcription complex, which promotes accurate transcription by RNA polymerase III. With the use of mutations that are predicted to disrupt the folding of a zinc finger, we have examined the roles of zinc fingers 1 through 7 of yeast TFIIIA in the establishment of a functional transcription complex both in vitro and in vivo. Our data indicate that, in addition to their role in DNA binding, the first and seventh zinc fingers contribute other essential roles in the assembly of an active transcription complex. Alanine-scanning mutagenesis identified residues within zinc finger 1 that are not required for DNA binding but are required for incorporation of TFIIIC into the TFIIIA–DNA complex. Although disruption of zinc finger 2 or 3 had a deleterious effect on the activity of TFIIIA both in vitro and in vivo, we found that increasing the level of their in vivo expression allowed these mutant proteins to support cell viability. Disruption of zinc fingers 4, 5 or 6 had minimal effect on the DNA binding and TF activities of TFIIIA.
INTRODUCTION
Promoter recognition by RNA polymerase III, which transcribes small non-coding genes such as tRNA genes and 5 S RNA genes, is mediated by the transcription factors (TFs) TFIIIA, TFIIIB and TFIIIC (1,2). These factors assemble in a stepwise manner, beginning with recruitment of the multi-subunit TFIIIC to the promoter of a tRNA gene and the single-subunit TFIIIA to the promoter of the 5 S RNA gene. The intragenic promoters of these genes, which are referred to as internal control regions (ICRs), are often multipartite, with the sub-elements contributing to the appropriate architecture of the final transcription complex (1,2). The protein–DNA and protein–protein interactions that promote assembly of a transcription complex on a tRNA gene have been extensively studied with TFs from Saccharomyces cerevisiae (3–5). Binding of the six-subunit yeast TFIIIC to the bipartite ICR of a tRNA gene protects the entire gene against cleavage by DNase I. TFIIIC then mediates stable binding of TFIIIB, which consists of three polypeptides including the TATA-binding protein, upstream of the start site of transcription. TFIIIB is responsible for recruitment of RNA polymerase III and is able by itself to direct multiple rounds of accurate transcription. In contrast to the tRNA gene, the 5 S RNA gene does not bind TFIIIC directly; rather, the TFIIIA–5 S DNA complex provides the platform for TFIIIC recruitment. Formation of the TFIIIC–TFIIIA–5 S DNA complex is a prerequisite for the subsequent recruitment of TFIIIB and RNA polymerase III. In this way, TFIIIA acts as an adaptor protein to assemble an initiation complex at the promoter of the 5 S RNA gene in which the placement of TFIIIC and TFIIIB is similar to that on a tRNA gene (6–8).
In most organisms, TFIIIA contains nine sequential Cys2His2 zinc fingers that are separated by short linker sequences. However, TFIIIAs with longer spacer regions and additional zinc fingers have been identified in some species (9,10). The interaction of TFIIIA from Xenopus with the tripartite ICR of the amphibian 5 S RNA gene has been studied in detail. The ICR consists of an A box (nt +50 to +64), an intermediate element (nt +67 to +72) and a C box (nt +80 to +97). The results of DNase I protection and chemical protection/interference studies with full-length and truncated polypeptides bound to DNA suggest that the three amino-terminal and the three carboxyl-terminal fingers of Xenopus TFIIIA wind around the major groove, making contacts with the C and A boxes, respectively. Fingers 4, 5 and 6 are proposed to bridge these two domains, with fingers 4 and 6 crossing the minor groove and finger 5 making contacts in the major groove of the intermediate element (11–18). Structures of polypeptides containing the amino-terminal 3 or 6 fingers bound to DNA have been determined by X-ray and NMR analyses and support the models derived from biochemical and genetic studies (13,16,17).
Because a polypeptide consisting of the three amino-terminal zinc fingers of Xenopus TFIIIA was found to bind to the 5 S RNA gene with an affinity similar to the full-length protein, this amino-terminal module was initially considered to be the main contributor to the affinity of the protein for DNA (19–21). However, additional studies with truncated proteins and full-length proteins that contained point mutations, ‘broken’ fingers and ‘swapped’ fingers, as well as insights derived from structural analyses, led to the conclusion that the carboxyl-terminal fingers also make a large contribution to the affinity of the protein–DNA complex (22–25). Accommodating the binding of the full-length Xenopus TFIIIA to DNA has been shown to involve both neighboring finger–finger interactions as well as longer range interactions that contribute both favorably and unfavorably to the overall affinity of TFIIIA for the promoter (22,23).
Once bound to DNA, TFIIIA directs assembly of the TFIIIB–TFIIIC–TFIIIA–DNA complex. The observation that the association of TFIIIA with DNA is much more stable in a fully assembled complex and in a TFIIIC–TFIIIA–DNA complex than in a TFIIIA–DNA complex has led to the suggestion that the rate of assembly of the complete transcription complex is largely independent of the equilibrium binding constant of TFIIIA for DNA as measured in the formation of a binary complex (24,26). For example, a version of Xenopus TFIIIA that contains a disruption of zinc finger 3 supports wild-type levels of 5 S RNA synthesis in vitro despite having a 27-fold reduction in its DNA-binding affinity (24).
The carboxyl-terminal portion of Xenopus TFIIIA, beginning at zinc finger 7, appears to have a role in the formation of the initiation complex that is distinct from its role in contacting the A box of the promoter (27). A mutation that prevents proper folding of any one of the first six zinc fingers of TFIIIA does not affect either the in vitro or in vivo TF activity of the protein (24,28). A mutation that disrupts the structure of zinc finger 7, 8 or 9 or deletion of a 14-residue sequence in the C-terminal extension of the protein leads to transcriptional defects that cannot be accounted for solely by effects on DNA binding (23–25,28–31). For example, disruption of zinc finger 7 or zinc finger 8 leads to a similar reduction in DNA-binding affinity of TFIIIA. However, disruption of zinc finger 7 leads to only a modest reduction in the activity of TFIIIA whereas disruption of zinc finger 8 abolishes the ability of the protein to support transcription of the 5 S RNA gene in vitro and greatly reduces its activity in vivo (23,24,28).
Like its Xenopus counterpart, yeast TFIIIA contains nine Cys2His2 zinc fingers. Yeast TFIIIA also contains a unique 81-amino-acid sequence between zinc fingers 8 and 9 that is essential for its TF activity (32). The ICR of the yeast 5 S RNA gene consists of only a C-box element, located between nucleotides +81 and +94 (33). DNase I footprinting and methylation protection studies indicate that the three amino-terminal zinc fingers span the ICR, contacting residues from +79 to +98 (34). Zinc finger 5 contacts residues 73 and 74 of the 5 S RNA gene, but this interaction does not appear to contribute to the affinity of the protein for the 5 S RNA gene (34). Although zinc fingers 6 through 9 are not in close contact with DNA in the TFIIIA–DNA complex (34), these fingers are in close proximity to DNA in the fully assembled initiation complex (6,8,35). A truncated version of yeast TFIIIA containing the three amino-terminal zinc fingers interacts with the ICR with high affinity. Although this TFIIIA–DNA complex is able to recruit TFIIIC, the resulting complex is non-functional (34,36). The recruitment of TFIIIC appears to require zinc finger 1, as a polypeptide that begins with finger 2 is able to bind to the 5 S RNA gene, albeit with reduced affinity, but does not recruit TFIIIC (36). A short leucine-rich segment within the 81-amino-acid region present between zinc fingers 8 and 9 of yeast TFIIIA is also required for the assembly of a functional transcription complex (36,37). This region might serve as a second docking site for TFIIIC and in combination with zinc fingers 8 and 9, which contribute in a redundant manner to the TF activity of TFIIIA, enforce a topography on the TFIIIC-TFIIIA-5 S DNA complex that promotes proper positioning of TFIIIB (1,36,37).
We have examined the roles of zinc fingers 1 through 7 of yeast TFIIIA in the establishment of a functional transcription complex on the 5 S RNA gene. We constructed versions of TFIIIA in which the folding of an individual zinc finger was disrupted by substitution of a zinc-coordinating histidine residue by an asparagine residue. In this study, we compare the ability of these mutant TFIIIAs to bind DNA and to support in vitro and in vivo transcription of the 5 S RNA gene.
MATERIALS AND METHODS
Site-directed mutagenesis
The construction of pXS-TFC2, which contains the coding region for TFIIIA downstream of a T7 RNA polymerase promoter, has been described elsewhere (37). This plasmid served as the parental plasmid for the introduction of mutations into the coding sequence of TFIIIA. Site-directed mutagenesis was achieved by recombinant PCR using the overlap extension procedure as described previously (37) with pXS-TFC2 as the template except as otherwise noted. All PCR amplifications were performed by using the high fidelity Vent DNA polymerase as instructed by the manufacturer (New England Biolabs), and the sequence of all amplified DNA was verified by DNA sequencing.
A series of pXS-TFC2-derived plasmids was constructed to code for broken-finger versions of TFIIIA containing a histidine-to-asparagine substitution in each of zinc fingers 1 through 7. The upstream and downstream primers used for the construction of pXS-TFC2(H69N), pXS-TFC2(H98N), pXS-TFC2(H126N) and pXS-TFC2(H154N), which encode versions of TFIIIA containing disruptions of zinc fingers 1 to 4, respectively, annealed to the DNA sequence just upstream of the TFIIIA-coding region and to the DNA sequence at the end of the coding region for zinc finger 4. The final PCR product in each case was digested with NcoI and HindIII, and the resulting ∼510-bp restriction fragments were gel-purified and used to replace the corresponding fragment of pXS-TFC2. The primers used for the construction of pXS-TFC2(H181N), which contains a disruption of zinc finger 5, annealed to the DNA sequence at the beginning of the coding region for zinc finger 4 and to the DNA sequence at the end of the coding region for zinc finger 8. The final PCR product was digested with HindIII and XbaI, and the resulting ∼280-bp restriction fragment was gel-purified and used to replace the corresponding fragment of pXS-TFC2. The primers used for the construction of pXS-TFC2(H214N) and pXS-TFC2(H240N), which encode versions of TFIIIA containing disruptions of zinc fingers 6 and 7, annealed to the DNA sequence just upstream of the TFIIIA-coding region and to the DNA sequence at the end of the coding region for zinc finger 8. The final PCR product in both cases was digested with NcoI and XbaI, and the resulting ∼850-bp restriction fragments were gel-purified and used to replace the corresponding fragment of pXS-TFC2.
Another series of plasmids encoding versions of TFIIIA with disruption of two zinc fingers was made by combining the appropriate restriction fragments from parental plasmids described above. The ∼510-bp NcoI–HindIII fragment from pXS-TFC2(H126N) (third finger disruption) or pXS-TFC2(H154N) (fourth finger disruption) was used to replace the corresponding fragment in plasmids pXS-TFC2(H181N) (fifth finger disruption), pXS-TFC2(H214N) (sixth finger disruption), pXS-TFC2(H240N) (seventh finger disruption) and pXS-TFC2(C367Y) [ninth finger disruption; (37)] to generate plasmids encoding versions of TFIIIA containing a disrupting mutation in both fingers 3 and 5, 3 and 6, 3 and 9, 4 and 5, 4 and 6, 4 and 7 and 4 and 9. The plasmid encoding the version of TFIIIA containing disruptions of zinc fingers 5 and 6 was constructed by replacing the ∼560-bp NcoI–Bsu36I fragment from pXS-TFC2(H214N) (sixth finger disruption) with the corresponding fragment from pXS-TFC2(H181N) (fifth finger disruption).
Another series of constructs, which was also made by overlap extension PCR with pXS-TFC2 as the template, directed the synthesis of versions of TFIIIA containing the following alanine-scanning mutations of the first finger of TFIIIA: Y49A, F50A, F50E, D52A, Y53A, Y53E, D54A, G55A, D57A, K58A, A59V, F60A, L71A, V73A and the double mutation Y53A/D54A. pXS-TFC2(F50A) was used as the template in the PCR construction of a version of TFIIIA containing the double mutation F50A/Y53A. pXS-TFC2(D54A) was used as the template in the construction of a version of TFIIIA containing the double mutation F50A/D54A, and pXS-TFC2(K58A) was used as the template in the construction of the versions of TFIIIA containing the double mutations F50A/K58A, Y53A/K58A and K58A/L71A. The final PCR product in all cases was digested with NcoI and HindIII, and the resulting ∼510-bp fragment was used to replace the corresponding fragment of pXS-TFC2.
Plasmids for bacterial expression of wild-type and mutant forms of yeast TFIIIA
The construction of a plasmid for bacterial expression of wild-type TFIIIA was described previously (34). Plasmids for the expression of mutant versions of TFIIIA described above were generated by inserting the 2.3-kb NcoI–BamHI fragment of the appropriate pXS-TFC2 plasmid between the corresponding sites of pET-11d (38). This places the coding region of TFIIIA under the control of a bacteriophage T7 RNA polymerase promoter.
Plasmids for in vivo analysis of mutant versions of TFIIIA
Two series of plasmids were used for in vivo expression of wild-type and mutant forms of TFIIIA. The yeast shuttle vector ΔpG-3, a pUC18-derived plasmid that contains a 2 μ origin of replication and the selectable marker TRP1, has been described previously (37). KpnI–BamHI fragments containing the open-reading frames of the wild-type and mutant versions of TFIIIA were purified from pXS-TFC2 plasmids and inserted between the KpnI and SalI sites of ΔpG-3 after the BamHI- and SalI-generated ends had been filled in by the Klenow form of DNA polymerase I in the presence of dNTPs. This placed the coding region of TFIIIA between the constitutive promoter of the glyceraldehyde-3-phosphate dehydrogenase (GPD) gene and the transcription terminator of the phosphoglycerate kinase (PGK) gene.
pRS314+-TFC2, a low-copy plasmid containing the wild-type coding sequence for TFIIIA under the control of its own promoter, was constructed as follows. The promoter sequence of the TFIIIA gene was amplified by PCR using pJA230 (39) as a template. The upstream primer annealed 230-bp upstream of the translation start site and introduced a KpnI site, whereas the downstream primer annealed at the translation start site and introduced an NcoI site. The 230-bp PCR product was digested with KpnI and inserted between the KpnI and EcoRI sites of the plasmid pRS314 after the EcoRI site had been blunted with Klenow in the presence of dNTPs. This generated plasmid pRS314+. This CEN/ARS-containing plasmid carries the TRP1 selectable marker and a yeast autonomously replicating sequence (ARS). NcoI–BamHI fragments containing the open reading frames of wild-type and mutant versions of TFIIIA were purified from pXS-TFC2 plasmids and inserted between the corresponding sites of pRS314+.
In vitro synthesis of TFIIIA
Wild-type and mutant versions of TFIIIA were synthesized in vitro using the TnT (Promega) coupled transcription–translation system essentially as described previously (34). pXS-TFC2 and its variants were used as template unless otherwise indicated. Five microlitrers of each 20 μl reaction was loaded on an SDS–15% polyacrylamide gel, transferred to nitrocellulose for western blotting and probed with anti-TFIIIA as described below to confirm that each protein was synthesized and stable. The remainder of the in vitro synthesized protein was used in electrophoretic mobility shift assays (EMSAs) and transcription assays.
Purification of full-length yeast TFIIIA from bacteria
The pET-11d-derived plasmids were transformed into the Escherichia coli strain BL21(DE3), and TFIIIA was purified as described (34).
Electrophoretic mobility shift and in vitro transcription assays
EMSAs were performed as described (37). A 20 μl reaction contained a 2 μl aliquot of TFIIIA, 4 μl of partially purified TFIIIC where indicated, and a 270-bp end-labeled DNA fragment, which was excised from p19-5 S and contains the yeast 5 S RNA gene (33). TFIIIA for these experiments was either purified from bacteria or synthesized in an in vitro transcription–translation reaction programmed to produce the indicated version of TFIIIA. The partially purified TFIIIC-containing fraction derived from yeast was prepared as described for fraction j in Taylor and Segall (40). In vitro transcription assays were performed as described (40) using the yeast 5 S RNA gene (p19-5 S) as template. A 50 μl reaction contained a 4.5 μl-aliquot of either bacterially expressed or in vitro synthesized TFIIIA and 12.5 μl of a yeast-derived heparin-agarose fraction (fraction h) (40) that contained TFIIIC, TFIIIB and RNA polymerase III.
In vivo analysis of the mutant versions of TFIIIA
The haploid yeast strain YRW1 (MATa can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 ade2-1 tfc2::LEU2 and harboring pJA230), which was used for in vivo analysis of wild-type and mutant versions of TFIIIA, has been described previously (34). pJA230 is a CEN/ARS plasmid with a URA3 selectable marker and a 10-kb insert of yeast DNA containing TFC2, the gene encoding TFIIIA (32). Since TFC2 is an essential gene, viability of YRW1 depends on the presence of pJA230 (32).
ΔpG-3 and pRS314+ plasmids directing expression of wild-type or mutant forms of TFIIIA were transformed into YRW1. After transformants had been selected on medium lacking uracil and tryptophan, the ΔpG-3- and pRS314+-containing strains were grown on medium lacking tryptophan and containing uracil to allow for loss of pJA230. Cells were then streaked on plates containing medium supplemented with 5-fluoro-orotic acid (5-FOA) and uracil and lacking tryptophan. Because 5-FOA kills cells containing the URA3 gene, only those cells which have lost pJA230 and contain a ΔpG-3 or pRS314+ derivative encoding a functional version of TFIIIA will grow on the 5-FOA-containing medium.
Western analysis of ΔpG-3-driven TFIIIA expression in vivo
YRW1 and strains of YRW1 containing ΔpG3-derived plasmids that directed expression of mutant versions of TFIIIA were grown overnight in minimal medium lacking tryptophan to an absorbance at 600 nm of ∼4.0. The cells from 10 ml of culture were harvested, washed twice with cold water and re-suspended in 200 μl of lysis buffer (30 mM HEPES, pH 7.9, 150 mM NaCl, 1 mM DTT and 0.1% NP-40). Cells were vortexed in the presence of 200 μl glass beads for six 1-min bursts interrupted by chilling on ice. Samples were spun at 4° for 10 min at 23 500 g, and the supernatant transferred to a fresh tube and re-spun for an additional 10 min. Protein concentration was determined using the Bradford assay with bovine serum albumin as the standard. Protein (3.75 μg) was loaded on an SDS 15%–polyacrylamide gel, transferred to nitrocellulose for western blotting and probed with anti-TFIIIA antibody as described previously (37).
RESULTS
DNA binding and TF activity of in vitro synthesized versions of yeast TFIIIA containing individual disruptions of zinc fingers 1 through 7
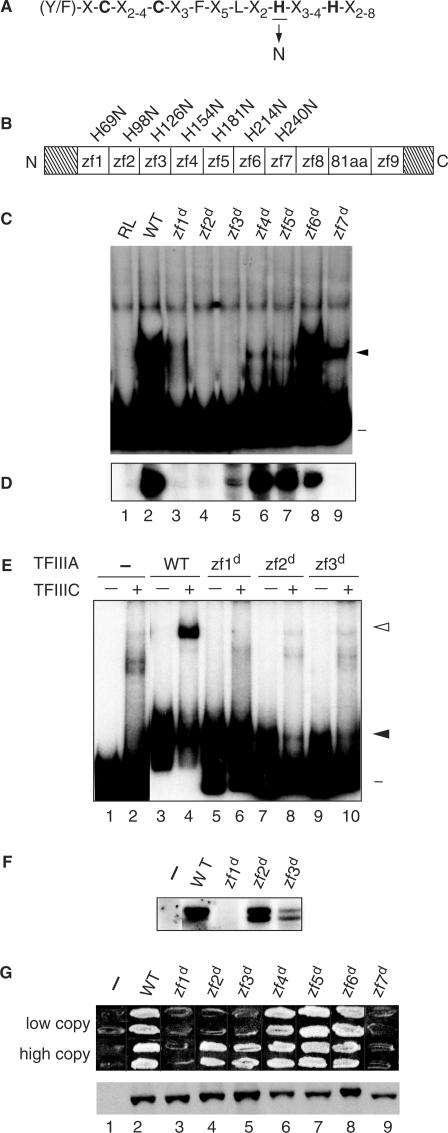
In this study, we have assessed the requirements for zinc fingers 1 through 7 of yeast TFIIIA in the establishment of a productive transcription complex on the 5 S RNA gene. A previous assessment of the activities of truncated versions of yeast TFIIIA suggested that the first zinc finger contributes to DNA binding and to recruitment of TFIIIC (36). Analysis of the effect of substitutions in zinc fingers 8 and 9 that are predicted to disrupt the folding of the finger indicated that these two zinc fingers contribute in a redundant manner to the TF activity of yeast TFIIIA (37). To extend this analysis, we generated a series of plasmids for in vitro and in vivo expression of proteins in which the second zinc-coordinating histidine residue of each of zinc fingers 1 through 7 was replaced by an asparagine residue (Figure 1A). We refer to this substitution, which is predicted to prevent stable folding of the finger, as a disrupting mutation and to each disrupted finger as zf#d, with # identifying the disrupted zinc finger. The substituted residue for disruption of zinc finger 1 through 7 was H69, H98, H126, H154, H181, H214 and H240, respectively (Figure 1B). As a first test of the effect of these mutations on the activity of TFIIIA, we compared the ability of approximately equal amounts of in vitro synthesized protein (data not shown) to bind to the 5 S RNA gene as assessed by an EMSA (Figure 1C). Because we have used this assay to give a qualitative assessment of protein–DNA interactions and have not determined Kd values, we use the term relative affinity in describing our interpretation of the EMSAs. TFIIIA(zf6d) was the only mutant protein that retained the ability to bind to the 5 S RNA gene with a relative affinity similar to the wild-type protein (Figure 1C, compare lanes 2 and 8). TFIIIA with a disruption of zinc finger 1, 4, 5 or 7 appeared severely compromised for DNA-binding activity, with disruption of zinc finger 7 being less deleterious than disruption of zinc finger 1, 4 or 5 (Figure 1C, lanes 3–7 and 9). TFIIIA(zf2d) and TFIIIA(zf3d) had no detectable DNA-binding activity (Figure 1C, lanes 4 and 5). Despite the qualitative nature of this assay, the data suggest that within the context of the entire protein, only zinc finger 6 of the first 7 fingers can be disrupted without compromising DNA-binding activity. This analysis also revealed the presence of a minor amount of protein–DNA complexes of higher mobility than typically observed for the TFIIIA–DNA complex, particularly for reactions containing TFIIIA(zf4d) and TFIIIA(zf5 d). It is possible that these complexes contained a truncated protein generated by proteolytic cleavage within the disrupted finger.
Figure 1.
Zinc finger 1 and zinc finger 7 are essential for the TF activity of TFIIIA. (A) The consensus amino acid sequence of the Cys2His2 zinc finger is given in single-letter code and the zinc-coordinating cysteines and histidines are in bold. The histidine substituted with asparagine to disrupt the structure of individual fingers is underlined. (B) Schematic representation of yeast TFIIIA. The boxes denoted zf1 to zf9 represent the nine zinc fingers; the boxed 81aa denotes the 81-amino-acid domain; and the diagonally stripped boxes represent the 48-amino-acid amino-terminal and the 35-amino-acid carboxyl-terminal regions. The histidine-to-asparagine substitution for each disruption is denoted above the zinc finger. zf = zinc finger. (C) Ability of mutant versions of TFIIIA to bind to the 5 S RNA gene as assessed by EMSA. A radioactively labeled DNA fragment containing the yeast 5 S RNA gene was incubated with in vitro synthesized versions of TFIIIA prior to electrophoresis on a non-denaturing polyacrylamide gel. Lane 1, RL (reticulocyte lysate; in vitro transcription–translation reaction that was not programmed to synthesize TFIIIA); lane 2, WT (wild-type) TFIIIA; lanes 3 through 9, versions of TFIIIA containing a histidine-to-asparagine mutation in the indicated zinc finger. Lane numbers are as given in panel D. The positions of the free DNA (minus sign) and the TFIIIA–DNA complex (closed arrowhead) are indicated on the right. (D) Abilities of mutant versions of TFIIIA to support in vitro transcription of the 5 S RNA gene. In vitro transcription reaction mixtures contained the yeast 5 S RNA gene as template, partially purified yeast TFIIIC, TFIIIB and RNA polymerase III and the version of in vitro synthesized TFIIIA indicated in the corresponding lanes of panel C. The RNAs synthesized in vitro were analyzed on a 7 M urea–10% polyacrylamide gel. The autoradiogram shows the portion of the gel containing 5 S RNA. (E) Abilities of mutant versions of TFIIIA purified from bacteria to bind to the 5 S RNA gene and to recruit TFIIIC to the TFIIIA–DNA complex as assessed by EMSA. A radioactively labeled DNA fragment containing the yeast 5 S RNA gene was incubated with protein extract containing the indicated version of bacterially produced TFIIIA in the absence (odd numbered lanes) or presence (even numbered lanes) of partially purified yeast TFIIIC prior to electrophoresis on a non-denaturing polyacrylamide gel. Lanes are labeled as in panel C with lanes 1 and 2 containing no added bacterial protein. The positions of the free DNA (minus sign), TFIIIA–DNA complexes (solid arrowhead) and TFIIIC–TFIIIA–DNA complexes (arrowhead) are indicated at the right. (F) Abilities of mutant versions of TFIIIA purified from bacteria to support in vitro transcription of the 5 S RNA gene. For details, see legend for Figure 1D. (G) Abilities of mutant versions of TFIIIA to support cell viability. Top panel: a plasmid shuffle assay to test the abilities of mutant versions of TFIIIA to replace wild-type TFIIIA in vivo. Cells of YRW1 that had been transformed with plasmids containing copies of the TFC2 gene were tested for their abilities to grow on medium containing 5-FOA (see Results Section). Each patch contains cells from a different transformant. Two series of plasmids were tested: in the pRS314+ series (labeled ‘low copy’), TFC2 is expressed under the control of its own promoter from a low-copy (CEN ARS) plasmid; in the ΔpG3 series (labeled ‘high copy’), TFC2 is expressed untrol of the strong GPD promoter from a high-copy (2 μ) plasmid. Cells containing the parental vectors not encoding a version of TFIIIA are indicated by the minus sign. Bottom panel: assessment by western blot of in vivo protein levels of mutant versions of TFIIIA. Aliquots of crude lysates prepared from representative YRW1 yeast cells containing the indicated ΔpG3-derived plasmids were separated on a 15% SDS polyacrylamide gel, transferred to a nitrocellulose filter, and probed with anti-TFIIIA antibody. Note that TFIIIA expressed from the pRS314+ series of plasmids is below the level of detection in this blot.
We next tested the TF activity of the in vitro synthesized proteins. TF activity, which refers to the ability of TFIIIA to assemble a functional transcription complex, was monitored in reactions that contained partially purified TFIIIC, TFIIIB, RNA polymerase III and 5 S DNA (see Materials and Methods section). Comparison of TF activity of the mutant TFIIIAs, as measured by the amount of 5 S RNA synthesized in in vitro transcription reactions (Figure 1D), with the EMSA results (Figure 1C) showed that there was little correlation between the relative ability of a mutant TFIIIA to bind to the 5 S RNA gene and to support transcription of the gene. For example, TFIIIA(zf4d) and TFIIIA(zf5d), which appeared severely compromised in their ability to interact with the 5 S RNA gene, were nonetheless able to support near wild-type levels of in vitro transcription (Figure 1C and D, lanes 6 and 7); similarly, TFIIIA(zf3d), which had no detectable DNA-binding activity, was able to support a low level of transcription (Figure 1C and D, lane 5). This suggested that in the presence of yeast TFIIIC, TFIIIB and RNA polymerase III, versions of TFIIIA with low affinity for the ICR could nonetheless be captured on DNA to form transcriptionally active complexes. Indeed, previous studies indicated that TFIIIC influences the stability of the TFIIIA–DNA complex (41–44). Detailed studies with Xenopus TFIIIA led to the conclusion that the activity of TFIIIA in an in vitro transcription reaction is independent of its apparent affinity for 5 S DNA as determined in a binary reaction and that TFIIIA molecules can be sequestered in higher order, essentially irreversible transcription complexes (24). It is also possible that the activity of TFIIIA(zf4d) and TFIIIA(zf5d) increased because potentially proteolytic sensitive sites in these proteins became protected. In contrast, TFIIIA(zf7d) did not support transcription of the 5 S RNA gene despite having a readily detectable DNA-binding activity (Figure 1C and D, compare lanes 9). TFIIIA(zf1d) and TFIIIA(zf2d), which had minimal or no DNA-binding activity, respectively, also appeared unable to support in vitro transcription of the 5 S RNA gene (Figure 1C and D, lanes 3 and 4). Finally, TFIIIA(zf6d), which had near wild-type DNA-binding activity, supported a high level of in vitro transcription (Figure1C and D, lane 8). In summary, in vitro synthesized TFIIIAs with a disruption of zinc finger 4, 5 or 6 retained a high level of in vitro TF activity whereas TFIIIAs with a disruption of zinc finger 1, 2, 3 or 7 appeared to have defects in TF activity.
DNA binding and TF activity of bacterially expressed versions of yeast TFIIIA containing a disruption of zinc finger 1, 2 or 3
We also expressed wild-type TFIIIA and TFIIIA with a disruption of zinc finger 1, 2 or 3 in bacteria so that we could test the in vitro activities of these proteins at higher concentrations than was possible with in vitro synthesized protein. We found that we could readily detect TFIIIA–5 S DNA complexes by EMSA with these three mutant TFIIIAs purified from bacteria. Relative to wild-type TFIIIA, however, all three mutant TFIIIAs showed reduced affinity for the 5 S RNA gene. The interaction of TFIIIA(zf2d) and TFIIIA(zf3d) with DNA appeared to be compromised to a greater extent than the interaction of TFIIIA(zf1d) (Figure 1E, lanes 3, 5, 7 and 9). We emphasize that we have not carried out quantitative measurements to determine Kd values and it is possible that these reactions contained close to saturating amounts of TFIIIA. Of note, however, is that TFIIIA(zf1d) was clearly able to bind to the 5 S RNA gene. We also monitored the effect of the addition of partially purified TFIIIC to the EMSA reactions. As observed previously (33), recruitment of TFIIIC to a wild-type TFIIIA–DNA complex generated a complex that had greatly reduced mobility (Figure 1E, lane 4). None of the mutant TFIIIA–DNA complexes appeared, however, to be able to efficiently recruit TFIIIC (Figure 1F, lanes 6, 8 and 9).
We next tested the ability of these bacterially expressed versions of TFIIIA to support in vitro transcription. TFIIIA(zf1d) was unable to support transcription (Figure 1F, lane 3) whereas TFIIIA-zf2d and TFIIIA-zf3d were able to support in vitro transcription of the 5 S RNA gene (Figure 1F, lanes 4 and 5). These results support the idea that zinc finger 1 plays an essential role in the assembly of an active initiation complex (36).
In vivo activity of mutant versions of yeast TFIIIA
We used a plasmid-shuffling protocol to test the mutant versions of TFIIIA for their ability to support cell viability. For this study we used two sets of plasmids: a high-copy ΔpG3-derived series of plasmids in which expression of TFC2 was directed by the strong, constitutive GPD promoter (37) and a low-copy CEN-ARS series of plasmids in which expression of TFC2 was under the control of its own promoter (see Materials and Methods section). Plasmids encoding the mutant proteins were introduced into a Δtfc2 ura3 strain that harbored pJA230. This is a low-copy plasmid that contains TFC2, which encodes TFIIIA and is an essential gene, and URA3 as a selectable marker (37). The transformed cells were then tested for their ability to grow on 5-FOA-containing medium; growth on this medium requires that the URA3-marked pJA230 plasmid has been lost from cells and is therefore diagnostic of the newly introduced mutant version of TFC2 being able to support expression of the 5 S RNA gene. The only essential role of TFIIIA is in directing expression of the 5 S RNA gene (45).
We found that TFIIIA containing a disruption of zinc finger 4, 5 or 6 supported cell growth when expressed at either a low or a high level. TFIIIA containing a disruption of zinc finger 2 or 3 was able to support cell viability, but only when expressed from the high-copy vector. TFIIIA containing a disruption of zinc finger 1 or 7 failed to support cell viability irrespective of expression level (Figure 1G, top panel). Our anti-TFIIIA antibody is insufficiently sensitive to detect endogenous TFIIIA or TFIIIA expressed from a low-copy vector in our standard western blot analysis (37) (Figure 1G, lane 1). However, TFIIIA expressed from the high-copy vector can be readily detected (37) (Figure 1G, lane 2). We therefore used western blot analysis to test for cellular accumulation of mutant TFIIIAs expressed from the high-copy plasmid in cells prior to growth on 5-FOA-containing medium. These cells were also expressing wild-type TFIIIA from the low-copy plasmid pJA230. We found that all mutant proteins were sufficiently stable to be readily detected (Figure 1G, bottom panel) and were thus present in cells at a higher level than chromosomally expressed TFIIIA.
In summary, this analysis indicated that zinc fingers 1 and 7 are essential for the in vivo assembly of a functional transcription complex on the 5 S RNA gene. The observation that the deleterious effect of disruption of zinc finger 2 or zinc finger 3 could be compensated for by a higher level of in vivo expression of the mutant protein was consistent with the observations made in comparing the activities of in vitro synthesized and bacterially expressed proteins (Figure 1C–F). Disruption of zinc finger 4, 5 or 6 did not appear to compromise the in vivo activity of TFIIIA.
Analysis of versions of TFIIIA with two disrupted zinc fingers
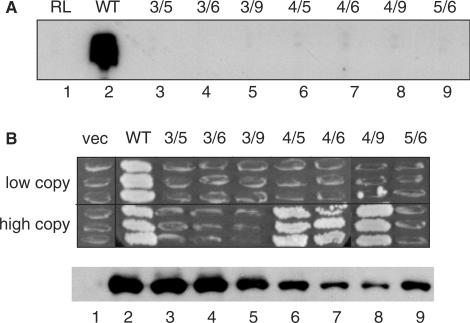
Because cells expressing TFIIIA with a disruption in any one of zinc fingers 4, 5 or 6 from a low-copy plasmid were viable, we investigated the possibility that the tertiary structure of this central trimeric unit of fingers might be dispensable. For example, this region of the molecule might serve as a flexible linker to allow appropriate positioning of the amino-terminal and carboxyl-terminal regions of the molecule. We therefore tested the effect of simultaneous disruptions of zinc fingers 4 and 5, 4 and 6, and 5 and 6 on the activity of TFIIIA. For comparison, we also monitored the activity of TFIIIA with simultaneous disruption of zinc fingers 3 and 5, 3 and 6, 3 and 9 and 4 and 9. When these double mutant versions of TFIIIA were synthesized in vitro and tested for their TF activity, all were found to be defective in the ability to promote 5 S RNA synthesis (Figure 2A). However, versions of TFIIIA that combined a disruption of zinc finger 4 with a disruption of zinc finger 5, 6 or 9 supported cell viability when expressed from the high-copy plasmid in vivo, although they failed to do so when expressed from the low-copy plasmid (Figure 2B, lanes 6–8). TFIIIAs that combined a disruption of zinc finger 3, which by itself reduced the activity of TFIIIA such that it could only support cell viability when expressed from a high-copy plasmid (Figure 1G), with a disruption of zinc finger 5, 6 or 9 were stable, but inactive in vivo (Figure 2B, lanes 3–5). TFIIIA(zf5d, zf6d) was also unable to support cell viability even when expressed at a high level (Figure 2B, lane 9). In summary, we found that the activity of TFIIIA(zf3d) was sufficiently compromised such that it lost all activity on disruption of a second zinc finger. In contrast, the structure of zinc finger 4 appears to be less important for the overall function of TFIIIA, and zinc fingers 5 and 6 may contribute in a redundant manner to an essential function of yeast TFIIIA.
Figure 2.
Abilities of versions of TFIIIA containing disruptions of two zinc fingers to support TF activity. (A) Abilities of mutant versions of TFIIIA to support in vitro transcription of the 5 S RNA gene. For simplicity, versions of TFIIIA containing histidine-to-asparagine substitutions in multiple fingers are named by the numbers of the disrupted fingers; for example, 3/5 represents TFIIIA with disruptions in zinc fingers 3 and 5 [TFIIIA(H126N/H181N)]. See Figure 1D for details. (B) Abilities of versions of TFIIIA containing disruptions of two zinc fingers to support cell viability. See Figure 1G for details.
Alanine-scanning mutagenesis of zinc finger 1 of TFIIIA
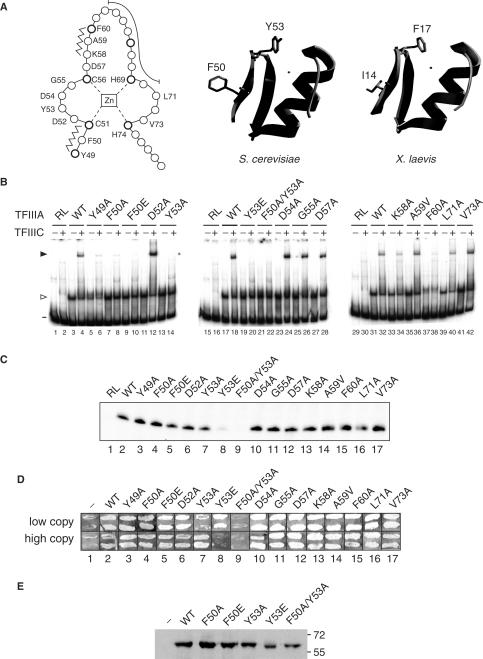
Our previous (36) and current data suggest that the first zinc finger of yeast TFIIIA contributes not only to DNA binding, but also serves a role in recruitment of TFIIIC to the TFIIIA–DNA complex. To identify residues in the first zinc finger that might contribute to an interaction with TFIIIC, we mutated amino acids that would be expected to be surface exposed and not directly involved in DNA binding. The solved structures of several zinc finger modules bound to DNA show that such surface-exposed residues are present in the short antiparallel β-sheets and in the β-turn connecting the β-strands (17,46–49). We therefore carried out alanine-scanning mutagenesis of the corresponding region of the first zinc finger of yeast TFIIIA. Residues from Y49 to F60 were mutated with the exception of the two zinc-liganding cysteines, C51 and C56 (Figure 3A). A59 was mutated to valine; all other substitutions were to alanine. For comparison, we also mutated residues L71 and V73, which are situated between the two zinc-coordinating histidine residues that follow the DNA-binding α-helix.
Figure 3.
TF activity of alanine-scanning mutants of the first zinc finger of TFIIIA. (A) A linear representation of the first zinc finger of TFIIIA from S. cerevisiae is on the left. Amino acids are represented by circles, with the position of conserved residues of the zinc finger motif shown in bold. The zinc ion, which is tetrahedrally coordinated by two cysteine and two histidine residues, is boxed in the center of the diagram. Residues that form the short anti-parallel β-sheet of the zinc finger domain are indicated by the zigzag lines, while residues that make up the DNA-binding α-helix are shown by the long curved line. Zinc-coordinating residues and residues that were mutated are identified by their single-letter codes. A ribbon representation of a 3D homology model of the first zinc finger of yeast TFIIIA (residues Y49 to A79) is in the middle. This modeled structure was obtained with the use of the SWISS-MODEL server (63,64) and with the coordinates of the first zinc finger of TFIIIA from X. laevis as the template (17; ExPDB entry 1tf3A; shown on the right). The side chains of F50 and Y53 of the yeast protein and the side chains of the corresponding residues, I14 and F17, of the amphibian protein are shown. The zinc ion, shown by the small circle, is positioned in the yeast structure according to its coordinates in the amphibian protein. (B) Abilities of versions of TFIIIA with alanine-scanning mutations to bind to the 5 S RNA gene and to recruit TFIIIC to the TFIIIA–DNA complex as assessed by EMSA. See Figure 2E for details. The version of TFIIIA used in each reaction is indicated below the gels; RL, reticulocyte transcription–translation reactions not programmed to synthesize TFIIIA; WT, wild-type TFIIIA. The presence (+) or absence (–) of TFIIIC is indicated above the gel. The positions of free DNA (minus sign), TFIIIA–DNA complexes (arrowhead) and TFIIIC–TFIIIA–DNA complexes (solid arrowhead) are shown on the left. (C) Abilities of mutant versions of TFIIIA to support in vitro transcription of the 5 S RNA gene. See Figure 1D for details. (D) Abilities of mutant versions of TFIIIA to support cell viability. See Figure 1G for details. (E) Assessment by western blot analysis of in vivo expression of selected mutant versions of TFIIIA from the ΔpG3 series of plasmids. See Figure 1G for details.
We first tested approximately equivalent amounts of each mutant TFIIIA that had been synthesized in vitro (data not shown) for its ability to bind to the 5 S RNA gene and to recruit TFIIIC. All mutant proteins bound to DNA with a relative affinity similar to wild-type TFIIIA with the exception of TFIIIA(Y49A) and TFIIIA(F60A), which appeared to have a modest reduction in affinity for DNA (Figure 3B, odd numbered lanes, open arrowhead). TFIIIA(K58A) was slightly compromised in its ability to recruit TFIIIC to the TFIIIA–DNA complex (Figure 3B, lane 34) and TFIIIA(Y49A), TFIIIA(F50A), TFIIIA(Y53A) and TFIIIA(F60A) were severely compromised in their ability to recruit TFIIIC (Figure 3B, lanes 6, 8, 14 and 38). As both Y49 and F60 are conserved residues of the zinc-finger motif and contribute to overall folding, the first zinc-finger of TFIIIA(Y49A) and TFIIIA(F60A) may be unable to adopt a stable tertiary structure and this may lead to the apparent defect in recruitment of TFIIIC. In the TFIIIA homologs of Xenopus laevis, Rana catesbeiana, Homo sapiens and other vertebrates, the two positions equivalent to F50 and Y53 are represented by isoleucine and phenylalanine, respectively (9,50). This conservation of hydrophobic residues in a region of the molecule that is predicted to be surface exposed is consistent with the notion that this region, and particularly F50 and Y53 of yeast TFIIIA, participates in a protein–protein interaction. In a homology model of the first zinc finger of yeast TFIIIA, these residues are indeed surface exposed (Figure 3A). Additionally, we found that reducing the charge in this region, by mutation of D52 or D54 to alanine, did not affect the ability of the TFIIIA–DNA complex to recruit TFIIIC (Figure 3B, lanes 12 and 24).
We next tested the in vitro synthesized proteins for their ability to support transcription of the 5 S RNA gene in the presence of a yeast fraction containing TFIIIC, TFIIIB and RNA polymerase III. All versions of TFIIIA were active in this assay (Figure 3C, lanes 1–4, 6, 7 and 10–17). Because both TFIIIA(F50A) and TFIIIA(Y53A) were defective in recruitment of TFIIIC as assessed by EMSA (Figure 3B, lanes 8 and 14), we next tested the activity of versions of TFIIIA in which these residues were mutated to glutamate, anticipating that the introduction of a charged residue might be more disruptive to the function of the putative hydrophobic interaction interface and thus have a more pronounced effect on the activity of TFIIIA than the alanine substitutions. We also examined the activity of TFIIIA in which the F50A and Y53A substitutions were combined. All three mutants retained the ability to bind to the 5 S RNA gene as assessed by EMSA (Figure 3B, lanes 9, 19 and 21). TFIIIA(F50E) retained the ability to support in vitro transcription of the 5 S RNA gene, but TFIIIA(Y53E) was severely compromised and TFIIIA(F50A/Y53A) did not (Figure 3C, lanes 5, 8 and 9).
With the use of the plasmid-shuffling protocol, we found that TFIIIA containing the double mutation F50A/Y53A was the only mutant from the collection of first finger mutants that was unable to support cell viability when expressed at either high or low levels in vivo (Figure 3D, lane 9). As assessed by western blot analysis, this mutant protein accumulated to approximately the same level as did wild-type TFIIIA expressed from the high-copy plasmid (Figure 3E, lanes 2 and 7). TFIIIA(Y53E), which was the only form of TFIIIA other than TFIIIA(F50A/Y53A) that was severely compromised for its ability to support in vitro transcription (Figure 4C, lane 8), was able to support cell viability when expressed from the low-copy plasmid, but not from the high-copy plasmid (Figure 3D, lane 8). This toxic effect of high-level expression of TFIIIA(Y53E) occurred only when cells were challenged to grow on 5-FOA-containing medium. This unusual observation is difficult to explain. It is possible that TFIIIA(Y53E) is modestly unstable and when present at high levels is inactivated by aggregation in a process that does not affect wild-type TFIIIA present in the same cells.
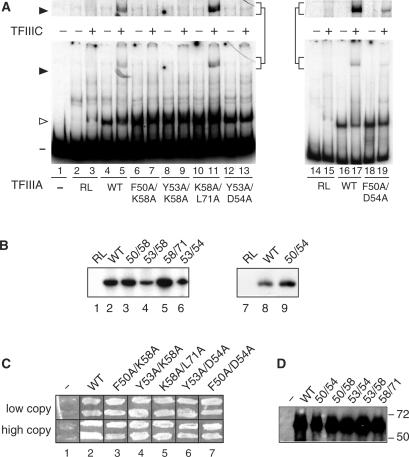
Figure 4.
Effects of alanine-scanning mutations within the first finger on the activity of TFIIIA. (A) Abilities of versions of TFIIIA with alanine-scanning mutations to bind to the 5 S RNA gene and to recruit TFIIIC to the TFIIIA–DNA complex as assessed by EMSA. See Figure 1E for details. The portion of the gel containing the TFIIIC–TFIIIA–DNA complex is shown at a higher exposure above the full-sized gel. (B) Abilities of mutant versions of TFIIIA to support in vitro transcription of the 5 S RNA gene. See Figure 1D for details. (C) Abilities of mutant versions of TFIIIA to support cell viability. See Figure 1G for details. (D) Assessment by western blot analysis of in vivo expression of mutant versions of TFIIIA from the ΔpG3 series of plasmids. See Figure 1G for details.
The ability of TFIIIA(Y53E) and TFIIIA(F50A/Y53A) to bind to the 5 S RNA gene, their inability to support in vitro transcription and the failure of TFIIIA(F50A/Y53A) to support cell viability suggested that residues F50 and Y53 were involved in the assembly of the transcription complex on the TFIIIA–DNA complex. For comparison, we also tested the effect of combining two other mutations, D54A and K58A, with F50A and Y53A; we also combined K58A with the more distant mutation L71A. These five double mutant versions of TFIIIA (F50A/K58A; Y53A/K58A; Y53A/D54A; F50A/D54A; K58A/L71A) retained the ability to bind to the 5 S RNA gene (Figure 4A, odd numbered lanes; TFIIIA shift indicated with open arrowhead). Although all proteins, with the exception of TFIIIA(K58A/L71A), were defective in recruitment of TFIIIC (Figure 4A, even numbered lanes; TFIIIC shift indicated by closed arrowhead and shown at higher exposure above the main figure), all of the double-mutant proteins were nonetheless able to direct in vitro transcription of the 5 S RNA gene in the presence of a yeast fraction containing TFIIIC, TFIIIB and RNA polymerase III (Figure 4B) and were able to support cell viability when expressed in vivo at either a high or low level (Figure 4C). The observation that the TFIIIA molecules containing these additional combinations of mutations retained in vivo activity further supported the conclusion that residues F50 and Y53 play a direct role in the recruitment of TFIIIC to the promoter.
We also carried out alanine-scanning mutagenesis of zinc finger 7 (data not shown). In this case, this approach did not identify any individual amino acid or combination of amino acids as being required for the essential TF activity of this finger.
DISCUSSION
Although it is well established that TFIIIA binds to the ICR of the 5 S RNA gene and recruits TFIIIC to the TFIIIA–DNA complex, the roles of individual zinc fingers in this process are less clear. Whereas specific contacts between zinc fingers and the ICR are a prerequisite for high-affinity, site-specific binding of TFIIIA to DNA, lower-affinity interactions could be important contributors to the overall architecture of the fully assembled transcription complex. Similarly, protein–protein interactions, in addition to being required for the recruitment of TFIIIC to the TFIIIA–DNA complex, could also promote conformational changes in the interacting proteins that are essential for the activity of the transcription complex. In this study, we have presented our initial analysis of the roles of individual zinc fingers of yeast TFIIIA in the assembly of an active transcription complex. We have used broken-finger versions of TFIIIA in which the third zinc ligand (i.e. the first of the two zinc-coordinating histidine residues) of a finger motif is mutated to asparagine in the context of the full-length protein (23). This mutation is predicted to disrupt the folding of the finger by interfering with its ability to coordinate zinc (51–53) and this would abolish its ability to bind to DNA (54). Examination of the products of partial proteolysis of broken-finger versions of Xenopus TFIIIA indicates that a finger containing such as histidine-to-asparagine mutation is indeed unstructured (23). In some cases, however, the fourth zinc-coordinating residue has been shown to be dispensable for folding. The ninth zinc finger of the recently characterized TFIIIA from Schizosaccharomyces pombe, which contains an unprecedented 10 potential zinc fingers, lacks the carboxyl-terminal Zn2+-coordinating histidine residue. This module is nonetheless able to fold and function in DNA binding and TF activity (10,55). Based on studies with synthetic, truncated zinc -finger polypeptides that showed that these peptides are able to fold in the absence of a carboxyl-terminal histidine residue (56), Schulman and Setzer (2003) speculate that a water molecule or a dithiothreitol-provided thiolate acts as the fourth zinc ligand in finger 9 of the S. pombe homolog of TFIIIA.
The effects of disrupting a zinc finger could in part reflect an influence of the mutation on the properties of neighboring zinc fingers. Studies with Xenopus TFIIIA suggest that disruption of any one zinc finger does not perturb the folding or function of adjacent fingers (22–24,28,57). However, NMR-derived structures of a polypeptide containing the first three zinc fingers of Xenopus TFIIIA bound to its cognate DNA site indicate that substantial packing interactions do occur between these zinc fingers when bound to DNA (13,17). Thus, without more rigorous analysis of the properties of the broken-finger versions of S. cerevisiae TFIIIA used in this study, we cannot be certain that the histidine-to-asparagine mutations are effectively and uniquely disrupting the structure and function of the mutated finger.
In this study, we compared the in vitro and in vivo activities of mutant TFIIIAs of S. cerevisiae. Both these assays showed that zinc fingers 1 and 7 play an essential role in assembly of an active transcription complex. Although our previous (36) and current analyses with in vitro synthesized proteins suggest that the absence of zinc finger 1 leads to a major reduction in affinity of the protein for the ICR, we were able to show with the use of bacterially expressed protein that TFIIIA(zf1d) does indeed bind the 5 S RNA gene. This finding supported the idea that the essential role of finger 1 is not its contribution to DNA binding, but rather its recruitment of TFIIIC (36). Indeed a comparison of bacterially expressed TFIIIA(zf1d), TFIIIA(zf2d) and TFIIIA(zf3d) suggested that TFIIIA(zf1d) bound DNA as efficiently as TFIIIA(zf2d) and TFIIIA(zf3d). The latter two versions of TFIIIA, however, but not TFIIIA(zf1d), were able to support in vitro transcription of the 5 S RNA gene and cell viability when expressed from a high-copy plasmid under the control of a strong, constitutive promoter. The observation that TFIIIA(zf2d) and TFIIIA(zf3d) were unable to support cell viability when expressed from a low-copy plasmid with the TFC2 gene under the control of its own promoter suggests that the function of TFIIIA that is compromised by disruption of finger 2 or finger 3 can be overcome by increasing the concentration of these proteins in the cell.
Our EMSA analysis of the bacterial proteins suggested that each of the first three zinc fingers contributed to recruitment of TFIIIC, with the contribution of the first finger being essential. Using an alanine-scanning mutagenesis approach, we identified residues within zinc finger 1 that are unlikely to participate in DNA binding but were necessary for the assembly of an active transcription complex. In particular, TFIIIA(F50A/Y53A) bound to the 5 S RNA gene, but did not recruit TFIIIC and did not support in vitro transcription nor cell viability, even when expressed at a high level. We speculate that the hydrophobic residues F50 and Y53 are surface-exposed, but become masked by an interaction with TFIIIC. Charged residues in this region of TFIIIA appeared less important for this predicted protein–protein interaction as the mutations D52A, D54A, D57A and K58A were without effect. Based on the known topography of the multisubunit TFIIIC on the 5 S RNA gene (7,8), the 138-kDa subunit of TFIIIC is a likely candidate for an interaction with the first zinc finger of TFIIIA. Other potential interacting partners are the 91-kDa subunit which has also been positioned at the 3′-end of the gene in close proximity to DNA around the transcription terminator, and the 60-kDa subunit, which has not been detected in photocrosslinking studies on either the tRNA gene or the 5 S RNA gene (8,58,59). Although a double mutation within finger 1 was required to completely eliminate the TF activity of TFIIIA, we note that several other mutations that were combined with F50A or Y53A were without effect.
The transcriptional defect of TFIIIA(H240N), containing a disruption of zinc finger 7, is unlikely to be in its binding affinity to 5 S DNA or in the initial recruitment of TFIIIC; in vitro synthesized TFIIIA(zf7d) showed some ability to bind to DNA and the first three zinc fingers of TFIIIA suffice for recruitment of TFIIIC (34,36). It is possible that a non-specific interaction of zinc finger 7 with DNA is essential for the appropriate positioning of TFIIIA in the fully assembled transcription complex. Site-specific DNA–protein photocrosslinking studies show that the interaction of TFIIIA with DNA is more extensive in the fully assembled transcription complex than in the TFIIIA–DNA complex itself (7,8). Another potential role for zinc finger 7 is in interactions with other TFs. TFIIIC can be recruited to the 5 S RNA gene by a TFIIIA that has mutations within the 81-amino-acid region that separates fingers 8 and 9, but the resulting TFIIIA–DNA complex is non-functional (37). We have previously suggested that the 81-amino-acid region provides a second docking site for TFIIIC and that this is important in adjusting the topography of the complex. Thus, it is possible that the essential function of zinc finger 7 from yeast TFIIIA is in proper presentation of the 81-amino-acid domain for its putative interaction with TFIIIC. However, zinc finger 7 could also be playing a more direct role in the TF activity of the protein. For example, it could interact directly with TFIIIC. Such an interaction would most likely be with the τA domain of TFIIIC, which consists of the 131- the 95- and the 55-kDa subunits. This domain is essential for transcription activation and start-site selection by virtue of its role in positioning TFIIIB upstream of the transcribed region (60,61).
We found that both the in vitro and in vivo TF activity of yeast TFIIIA was tolerant to disruption of zinc finger 4, 5 or 6. Thus, as suggested previously (34), the contacts that are made between zinc finger 5 and guanines +73 and +74 of the 5 S RNA gene are not essential. TFIIIA containing disruptions of both zinc fingers 4 and 5, zinc fingers 4 and 6 or zinc fingers 4 and 9 also supported cell viability, albeit only when expressed at high copy. In contrast, TFIIIA with a disruption of zinc fingers 5 and 6 as well as TFIIIAs that combined disruption of zinc finger 3 with disruption of finger 5, 6 or 9 were unable to support cell viability even when expressed at a high level. Although it is possible that some of these deficiencies reflect redundant functions shared between fingers, we consider it more likely that the inactivity of these proteins reflects a cumulative crippling effect.
In summary, our analysis of the effect of mutations in yeast TFIIIA on its activities in vitro and in vivo have shown that F50 and Y53 of the first zinc finger play a role in recruitment of TFIIIC and have revealed a requirement for the seventh zinc finger in assembly of a functional transcription complex. The fact that neither of these zinc fingers has an essential role in the amphibian TFIIIA (23,28) is consistent with the high rate of sequence divergence of TFIIIA (32) and the observed species specificity in the RNA polymerase III transcriptional machinery (62).
ACKNOWLEDGEMENTS
We thank Joshua Silver and Adelene Tan for assistance in the construction of several of the plasmids used in this study and Dr David Isenman for assistance with the homology modeling. This work was supported by a grant (MOP-6826) from the Canadian Institutes of Health Research to J.S. Funding to pay the Open Access publication charges for this article was provided by the Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Geiduschek EP, Kassavetis GA. RNA polymerase III transcription complexes. In: McKnight SL, Yamamoto KR, editors. Transcriptional Regulation. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 2.White RJ. RNA Polymerase III Transcription. Austin: R. G. Landes Company; 1994. [Google Scholar]
- 3.Camier S, Gabrielsen O, Baker R, Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985;4:491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kassavetis GA, Riggs DL, Negri R, Nguyen LH, Geiduschek EP. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stillman DJ, Geiduschek EP. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984;3:847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 7.Bartholomew B, Braun BR, Kassavetis GA, Geiduschek EP. Probing close DNA contacts of RNA polymerase III transcription complexes with the photoactive nucleoside 4-thiodeoxythymidine. J. Biol. Chem. 1994;269:18090–18095. [PubMed] [Google Scholar]
- 8.Braun BR, Bartholomew B, Kassavetis GA, Geiduschek EP. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 9.Mathieu O, Yukawa Y, Prieto JL, Vaillant I, Sugiura M, Tourmente S. Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Res. 2003;31:2424–2433. doi: 10.1093/nar/gkg335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulman DB, Setzer DR. Identification and characterization of transcription factor IIIA from Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:2772–2781. doi: 10.1093/nar/gkf385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens KR, Liao X, Wolf V, Wright PE, Gottesfeld JM. Definition of the binding sites of individual zinc fingers in the transcription factor IIIA-5S RNA gene complex. Proc. Natl Acad. Sci. USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairall L, Rhodes D. A new approach to the analysis of DNase I footprinting data and its application to the TFIIIA/5S DNA complex. Nucleic Acids Res. 1992;20:4727–4731. doi: 10.1093/nar/20.18.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster MP, Wuttke DS, Radhakrishnan I, Case DA, Gottesfeld JM, Wright PE. Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA. Nat. Struct. Biol. 1997;4:605–608. doi: 10.1038/nsb0897-605. [DOI] [PubMed] [Google Scholar]
- 14.Hansen PK, Christensen JH, Nyborg J, Lillelund O, Thogersen HC. Dissection of the DNA-binding domain of Xenopus laevis TFIIIA. Quantitative DNase I footprinting analysis of specific complexes between a 5 S RNA gene fragment and N-terminal fragments of TFIIIA containing three, four or five zinc-finger domains. J. Mol. Biol. 1993;233:191–202. doi: 10.1006/jmbi.1993.1499. [DOI] [PubMed] [Google Scholar]
- 15.Hayes JJ, Clemens KR. Locations of contacts between individual zinc fingers of Xenopus laevis transcription factor IIIA and the internal control region of a 5S RNA gene. Biochemistry. 1992;31:11600–11605. doi: 10.1021/bi00161a045. [DOI] [PubMed] [Google Scholar]
- 16.Nolte RT, Conlin RM, Harrison SC, Brown RS. Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl Acad. Sci. USA. 1998;95:2938–2943. doi: 10.1073/pnas.95.6.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wuttke DS, Foster MP, Case DA, Gottesfeld JM, Wright PE. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity. J. Mol. Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 18.McBryant SJ, Meier E, Leresche A, Sharp SJ, Wolf VJ, Gottesfeld JM. TATA-box DNA binding activity and subunit composition for RNA polymerase III transcription factor IIIB from Xenopus laevis. Mol. Cell. Biol. 1996;16:4639–4647. doi: 10.1128/mcb.16.9.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen JH, Hansen PK, Lillelund O, Thogersen HC. Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 1991;281:181–184. doi: 10.1016/0014-5793(91)80388-j. [DOI] [PubMed] [Google Scholar]
- 20.Darby MK, Joho KE. Differential binding of zinc fingers from Xenopus TFIIIA and p43 to 5S RNA and the 5S RNA gene. Mol. Cell. Biol. 1992;12:3155–3164. doi: 10.1128/mcb.12.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao XB, Clemens KR, Tennant L, Wright PE, Gottesfeld JM. Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5 S RNA gene. J. Mol. Biol. 1992;223:857–871. doi: 10.1016/0022-2836(92)90248-i. [DOI] [PubMed] [Google Scholar]
- 22.Kehres DG, Subramanyan GS, Hung VS, Rogers GW, Jr, Setzer DR. Energetically unfavorable interactions among the zinc fingers of transcription factor IIIA when bound to the 5 S rRNA gene. J. Biol. Chem. 1997;272:20152–20161. doi: 10.1074/jbc.272.32.20152. [DOI] [PubMed] [Google Scholar]
- 23.Del Rio S, Menezes SR, Setzer DR. The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J. Mol. Biol. 1993;233:567–579. doi: 10.1006/jmbi.1993.1536. [DOI] [PubMed] [Google Scholar]
- 24.Del Rio S, Setzer DR. The role of zinc fingers in transcriptional activation by transcription factor IIIA. Proc. Natl Acad. Sci. USA. 1993;90:168–172. doi: 10.1073/pnas.90.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vrana KE, Churchill ME, Tullius TD, Brown DD. Mapping functional regions of transcription factor TFIIIA. Mol. Cell. Biol. 1988;8:1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brady KL, Ponnampalam SN, Bumbulis MJ, Setzer DR. Mutations in TFIIIA that increase stability of the TFIIIA-5 S rRNA gene complex: unusual effects on the kinetics of complex assembly and dissociation. J. Biol. Chem. 2005;280:26743–26750. doi: 10.1074/jbc.M502677200. [DOI] [PubMed] [Google Scholar]
- 27.Hayes J, Tullius TD, Wolffe AP. A protein-protein interaction is essential for stable complex formation on a 5 S RNA gene. J. Biol. Chem. 1989;264:6009–6012. [PubMed] [Google Scholar]
- 28.Rollins MB, Del Rio S, Galey AL, Setzer DR, Andrews MT. Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol. Cell. Biol. 1993;13:4776–4783. doi: 10.1128/mcb.13.8.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing YY, Worcel A. The C-terminal domain of transcription factor IIIA interacts differently with different 5S RNA genes. Mol. Cell. Biol. 1989;9:499–514. doi: 10.1128/mcb.9.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao X, Darby MK. A position-dependent transcription-activating domain in TFIIIA. Mol. Cell. Biol. 1993;13:7496–7506. doi: 10.1128/mcb.13.12.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith DR, Jackson IJ, Brown DD. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984;37:645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- 32.Archambault J, Milne CA, Schappert KT, Baum B, Friesen JD, Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J. Biol. Chem. 1992;267:3282–3288. [PubMed] [Google Scholar]
- 33.Challice JM, Segall J. Transcription of the 5 S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J. Biol. Chem. 1989;264:20060–20067. [PubMed] [Google Scholar]
- 34.Rowland O, Segall J. Interaction of wild-type and truncated forms of transcription factor IIIA from Saccharomyces cerevisiae with the 5 S RNA gene. J. Biol. Chem. 1996;271:12103–12110. doi: 10.1074/jbc.271.20.12103. [DOI] [PubMed] [Google Scholar]
- 35.Braun BR, Riggs DL, Kassavetis GA, Geiduschek EP. Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc. Natl Acad. Sci. USA. 1989;86:2530–2534. doi: 10.1073/pnas.86.8.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milne CA, Segall J. Mapping regions of yeast transcription factor IIIA required for DNA binding, interaction with transcription factor IIIC, and transcription activity. J. Biol. Chem. 1993;268:11364–11371. [PubMed] [Google Scholar]
- 37.Rowland O, Segall J. A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol. 1998;18:420–432. doi: 10.1128/mcb.18.1.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Archambault J, Schappert KT, Friesen JD. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol. Cell. Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor MJ, Segall J. Characterization of factors and DNA sequences required for accurate transcription of the Saccharomyces cerevisiae 5 S RNA gene. J. Biol. Chem. 1985;260:4531–4540. [PubMed] [Google Scholar]
- 41.Bieker JJ, Martin PL, Roeder RG. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985;40:119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- 42.Bogenhagen DF, Wormington WM, Brown DD. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982;28:413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 43.Lassar AB, Martin PL, Roeder RG. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 44.Segall J. Assembly of a yeast 5 S RNA gene transcription complex. J. Biol. Chem. 1986;261:11578–11584. [PubMed] [Google Scholar]
- 45.Camier S, Dechampesme AM, Sentenac A. The only essential function of TFIIIA in yeast is the transcription of 5S rRNA genes. Proc. Natl Acad. Sci. USA. 1995;92:9338–9342. doi: 10.1073/pnas.92.20.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 47.Houbaviy HB, Usheva A, Shenk T, Burley SK. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl Acad. Sci. USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim CA, Berg JM. A 2.2 Å resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 49.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 50.Gaskins CJ, Hanas JS. Sequence variation in transcription factor IIIA. Nucleic Acids Res. 1990;18:2117–2123. doi: 10.1093/nar/18.8.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diakun GP, Fairall L, Klug A. EXAFS study of the zinc-binding sites in the protein transcription factor IIIA. Nature. 1986;324:698–699. doi: 10.1038/324698a0. [DOI] [PubMed] [Google Scholar]
- 52.Frankel AD, Berg JM, Pabo CO. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc. Natl Acad. Sci. USA. 1987;84:4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parraga G, Horvath SJ, Eisen A, Taylor WE, Hood L, Young ET, Klevit RE. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988;241:1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- 54.Hanas JS, Hazuda DJ, Bogenhagen DF, Wu FY, Wu CW. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J. Biol. Chem. 1983;258:14120–14125. [PubMed] [Google Scholar]
- 55.Schulman DB, Setzer DR. Functional analysis of the novel C-terminal domains of S. pombe transcription factor IIIA. J. Mol. Biol. 2003;331:321–330. doi: 10.1016/s0022-2836(03)00730-7. [DOI] [PubMed] [Google Scholar]
- 56.Merkle DL, Berg JM. Metal requirements for nucleic acid binding proteins. Methods Enzymol. 1991;208:46–54. doi: 10.1016/0076-6879(91)08006-4. [DOI] [PubMed] [Google Scholar]
- 57.Setzer DR, Menezes SR, Del Rio S, Hung VS, Subramanyan G. Functional interactions between the zinc fingers of Xenopus transcription factor IIIA during 5S rRNA binding. RNA. 1996;2:1254–1269. [PMC free article] [PubMed] [Google Scholar]
- 58.Bartholomew B, Meares CF, Dahmus ME. Photoaffinity labeling of RNA polymerase III transcription complexes by nascent RNA. J. Biol. Chem. 1990;265:3731–3737. [PubMed] [Google Scholar]
- 59.Bartholomew B, Kassavetis GA, Geiduschek EP. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gabrielsen OS, Marzouki N, Ruet A, Sentenac A, Fromageot P. Two polypeptide chains in yeast transcription factor tau interact with DNA. J. Biol. Chem. 1989;264:7505–7511. [PubMed] [Google Scholar]
- 61.Joazeiro CA, Kassavetis GA, Geiduschek EP. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 62.Paule MR, White RJ. Survey and summary. Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–1723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]