Abstract
The optional Escherichia coli restriction tRNase PrrC represents a family of potential antiviral devices widespread among bacteria. PrrC comprises a functional C-domain of unknown structure and regulatory ABC/ATPase-like N-domain. The possible involvement of a C-domain sequence in tRNALys recognition was investigated using a matching end-protected 11-meric peptide. This mimic, termed here LARP (Lys-anticodon recognizing peptide) UV-cross-linked tRNALys anticodon stem-loop (ASL) analogs and inhibited their PrrC-catalyzed cleavage. Trimming LARP or introducing in it inactivating PrrC missense mutations impaired these activities. LARP appeared to mimic its matching protein sequence in ability to dimerize in parallel, as inferred from the following results. First, tethering Cys to the amino- or carboxy-end of LARP dramatically enhanced the ASL-cross-linking and PrrC-inhibiting activities under suitable redox conditions. Second, Cys-substitutions in a C-domain region containing the sequence corresponding to LARP elicited specific intersubunit cross-links. The parallel dimerization of PrrC's C-domains and expected head-to-tail dimerization of its N-domains further suggest that the NTPase and tRNALys-binding sites of PrrC arise during distinct assembly stages of its dimer of dimers form.
INTRODUCTION
PrrC, the optional Escherichia coli anticodon nuclease (ACNase) (1,2) represents potential antiviral devices widespread among bacteria (3). PrrC's activity is silenced by the genetically linked type Ic DNA restriction endonuclease EcoprrI (4–7) and is unleashed by Stp, the phage T4-encoded peptide inhibitor of EcoprrI (8). The activation of PrrC causes specific cleavage of tRNALys 5′ to the wobble base (2) and, consequently, could block T4-late translation and contain the infection (9). However, the T4-coded RNA healing and sealing enzymes 3′-phosphatase/5′-polynucleotide kinase (Pnk) and RNA ligase 1 (Rnl1) (10) normally restore the intact form of tRNALys (1,2), exercising perhaps their intended functions (11,12). Known PrrC homologs are invariably linked to EcoprrI homologs and, hence, could also act as secondary defenses mobilized when an associated DNA restriction endonuclease is compromised (3,8,13).
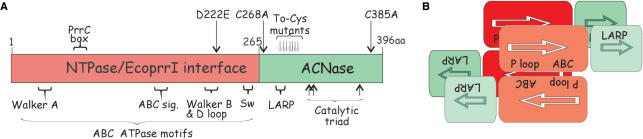
When PrrC is expressed by itself it exhibits overt (core) ACNase activity (14) that purifies with a homo-oligomeric PrrC form, possibly a dimer of dimers (3). PrrC's N-proximal ∼265 amino acids are thought to constitute an NTPase domain that mediates the activation of the latent ACNase (3,13) (Figure 10A). This region contains ABC ATPase-like motifs (15), albeit, sufficiently different from the typical to justify classifying PrrC's N-domain as a distinct subtype. The divergent sequence of PrrC's N-domain could account for the unusual nucleotide requirements of the ACNase activation reaction and PrrC's idiosyncratic nucleotide binding attributes. Namely, the activation depends on the cooperation of GTP and dTTP and is inhibited by ATP. Moreover, dTTP exhibits higher affinity for PrrC than GTP or ATP (μM versus mM-range); and dTTP but not GTP or ATP stabilizes PrrC's core ACNase activity. PrrC differs from the typically dimeric ABC ATPases (16) also in its apparent dimer of dimers structure (3). The remaining ∼130 amino acid region of PrrC harbors residues implicated in tRNALys recognition (17–19) and cleavage (3) and does not resemble a known protein structure. The main cues PrrC recognizes in tRNALys map to the anticodon stem-loop (ASL). They comprise the anticodon sequence, base modifications and base-pairing interactions (17–20).
Figure 10.
PrrC's functional organization and possible quaternary topology. (A) Functional organization of PrrC. The crimson bar represents the N-proximal NTPase domain, the green bar the C-proximal ACNase domain. The ABC ATPase-like motifs indicated below the bar are, from left to right, the Walker A (P-loop), ABC signature, Walker B/D loop and switch region motifs (15). The PrrC-box motif indicated above the bar is unique to the PrrC proteins. The D222E mutation in this domain attenuates ACNase activity and facilitates the overproduction and isolation of PrrC (3). In the C-proximal ACNase domain are indicated the sequence matching LARP thought to partake in tRNALys recognition, the conserved putative catalytic residues Arg320, Glu324 and His356 (3), the seven to-Cys mutations in the 288–306 range and the wild-type Cys residues at the edges of the domain that were replaced by Ala in PrrC*. (B) Proposed quaternary topology of PrrC. According to this model, the nucleotide binding pockets arise by head-to-tail interaction of the regulatory N-domains (crimson rectangles), as in ABC/ATPases (15). The tRNALys-sites are formed at parallel dimer–dimer interfaces between the ACNase C-domains (green squares). The upper dimer (darker forms) obscures part of the lower dimer (paler forms). P-loop, Walker A motif, ABC, ABC signature motif.
Peptide mimicry and Cys-mediated intersubunit cross-linking data reported here suggest that the tRNALys-binding motif of PrrC is shared by C-domain portions interfacing in parallel. The proposed parallel dimerization of the C-domains can be reconciled with the expected head-to-tail dimerization of the N-domains (15) by further suggesting that the regulatory and functional sites of PrrC arise at distinct assembly stages of its dimer of dimers form.
MATERIALS AND METHODS
Materials
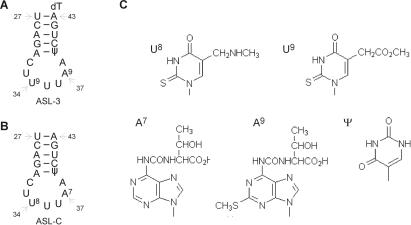
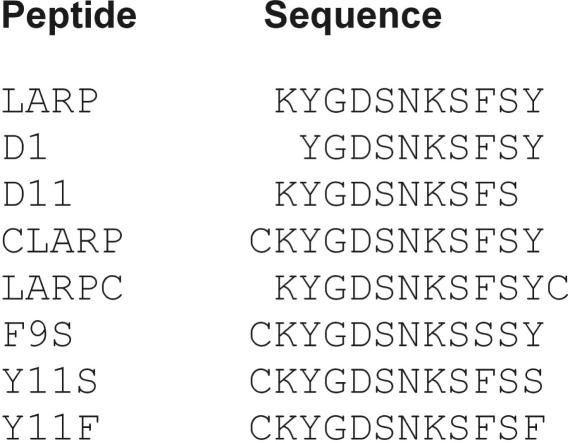
Purified E. coli tRNALys labeled with 32P at the 33p34 junction (13,21) and synthetic tRNALys ASL analogs (22,23) were prepared as previously described. The analogs included ASL-3 that matches mammalian tRNALys3 in RNA sequence and base modifications but contains an extra 3′-dT that facilitated its synthesis; and ASL-C that has the same RNA sequence but base modifications of E. coli tRNALys (Figure 1); both were [5′-32P] labeled as described (19). The synthetic end-protected peptides used (Figure 2) were purchased from Genscript Corporation and were over 80% pure. Diazenedicarboxylic acid (diamide) was purchased from Sigma.
Figure 1.
ASL analogs of tRNALys and their modified bases. (A and B) Secondary structure presentations and indicated tRNA positions of ASL-3 and ASL-C, respectively. (C) Notations and structures of modified ASL bases. U9 (mcm5s2U), A9 (ms2t6A) appear in human tRNALys3, U8 (mnm5s2U) and A7 (t6A) in corresponding positions of E. coli tRNALys and Ψ appears in both.
Figure 2.
Sequences of LARP and LARP derivatives used in this work. All the peptides used were protected at their N- and C-ends by respective acetyl and amide groups.
PrrC mutagenesis
The PrrC forms used contained a C-terminal His6 tag and, except where indicated, also the leaky D222E mutation that allows high level expression of the protein (3). The triple mutant D222E/C268A/C385A termed PrrC*, its Cys replacement derivatives and the H356A/F292S double mutant were generated by Quick Change (24). Other PrrC mutants have been described (3,17).
PrrC-expression plasmids and bacterial hosts
The PrrC proteins were expressed under the control of the T7-Lac promoter and Shine–Dalgarno sequence of plasmid pRRC11 (17) in E. coli Rosetta (DE3)pLysS (Novagen, UK) encoding T7 RNA polymerase, T7 lyzozyme (25) and rare tRNAs from plasmid pRARE. The cells were grown in LB medium at 37°C to a density of ∼6.108 cells/ml. They were shifted then to 30°C and PrrC's expression induced by adding 1 mM IPTG. After further incubation for 2 h, the cells were harvested and the PrrC proteins purified by immobilized metal affinity chromatography as described (3).
Diamide treatment of cells expressing PrrC to-Cys mutants
Escherichia coli cells induced to express the indicated PrrC forms were harvested by high-speed centrifugation, re-suspended in 0.01 vol of 10 mM diamide in water and incubated at 10°C for the indicated time. The cells were then lyzed by heating them for 1 min at 100°C in SDS–PAGE sample buffer, their proteins were separated by SDS–PAGE and PrrC monitored by immunoblotting using purified polyclonal anti-PrrC antibodies (17).
ACNase isolation and assays
The standard core ACNase form used in the in vitro assays was a His6-tagged derivative of the leaky PrrC mutant D222E. It was purified by TALON® (Clontech) affinity chromatography followed by Superdex-200 gel filtration (3). The standard ACNase reaction mixture (10 μl) contained 1 fmol of the 32P-labeled E. coli tRNALys or the indicated [5′-32P] labeled ASL, both at 3000 Ci/mmol; 2 μM dTTP, 4 mM Na-HEPES buffer, pH 7.5; 0.5 mM MgCl2, 15 mM NaCl, 5% glycerol and 0.5 M trimethylamine-N-oxide (TMAO). The reaction was performed at 10°C. It was initiated by adding the enzyme and terminated at the indicated time by adding 1.5 vol of 10 M urea, 0.01% bromphenol blue and 0.01% xylene cyanol. The products were separated by polyacrylamide–urea gel electrophoresis and quantified by densitometry or counting.
Peptide-RNA UV-cross-linking
Peptide-RNA UV-cross-linking was performed in mixtures (10 μl) containing ∼0.1 pmol of [5′-32P] labeled ASL-3, 0.1–2.0 nmol of Lys-anticodon recognizing peptide (LARP) or indicated LARP derivative, 5 mM Na-HEPES buffer, pH 7.5; 1 mM MgCl2, 30 mM NaCl, 0.5–1.0 M TMAO and 10% glycerol. The mixture was placed in a 35-mm polystyrene culture dish and irradiated for 5 min at 4°C over a TFX-20 M transilluminator providing radiation spectrum with a peak at 312 nm. Aliquots were mixed with 1.5 vol of 10 M urea containing 0.01% each of xylene cyanol and bromphenol blue and separated by denaturing gel electrophoresis.
PrrC-RNA UV-cross-linking
ThePrrC mutant proteins H356A and H356A/F292S were cross-linked to ASL-3 in 12 μl ACNase reaction mixtures essentially as described above but containing 0.3 pmol of ASL-3 and ∼1 μg of the indicated PrrC allele purified by TALON cobalt affinity chromatography. The mixture was irradiated for 5 min at 4°C as described for the LARP-ASL-3 cross-linking. An equal volume of 2× SDS–PAGE gel sample buffer devoid of reducing agent was added and the products separated by 10% SDS–PAGE. The proteins were transferred then to a nitrocellulose membrane, PrrC monitored by immunoblotting using anti-PrrC antibodies (17) and the PrrC/ASL-3 cross-linking product detected by autoradiography.
Glutaraldehyde (GA)-mediated protein–protein cross-linking
The indicated PrrC forms were subjected to GA-mediated protein–protein cross-linking as previously described and the products monitored by immunoblotting (3).
RESULTS AND DISCUSSION
The synthetic 11-mer LARP matching PrrC residues 284–294 inhibits ACNase activity
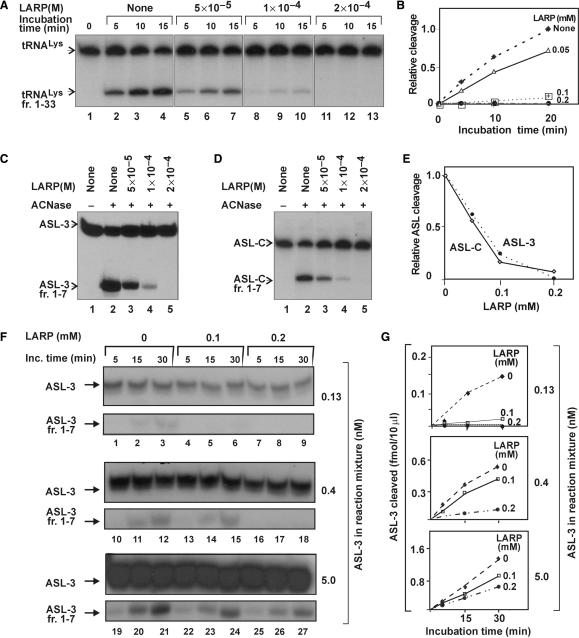
Mutations that alter ACNase cleavage specificity have implicated PrrC's Asp287 in tRNALys recognition (17–19). Asp287 maps to a consensus sequence shared by a subset of the known PrrC homologs. This sequence comprises a predicted coil region (284–92) and the essential aromatic residues Phe292 and Tyr294 (Figure 3) (3,17). Its possible role in tRNALys recognition was investigated using an end-protected mimic (Ac-KYGDSNKSFSY-NH2). This mimic was termed LARP to indicate its anticipated function. LARP inhibited PrrC's ACNase activity in a dose-dependent manner when assayed with E. coli tRNALys (Figure 4A and B) or ASL analogs (Figure 4C–E). An IC50 value of ∼10−4M was obtained in all cases. The inhibition decreased with increasing substrate level within the range examined (Figure 4F and G). These results hinted that LARP acted by occluding the RNA substrate and/or competing with the substrate over the protein.
Figure 3.
Multiple sequence alignment of a region containing a residue implicated in tRNALys recognition. Escherichia coli PrrC (indicated by Eco) residues 279–330 were aligned using ClustalW (31) with corresponding portions of representative homologs detected by BLAST (32). Secondary structure predictions according to SSpro (33) and Jpred (34) are indicated above the alignment. The region corresponding to LARP in a subset of the homologs is shaded. The arrows indicate residues whose replacement by Cys triggered intersubunit cross-links (Figure 9). The PrrC homologs indicated by the three-letter abbreviations are encoded by the following bacteria and indicated by the respective accession numbers: Yfr—Yersinia frederiksenii ATCC 33641, ZP_00829622.1; Nme—Neisseria meningitidis MC58, Hin—Haemophilus influenzae R2846, ZP_00154666.1; Lin—Leptospira interrogans serovar Copenhageni str. (Lin.), NC_005823.1; YP_000904.1; Photorhabdus luminescens subsp. laumondii TTO1 (Plu), NP_931496.1; RhpRhodopseudomonas palustris HaA2, NZ_AALQ01000003.1; Smu—Streptococcus mutans, NC_004350.1; Bli—Brevibacterium linens BL2 H, NZ_AAGP01000001.1. Identical residues shared by all homologs are marked in red, conserved residues in green.
Figure 4.
LARP inhibits ACNase activity. (A) Effect of LARP on the PrrC-catalyzed cleavage of E. coli tRNALys. The ACNase assay was performed in the presence of the indicated LARP levels and aliquots analyzed at the indicated time points. (B) Time course of the reactions of panel A. (C and D) Effect of LARP on the respective PrrC-catalyzed cleavages of ASL-3 or ASL-C. Only 15-min time points are shown. (E) ASL-3 or ASL-C cleavage by PrrC versus LARP's level. (F and G) Effect of LARP on ACNase activity at different ASL-3 levels. tRNALys fr. 1–33, ASL-3 fr. 1–7 and ASL-C fr. 1–7, respective labeled ACNase cleavage products of tRNALys, ASL-3 and ASL-C.
LARP UV-cross-links ASL-3
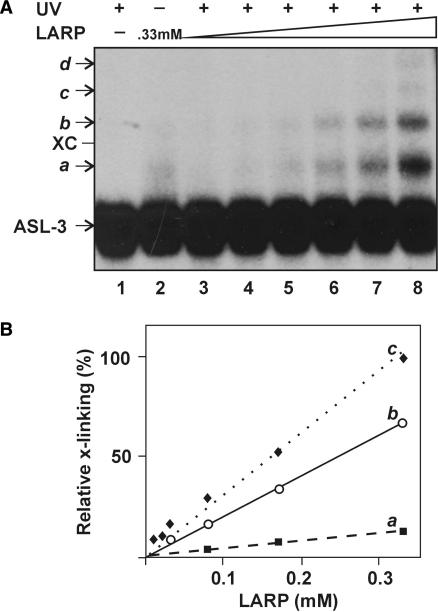
The possible interaction of LARP with the ACNase substrate was tested by attempting to UV-cross-link the peptide to ASL-3. The cross-linking was performed at 312 nm where the tRNALys wobble base is highly photoreactive (26). Several cross-linking products were obtained in amounts proportional to the dose of LARP (Figure 5A and B). The most abundant (designated a) was retarded in gel electrophoresis relative to ASL-3 by the equivalent of ∼4 nt, possibly due to a single LARP adduct. Slower migrating products designated b-d could contain additional LARP moieties, judged from their incremental retardations and dependence on LARP. Presumably, they arose through peptide–peptide cross-links since data shown below suggested that LARP could also self-interact (Figures 8 and 9). The non-irradiated mixture yielded weaker bands over a continuous distribution trailing behind ASL-3 (lane 2), probably non-covalent complexes that partially dissociated during the fractionation.
Figure 5.
LARP UV-cross-links ASL-3. (A) LARP was cross-linked to ASL-3 by irradiating their mixture at 312 nm (UV) as detailed in the Methods section. Lanes: 1, ASL-3 irradiated alone; 2, Non-irradiated mixture of ASL-3 and LARP (330 μM); 3–8, Irradiated mixtures of ASL-3 with the respective levels of LARP: 8.25, 16.5, 33, 82.5, 165 or 330 μM. XC, dye marker corresponding in position to a 22 nt oligoribonucleotide; a–d indicate cross-linking products assumed to contain one, two, three or four LARP moieties per ASL-3, respectively. (B) Relative levels of the primary (a), secondary (b) and tertiary (c) conjugates versus that of LARP. The amount of primary conjugate formed at the highest LARP level is assigned a value of 1.0.
Figure 8.
Cys extensions enhance LARP activities under suitable redox conditions. (A) Terminal Cys extensions enhance the UV-cross-linking of LARP to ASL-3. The indicated peptides employed each at 0.2 mM were UV-cross-linked to ASL-3 in the presence of 1mM DTT. (B–D) Effect of the DTT level on the cross-linking efficiencies of LARP, CLARP and LARPC on their ability to form UV-cross-links to ASL-3. LARP, CLARP and LARPC employed at the indicated concentrations were UV-cross-linked to ASL-3 at 1 mM DTT (panel B), without DTT (panel C) or at 10 mM DTT (panel D). (E) Effect of LARP, CLARP and LARPC on ACNase activity at 1 mM DTT. The incubation times were 2 (lanes 2, 5, 8, 11, 14 and 17), 5 (lanes 3, 6, 9, 12, 15 and 18) and 10 min (lanes 4, 7, 10, 13, 16 and 19). (F) Effect of 10 mM DTT on the ACNase-inhibiting potential of LARP, CLARP or LARPC. The incubation times were 2 (lanes 2, 5, 8, 11, 14, 17 and 20), 5 (lanes 3, 6, 9, 12, 15, 18 and 21) and 10 min (lanes 4, 7, 10, 13, 16, 19 and 22). a–c indicate respective cross-linking products assumed to contain one, two or three LARP moieties per ASL-3; a′-c′, corresponding products of CLARP and LARPC. ASL-3 fr. 1–7, labeled ACNase cleavage product of ASL-3.
Figure 9.
Cys substitutions within or near the PrrC region corresponding to LARP trigger inter-subunit cross-links. (A) In vitro ACNase activity of PrrC* and the indicated to-Cys derivatives of PrrC*. These PrrC forms were isolated by affinity chromatography and assayed for ACNase activity as detailed in the Methods section. (B) Escherichia coli cells expressing the indicated PrrC forms were suspended in 10 mM diamide and incubated at 10°C for the indicated time. The cells were lyzed, their proteins separated in SDS–PAGE without a reducing agent and PrrC visualized by immunoblotting. (C) The indicated PrrC forms were purified by TALON® immobilized-metal affinity-chromatography and further fractionated by SDS–PAGE, as such (lanes 1, 4, 7 and 10), after treatment with 1 mM DTT (lanes 2, 5, 8 and 11) or further treatment with 2 mM diamide (lanes 3, 6, 9 and 12) and then monitored by immunoblotting. (D) The indicated PrrC forms were treated as in panel C and monitored by protein staining. (E) The indicated PrrC forms were subjected to GA-mediated protein–protein cross-linking. (F) The indicated PrrC mutant forms were analyzed as in panel B: L298C (lanes 1–3), Q300C (lanes 4–6), I302C (lanes 7–9), I306C (lanes 10–12), PrrC* (lanes 13–15), PrrC-D222E (lanes 16–18). The bands indicated as Xa and Xb are in respective order the common form of band X generated by the majority of the to-Cys mutants and the faster migrating form generated only by Q300. ASL-3 fr. 1–7, labeled ACNase cleavage product of ASL-3 containing residues 1–7; kDa, protein size markers; 2 mers, 3 mers and 4 mers, indicate cross-linking forms thought to contain 2, 3 or 4 PrrC subunits, respectively; M, PrrC monomer; X, the major S-S cross-linking product.
Mutating LARP impairs the ASL-cross-linking and ACNase-inhibiting activities
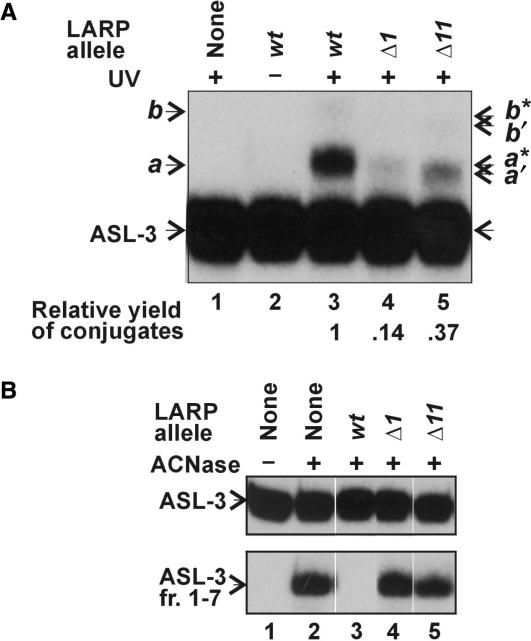
LARP was mutated to evaluate the importance of its sequence to the ASL-cross-linking and ACNase-inhibiting activities. Trimmed derivatives lacking Lys1 (Δ1) or Tyr11 (Δ11) formed UV-cross-links to ASL-3 ∼ 7- or 3-fold less efficiently than LARP, respectively (Figure 6A). The conjugates obtained with the trimmed mutants (designated a′, a*, b′ and b*) were less retarded than those of LARP, possibly due to their lower mass. Δ1 and Δ11 also hardly inhibited ACNase (Figure 6B).
Figure 6.
Effect of trimming LARP on its ASL-3 cross-linking and ACNase-inhibiting activities. (A) The UV-cross-linking of the indicated peptides (employed each at 200 μM) to ASL-3 was performed essentially as in Figure 3A. (B) Effect of the indicated peptides on the PrrC-catalyzed cleavage of ASL-3. The ACNase assay was performed essentially as in Figure 2C. a and b indicate respective cross-linking products assumed to contain one or two LARP moieties per ASL-3; a′ and b′ indicate corresponding cross-linking conjugates of Δ1, a* and b* indicate cross-linking conjugates of Δ11.
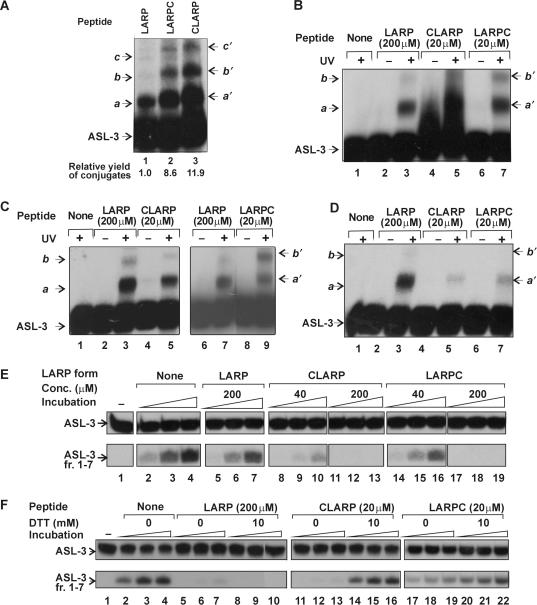
LARP's sequence was also changed by PrrC missense mutations that confer an ACNase null phenotype. These mutations: F292S (17), Y294S or Y294F (3) were respectively termed F9S, Y11S and Y11F in the peptide context. They were introduced in a LARP derivative containing an extra N-terminal Cys (CLARP, described in more detail later). CLARP was far more efficient than LARP both in cross-linking ASL-3 and inhibiting ACNase (Figure 8) and using it as the reference peptide facilitated the detection of residual mutant activities.
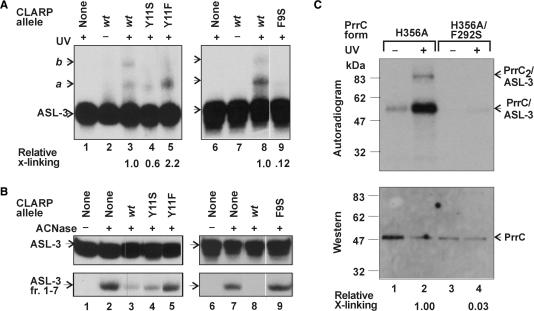
F9S and Y11S impaired the two activities in a correlated manner. The milder Y11S mutation reduced the yield of the primary UV-cross-linking conjugate with ASL-3 nearly 2-fold (Figure 7A, lane 3 versus 4) and partially impaired the inhibition of ACNase (Figure 7B, lane 3 versus 4). It is noteworthy that this mutation also prevented the formation of a stable secondary conjugate, as inferred from the absence of the expected band b and appearance instead of a smear trailing behind band a. The more drastic mutation F9S reduced the cross-linking to ASL-3 8-fold (Figure 7A, compare lanes 8 and 9) and severely impaired ACNase inhibition (Figure 7B, compare lanes 8 and 9). The corresponding PrrC mutation F292S could also abrogate the UV-cross-linking of PrrC to ASL-3. This was inferred from the observation that the active site PrrC mutant H356A, which lacks ACNase activity (3) but efficiently binds tRNALys (data not shown), formed UV-cross-links to ASL-3 ∼ 30-fold more efficiently than F292S/H356A (Figure 7C).
Figure 7.
ASL-3 cross-linking and ACNase-inhibiting activities of missense mutants of CLARP. (A) The UV-cross-linking of CLARP and its indicated missense mutants to ASL-3 was performed essentially as in Figure 3A except that the peptide level was only 20 μM. (B) The effect of CLARP and its indicated missense mutants on PrrC-catalyzed cleavage of ASL-3 was determined essentially as in Figure 2C except that the peptide level was 20 μM. (C) The F292S mutation of PrrC inhibits the UV-cross-linking of the protein to ASL-3. ASL-3 was UV-cross-linked to the indicated alleles of PrrC. The products were separated by SDS–PAGE, transferred to a nitrocellulose membrane and detected by immunoblotting and autoradiography as detailed in the Methods section. a and b indicate respective cross-linking products assumed to contain one or two LARP moieties per ASL-3. ASL-3 fr. 1–7; labeled ACNase cleavage product of ASL-3; PrrC/ASL-3 and PrrC2/ASL-3, respective UV-cross-linking products of ASL-3 with a PrrC monomer or PrrC dimer. X-linking, cross-linking.
The Y11F mutation conferred a different phenotype. It nearly abolished the ACNase-inhibiting potential of CLARP (Figure 7B, compare lanes 2, 3 and 5) yet doubled the yield of the primary UV-cross-linking conjugate obtained with ASL-3 (Figure 7A, compare lanes 3 and 5). In part this increase could be ascribed to the failure of Y11F to form stable higher conjugates, as with Y11S. Nonetheless, the overall cross-linking yield obtained with Y11F suggested that this mutant bound ASL-3 at least as efficiently as CLARP. The discrepant behavior of Y11F hinted that the ACNase-inhibiting forms of LARP exerted their effect not only by occluding the RNA substrate but also by forming a binary complex with PrrC. Since LARP was employed in excess over PrrC and ASL-3, it could exert these two ACNase-inhibiting modes independently. The possibility that LARP interacted with PrrC directly was reinforced by data shown below suggesting that both LARP and its matching PrrC sequence can self-interact (Figures 8 and 9). The failure of the Y11S and Y11F mutants to form stable conjugates containing additional peptide moieties could not be ascribed to a critical role of Tyr11 in peptide–peptide UV-cross-linking since deleting this residue did not elicit such an effect (Figure 6A, lane 5).
Extending LARP with Cys augments its activities under suitable redox conditions
LARP derivatives extended at the N- or C-end with Cys (CLARP and LARPC, respectively) were intended for further modification with Fe-EDTA that rendered them artificial nucleases (M. Amitsur, D. Klaiman and G. Kaufmann; unpublished data). Unexpectedly, CLARP and, to a lesser extent, LARPC were far more potent than LARP in UV-cross-linking ASL-3 and inhibiting ACNase. Thus, when CLARP, LARPC or LARP were UV-cross-linked to ASL-3 at 1 mM DTT and identical peptide levels, similar product patterns were obtained. However, the product yields with CLARP or LARPC were about an order of magnitude higher than with LARP (Figure 8A). Employing the Cys-containing peptides at a level 10-fold lower than that of LARP resulted in comparable yields (Figure 8B). CLARP and LARPC UV-cross-linked ASL-3 more efficiently than LARP also without DTT, LARPC yielding under these conditions a relatively high proportion of the secondary conjugate (Figure 8C). However, the cross-linking efficiency of CLARP and LARPC to ASL-3 was relatively weak at 10 mM DTT (Figure 8D). CLARP and LARPC inhibited ACNase more strongly than LARP at 1 mM DTT (Figure 8E) but were less effective than LARP at 10 mM DTT (Figure 8F).
Thus, tethering Cys to one or the other end of LARP enhanced the ACNase-inhibiting and ASL-3 cross-linking activities of the peptide although not under highly reducing conditions. Moreover, LARPC yielded under oxidizing conditions higher proportions of conjugates likely to contain two peptide moieties per ASL-3. These facts may be accounted for by the stabilization of a common functional form of LARP both by a C-terminal or N-terminal S-S bond, a requirement satisfied by a parallel but not a head-to-tail LARP dimer. An alternative explanation is that mere dimerization of LARP by a disulfide bond at either end increased the probability of non-specific electrostatic, hydrogen bonding and stacking interactions between the peptide and the RNA.
The tethered Cys residue enhanced the UV-cross-linking efficiency of LARP to ASL-3 optimally at intermediate redox conditions (1 mM) rather than oxidizing conditions. This outcome suggested that reversible formation of the disulfide link facilitated the fixation of original LARP–LARP and LARP–RNA contacts by UV-cross-linking. In other words, a stable S-S bond could impede the UV-cross-linking by constraining the dimer.
Placing Cys at or near the PrrC region matching LARP triggers intersubunit cross-links
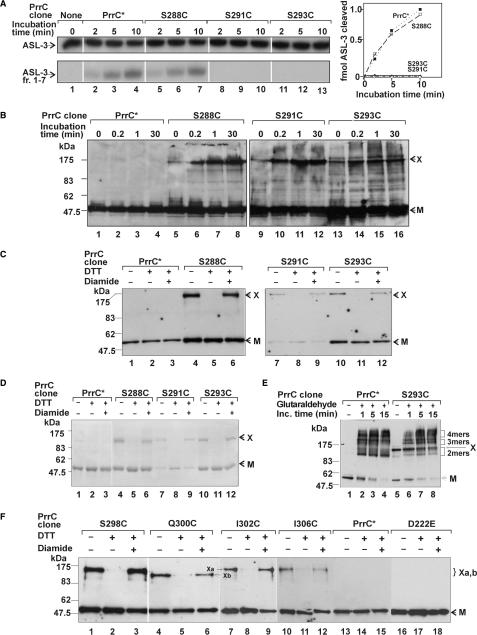
The suspicion that LARP exerted its ASL-3 cross-linking and ACNase inhibiting activities as a parallel dimer prompted us to examine if the matching PrrC sequence dimerizes similarly. To this end, a Cys residue was introduced in this sequence instead of Ser288, Ser291 or Ser293. We expected that the mutant Cys residues will self-interact and form intersubunit disulfide cross-links if the region containing them dimerizes in parallel. Each of the three mutations was placed over the D222E/C268A/C385A background termed PrrC*. This facilitated the isolation of the mutant proteins (3,17) and precluded the formation of non-specific S-S cross-links due to the wild-type Cys residues. The PrrC* control and to-Cys derivatives exhibited comparable in vivo ACNase activities and protein levels (data not shown). However, only PrrC* and S288C/PrrC* retained ACNase activity in vitro (Figure 9A).
To induce the formation of S-S cross-links the cells expressing the to-Cys mutants or the PrrC* control were exposed to diamide (27). The cellular proteins were separated then by SDS–PAGE under non-reducing conditions and PrrC visualized by immunoblotting. As shown, the three mutants, but not PrrC* yielded products that migrated in SDS–PAGE slower than the ∼47.5-kDa PrrC monomer (Figure 9B). These products included a major form designated X that migrated below the 175-kDa size marker as well as several less pronounced and faster migrating forms. These various products appeared also without diamide, perhaps due to partial in vitro oxidation. However, their amounts increased considerably with diamide, consistent with their formation by disulfide cross-linking.
To determine if any of the diamide-dependent products arose by intra-PrrC cross-links, the four PrrC forms were isolated by immobilized metal affinity chromatography in the absence of a reducing agent. Subsequently they were fractionated by SDS–PAGE as such, after prior treatment with 1 mM DTT or also with 2 mM diamide. Western analysis revealed that the purified Ser → Cys mutants, but not PrrC*, yielded a single product with mobility similar to that of band X of the diamide treated cells (compare Figure 9C, lanes 1, 4, 7 and 10 to Figure 9B lanes 4, 8, 12 and 16). Moreover, this band was abolished by DTT (Figure 9C. lanes 2, 5, 8 and 11) and restored by excess diamide (lanes 3, 6, 9 and 12), indicating it depended on disulfide cross-linking. Corresponding protein staining (Figure 9D) ascertained that the various PrrC forms were not contaminated by significant amounts of other proteins and, consequently, that band X was unlikely to contain proteins other than PrrC. The failure of a considerable fraction of the to-Cys mutant proteins to generate band X could be attributed to its misfolding since PrrC is thermally unstable. Alternatively, this fraction could represent a functionally relevant conformation incompatible with the S-S cross-linking; e.g. due to a substrate or inhibitor bound to the tRNALys site.
Band X migrated in SDS–PAGE less than expected of a cross-linked PrrC dimer of ∼95 kDa. This fact could be attributed to its particular shape or irreversible entanglement of two such dimers within the PrrC tetramer. To distinguish between these possibilities, we subjected PrrC* and S293C/PrrC* to GA-mediated protein–protein cross-linking. PrrC* yielded the familiar pattern (3) where the monomer is gradually converted into apparent GA-cross-linked tetramer forms via dimeric and less pronounced trimeric intermediates (Figure 9E, lanes 1–4). The monomeric fraction of the mutant behaved similarly (lanes 5–8). Band X, which coincided with the most retarded GA-cross-linked dimers, was also gradually converted into higher forms superimposed over those generated by the fraction that failed to form the S-S cross-links. These data suggested that band X was a dimer that was retarded due to its particular shape.
We analyzed in a similar fashion PrrC* derivatives containing Cys in the predicted α-helix found immediately downstream to the LARP-like region (Figure 3). It replaced L298, I302 or I306 of the α-helix hydrophobic face or the hydrophilic Q300. These mutants also yielded the S-S cross-linked form designated X, L298C yielding the highest proportion (Figure 9F, lanes 1–3), possibly due to closer contact of the dimerizing α-helices at position 298. Q300C yielded a variant (designated Xb) that migrated slightly faster than the form X generated by the other mutants designated here Xa (Figure 9F, compare lanes 3 and 4, 6 and 7). The slight mobility difference may be attributed to the presence of Cys300 at the hydrophilic and, hence, outward-pointing face of the dimerizing α-helix. Consequently, an S-S cross-link mediated by it could constrain the dimer into a more compact, faster migrating form. The PrrC mutant D222E containing the wild-type residues Cys268 and Cys385 failed to generate band X (Figure 9F, lanes 16–18). This result underscored the specificity of the intersubunit cross-links triggered by the mutant Cys residues placed in the 288–306 range.
Peptide mimicry of tRNALys recognition by PrrC
LARP formed UV-cross-links to ASL analogs of tRNALys and inhibited their PrrC-catalyzed cleavage (Figures 4 and 5). Moreover, the two activities were impaired by trimming LARP (Figure 6) or placing in it missense mutations that confer a null ACNase phenotype in the PrrC context (Figure 7). Importantly, the CLARP mutation F9S and the corresponding inactivating PrrC mutation F292S severely inhibited the respective UV-cross-linking of ASL-3 to the peptide (Figure 7A) or protein (Figure 7C). These results could be taken to indicate that LARP mimics its matching PrrC sequence in tRNALys recognition. However, even in that case LARP would probably represent only part of PrrC's tRNALys-binding motif since the level at which it effectively inhibited ACNase (Figure 4) far exceeds the Km of efficient ACNase substrates (19). Moreover, known RNA-binding motifs including that of colicin E5-ACNase are several fold longer than LARP (28,29).
The tRNALys site of PrrC may arise between C-domains interfacing in parallel
The functional relevance of LARP to its matching PrrC region was suggested also by the apparent ability of both to dimerize in parallel. We deduced that LARP acted as a parallel dimer in forming UV-cross-links to ASL-3 and inhibiting ACNase from the ability of a single Cys residue tethered to either the N- or C-end of LARP to dramatically enhance these activities and abolition of these enhancements under highly reducing conditions (Figure 6). These coincidences could be accounted for by the stabilization of a common functional dimeric structure of LARP by a disulfide bond formed at one or the other end of its protomers, a requirement met by a parallel but not anti-parallel dimer. Alternatively, the similar effects of the N- and C-proximal Cys extensions reflected the increased probability of non-specific peptide–RNA interactions caused by mere dimerization of the peptide, whether caused by the N- or C-terminal S-S bond. However, we favor the first explanation in view of the intersubunit S-S cross-links triggered by all seven Cys substitutions placed along the C-proximal PrrC 288–306aa region overlapping the sequence matching LARP (Figure 9). Such an outcome is consistent with parallel dimerization of the mutated region and lends support to the assumption that LARP could also dimerize in this orientation. The PrrC region that dimerizes in parallel could be confined to a portion of the ACNase domain since the regulatory N-domain is expected to dimerize in a head-to-tail fashion, like typical ABC/ATPases where the nucleotide binding pocket arises by the interaction of the Walker A motif of one subunit with the ABC signature motif of the second (15). Moreover, the wild-type Cys268 and 385 at the edges of the C-domain (Figure 10A) did not trigger intersubunit cross-links (Figure 9F). The relation of the PrrC region that dimerizes in parallel to the tRNALys-binding motif may be revealed by accurately mapping and functionally characterizing it.
Finally, as illustrated in Figure 10B, the opposite orientations in which PrrC's N- and C-domains dimerize dictate a phosphofructokinase-like topology (30) where the NTPase and tRNALys sites arise at distinct assembly stages of the PrrC dimer of dimers (3).
ACKNOWLEDGEMENTS
We thank Ezra Yagil for comments and Elena Davidov for PrrC mutant proteins. This work was supported by grants from the Israel Science Foundation, and the German-Israeli Foundation for Scientific Research and Development to G.K. and the United-States-Israel Binational Science Foundation to G.K. and D.R.D. Funding to pay the Open Access publication charges for this article was provided by the Israel Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.David M, Borasio GD, Kaufmann G. T4 bacteriophage-coded polynucleotide kinase and RNA ligase are involved in host tRNA alteration or repair. Virology. 1982;123:480–483. doi: 10.1016/0042-6822(82)90284-7. [DOI] [PubMed] [Google Scholar]
- 2.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanga-Kanfi S, Amitsur M, Azem A, Kaufmann G. PrrC-anticodon nuclease: functional organization of a prototypical bacterial restriction RNase. Nucleic Acids Res. 2006;34:3209–3219. doi: 10.1093/nar/gkl415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linder P, Doelz R, Gubler M, Bickle TA. An anticodon nuclease gene inserted into a hsd region encoding a type I DNA restriction system. Nucleic Acids Res. 1990;18:7170. doi: 10.1093/nar/18.23.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amitsur M, Morad I, Chapman-Shimshoni D, Kaufmann G. HSD restriction-modification proteins partake in latent anticodon nuclease. EMBO J. 1992;11:3129–3134. doi: 10.1002/j.1460-2075.1992.tb05385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyndall C, Meister J, Bickle TA. The Escherichia coli prr region encodes a functional type IC DNA restriction system closely integrated with an anticodon nuclease gene. J. Mol. Biol. 1994;237:266–274. doi: 10.1006/jmbi.1994.1230. [DOI] [PubMed] [Google Scholar]
- 8.Penner M, Morad I, Snyder L, Kaufmann G. Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 1995;249:857–868. doi: 10.1006/jmbi.1995.0343. [DOI] [PubMed] [Google Scholar]
- 9.Sirotkin K, Cooley W, Runnels J, Snyder L. A role in true-late gene expression for the T4 bacteriophage 5′-polynucleotide kinase 3′-phospatase. J. Mol. Biol. 1978;123:221–233. doi: 10.1016/0022-2836(78)90322-4. [DOI] [PubMed] [Google Scholar]
- 10.Ho CK, Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galburt EA, Pelletier J, Wilson G, Stoddard BL. Structure of a tRNA Repair Enzyme and Molecular Biology Workhorse: T4 Polynucleotide Kinase. Structure. 2002;10:1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 12.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, Legrice SF, Stammers DK. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 13.Amitsur M, Benjamin S, Rosner R, Chapman-Shimshoni D, Meidler R, Blanga S, Kaufmann G. Bacteriophage T4-encoded Stp can be replaced as activator of anticodon nuclease by a normal host cell metabolite. Mol. Microbiol. 2003;50:129–143. doi: 10.1046/j.1365-2958.2003.03691.x. [DOI] [PubMed] [Google Scholar]
- 14.Morad I, Chapman-Shimshoni D, Amitsur M, Kaufmann G. Functional expression and properties of the tRNALys-specific core anticodon nuclease encoded by Escherichia coli prrC. J. Biol. Chem. 1993;268:26842–26849. [PubMed] [Google Scholar]
- 15.Moody JE, Thomas PJ. Nucleotide binding domain interactions during the mechanochemical reaction cycle of ATP-binding cassette transporters. J. Bioenerg. Biomembr. 2005;37:475–479. doi: 10.1007/s10863-005-9494-8. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Meidler R, Morad I, Amitsur M, Inokuchi H, Kaufmann G. Detection of Anticodon Nuclease Residues Involved in tRNALys Cleavage Specificity. J. Mol. Biol. 1999;287:499–510. doi: 10.1006/jmbi.1999.2634. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Meidler R, Amitsur M, Kaufmann G. Specific Interaction between Anticodon Nuclease and the tRNALys Wobble Base. J. Mol. Biol. 2001;305:377–388. doi: 10.1006/jmbi.2000.4282. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Blanga S, Amitsur M, Meidler R, Krivosheyev E, Sundaram M, Bajji AC, Davis DR, Kaufmann G. Structural features of tRNALys favored by anticodon nuclease as inferred from reactivities of anticodon stem and loop substrate analogs. J. Biol. Chem. 2002;277:3836–3841. doi: 10.1074/jbc.M110072200. [DOI] [PubMed] [Google Scholar]
- 20.Shterman N, Elroy-Stein O, Morad I, Amitsur M, Kaufmann G. Cleavage of the HIV replication primer tRNALys,3 in human cells expressing bacterial anticodon nuclease. Nucleic Acids Res. 1995;23:1744–1749. doi: 10.1093/nar/23.10.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amitsur M, Morad I, Kaufmann G. In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J. 1989;8:2411–2415. doi: 10.1002/j.1460-2075.1989.tb08371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajji AC, Sundaram M, Myszka DG, Davis DR. An RNA complex of the HIV-1 A-loop and tRNALys,3 is stabilized by nucleoside modifications. J. Am. Chem. Soc. 2002;124:14302–14303. doi: 10.1021/ja028015f. [DOI] [PubMed] [Google Scholar]
- 23.Sundaram M, Crain PF, Davis DR. Synthesis and characterization of the native anticodon domain of E. coli tRNALys: simultaneous incorporation of modified nucleosides mnm(5)s(2)U, t(6)A, and pseudouridine using phosphoramidite chemistry. J. Org. Chem. 2000;65:5609–5614. doi: 10.1021/jo000338b. [DOI] [PubMed] [Google Scholar]
- 24.Ansaldi M, Lepelletier M, Mejean V. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal. Biochem. 1996;234:110–111. doi: 10.1006/abio.1996.0060. [DOI] [PubMed] [Google Scholar]
- 25.Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 26.Thomas G, Thiam K, Favre A. tRNA thiolated pyrimidines as targets for near-ultraviolet- induced synthesis of guanosine tetraphosphate in Escherichia coli. Eur. J. Biochem. 1981;119:381–387. doi: 10.1111/j.1432-1033.1981.tb05619.x. [DOI] [PubMed] [Google Scholar]
- 27.Kosower NS, Kosower EM. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 28.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res. 2006;34:6074–6082. doi: 10.1093/nar/gkl729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schirmer T, Evans PR. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 34.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]