Abstract
For production of proteins that are encoded by the mitochondrial genome, mitochondria rely on their own mitochondrial translation system, with the mitoribosome as its central component. Using extensive homology searches, we have reconstructed the evolutionary history of the mitoribosomal proteome that is encoded by a diverse subset of eukaryotic genomes, revealing an ancestral ribosome of alpha-proteobacterial descent that more than doubled its protein content in most eukaryotic lineages. We observe large variations in the protein content of mitoribosomes between different eukaryotes, with mammalian mitoribosomes sharing only 74 and 43% of its proteins with yeast and Leishmania mitoribosomes, respectively. We detected many previously unidentified mitochondrial ribosomal proteins (MRPs) and found that several have increased in size compared to their bacterial ancestral counterparts by addition of functional domains. Several new MRPs have originated via duplication of existing MRPs as well as by recruitment from outside of the mitoribosomal proteome. Using sensitive profile–profile homology searches, we found hitherto undetected homology between bacterial and eukaryotic ribosomal proteins, as well as between fungal and mammalian ribosomal proteins, detecting two novel human MRPs. These newly detected MRPs constitute, along with evolutionary conserved MRPs, excellent new screening targets for human patients with unresolved mitochondrial oxidative phosphorylation disorders.
INTRODUCTION
Mitochondria are primary ATP-producing organelles which originated from an ancestral alpha-proteobacterial endosymbiont (1,2). The oxidative phosphorylation (OXPHOS) pathway, consisting of the mitochondrial respiratory chain and the ATP synthase complex, is the final biochemical pathway in energy conversion (3). However, mitochondria have been found to perform pivotal roles in many other metabolic, regulatory and developmental processes. During the evolution of the mitochondria, most genes of the ancestral endosymbiont have been transferred from the mitochondrial to the nuclear genome. The genes that reside on the mitochondrial DNA (mtDNA), e.g. 13 OXPHOS-related genes in human, are translated into their respective protein products by the mitochondrial translation machinery, which resembles the prokaryotic translation system (4), comprising a mitochondrial ribosome (mitoribosome) and several translation factors. In mammals, all proteins that constitute the mitochondrial translation machinery are encoded by the nuclear genome. Also the mitochondrial genomes of fungi generally encode only very few mitochondrial ribosomal proteins (MRPs). In contrast, the mitochondrial genome of many protozoa and plants encode numerous MRPs. Most notably, the ‘primitive’ mitochondrial genome of the freshwater protist Reclinomonas americana was found to contain as many as 27 MRP-encoding genes (5).
Mitoribosomes have undergone major remodeling during their evolution. For instance, it has been found that, despite the fact that their rRNA content is only half the content of bacterial ribosomes (6), mitoribosomes generally exceed the bacterial ribosomes both in molecular mass as well as in physical dimensions (6–8). Mass spectrometry studies of the bovine and yeast mitoribosomes, which have served as model systems during the past years, revealed that mammalian mitoribosomes comprise about 80 MRPs (9–13). In yeast, about 70 MRPs have been identified thus far, although protein–protein interaction data and mutational analysis suggest that this number might be substantially higher (14–16). Interestingly, a recent proteomics survey of mitochondrial ribosome-related complexes in the kinetoplast-mitochondria of Leishmania tarentolae revealed an additional protein complex which was found to be specifically associated with the small subunit (SSU), but not with the large subunit (LSU) of the mitoribosome (17). The biological role of the additional SSU-associated complex, which mostly consists of proteins unique to kinetoplastida, remains unknown.
Apparently, mitoribosomes have expanded their protein content in the course of evolution by acquiring numerous extra ‘supernumerary’ MRPs. Currently information regarding the functions of these supernumerary MRPs is rather limited. In addition, loss of MRPs that are normally part of the ‘bacterial core ribosome’ is also observed.
Recently, several attempts have been made to identify the MRPs in other eukaryotes, such as Neurospora crassa (18), Arabidopsis thaliana (19) and L. tarentolae (20), however, a comprehensive study of the evolution of the mitoribosomal proteome remains to be established. Such a study bears relevance for the potential identification of genes that cause mitochondrial dysfunctions in human patients. Since mitochondria perform many fundamental functions, mitochondrial dysfunction results in a wide variety of multisystemic diseases, predominantly affecting tissues with high metabolic energy rates (21). These disorders are mostly caused by the dysfunction of one or more enzyme complexes of the OXPHOS system and several mutations have been identified in mtDNA (22) as well as in nuclear DNA (23). Not only mutations in structural OXPHOS genes, but also mutations in genes involved in the mitochondrial transcription or translation, or in the assembly of OXPHOS complexes can result in OXPHOS disease. Although the vast majority of components of the mitochondrial translation system are nuclear encoded, thus far most mutations associated with mitochondrial translation defects have been reported in mtDNA-encoded tRNAs and rRNAs (22,24). Recently, mutations in four different nuclear gene products involved in mitochondrial protein synthesis have been reported in patients with mitochondrial disease: elongation factors EFG1 (25), EFTs (26) and EFTu (27), and small ribosomal subunit protein MRPS16 (28). Additional information regarding which MRPs could be implicated in disease could be obtained by studying the evolutionary conservation of the MRPs.
The aim of the present study is 2-fold, from an evolutionary and a disease point of view: (i) gain insight into the evolution of the mitoribosome and its protein content in various eukaryotic species; (ii) prioritize MRPs as candidates for their involvement in mitochondrial disease. We have performed a comprehensive comparative genomics analysis of the MRPs in 18 eukaryotic species from which both the complete nuclear and the organellar genome sequences were available. Representatives are included from different phylogenetic groups: six metazoa (Homo sapiens, Mus musculus, Tetraodon nigroviridis, Drosophila melanogaster, Anopheles gambiae and Caenorhabditis elegans), two fungi (Saccharomyces cerevisiae and N. crassa), one microsporidian (Encephalitozoon cuniculi), one mycetozoan (Dictystelium discoideum), one plant (A. thaliana), one alga (Chlamydomonas reinhardtii), one apicomplexa (Plasmodium falciparum), one ciliophora (Tetrahymena thermophila), two kinetoplastida (Trypanosoma brucei and Leishmania major), one diplomonad (Giardia lamblia) and the mitochondrial genome of the freshwater protist Reclinomonas americana. In addition, we included a balanced, phylogenetically diverse subset of completely sequenced prokaryotic genomes in our survey.
Our analysis retrieved multiple previously unidentified MRPs in several species, two of which constitute potential novel human MRPs. Furthermore, the current study established orthology relationships between seven MRPs reported to be fungi specific and ribosomal proteins from other eukaryotes. The establishment of these homology relationships enabled us to trace the origins of some of the supernumerary MRPs and to predict their molecular functions. In addition, we have investigated all MRPs in the presence of additional, newly acquired protein domains that might point at mitochondria-specific adaptations of the mitoribosome, and we have reconstructed the evolutionary history of the mitoribosomal proteome in terms of gains and losses of the MRPs along the different eukaryotic lineages. The newly detected mitochondrial ribosomal genes constitute, in addition to the set of evolutionary conserved MRPs, excellent new screening targets for human patients with unresolved mitochondrial oxidative phosphorylation disorders.
MATERIALS AND METHODS
Use of genomic data
Genome data (both proteomes and genomes) of the eukaryotes described in this study were obtained from the Integr8 database (http://www.ebi.ac.uk/integr8/, Release 41) at EBI (29), except for the genomes of Tetrahymena thermophila (version 2 obtained from TIGR, ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/t_thermophila/), Trypanosoma brucei (version 4 obtained from Sanger, ftp://ftp.sanger.ac.uk/pub/databases/T.brucei_sequences/), N. crassa (version 7.0 obtained from Broad institute at MIT, http://www.broad.mit.edu/annotation/genome/neurospora/Downloads.html) and Chlamydomonas reinhardtii (version 3.0 obtained from JGI, ftp://ftp.jgi-psf.org/pub/JGI_data/Chlamy/v3.0/). For most eukaryotic species, the organelle genomes were included in the gene set per species, otherwise those were downloaded separately from GOBASE (30) (http://megasun.bch.umontreal.ca/ogmp/projects/other/mt_list.html).
In addition to the eukaryotic genomes mentioned above, we have also included a balanced, phylogenetically diverse subset of completely sequenced prokaryotic genomes. We have explicitly chosen not to include the complete set of completely sequenced prokaryotic genomes available, since the large number of prokaryotic proteins might cause an imbalance in the PSI-BLAST analysis described below. Sequence data (proteomes and genomes) of the selected prokaryotic genomes were all downloaded from the Integr8 database at EBI, and included: Escherichia coli, Pseudomonas aeruginosa, Vibrio cholerae (gamma-proteobacteria), Bacillus subtilis, Clostridium acetobutylicum (firmicutes), Borrellia burgdorferi (spirochetes), Agrobacterium tumefaciens WashU, Brucella suis, Rickettsia prowazeckii, Caulobacter crescentus, Rhizobium meliloti, Rhodopseudomonas palustris (alpha-proteobacteria), Nostoc sp. PCC 7920, Synechocystis sp. PCC 6803, Prochlorococcus marinus SS120 (cyanobacteria), Deinococcus radiodurans (Deinococcus-Thermus group), Aquifex aeoliticus (aquificae), Sulfolobus solfataricus (crenarchaea), Pyrococcus furiosus (euryarchaea).
Detection of MRPs in eukaryotic genomes
For each of the experimentally verified proteins of the Leishmania (17,20), fungal (14–16) and mammalian (6) mitochondrial ribosomal proteins we have performed iterative PSI-BLAST searches (31) against a locally assembled protein database comprised of the proteomes of the organisms indicated above. For each of these searches, an E-value inclusion threshold of 0.005 was applied, and the proteins that were retrieved after five iterations were aligned using MUSCLE (32).
For those cases where a candidate MRP, from a species from which the query MRP was still missing, appeared in the PSI-BLAST hit list, but with an E-value between 0.005 and 10, a reciprocal search was performed. If this search successfully retrieved a bona fide MRP within five iterations (E-value inclusion threshold of 0.005), it was added to the initial set of homologous proteins, and a new alignment was created. Whenever a given MRP was still not found in the predicted proteome of a eukaryotic organism, a hidden Markov model (HMM) of the respective protein was created from the above-mentioned alignments using the HMMER program (33). Subsequently, the HMM was used to screen the genomic sequences at the DNA level. The latter screen served as a final control for missing MRPs that represent unpredicted genes in the genome annotation process. To delineate orthologous groups, neighbor-joining trees were derived from these alignments using Kimura distances (34). A bootstrap analysis using 1000 samples was conducted for each phylogenetic tree. The selection of the orthologous group for each MRP was subsequently done by manually examining all trees. Assignment of orthologous relationships among the eukaryotic sequences was based on the species-phylogeny for eukaryotes (35,36) and the presence of these sequences within a monophyletic branch of the tree that contained known MRPs. Homologies between different families of MRPs, if not already found during the PSI-BLAST searches described above, were detected using the hhsearch algorithm (37), by comparing HMM profiles constructed from aligned MRPs with each other, using an E-value cut-off of 1e−4.
Phylogenetic analyses
Maximum-likelihood (ML) trees were created using PHYML (version 2.2.4) (38) based on alignments of the above-mentioned orthologous groups. Prior to the analyses, global and pairwise gaps were removed from the alignments. The appropriate model of protein evolution was selected using MODELSELECTOR (39). Bootstrap support values were derived from 100 replicates.
Detection of newly evolved MRP domains
All MRPs homologous to bacterial core RPs that were identified in the current research were examined for additional, potentially newly evolved domains. Using HMMER (33), each MRP was analyzed with an HMM profile that was built from an alignment of its ancestral bacterial counterpart. These parts of the MRPs that were not covered by the respective HMM profile (minimum length 30 amino acids), were searched against the PFAM (40) and CDD (41) databases using HMMER (33) and RPS-BLAST (41) respectively. Hits below an E-value threshold of 0.01 were regarded significant.
RESULTS
Detection of MRPs in eukaryotic genomes
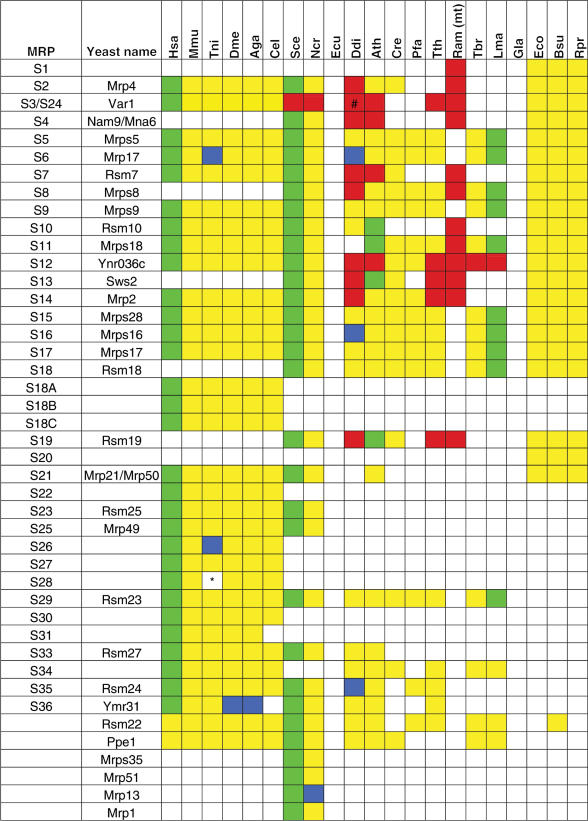
We have employed a systematic analysis of nuclear and mitochondrial genomes in order to identify the mitoribosomal proteome content encoded in a wide variety of eukaryotic species. To avoid confusion regarding different nomenclatures that are used for ribosomal proteins, we have adopted the nomenclature for human MRPs, approved by the Human Genome Nomenclature Committee (http://www.gene.ucl.ac.uk/nomenclature/). All proteins are designated MRPSxx or MRPLxx, where S and L stand for small and large subunit, respectively, and xx is the number of the corresponding bacterial ribosomal protein. The MRPs lacking bacterial orthologs (supernumerary proteins) are assigned higher numbers, starting at MRPS22 and MRPL37 for the small and large subunit, respectively. In yeast, unfortunately, the nomenclature is as variable as the methods employed for their characterization. Therefore, in order to prevent further confusion about proteins with the same name in human and yeast that are not part of the same orthologous group, we have indicated all human MRPs in capital letters throughout the manuscript (e.g. MRPS28), whereas for the yeast proteins we only capitalized the first letter (e.g. Mrps28, which corresponds to MRPS15 in human, also see Table 1 and Supplementary Data).
Table 1.
Phylogenetic distribution of SSU mitochondrial ribosomal proteins and their bacterial counterparts
The first and second column contain, if applicable, the MRP identifiers according to the human and yeast nomenclature, respectively. MRPs and bacterial RPs that were identified in the predicted proteome are indicated in yellow, MRPs that were not found in the predicted proteome, but for which the unpredicted gene could be identified on the genomic DNA are indicated in blue. MRPs that have been experimentally verified as being associated with the mitoribosome by means of large-scale proteomics interaction studies or small-scale functional studies are indicated in green (except for the human MRPs, for which mostly the bovine counterparts were experimentally verified). MRPs that are encoded by the mitochondrial genome are indicated in red. A bracket (#) denotes a peculiar case of a split MRPS3 gene in D. discoideum, which is encoded by its mitochondrial genome. The 5′-and 3′ ends are fused to genes of unknown function (ORF425 and ORF1740, respectively) (79,80). It is unknown if the split MRPS3 genes results in a functional protein. An asterisk (*) denotes cases where a given MRP was not identified most probably as a result of incompleteness of the genome sequence, i.e. the MRP was not found in the predicted proteome, nor was it found by analyzing the genomic DNA, but could be identified in a closely related species. See Supplementary for protein identifiers.
Species abbreviations: Hsa: Homo sapiens; Mmu: Mus musculus; Tni: Tetraodon nigroviridis; Dme: Drosophila melanogaster; Aga: Anopheles gambiae; Cel: Caenorhabditis elegans; Sce: Saccharomyces cerevisiae; Ncr: Neurospora crassa; Ecu: Encephalitozoon cuniculi; Ddi: Dictyostelium discoideum; Ath: Arabidopsis thaliana; Cre: Chlamydomonas reinhardtii; Pfa: Plasmodium falciparum; Tth: Tetrahymena thermophila; Ram (mt): Reclinomonas americana (mitochondrial genome); Tbr: Trypanosoma brucei; Lma: Leishmania major; Gla: Giardia lamblia; Eco: Escherichia coli; Bsu: Brucella suis; Rpr: Rickettsia prowazekii.
Starting from a curated list of MRPs that have been identified in various studies, we first searched for homologs using several search algorithms (see Materials and Methods). Subsequently, the list of homologs was subjected to a phylogenetic analysis in order to define orthologous groups of MRPs. Not only does our analysis distinguish MRPs from their cytosolic counterparts, it also enabled for differentiation between ‘true’ MRPs and homologous ribosomal proteins of other organelles, such as the chloroplast (in plants) or apicoplast (in the apicomplexa P. falciparum). We realize that this approach has its limitations, as it does not account for potential retargeting events that have been shown to occur, such as in A. thaliana, where a chloroplast-type S13 protein encoded by a diverged duplicated nuclear gene has been demonstrated to be targeted to the mitochondrion (42). In order to account for such cases, we included such an experimentally verified protein in our list, whenever our analysis did not suggest a candidate MRP.
Altogether, our survey resulted in the identification of highly variable numbers of MRPs across the genomes of the investigated eukaryotic species, several of which had not been identified as MRPs previously (see Table 1, Table 2 and Supplementary Data). The total number of identified MRPs ranged from 81 in most metazoan species, to 80 in yeast, to 63 in plants, down to a mere 39 MRPs in the apicomplexan P. falciparum. We recognize the fact that the latter numbers are probably an underestimate of the actual numbers of MRPs in these species, as there are no experimental studies to identify potential lineage-specific supernumerary MRPs. Moreover, since protein sequences of these species have been found to evolve relatively fast (43), it is at least feasible that we failed to identify some MRPs that have evolved beyond the detection limits of our search algorithms.
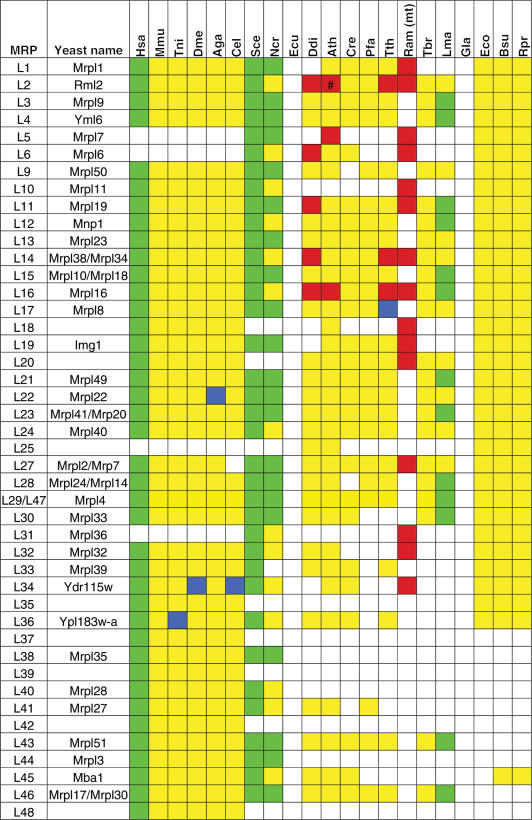
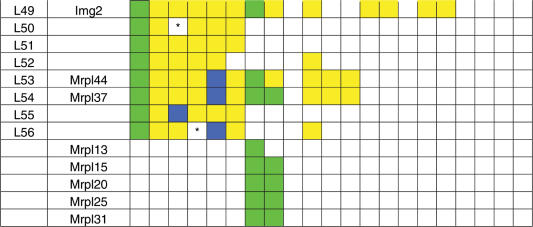
Table 2.
Phylogenetic distribution of LSU mitochondrial ribosomal proteins and their bacterial counterparts
For explanation of used MRP protein names, color coding and species abbreviations, see comments below Table 1. Subunits that are indicated to be experimentally characterized for L. major were in fact identified to be part of the mitoribosome of the closely related species L. tarentolae (17). A bracket (#) denotes a special case of the A. thaliana MRPL2, of which the N-terminal half is encoded by the mitochondrial genome and the C-terminal half is nuclear encoded. See Supplementary Data for protein identifiers. An asterisk (*) denotes cases where a given MRP was not identified as a result of incompleteness of the genome sequence, i.e. the MRP was not found in the predicted proteome, nor was it found by analyzing the genomic DNA, but could be identified in a closely related species. For example, MRPS28 and MRP10 were not detected in T. nigroviridis, but could be found in Danio rerio. For example, MRPL56 was not detected in D. melanogaster, but was found in the genomes of D. willistoni and A. gambiae. Originally, MRPS32 was identified as a protein in the small subunit (9) and MRPL42 as a protein in the large subunit [MRPL31 in (89)]. However, the protein sequences of these proteins were found to be identical. Later, MRPS32 was omitted since its identification was probably caused by contamination in the preparation of small subunits, and thus was in fact MRPL42 (Thomas O’Brien, personal communication).
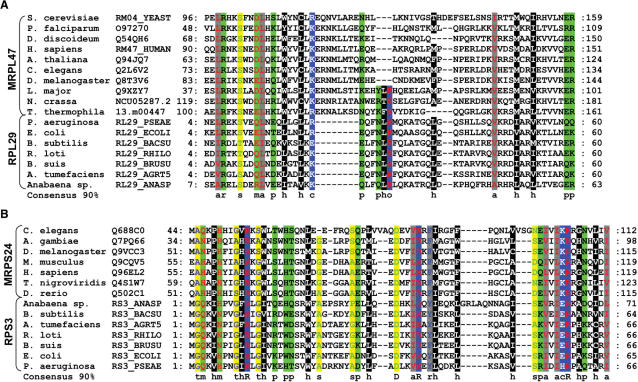
Apart from mapping MRPs that are encoded by eukaryotic genomes, we were able to find hitherto undetected homology between experimentally verified supernumerary MRPs and bacterial ribosomal proteins that were presumed to have been lost from the primitive eukaryotic mitoribosome. Using sensitive profile–profile searches for detection of distant homology between protein families, we connected the MRPS24 orthologous group to the bacterial RPS3 ribosomal protein family (E-value 2.2e−5) and similarly, we connected the MRPL47 orthologous group to the bacterial RPL29 ribosomal protein family (E-value 8.6e−7) (see Figure 1). The complementary phylogenetic distribution patterns of the respective genes are in support of this suggestion. For example, MRPL47 is present in all eukaryotes analyzed (except for E. cuniculi and G. lamblia), but not in bacteria. The opposite phylogenetic pattern is observed for bacterial ribosomal protein RPL29, for which no homolog was identified in any eukaryotic genome. Apparently, these MRPs have diverged almost beyond homology detection limits during the course of evolution (see Figure 1), possibly as an adaptation to specific mitoribosomal functionality. Given the detected homology between the above-mentioned MRPs and bacterial ribosomal proteins, we have placed the MRPs in the orthologous group of their respective bacterial counterparts (see Table 1 and 2).
Figure 1.
Distant homology between mitochondrial and bacterial ribosomal protein families detected in this study. Multiple sequence alignments of MRPL47 and RPL29 proteins (A) and of MRPS24 and RPS3 proteins (B). Amino acid residues are shaded according to the following physicochemical properties: t: tiny (blue font on yellow shading); m: amphoteric (red font on green shading); h: hydrophobic (white font on black shading); o: positive (red font on blue shading); p: polar (black font on green shading); s: small (green font on yellow shading); a: aliphatic (red font on gray shading), r: aromatic (blue font on gray shading); c: charged (white font on blue shading). Sequences are annotated by the organism name and Swiss-Prot protein identifier and the boundaries of the segment that is used in the alignment are indicated. A 90% consensus sequence is depicted below the alignments using the same classification, with invariant residues being indicated in capitals.
We also encountered some cases where we could not detect an expected MRP ortholog in some species, which might be caused by incompleteness of the respective genomes. For example, despite the fact that we could identify orthologs of MRPL50, MRPS28 and Mrp10 in all metazoa analyzed in this survey, as well as in the Danio rerio genome (data not shown), we were unable to detect these MRPs in the genome of T. nigroviridis. Likewise, MRPL56 was found in all metazoa, including A. gambiae, but not in D. melanogaster (see Table 2). In addition, in some instances our analysis proposed different proteins than have been reported in earlier reports. The discrepancies between our study and that of others are outlined in the Supplementary Data.
Not unexpectedly, MRPs were not detected in the genomes of the microsporidian E. cuniculi and the diplomonad G. lamblia. These organisms do not contain full-fledged mitochondria, but instead they contain mitosomes (E. cuniculi) or mitochondrial remnants (G. lamblia), which typically lack organellar DNA. Consequently, these organisms have no need for mitochondrial translation machineries, explaining the observed loss of MRP-encoding genes.
Identification of novel (human) MRPs
Our analysis expanded orthologous groups that were previously reported to contain fungi-specific MRPs to include other eukaryotic proteins, resulting in the identification of two potentially novel human MRPs, namely the human orthologs of yeast SSU proteins Rsm22 and Mrp10, extending the total number of human MRPs to 81. The detection of human orthologs of yeast MRP proteins Rsm22 and Mrp10 is of particular interest, as defects within these genes might be linked to mitochondrial dysfunctions in human. Interestingly, extensive biochemical data is available for the yeast gene products, enabling to speculate about their respective molecular functions.
Yeast Rsm22 is homologous to an rRNA methyltransferase and has been shown to be part of the SSU of the yeast mitochondrial ribosome (44). The association of Rsm22 with the SSU has also been confirmed in a large-scale affinity-capture mass spectrometry study (45). We have detected an ortholog of Rsm22 in most eukaryotes, as well as in some prokaryotes. Interestingly, the STRING database (46) shows that the gene encoding the Rsm22 ortholog in some prokaryotes is clustered on the genome with genes that encode ribosomal proteins (not shown), suggesting that the Rsm22 orthologs in these species are also associated with the ribosome. In Schizosaccharomyces pombe, the genes encoding Rsm22 and Cox11 are fused. Cox11 is an assembly factor of the OXPHOS enzyme complex cytochrome c oxidase and in S. cerevisiae, Cox11 has been shown to associate with the mitoribosome and is postulated to function in co-translational insertion of copper ions into Cox1, a subunit of cytochrome c oxidase (47). Finally, Rsm22-deficient yeast strains exhibit a growth defect on non-fermentable (respiratory) carbon sources, indicative of mitochondrial respiratory dysfunction (44). All these lines of evidence suggest that Rsm22 is a protein of the mitoribosomal small subunit that is essential for proper mitochondrial function. As Rsm22 is a predicted rRNA methyltransferase, it might be involved in methylation of mitochondrial rRNA, and as such play a role in the regulation of protein translation efficiency.
Yeast Mrp10 was shown to be a component of the SSU of mitoribosomes as well and disruption of the gene resulted in a mitochondrial translation defect and a tendency to accumulate deletions in mtDNA (48). Jin et al. observed low sequence similarity to yeast Mrpl37, a constituent of the LSU (49). However, we identified yeast Mrpl37 as an ortholog of human MRPL54 and did not detect any significant sequence homology between Mrp10 and Mrpl37. Nevertheless, we did find Mrp10 orthologs in many eukaryotes, all of which were found to contain a CHCH domain (Coiled coil 1—Helix 1—Coiled coil 2—Helix 2, PFAM domain 06747). Within each helix of this CHCH domain, two invariant cysteine residues are present in a C–X9–C motif. Besides in Mrp10, CHCH domains have also been detected in Cox19 and in the respiratory Complex I (NADH-ubiquinone oxidoreductase) 19 kDa subunit (NDUFA8) (50). In yeast, Cox19 is present in the cytoplasm as well as in the inter membrane space of the mitochondria, where it is suggested to play a posttranslational role in the assembly of cytochrome c oxidase through copper transport to mitochondria. It has been proposed that the conserved cysteine residues of the CHCH motif in Cox19 mediate as ligands for the copper ions (51,52). Most likely, yeast Mrp10 and their orthologs in other eukaryotes, including human, constitute an essential component of the mitochondrial translation machinery and we anticipate that the CHCH domains that are present in this orthologous group, as well as in Complex I subunit NDUFA8, are involved in trafficking of metal ions.
We also detected a potential third novel human MRP, Ppe1, however this case is less clear-cut than these described above. Ppe1 is a protein, which has been shown to be part of the SSU of the yeast mitoribosome (53). We detected Ppe1 orthologs in all eukaryotes analyzed, except for P. falciparum and T. thermophila (and the mtDNA lacking eukaryotes). However, the localization of Ppe1 in the mitochondrion is questionable, as the report by Kitakawa et al. is the only indication for this (53). Moreover, the yeast and human Ppe1 have previously been reported to function as protein phosphatase methylesterases of Protein phosphatase 2A (54,55), which are mainly localized in the nucleus and cytoplasm (56). Additionally, yeast Ppe1 mutant strains do not exhibit any growth defects (55), while mutations affecting synthesis or function of respiratory chain components generally result in growth impairment on a non-fermentable carbon source. Thus currently there is, besides the reported association with SSU of the mitoribosome in yeast, no clear indication that Ppe1 is implicated in mitochondrial translation. Therefore, additional research is needed to corroborate the cellular localization and function of yeast Ppe1 and its orthologs in other eukaryotes.
The evolution of the mitochondrial ribosomal proteome along eukaryotic lineages
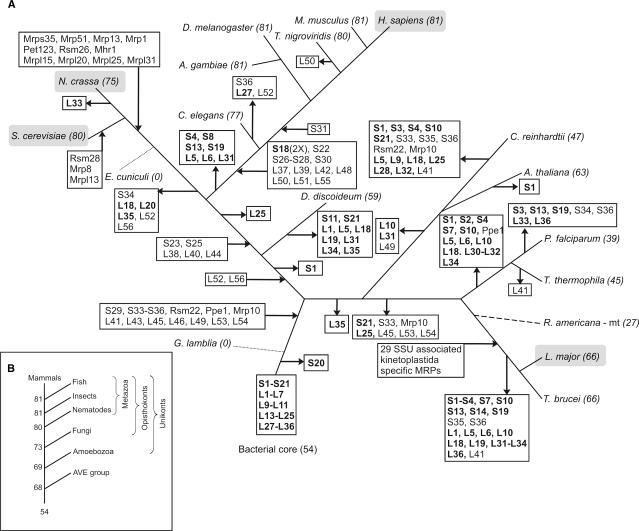
The protein composition of mitoribosomes is variable, not only in terms of the number of MRPs, but also in terms of the specific MRPs contained (see Tables 1 and 2). By mapping the distribution of the orthologous groups of MRPs onto a reconstructed tree of the eukaryotes (35) while always considering the most parsimonious scenario, we obtained an overview of the history of gains and losses of MRPs during the divergence of eukaryotes (see Figure 2). Mitoribosomes originated from the bacterial ribosome present in the alpha-proteobacterial ancestor of mitochondria, consisting of a microbial core of 54 ribosomal proteins in total. Already in the earliest stage of eukaryotic evolution, prior to the major divergence of the fungal, plant and metazoan lineages, several supernumerary proteins were recruited to the mitoribosome, while only one bacterial core protein was lost at this stage (RPS20, see Figure 2), resulting in a primitive eukaryotic mitoribosome of 68 MRPs. Subsequent to the gain of this primary set of 15 proteins, the mitoribosome diversified along the various eukaryotic lineages and a number of lineage-specific gain and loss events can be observed. Another gain of MRPs occurred at the level of the metazoa, prior to the bifurcation of nematodes and eumetazoa. At this stage, 14 proteins were recruited to the metazoan mitoribosome, whereas seven bacterial core proteins were lost. Major protein gains at these two time points have also been observed in the evolutionary history of Complex I (57). Additionally, 11 fungi-specific MRPs were recruited after the animal-fungi radiation. In the lineage towards the kinetoplastida, a major surge of 29 proteins is observed. These proteins have been shown to comprise a protein complex that is associated with the SSU of kinetoplastid-mitoribosomes (17). The role of this additional protein complex and its association with the SSU is still unclear. Apart from this major gain in the kinetoplastida, we observe loss of several proteins in the AVE (alveolates, viridiplantae and excavates) lineage, predominantly consisting of bacterial core proteins, but also including supernumerary MRPs in some lineages. Possibly, some of these proteins remained undetected due to sequence divergence. The fact that the AVE lineages are dominated by loss events of MRPs is likely due to the lack of experimental data of mitoribosomal content in these species.
Figure 2.
Reconstruction of the evolutionary history of the mitochondrial proteome across the different eukaryotic lineages. (A) Incoming and outgoing arrows indicate the gains and losses of the MRPs that are indicated in the box. Genes encoding proteins of the bacterial core are shown in bold. The scenario of gene gain and loss was adjusted according to the MRP content encoded by the mitochondrial genomes of rice and R. americana (5) [dashed line, position in tree inferred from (89)]. Dotted lines indicate lineages with highly reduced mitochondria in which all MRP-encoding genes have been lost. Species for which the mitoribosomal proteome has been experimentally investigated are highlighted in a gray shaded box. Numbers following the species names denote the total number of MRPs identified in that species. Note that several MRPs have been lost independently in different lineages. For example, MRPS1 is lost independently in C. reinhardtii, A. thaliana (MRPS1 is encoded on the mitochondrial genomes of Oryza sativa and R. americana) and in the kinetoplastida and alveolata lineages. (B) Evolutionary trajectory towards the human mitochondrial ribosomal proteome. Main gene gain events can be observed at major branching points of the tree, such as in the ancestral eukaryote and at the origin of the metazoa, after which the mitoribosomal proteome is maintained at a constant level. AVE: alveolates, viridiplantae and excavates branch.
Compared to the 54 ribosomal proteins present in the alpha-proteobacterial ancestor, 81 MRPs in human represent a substantial gain in complexity (see Figure 2B). However, the observed increase in complexity following the endosymbiotic event is not uncommon, as it has also been reported for electron transport chain complexes (57). It has been proposed that the recruited supernumerary proteins are mainly involved in the assembly and stabilization of the complexes (57,58). The mammalian supernumerary MRPs are shown to be mostly localized to the peripheral regions of the mitoribosome (59–61). Since all proteins synthesized by metazoan mitoribosomes are inserted into the inner mitochondrial membrane, at least some of the metazoa-specific supernumerary MRPs are presumed to be associated with positioning the mitoribosome during co-translational insertion of nascent polypeptides into the membrane. Interestingly, a protein that has been proposed to be essential for co-translational insertion in yeast, Mba1 (MRPL45, see ‘Origins from outside of the mitoribosomal proteome—MRP recruitment’), was already part of the primitive eukaryotic mitoribosome and has been recruited from the alpha-proteobacterial ancestor. Similar to the evolution of Complex I, we observe the recruitment of proteins of bacterial origin to the mitoribosome (MRPL45 and Rsm22), however, this number is considerably lower (only two, compared to six in Complex I), especially considering that in the evolution of the mitoribosome more proteins were added compared to Complex I (37 and 32 in the human mitoribosome and Complex I, respectively).
Tracing the origins of supernumerary MRPs
As outlined above, the protein content of the mitoribosome has significantly increased in most species during the evolution of the mitochondria. Our analysis indicates that in several stages of the mitoribosomal evolution various extra proteins, the supernumerary MRPs, have been added to the mitoribosomal proteome. Some of these supernumerary MRPs were found to be present in all eukaryotes that were investigated, but not in bacteria, such as MRPS29 (see Table 1). Clearly, these MRPs have been recruited in an early stage of the evolution of the eukaryotes, before the formation of major eukaryotic kingdoms. In contrast, other MRPs have originated relatively recently, such as MRPL37, which is specific for the metazoa. However, it is not only interesting to know ‘when’ certain MRPs were added to the mitoribosome, it is also enticing to trace their origins. We found that several of the supernumerary MRPs are homologous to other proteins, some of which are also part of the mitoribosome (see Table 3). The latter finding implies that gene duplication events have played a substantial role in the expansion of the mitoribosomal proteome (see Table 3). Below we will discuss some of the most protruding cases.
Table 3.
Paralogous relationships involving MRPs
| MRP | Mitochondrial [cytosolic] ribosome homolog | Other |
|---|---|---|
| SSU | ||
| S10 | L48, [S20] | |
| S18 | S18A, S18B, S18C | |
| S26 | Several coiled coil containing proteins, including subunit 10 theta of the eukaryotic translation initiation factor 3 | |
| S27 | TPR/PPR repeat proteins/salt-inducible proteins (Arabidopsis) | |
| S29 | DAP3 | |
| S30 | L37 | PDCD9 |
| Mrp1 | Rsm26 (yeast) | Superoxide dismutase |
| Pet123 | Several coiled coil containing proteins | |
| Mrp10 | COX19, NDUFA8 | |
| Rsm28 | Kinases | |
| LmjF30.3530 | Rhodanese, inactive domain | |
| LmjF25.2160 | Ubiquitin | |
| LmjF32.0650 | AKAP7 | |
| LSU | ||
| L2 | [L8] | NUZM (fungi-specific OXPHOS complex I subunit) |
| L38 | Phosphatidylethanolamine-binding protein 1 (PEBP-1) and Flowering Locus T | |
| L39 | Threonyl- and alanyl-tRNA synthetase | |
| L43 | S25 | Respiratory Complex I subunit B8 |
| L44 | Mrpl15, Mrpl3 (yeast) | Ribonuclease III and other ribonuclease_3 domain containing proteins |
| L45 | Tim44 | |
| L46 | mRNA decapping enzyme 2 (DCP2) and Nudix hydrolase (NUDT) | |
| L56 | Beta-lactamase | |
| Mrpl20 | Mesenchymal stem cell protein |
Names of MRPs are indicated in the first column and supernumerary MRPs and proteins from outside the ribosome are indicated in the second and third column, respectively. In addition, if a homologous cytosolic ribosomal protein is present, it is indicated between square brackets in the second column.
Origins from within the mitoribosomal proteome—MRP duplication
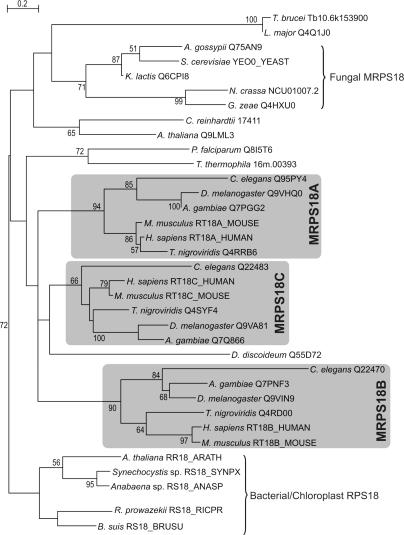
Some supernumerary MRPs are homologous to MRPs that are part of the bacterial core of the mitoribosome. These proteins have seemingly been duplicated from within the mitoribosomal proteome. One of the most straightforward examples is the emergence of three variants of MRPS18 in the metazoan lineage resulting from gene duplication events (see Figure 3). In contrast with earlier reports (9), we identified three MRPS18 variants in C. elegans and we found that all three metazoan variants emerged from the same ancestral metazoan sequence, which is unlikely to be derived from chloroplast S18, as was reported by Cavdar Koc and co-workers (9) (see Figure 3). All three C. elegans MRPS18 variants seem to be essential in embryonal development, as targeted disruption of these genes result in an embryonic lethal phenotype (62). Independent from the observed MRPS18 duplications in metazoa, it is interesting to note that most sequenced actinobacterial genomes encode two variants of the S18 gene, probably the result of a gene duplication event in these species. Currently, experimental evidence regarding the functions of the different MRPS18 versions in metazoa is lacking, but it is believed that each mitoribosome contains only one copy of MRPS18, giving rise to a heterogeneous population of mitoribosomes (9).
Figure 3.
Maximum-likelihood tree of the MRPS18 orthologous group. The metazoa contain three copies of this family (MRPS18A, MRPS18B and MRPS18C) as a result of two consecutive duplication events at the root of the metazoan radiation. Proteins are indicated by a species name and a protein identifier. Bootstrap values for partitions that are supported with values above 50% (out of 100 replicates) are displayed.
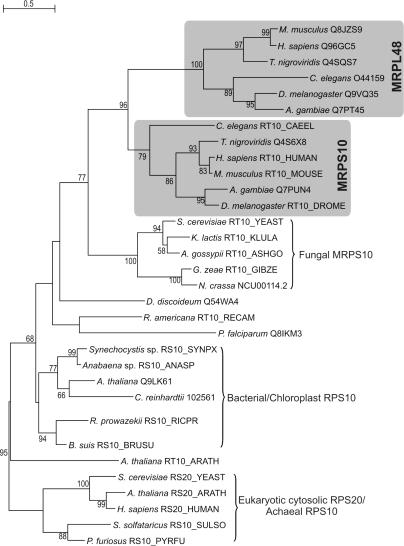
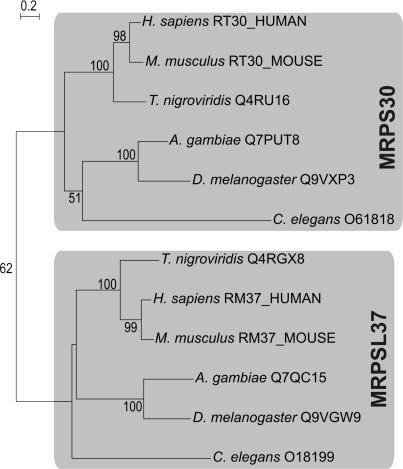
Another example can be found in the duplication of MRPS10 in the lineage of the metazoa, which gave rise to MRPL48 (see Table 3 and Figure 4). This duplication event constitutes one of the cases where the duplicate gene product has become part of the other mitoribosomal subunit, in this case from the SSU (MRPS10) to the LSU (MRPL48). Likewise, a duplication event at the base of the metazoan radiation seems to have given rise to the supernumerary MRPs MRPS30 and MRPL37, both of which are only present in metazoa (see Figure 5). Interestingly, MRPS30 is also known as programmed cell death protein 9 (PDCD9 or p52) (63,64). Another supernumerary protein, MRPS29, is also implicated in apoptosis, as it has been shown to be identical to death-associated protein 3 (DAP3) (63,65). Given the dual roles of these MRPs, the processes of mitochondrial translation and apoptosis are apparently closely coupled (66).
Figure 4.
Maximum-likelihood tree showing the origin of supernumerary MRPL48 as a result of a duplication event at the base of the metazoan radiation within the MRPS10 orthologous group. All depicted proteins were connected using PSI-BLAST (see Materials and Methods for details). For instance, a search started with human MRPS10 retrieved both the human MRPL48 (Q96GC5) and the cytosolic SSU ribosomal protein RPS20 (RS20_HUMAN) in the second iteration with E-values of 1e−9 and 0.001, respectively. Proteins are indicated by a species name and a protein identifier. Bootstrap values for partitions that are supported with values above 50% (out of 100 replicates) are displayed.
Figure 5.
Maximum-likelihood tree showing that metazoan-specific supernumerary MRPs MRPS30 and MRPL37 originated as a result of a duplication event at the base of the metazoan radiation. The depicted proteins were connected using PSI-BLAST (see Materials and Methods for details). For instance, a search started with human MRPS30 retrieves human MRPL37 within the first iteration with an E-value of 0.001. All proteins are indicated by a species name and a protein identifier. Bootstrap values for partitions that are supported with values above 50% (out of 100 replicates) are displayed.
Origins from outside of the mitoribosomal proteome—MRP recruitment
We found that some supernumerary MRPs have probably been recruited to the mitoribosome, as they display significant homology to other mitochondrial proteins, some of which are also implicated to play a role in mitochondrial translation (see Table 3). Of course, it cannot always be resolved if an MRP has been part of the mitoribosome prior to the duplication event. For some instances, we can use the phylogenetic distribution patterns of the copies in order to point out which was first. One of such examples, where a pre-existing protein has been recruited to the mammalian mitoribosome, is the acquisition of MRPL39, which is homologous to the N-terminal domain of threonyl-tRNA synthetases and which has a universal phylogenetic distribution. This homology has been noticed before and it is suggested that MRPL39 can bind tRNA in a similar manner as the E. coli threonyl-tRNA synthetase (67). Conceivably, the N-terminal tRNA-binding domain of a mitochondrial threonyl-tRNA synthetase was recruited to the mitoribosome in order to compensate for the loss of bacterial ribosomal proteins that are involved in tRNA binding, such as RPS13 and RPL5 (see Tables 1 and 2).
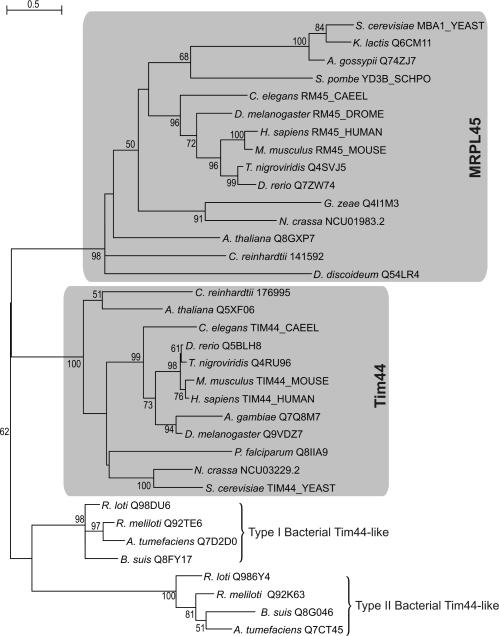
The most striking example of MRP recruitment is that of MRPL45 (see Figure 6). First we found that MRPL45 and yeast protein Mba1 belong to the same orthologous group, which in addition comprises proteins from all metazoa, the plant A. thaliana, the alga C. reinhardtii as well as from the mycetozoan D. discoideum. Mba1 is a mitochondrial protein associated with the matrix face of the inner membrane, which associates with the highly conserved inner membrane protein Oxa1. Mba1 and Oxa1 orchestrate the insertion of both mitochondrial- and nuclear-encoded proteins from the mitochondrial matrix into the inner membrane (68,69). Moreover, we detected homology between MRPL45 (including Mba1) and Tim44 (see Figure 6), a peripheral mitochondrial inner membrane protein that is an essential component of the import motor PAM of the TIM23 complex, which is implicated in the translocation of proteins from the inter membrane space to the mitochondrial matrix [for reviews see (70,71)]. The apparent sequence homology, as well as functional homology (i.e. protein translocation), suggests that MRPL45 and Tim44 share a common alpha-proteobacterial origin. Many alpha-proteobacteria contain two distinct types of Tim44/MRPL45-like genes (see Figure 6).
Figure 6.
Maximum-likelihood tree indicating common ancestry of supernumerary mitoribosomal protein MRPL45 and Tim44, an essential component of the TIM23/PAM complex, which mediates the translocation of nuclear-encoded proteins across the mitochondrial inner membrane. The tree contains alpha-proteobacterial sequences from which mitochondrial MRPL45 and Tim44 have evolved. Note that there are two paralogous Tim44-like families within the alpha-proteobacteria, resulting from a duplication within that lineage. The depicted proteins were connected using PSI-BLAST as outlined in the Materials and Methods. For instance, a search started with human MRPL45 retrieves the Rhizobium meliloti Type I Tim44-like protein (Q92TE6) in the second iteration with an E-value of 1e−7. The yeast Mba1 and human Tim44 proteins were both retrieved in the third iteration (with E-values of 2e−13 and 2e−10, respectively). Proteins are indicated by a species name and a protein identifier. Bootstrap values for partitions that are supported with values above 50% (out of 100 replicates) are displayed.
A close examination of MRPL45 and Tim44 proteins reveals that the N-terminal region of Tim44 is absent in MRPL45 (not shown). This region is supposed to be located in the mitochondrial matrix where it interacts with mitochondrial Hsp70 chaperone, which is also a component of the PAM complex (72). Apart from its association with Oxa1, MRPL45 has been shown to be a constituent of the mitoribosomal LSU in multiple studies (10,73). Moreover, it has been shown that in yeast, transcription of MBA1 is tightly co-regulated with that of genes encoding MRPs (74), implying a functional link between the respective gene products. The anticipated association of the mitoribosome and membrane-bound constituents of the mitochondrial protein insertion machinery would explain the fact that up to 50% of the bovine mitoribosomes are found to associate with the inner membrane fraction of mitochondria (75). Taking all this in consideration, we postulate that MRPL45 functions as a bridging factor between the mitoribosome and Oxa1, and as such constitutes an essential component for successful co-translational insertion of proteins into the inner membrane of mitochondria in eukaryotes. Interestingly, the MRPL45 orthologous group, which also includes the bacterial Tim44-like proteins, shows a similar phylogenetic distribution pattern as the orthologous group of Cox11, an assembly factor of cytochrome oxidase, which is involved in the co-translational insertion of copper ions into the nascent Cox1 protein (47). The observed pattern of co-presence and absence of these genes across a phylogenetically diverse set of species indicates that their gene products might be involved in the same process (76). As Cox11 has been shown to associate transiently with the mitoribosome as well, it is feasible that MRPL45 and Cox11 act together in order to functionally insert Cox1 into the inner mitochondrial membrane (47).
Other examples of MRPs that have been recruited to the mitoribosome are the supernumerary proteins MRPL43 and MRPS25 (see Table 3), which are homologous to each other as well as to the B8 subunit (NI8M) of respiratory Complex I (NADH-ubiquinone oxidoreductase) of the OXPHOS system (57,77). Presumably, MRPL43 and the B8 subunit of Complex I originated from a single gene that was present in the primitive eukaryote, as they are present in a phylogenetically wide range of organisms. Most likely, another duplication of MRPL43 then gave rise to MRPS25, which is only found in metazoa and fungi. More examples of MRP recruitment include yeast MRPs Mrp1 and Rsm26, which are both part of the iron/manganese superoxide dismutase family (44), and MRPL44, which is homologous to a wide range of double-stranded RNA-binding proteins (see Table 3).
Detection of orthologs of MRPs previously reported to be fungi specific
Several comprehensive experimental studies on the yeast mitochondrial translation machinery have significantly extended its mitoribosomal proteome (14–16). However, for a large number of MRPs that have been characterized in yeast thus far, no significant or conclusive sequence similarity has been reported to ribosomal proteins from other sources. In the current study, we were able to link seven of these seemingly fungi-specific MRPs to known ribosomal proteins in other species, as will be outlined below.
Apart from the homology between yeast Mba1 and MRPL45, which has already been discussed above, we identified Mrp49 as the ortholog of MRPS25. It is, however, surprising that the orthologs of MRPS25 found in S. cerevisiae as well as in N. crassa are reported to be located in the large instead of the small subunit of the mitoribosome (18,78), in contrast with the location of bovine MRPS25 (59). It is possible that the subunit localization of the MRPS25 orthologs is different between metazoa and fungi; however, further experimental evidence is needed in order to clarify this.
We also connected Rsm27, a yeast protein of the small subunit (44), to the MRPS33 protein family. In addition, we detected MRPS33 orthologs in all metazoa as well as D. discoideum and A. thaliana. Moreover, we found that yeast MRPs Mrpl28 (53), Mrpl44 (79), Mrpl50 (53) and Mrpl40 (53) are in fact the orthologs of MRPL40, MRPL53, MRPL9 and MRPL24, respectively. Both MRPL24 and Mrpl40 were found to contain a KOW motif (PFAM domain pfam00467), which is present in a variety of ribosomal proteins as well as in the bacterial transcription elongation factor NusG (80). The homology between Mrpl50 and bacterial RPL9 has been noted before (14), but it was dismissed as being insignificant, since the sequence similarity was not conserved throughout the full length of Mrpl50. In our analysis, however, we did detect significant homology between Mrpl50 and the MRPL9 family (including bacterial RPL9). Our finding is supported by the fact that Mrpl50 and MRPL9 representatives reside within the same protein superfamily (SSF55658, ‘L9 N-domain-like’) (81).
Finally, we were able to confirm the distant homology between Var1 and Rps5, two mtDNA-encoded homologs of bacterial RPS3 in fungi, as was previously reported by Lang and colleagues (82). Var1 and Rps5, which are the only mtDNA-encoded MRPs in yeast and N. crassa, respectively, have both been identified as essential components of the SSU of the mitoribosome, showing similar phenotypes in mutant strains (82). Despite the fact that RPS3 has been shown to play an essential role in bacterial ribosome assembly, and that Var1 and Rps5 are both essential for assembly of the mitochondrial SSU in fungi, we were unable to detect candidate mitochondrial RPS3 homologs in the metazoa. In the latter organisms, this essential role in ribosome assembly could be performed by MRPS24, which displays distant homology to RPS3 (see Figure 1). Moreover, we could detect mtDNA-encoded RPS3 homologs in T. thermophila and D. discoideum (82,83), the latter of which constitutes a peculiar case where the RPS3 encoding gene is split and both the 5′ and 3′ parts are fused to an ORF of unknown function (ORF425 and ORF1740, respectively). In any case, the fact that genes encoding RPS3 homologs are present on several mitochondrial genomes implies an alpha-proteobacterial origin of this protein.
Adaptive evolution of core MRPs
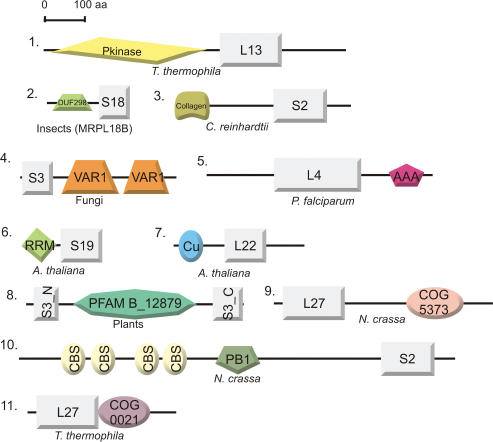
Compared to bacterial ribosomes, mammalian mitoribosomes contain scarcely half as much rRNA and over twice as much protein, due to the presence of enlarged bacterial core proteins and supernumerary proteins (6). Previously, it has been hypothesized that these proteins might compensate for the loss of rRNA segments in the mitoribosome (6,84). Recently however, it was shown that many supernumerary MRPs occupy new quaternary positions in both the large and small subunits of the mitoribosome (61), inconsistent with this hypothesis. We compared the sequences of all core MRPs with their bacterial counterparts in order to explore the possibility that core MRPs contain additional domains that perform extra, mitochondria-specific functions. Indeed, we observe that MRPs are significantly larger (on average almost twice as large) than their bacterial ancestral sequences (see Supplementary Data, Figure S8). In some cases, we could identify functional domains within these newly evolved regions, which we expect to be linked to ribosome assembly or function (see Figure 7). For instance, we identified a RNA recognition motif (RRM_1, pfam00076) and a copper-binding domain (Cu_bind_like, pfam02298) in the N-terminal regions of A. thaliana MRPS19 and MRPL22, respectively (see Figure 7). The RRM motif that is present in MRPS19 (85) is found in a wide variety of RNA-binding proteins, which are implicated in a wide variety of cellular processes. Here, the motif is most likely involved in increasing the binding affinity to and/or stabilization of the tertiary structure of rRNA. The role of the copper-binding domain in MRPL22 is less clear. It might be involved in the co-translational insertion of copper ions into proteins that require copper for activity, in a similar way as has been observed for the cytochrome c oxidase assembly factor in yeast, Cox11 (47) (also see above). The functions of other functional domains that are fused to MRPs are still obscure, such as a bacterial-type membrane protein (COG5373) to MRPL27 in N. crassa (see Figure 7).
Figure 7.
Domain architectures of MRPs to which additional functional domains have been added. Protein identifiers are depicted as in Table 1 or 2 and species distribution is indicated. Proteins are drawn approximately according to scale. Domain abbreviations: (1) Pkinase (pfam00069): Protein kinase catalytic domain. (2) DUF298 (pfam03556): Domain of unknown function, containing a basic helix-loop-helix leucine zipper motif. (3) Collagen (pfam01391): Collagen triple helix repeat. (4) VAR1 (pfam05316): This domain is specific for some fungal mitochondrial ribosomal S3 proteins and is often present in two copies (also see text). (5) AAA (pfam00004): ATPase family associated with various cellular activities. Proteins containing AAA domains often perform chaperone-like functions that assist in the assembly, operation or disassembly of protein complexes. (6) RRM (pfam00076): RNA-binding domain, found in a variety of RNA-binding proteins (also see text). (7) Cu (pfam02298): Copper-binding domain found in various proteins (also see text). (8) In plants, the N-terminus (S3_N, pfam00417) and C-terminus (S3_C, pfam00189) of MRPS3 are separated as a result of an insertion of a protein domain of unknown function (pfam B domain PB012879). (9) COG5373: Predicted membrane protein present in prokaryotes. (10) CBS (pfam00571) and PB1 (pfam00564) domains: CBS domains are small intracellular modules, which have a regulatory role in making proteins sensitive to adenosyl carrying ligands. PB1 is present in many eukaryotic cytoplasmic signaling proteins. (11) COG0021: Transketolase (1-deoxyxylulose-5-phosphate synthase).
DISCUSSION
In the present study, we have mapped the evolution of the mitochondrial ribosomal proteome using a comparative genomics approach that combined the most recent experimental data and computational techniques. The results of our study hint at a complex evolutionary scenario in which an ancestral ribosome of alpha-proteobacterial descent doubled its amount of protein in most eukaryotic lineages by elongation of its proteins and by recruiting new proteins of diverse origins. We observe that the mitoribosome has a complex history in terms of MRP gene gain and loss events during evolution. A major protein gain of the mitoribosome in eukaryotic evolution occurred prior to the divergence of the main eukaryotic domains, resulting in a primitive eukaryotic mitoribosome of 68 MRPs. Subsequently, the mitoribosome diversified along the different eukaryotic lineages with a continuous recruitment of new proteins, including a large gain after the bifurcation of the animal-fungi clade in each lineage. Another notable surge has taken place in the kinetoplastida lineage, where 29 new proteins were found to form a new, distinct protein complex with unknown function, which associates with the SSU of the Leishmania mitoribosomes (17).
In addition to the gain of MRPs, we also observe that numerous MRPs have been lost during the course of evolution, some of which have occurred independently in different lineages, as guided from ribosomal protein content that is encoded by the ‘ancestral’ mitochondrial genome of R. americana (5). The extensive loss of genes encoding core MRPs in some lineages contrasts with the evolutionary scenario that is observed for another mitochondrial protein complex with an alpha-proteobacterial ancestry, respiratory Complex I (NADH:Ubiquinone oxidoreductase). During the evolution of this protein complex, which has approximately tripled its size in most eukaryotes, hardly any gene loss was observed, except for these lineages in which the complete respiratory complex was lost (57). Moreover, the bacterial core of Complex I has been restricted from gene loss events in all eukaryotic genomes analyzed thus far, which contrasts with the scenario of the mitoribosome. What is the underlying reason for the observed differences in the evolutionary trajectories? Evidently, some of the core ribosomal proteins are not as essential as the core subunits of Complex I for proper functioning of the protein complex, and are therefore expendable. This is the case for RPS20 and RPL25, which are absent from most mitoribosomes, for example. RPS20-deficient E. coli strains were found to be viable, but showed an increased misreading ability of nonsense codons (86); RPL25-defective and wild-type E. coli ribosomes were found to translate at the same rate in vitro, albeit that the mutant type was less efficient (87). In contrast, some MRPs that are missing in most eukaryotic species, such as MRPS1 and MRPL5, are indispensable for translation in E. coli and result in lethal phenotypes when mutated (87,88). Moreover, MRPL5-deficient yeast strains display a respiratory-deficient phenotype, probably caused by a defect in the mitochondrial translation machinery (15). Apparently, the evolution of the mitoribosomal proteome involved several drastic events that altered the absolute necessity of some ribosomal proteins.
In addition, the present study was performed in order to prioritize MRPs for their potential involvement in mitochondrial respiratory diseases. Based on the fact that essential genes appear more evolutionary conserved than non-essential genes, the bacterial core ribosomal proteins and translation factors that are conserved in most eukaryotes constitute excellent screening targets for human mitochondrial disorders. Mutations in non-essential MRPs could be risk factors for these diseases. Thus far, mutations in patients with mitochondrial respiratory disorders have been reported for the gene encoding small ribosomal subunit protein MRPS16 (28). But what about the supernumerary MRPs? In general, they are phylogenetically less conserved than the bacterial core proteins; however, their recruitment bears functional relevance, as is indicated by knock-out studies in yeast and C. elegans. As such, the two novel human MRPs, orthologs of yeast Rsm22 and Mrp10, are good candidates for unresolved mitochondrial disorders, since they show a wide phylogenetic distribution, have not been investigated before and yeast mutants show clear respiratory-deficient phenotypes. Translation-related mitochondrial disorders are not solely caused by mutations in mitoribosomal genes, but also by mutations in genes encoding factors that functionally interact with the mitoribosome, such as mitochondrial translation elongation factors EFG1 (25) and EFTs (26). In addition to the newly discovered human MRPs, these interactors constitute excellent screening targets for human patients with unresolved mitochondrial OXPHOS system disorders.
Altogether, our study reveals that the mitoribosomal proteome has, after initially having been acquired via the alpha-proteobacterial endosymbiont, been subjected to a complex evolution that involved numerous gain and loss events, resulting in a large variety in mitoribosomal protein content between different eukaryotic lineages. Potentially, the increased amount of proteins that seem to be associated with all modern eukaryotic mitoribosomes might reflect specific adaptations to mitochondria-specific functions, or, it might even reflect the increased complexity of eukaryotes themselves. Finally, we expect that additional experimental studies of mitoribosomal proteomes, such as the one that was recently performed on Leishmania mitoribosomes, will reveal an even more dramatic picture of the evolutionary trajectory of this protein complex. It will be a major challenge to gain more insight in the functional roles of the newly acquired MRPs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by a Rubicon grant from The Netherlands Organization for scientific research (NWO) and by the European Union's 6th Framework Program, contract number LSHM-CT-2004-503116 (EUMITOCOMBAT) and contract number LSHM-CT-2004-005260 (MITOCIRCLE). We thank Siv Andersson (Evolutionary Biology Center, Uppsala University, Sweden) for reading the manuscript and for helpful suggestions prior to submission. Funding to pay the Open Access publication charges for this article was provided by the European Union's 6th framework program EUMITOCOMBAT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Margulis L. The origin of the eukaryotic cell. New Haven: Yale University Press; 1970. [Google Scholar]
- 3.Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 4.Pel HJ, Grivell LA. Protein synthesis in mitochondria. Mol. Biol. Rep. 1994;19:183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- 5.Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003;55:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- 7.Chacinska A, Boguta M. Coupling of mitochondrial translation with the formation of respiratory complexes in yeast mitochondria. Acta Biochim. Pol. 2000;47:973–991. [PubMed] [Google Scholar]
- 8.Leaver CJ, Harmey MA. Isolation and characterization of mitochondrial ribosomes from higher plants. Biochem. J. 1972;129:37P–38P. doi: 10.1042/bj1290037p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 10.Cavdar Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien TW, Fiesler SE, Denslow ND, Thiede B, Wittmann-Liebold B, Mougey EB, Sylvester JE, Graack HR. Mammalian mitochondrial ribosomal proteins (2). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 1999;274:36043–36051. doi: 10.1074/jbc.274.51.36043. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien TW, Liu J, Sylvester JE, Mougey EB, Fischel-Ghodsian N, Thiede B, Wittmann-Liebold B, Graack HR. Mammalian mitochondrial ribosomal proteins (4). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 2000;275:18153–18159. doi: 10.1074/jbc.M909762199. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem. 2001;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 14.Gan X, Kitakawa M, Yoshino K, Oshiro N, Yonezawa K, Isono K. Tag-mediated isolation of yeast mitochondrial ribosome and mass spectrometric identification of its new components. Eur. J. Biochem. 2002;269:5203–5214. doi: 10.1046/j.1432-1033.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 15.Graack HR, Wittmann-Liebold B. Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem. J. 1998;329(Pt 3):433–448. doi: 10.1042/bj3290433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towpik J. Regulation of mitochondrial translation in yeast. Cell. Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- 17.Maslov DA, Spremulli LL, Sharma MR, Bhargava K, Grasso D, Falick AM, Agrawal RK, Parker CE, Simpson L. Proteomics and electron microscopic characterization of the unusual mitochondrial ribosome-related 45S complex in Leishmania tarentolae. Mol. Biochem. Parasitol. 2007;152:203–212. doi: 10.1016/j.molbiopara.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan X, Arita K, Isono S, Kitakawa M, Yoshino K, Yonezawa K, Kato A, Inoue H, Isono K. Identification and comparative analysis of the large subunit mitochondrial ribosomal proteins of Neurospora crassa. FEMS Microbiol. Lett. 2006;254:157–164. doi: 10.1111/j.1574-6968.2005.00024.x. [DOI] [PubMed] [Google Scholar]
- 19.Bonen L, Calixte S. Comparative analysis of bacterial-origin genes for plant mitochondrial ribosomal proteins. Mol. Biol. Evol. 2006;23:701–712. doi: 10.1093/molbev/msj080. [DOI] [PubMed] [Google Scholar]
- 20.Maslov DA, Sharma MR, Butler E, Falick AM, Gingery M, Agrawal RK, Spremulli LL, Simpson L. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol. Biochem. Parasitol. 2006;148:69–78. doi: 10.1016/j.molbiopara.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 21.Smeitink J, van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 22.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoubridge EA. Nuclear genetic defects of oxidative phosphorylation. Hum. Mol. Genet. 2001;10:2277–2284. doi: 10.1093/hmg/10.20.2277. [DOI] [PubMed] [Google Scholar]
- 24.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 25.Coenen MJ, Antonicka H, Ugalde C, Sasarman F, Rossi R, Heister JG, Newbold RF, Trijbels FJ, van den Heuvel LP, et al. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N. Engl. J. Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- 26.Smeitink JA, Elpeleg O, Antonicka H, Diepstra H, Saada A, Smits P, Sasarman F, Vriend G, Jacob-Hirsch J, et al. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am. J. Hum. Genet. 2006;79:869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valente L, Tiranti V, Marsano RM, Malfatti E, Fernandez-Vizarra E, Donnini C, Mereghetti P, De Gioia L, Burlina A, et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am. J. Hum. Genet. 2007;80:44–58. doi: 10.1086/510559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller C, Saada A, Shaul N, Shabtai N, Ben-Shalom E, Shaag A, Hershkovitz E, Elpeleg O. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 29.Pruess M, Kersey P, Apweiler R. The Integr8 project – a resource for genomic and proteomic data. In Silico Biol. 2005;5:179–185. [PubMed] [Google Scholar]
- 30.O'Brien EA, Zhang Y, Yang L, Wang E, Marie V, Lang BF, Burger G. GOBASE – a database of organelle and bacterial genome information. Nucleic Acids Res. 2006;34:D697–D699. doi: 10.1093/nar/gkj098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST–a tool for discovery in protein databases. Trends Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 32.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy SR, Mitchison G, Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J. Comput. Biol. 1995;2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. PHYLIP (PHYLogeny Inference Package) version 3.6, Distributed by the author. Seattle: Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- 35.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 36.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 38.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A, Anderson JB, Derbyshire MK, Deweese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams KL, Daley DO, Whelan J, Palmer JD. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 2002;14:931–943. doi: 10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Muller M, Le Guyader H. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc. Biol. Sci. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saveanu C, Fromont-Racine M, Harington A, Ricard F, Namane A, Jacquier A. Identification of 12 new yeast mitochondrial ribosomal proteins including 6 that have no prokaryotic homologues. J. Biol. Chem. 2001;276:15861–15867. doi: 10.1074/jbc.M010864200. [DOI] [PubMed] [Google Scholar]
- 45.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 46.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7 – recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khalimonchuk O, Ostermann K, Rodel G. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr. Genet. 2005;47:223–233. doi: 10.1007/s00294-005-0569-1. [DOI] [PubMed] [Google Scholar]
- 48.Jin C, Myers AM, Tzagoloff A. Cloning and characterization of MRP10, a yeast gene coding for a mitochondrial ribosomal protein. Curr. Genet. 1997;31:228–234. doi: 10.1007/s002940050199. [DOI] [PubMed] [Google Scholar]
- 49.Grohmann L, Graack HR, Kruft V, Choli T, Goldschmidt-Reisin S, Kitakawa M. Extended N-terminal sequencing of proteins of the large ribosomal subunit from yeast mitochondria. FEBS Lett. 1991;284:51–56. doi: 10.1016/0014-5793(91)80759-v. [DOI] [PubMed] [Google Scholar]
- 50.Triepels R, van den Heuvel L, Loeffen J, Smeets R, Trijbels F, Smeitink J. The nuclear-encoded human NADH:ubiquinone oxidoreductase NDUFA8 subunit: cDNA cloning, chromosomal localization, tissue distribution, and mutation detection in complex-I-deficient patients. Hum. Genet. 1998;103:557–563. doi: 10.1007/s004390050869. [DOI] [PubMed] [Google Scholar]
- 51.Nobrega MP, Bandeira SC, Beers J, Tzagoloff A. Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 2002;277:40206–40211. doi: 10.1074/jbc.M207348200. [DOI] [PubMed] [Google Scholar]
- 52.Sacconi S, Trevisson E, Pistollato F, Baldoin MC, Rezzonico R, Bourget I, Desnuelle C, Tenconi R, Basso G, et al. hCOX18 and hCOX19: two human genes involved in cytochrome c oxidase assembly. Biochem. Biophys. Res. Commun. 2005;337:832–839. doi: 10.1016/j.bbrc.2005.09.127. [DOI] [PubMed] [Google Scholar]
- 53.Kitakawa M, Graack HR, Grohmann L, Goldschmidt-Reisin S, Herfurth E, Wittmann-Liebold B, Nishimura T, Isono K. Identification and characterization of the genes for mitochondrial ribosomal proteins of Saccharomyces cerevisiae. Eur. J. Biochem. 1997;245:449–456. doi: 10.1111/j.1432-1033.1997.t01-2-00449.x. [DOI] [PubMed] [Google Scholar]
- 54.Ogris E, Du X, Nelson KC, Mak EK, Yu XX, Lane WS, Pallas DC. A protein phosphatase methylesterase (PME-1) is one of several novel proteins stably associating with two inactive mutants of protein phosphatase 2A. J. Biol. Chem. 1999;274:14382–14391. doi: 10.1074/jbc.274.20.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J, Tolstykh T, Lee J, Boyd K, Stock JB, Broach JR. Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J. 2000;19:5672–5681. doi: 10.1093/emboj/19.21.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turowski P, Fernandez A, Favre B, Lamb NJ, Hemmings BA. Differential methylation and altered conformation of cytoplasmic and nuclear forms of protein phosphatase 2A during cell cycle progression. J. Cell. Biol. 1995;129:397–410. doi: 10.1083/jcb.129.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabaldon T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) J. Mol. Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 58.Berry S. Endosymbiosis and the design of eukaryotic electron transport. Biochim. Biophys. Acta. 2003;1606:57–72. doi: 10.1016/s0005-2728(03)00084-7. [DOI] [PubMed] [Google Scholar]
- 59.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Koc H, Spremulli LL. A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J. Biol. Chem. 2000;275:32585–32591. doi: 10.1074/jbc.M003596200. [DOI] [PubMed] [Google Scholar]
- 60.Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, Agrawal RK, Harvey SC. A structural model for the large subunit of the mammalian mitochondrial ribosome. J. Mol. Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 62.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 63.Cavdar Koc E, Ranasinghe A, Burkhart W, Blackburn K, Koc H, Moseley A, Spremulli LL. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 64.Sun L, Liu Y, Fremont M, Schwarz S, Siegmann M, Matthies R, Jost JP. A novel 52 kDa protein induces apoptosis and concurrently activates c-Jun N-terminal kinase 1 (JNK1) in mouse C3H10T1/2 fibroblasts. Gene. 1998;208:157–166. doi: 10.1016/s0378-1119(97)00626-4. [DOI] [PubMed] [Google Scholar]
- 65.Kissil JL, Cohen O, Raveh T, Kimchi A. Structure-function analysis of an evolutionary conserved protein, DAP3, which mediates TNF-alpha- and Fas-induced cell death. EMBO J. 1999;18:353–362. doi: 10.1093/emboj/18.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Brien TW, O’Brien BJ, Norman RA. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–151. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 67.Spirina O, Bykhovskaya Y, Kajava AV, O'Brien TW, Nierlich DP, Mougey EB, Sylvester JE, Graack HR, Wittmann-Liebold B, et al. Heart-specific splice-variant of a human mitochondrial ribosomal protein (mRNA processing; tissue specific splicing) Gene. 2000;261:229–234. doi: 10.1016/s0378-1119(00)00504-7. [DOI] [PubMed] [Google Scholar]
- 68.Herrmann JM, Neupert W. Protein insertion into the inner membrane of mitochondria. IUBMB Life. 2003;55:219–225. doi: 10.1080/1521654031000123349. [DOI] [PubMed] [Google Scholar]
- 69.Preuss M, Leonhard K, Hell K, Stuart RA, Neupert W, Herrmann JM. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell. Biol. 2001;153:1085–1096. doi: 10.1083/jcb.153.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 71.Wiedemann N, Frazier AE, Pfanner N. The protein import machinery of mitochondria. J. Biol. Chem. 2004;279:14473–14476. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 72.Pavlov PF, Glaser E. Probing the membrane topology of a subunit of the mitochondrial protein translocase, Tim44, with biotin maleimide. Biochem. Biophys. Res. Commun. 2002;293:321–326. doi: 10.1016/S0006-291X(02)00221-8. [DOI] [PubMed] [Google Scholar]
- 73.Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 75.Liu M, Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J. Biol. Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- 76.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 77.Ton C, Hwang DM, Dempsey AA, Liew CC. Identification and primary structure of five human NADH-ubiquinone oxidoreductase subunits. Biochem. Biophys. Res. Commun. 1997;241:589–594. doi: 10.1006/bbrc.1997.7707. [DOI] [PubMed] [Google Scholar]
- 78.Fearon K, Mason TL. Structure and function of MRP20 and MRP49, the nuclear genes for two proteins of the 54 S subunit of the yeast mitochondrial ribosome. J. Biol. Chem. 1992;267:5162–5170. [PubMed] [Google Scholar]
- 79.Matsushita Y, Kitakawa M, Isono K. Cloning and analysis of the nuclear genes for two mitochondrial ribosomal proteins in yeast. Mol. Gen. Genet. 1989;219:119–124. doi: 10.1007/BF00261166. [DOI] [PubMed] [Google Scholar]
- 80.Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- 81.Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 2004;32:D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bullerwell CE, Burger G, Lang BF. A novel motif for identifying rps3 homologs in fungal mitochondrial genomes. Trends Biochem. Sci. 2000;25:363–365. doi: 10.1016/s0968-0004(00)01612-1. [DOI] [PubMed] [Google Scholar]
- 83.Iwamoto M, Pi M, Kurihara M, Morio T, Tanaka Y. A ribosomal protein gene cluster is encoded in the mitochondrial DNA of Dictyostelium discoideum: UGA termination codons and similarity of gene order to Acanthamoeba castellanii. Curr. Genet. 1998;33:304–310. doi: 10.1007/s002940050341. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 2001;276:21724–21736. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 85.Sanchez H, Fester T, Kloska S, Schroder W, Schuster W. Transfer of rps19 to the nucleus involves the gain of an RNP-binding motif which may functionally replace RPS13 in Arabidopsis mitochondria. EMBO J. 1996;15:2138–2149. [PMC free article] [PubMed] [Google Scholar]
- 86.Ryden-Aulin M, Shaoping Z, Kylsten P, Isaksson LA. Ribosome activity and modification of 16S RNA are influenced by deletion of ribosomal protein S20. Mol. Microbiol. 1993;7:983–992. doi: 10.1111/j.1365-2958.1993.tb01190.x. [DOI] [PubMed] [Google Scholar]
- 87.Korepanov AP, Gongadze GM, Garber MB, Court DL, Bubunenko MG. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J. Mol. Biol. 2007;366:1199–1208. doi: 10.1016/j.jmb.2006.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sorensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 89.Simpson AG, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol. Biol. Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.