Abstract
BsrDI and BtsI restriction endonucleases recognize and cleave double-strand DNA at the sequences GCAATG (2/0) and GCAGTG (2/0), respectively. We have purified and partially characterized these two enzymes, and analyzed the genes that encode them. BsrDI and BtsI are unusual in two respects: each cleaves DNA as a heterodimer of one large subunit (B subunit) and one small subunit (A subunit); and, in the absence of their small subunits, the large subunits behave as sequence-specific DNA nicking enzymes and only nick the bottom strand of the sequences at these respective positions: GCAATG (−/0) and GCAGTG (−/0). We refer to the single subunit, the bottom-strand nicking forms as ‘hemidimers’. Amino acid sequence comparisons reveal that BsrDI and BtsI belong to a family of restriction enzymes that possess two catalytic sites: a canonical PD-Xn-EXK and a second non-canonical PD-Xn-E-X12-QR. Interestingly, the other family members, which include BsrI (ACTGG 1/−1) and BsmI/Mva1269I (GAATGC 1/−1) are single polypeptide chains, i.e. monomers, rather than heterodimers. In BsrDI and BtsI, the two catalytic sites are found in two separate subunits. Site-directed mutagenesis confirmed that the canonical catalytic site located at the N-terminus of the large subunit is responsible for the bottom-strand cleavage, whereas the non-canonical catalytic site located in the small subunit is responsible for hydrolysis of the top strand. Top-strand specific nicking variants, Nt.BsrDI and Nt.BtsI, were successfully engineered by combining the catalytic-deficient B subunit with wild-type A subunit.
INTRODUCTION
Many restriction endonucleases (REases) bind to their target sequences and hydrolyze both strands of the duplex DNA simultaneously, cleaving the molecule in a single catalytic cycle. In general, hydrolysis reactions for the two strands proceed simultaneously, thus requiring the presence of two catalytic sites. Evolutionarily, the simplest way for a protein with one catalytic site to acquire the second one is to dimerize with one another—as a homodimer—and this is the strategy that many REases appear to have adopted. As a result, and in spite of their considerable diversity, REases usually accomplish double-stranded (ds) cleavage of DNA using two identical catalytic sites. Not all do so, however. Some enzymes use a single catalytic site to cleave the two strands of DNA sequentially in a single binding event (e.g. BfiI) or in consecutive binding events (e.g. MspI and HinP1I) (1–3). Here we describe a family of unusual REases from the bacterial genus Bacillus that accomplish ds cleavage using two different catalytic sites. Some members of this family are heterodimers with the two catalytic sites located in separate polypeptide chains. Other members are monomeric with the two catalytic sites located in a single polypeptide chain.
One common mode of homodimerization is exhibited by the familiar ‘Type IIP’ REases such as EcoRI (GAATTC −5/−1) and BglI (GCCNNNNNGGC −4/−7) that recognize palindromic sequences. These enzymes bind to ds DNA as homodimers and recognize their target sequences by a concerted process in which both subunits participate in equal and opposite measure: one subunit recognizes certain features of the sequence in one orientation, while the other subunit recognizes the same features but in the other orientation. In consequence, the overall recognition sequence is symmetric, and so are the cleavage positions within the sequence. Heterodimeric REases have been discovered in the forms of Bpu10I (CCTNAGC −5/−2), BbvCI (CCTCAGC −5/−2) and BspD6I (GAGTC 4/?), but they are relatively rare. Bpu10I and BbvCI each comprise two similar but non-identical subunits. Both subunits contain a single catalytic site, and both are required for ds cleavage activity. Mutating either catalytic site produces ‘half-active’ heterodimers that cleave one strand of the recognition sequence only; that is, ‘nick’ the DNA instead of cleaving it (Janulaitis,A. et al., 2005, US patent 6867028) (4). Another example of a heterodimeric REase is BseYI (CCCAGC −5/−1) consisting of two non-identical subunits that can be expressed separately in Escherichia coli. BseYI restriction activity can be reconstituted by mixing the two subunits (Nkenfou,C., Morgan,R. and S.-Y.X., unpublished data).
An alternative mode of homodimerization is exemplified by FokI (GGATG 9/13), the archetypal Type IIS REase. FokI binds to DNA as a monomer but cleaves cooperatively with another monomer (5–8) because its recognition sequence is asymmetric, it binds in one orientation only and cleaves asymmetrically, to one particular side of the recognition sequence. FokI comprises an N-terminal DNA-binding domain and a discrete C-terminal catalytic domain with a single catalytic site. Cleavage is accomplished by transient homodimerization between the catalytic domains of neighboring DNA-bound monomers (6). At low enzyme concentrations, solitary DNA-bound FokI molecules neither nick nor cleave the DNA. The truncated catalytic domain of FokI can dimerize transiently with such solitary molecules in vitro to stimulate ds cleavage, although this domain on its own is inactive (7). Cooperativity implies that the extent of cleavage increases sigmoidally with increasing enzyme concentration rather than monotonically, and that DNA molecules with a single recognition sequence are cleaved poorly compared with those containing multiple recognition sequences. These two mechanistic characteristics have been shown experimentally for FokI, suggesting that transient dimerization is a strategy commonly adapted by these REases. Steric considerations suggest that this form of dimerization is feasible only for enzymes in which the amino acid residues responsible for catalysis and DNA recognition are distinct and well separated; for enzymes that cleave well outside of their recognition sequences.
A number of REases cleave within asymmetric sequences or very close to the recognition sequences. These enzymes do not appear to engage in transient homodimerization. Instead these enzymes use two different catalytic sites for cleavage (9–11). We describe here four enzymes of this kind from thermophilic isolates of Bacillus: BsrDI, BtsI, BsmI and BsrI. The four appear to be distantly related: they exhibit limited amino acid sequence similarities; they recognize similar asymmetric five- and six-base sequences in ds DNA; and they all cleave very close to their recognition sequences and produce fragments with 2-base, 3′-overhangs.
Of interest is our finding that these enzymes possess two oligomeric organizations: BsrDI and BtsI endonucleases are heterodimers; BsmI, Mva1269I (a BsmI isoschizomer) and BsrI are monomers that might have evolved from heterodimeric ancestors by gene fusion. Of interest, too, is our finding that BsrDI and BtsI act as sequence-specific DNA nicking enzymes, or DNA cleaving enzymes, depending on their subunit compositions. Each enzyme comprises one large subunit and one small subunit. The large subunits, reminiscent of Type IIS enzymes, contain one catalytic site and all the elements for DNA recognition. On their own, they bind to DNA and specifically nick the bottom strand of their recognition sequence. We found that the small subunit of BsrDI and BtsI contain a catalytic site for top-strand cleavage. The small subunit is inactive on its own but in combination with the large subunit the heterodimer becomes a REase that cleaves both strands. We refer to the large subunits of BsrDI (Nb.BsrDI) and BtsI (Nb.BtsI)—natural, as opposed to mutationally engineered, DNA nicking enzymes—as ‘hemidimers’. By combining the catalytic-deficient large subunit of BsrDI and BtsI with its small subunit partner, we have successfully created top-strand-specific nicking enzymes Nt.BsrDI and Nt.BtsI.
MATERIALS AND METHODS
Strains and plasmid vectors
The BsrDI- and BtsI-producing strains were from NEB's strain collection. Escherichia coli strains ER2502, ER2566, ER2683 (KmR, lacIq) and ER2848 (TcR, lacIq) were provided by M. Sibley and E. Raleigh (NEB). Plasmids pRRS (ApR, a pUC19 derivative), pLG339 [pSC101 origin, KmR, TcR (12)], pAGR3 [ApR, a pBR322 derivative with Ptac promoter, (13)] and pAII17 (ApR, a pBR322 derivative with T7 promoter, Bill Jack, NEB) were used as the expression vectors.
Genomic DNA library construction and methylase selection
Bacillus stearothermophilus D70 genomic DNA was digested with REases and ligated into pRRS with compatible ends. Plasmid library was challenged with BsrDI and the digested DNA was used to transform ER2683 competent cells. Plasmids from cultures of individual transformants were screened for resistance to BsrDI digestion. The inserts in the methylase-positive clone and subclones were sequenced using a kit from ABI. A DNA fragment carrying bsrDIM1M2 genes were amplified in PCR, digested with BamHI and SphI and ligated to pLG339 with compatible ends.
Inverse PCR amplification, sequencing and expression of the REase genes
Inverse PCR was carried out to amplify the DNA adjacent to the M genes. Inverse PCR products were purified and directly sequenced by primer walking. The pre-modified host ER2848 (pLG-bsrDIM1M2) was used for endonuclease expression. Cells were cultured to late-log phase at 37°C and IPTG induction (0.5 mM) was carried out for 3 h. Cell extracts were prepared by sonication. In order to improve the expression level, bsrDIA and bsrDIB genes were amplified in PCR and independently cloned into the expression vector pAII17. Recombinant BsrDI activity was reconstituted by mixing cell extracts containing BsrDI A and B subunits or by mixing purified BsrDI A and B subunits. IPTG-induced cell extracts were used to digest λ or pUC19 DNA. Nicking activity was assayed on a single-site substrate pGPS2.1 or pUC-Km (a pUC19 derivative with a KmR gene).
Purification of BsrDI A and B subunits
Cell extract containing BsrDI A subunit was heated at 65°C for 30 min and the denatured E. coli proteins were removed by centrifugation. The BsrDI A subunit in the supernatant was further purified by chromatography through Heparin Sepharose FF, DEAE Sepharose and Q Sepharose columns (GE HealthCare). Cell extracts containing BsrDI B subunit were prepared from ∼20 g of IPTG-induced cells. BsrDI B subunit (Nb.BsrDI) was purified by chromatography through a Heparin Hyper D column (BioSepra), a Q Sepharose column and a Heparin column (Tosoh Bioscience).
Cloning of the BtsI R-M system
ApoI, NlaIII and Sau3AI partially digested genomic fragments from Bacillus thermoglucosidasius were ligated to EcoRI, SphI and BamHI digested, CIP-treated pUC19, respectively. The primary DNA libraries were challenged by BtsI digestion. After retransformation, plasmids were isolated and screened for resistance to BtsI digestion. The inserts in the resistant plasmids (M+ clones) were sequenced with pUC universal primers and custom-made primers. ORFs adjacent to btsIM genes were obtained by inverse PCR and direct sequencing of the PCR products. The btsIM1 and btsIM2 genes were amplified in PCR and cloned in two steps into pACYC184 to generate pre-modified host ER2683 (pACYC-btsIM1M2). The btsIRA and btsIRB genes were amplified in PCR, digested with appropriate REases and ligated to pUC19 separately and transferred into pre-modified host ER2683 (pACYC-btsIM1M2).
Purification of BtsI A and B subunits
BtsI A subunit was purified by heat denaturation of E. coli proteins and chromatography through a hydroxyapatite column. Cell extracts containing BtsI B subunit were heated at 65°C for 30 min and then centrifuged at 26 000 g for 20 min at 4°C to remove most of E. coli proteins. BtsI B subunit was purified by Heparin and hydroxyapatite column chromatography.
Reconstitution of BtsI endonuclease activity
Cell extracts containing BtsI A (small) and B (large) subunits or purified A and B subunits were mixed and incubated with 1 μg of ϕX174 DNA (RF form with a single BtsI site) at 55°C for 1 h. The cleavage products were analyzed by agarose gel electrophoresis and compared with the cleavage pattern of the native BtsI REase. The nicking activity was also assayed on ϕX174 ds DNA. To confirm the nicking strand specificity of the BtsI B subunit, the nicked circular product was subjected to run-off sequencing.
Site-directed mutagenesis of catalytic residues
The Asp, Glu and Lys catalytic residues in the active site for top-strand nicking (PDX12–17EXK) and the Asp, Glu, Gln and Arg residues in the active site for bottom-strand nicking (PDX13–18EX12QR) were mutated to Ala residue by site-directed mutagenesis with Phusion DNA polymerase and inverse PCR. Following DpnI digestion, plasmids coding for inactive proteins (inverse PCR products) were transformed into methylase-protected host for expression. The entire alleles were sequenced to confirm the desired mutation. Cell extracts were prepared from IPTG-induced cultures. Catalytically inactive A subunit was combined with wild-type (wt) B subunit or catalytic-deficient B subunit was mixed with wt A subunit. Reconstituted restriction or nicking activity was examined using the appropriate DNA substrates.
RESULTS
Cloning and expression of BsrDI R-M system
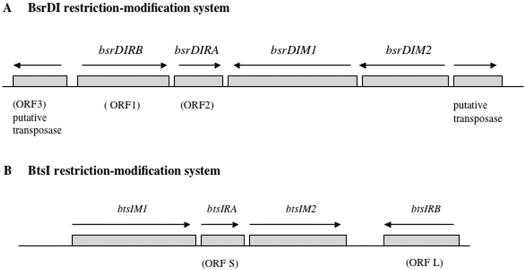
Several BsrDI-resistant clones were isolated following the methylase selection procedure (14). An M+ clone with a 4-kb insert was sequenced and three potential ORFs were found (Figure 1A). The aa sequence of one ORF was found to resemble a putative DNA transposase. The other two ORFs contain conserved motifs of adenine N6 and cytosine N4 methyltransferases and were named M1.BsrDI and M2.BsrDI, respectively. The two M genes were amplified in PCR and cloned into the plasmid vector pLG339 to produce pre-modified host ER2566 (pLG-bsrDIM1M2). After several rounds of inverse PCR and sequencing, three additional ORFs (ORF1, ORF2 and ORF3) were found (Figure 1A).
Figure 1.
Gene organization of BsrDI and BtsI R-M systems. ORF3 upstream of BsrDI R-M genes encodes a putative transposase. The ORF lengths are not drawn to scale. ORF1 = bsrDIRB, encoding BsrDI B subunit (Nb.BsrDI, large subunit); ORF2 = bsrDIRA, encoding BsrDI A subunit (small subunit). ORF S = btsIRA, encoding BtsI A subunit (small subunit); ORF L = btsIRB; encoding BtsI B subunit (Nb.BtsI, large subunit).
We fortuitously discovered that the gpORF1 is a strand-specific DNA nicking endonuclease. When IPTG-induced cell extracts containing gpORF1 was used to digest λ DNA at 65°C, no apparent ds DNA cleavage activity was detected. However, when the cell extracts of gpORF1 were incubated with supercoiled pUC19, the DNA was converted to nicked-circular form. The nicking is site specific and not due to contaminating E. coli nucleases since a pBR322 derivative with BsrDI site deletion was not nicked (data not shown). ORF2 was amplified in PCR and cloned into pAII17. IPTG-induced cell extracts lack site-specific restriction or nicking activity (data not shown). The negative result was not due to the absence of the expressed protein since a ∼25-kDa protein band was clearly detected by SDS–PAGE. In addition, no mutations had been introduced into the cloned insert.
Characterization of BsrDI large and small subunit activity
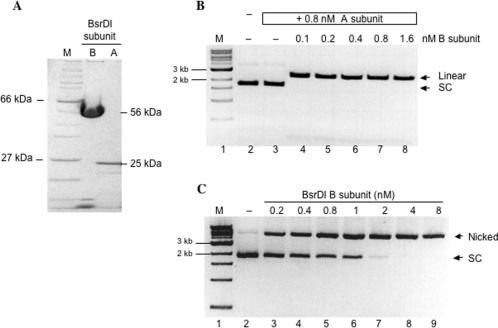
We confirmed that both the BsrDI large and small subunits are required for restriction activity. The BsrDI small subunit (A subunit) was partially purified and its purity was estimated to be 90% by SDS–PAGE analysis (Figure 2A, lane 3). BsrDI REase activity was reconstituted by mixing the cell extracts containing gpORF1 and gpORF2 (data not shown) or by mixing purified large and small subunits (Figure 2B, lanes 4–8). When 0.8 nM BsrDI A subunit was incubated with 8 nM pUC19 (lane 3) there was no change in DNA mobility. However, when 0.1 nM of the large subunit (B subunit) was added to the same reaction containing A subunit, complete cleavage of pUC19 was achieved (lane 4). The data from Figure 2B demonstrate that restriction activity requires the presence of both subunits. Thus, BsrDI is a heterodimeric REase encoded by bsrDIRB (ORF1) and bsrDIRA (ORF2).
Figure 2.
Characterization of BsrDI A/B subunits. (A) Purified recombinant BsrDI A and B subunits. The calculated molecular weight of BsrDI A and B subunits are 25 and 56 kDa, respectively. Lane 1, protein size marker; lane 2, BsrDI B subunit (Nb.BsrDI); lane 3, BsrDI A subunit. (B) Reconstituted BsrDI double-strand cleavage activity by mixing purified BsrDI A and B subunits. Lane 1, 1-kb DNA marker; lane 2, pUC19; lane 3, 8 nM pUC19 treated with A subunit; lanes 4–8, 8 nM pUC19 digested with A and B subunits (linear pUC19 = 2.7 kb); sc, supercoiled DNA. (C) Nb.BsrDI DNA nicking activity on 8-nM pUC19. Lane 1, 1-kb DNA marker; lane 2, pUC19; lanes 3–9, pUC19 digested with BsrDI B subunit, enzyme concentration as indicated on the top of each lane.
We further demonstrated that BsrDI large subunit (B subunit) is a bottom-strand specific DNA nicking endonuclease. The BsrDI large subunit (B subunit) was purified by chromatography such that >95% homogeneity was achieved (Figure 2A, lane 2). Most striking is the nicking activity displayed by the large subunit (Figure 2C, lanes 3–9). Complete conversion of 8 nM pUC19 to open circular form can be achieved with 4 nM of the large subunit in 30 min at 65°C. To determine the strand and sequence specificity, the plasmid pGPS2.1 with a single BsrDI site was treated with the large subunit. The nicked DNA product was purified and subjected to run-off sequencing. The sequencing peaks drop off at 5′ GCAATG(a) 3′, indicating a nick at 5′↓CATTGC 3′. The sequence was continuous on the opposite strand, indicating that no nicks have been introduced to the top-strand template (data not shown). The gene product of ORF1 was thus named Nb.BsrDI (Nb for bottom-strand nicking) with the specificity of 5′ ↓CATTGC 3′ (↓ indicating the nicking site). These results indicate that in addition to residues responsible for binding specifically to the recognition sequence, all catalytic residues necessary for bottom-strand cleavage (Cb) are located in the BsrDI large subunit (B subunit). In contrast, top-strand nicking and thus ds cleavage activity requires the assembly of A and B subunits. Amino acid sequence alignment of BsrDI large subunit with the N-terminus of BsmI/Mva1269I, and BsrI revealed a non-canonical catalytic site Cb with conserved residues P/A/F D-X13–17-E-X12–QR, whereas the BsrDI small subunit contains a canonical Type II catalytic site Ct with conserved amino acid residues PD-Xn-EXK. The BsmI/Mva1269I and BsrI REases are monomeric proteins with large molecular weights (78.1, 78.4 and 69.4 kDa, respectively) (15) (S.-Y.X., Z.Z. and G.G.W., unpublished data).
Cloning and expression of BtsI R-M system
The BtsI M genes were successfully cloned by the methylase selection procedure. The inserts of the M+ clones were sequenced and the assembled sequence generated a 4986-bp sequence with four potential ORFs. The predicted aa sequences of two ORFs contain conserved motifs of amino-methyltransferases and were named M1.BtsI and M2.BtsI. The two M genes were amplified separately in PCR and cloned in two steps into the plasmid vector pACYC184 to generate a pre-modified expression host ER2683 (pACYC-btsIM1M2). We surmised that the third ORF (ORF S, 495 bp) flanked by the two M genes might encode a regulatory protein or one subunit of the BtsI REase. A partial ORF of 657 bp was also found adjacent to the btsIM2 gene. Following inverse PCR and direct sequencing, a complete ORF of 987 bp was derived (ORF L, Figure 1B).
Expression of ORF S (btsIRA) and ORF L (btsIRB)
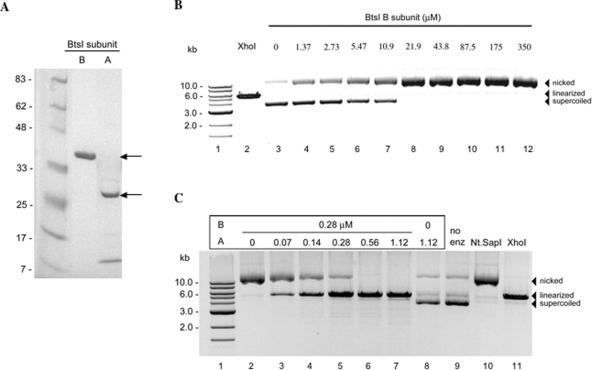
Similar to BsrDI, we found that the BtsI large subunit (B subunit, gpORF L) is a strand-specific DNA nicking endonuclease. Both the large and small subunits are required for BtsI restriction activity. The four genes in the BtsI R-M system are organized in the order of btsIM1, ORF S, btsIM2 and ORF L (Figure 1B). ORF S and ORF L are oriented in the opposite direction and are separated by the btsIM2 gene. The ORF S and ORF L were independently expressed in E. coli. No specific endonuclease activity was detected on λ or supercoiled ϕX174 DNA with cell extracts of gpORF S. However, cell extracts containing the large subunit prepared from ER2683 (pACYC-btsIM1M2, pUC19-ORF L) displayed DNA nicking activity when ϕX174 RF was used as the substrate (data not shown). Figure 3A shows the purified BtsI A/B subunits. The A subunit displayed aberrant migration on the protein gel. The apparent molecular mass is 27 kDa, while the predicted size is 18.6 kDa. Purified BtsI A subunit forms oligomers with three distinct species in native PAG electrophoresis (data not shown). The significance of this self-assembly is not known. The apparent molecular mass of the B subunit on the protein gel was as predicted (38 kDa). DNA nicking activity was detected when purified BtsI B subunit was incubated with ϕX174 ds DNA (Figure 3B, lanes 4–12). Ds cleavage activity of BtsI was reconstituted by mixing the two purified subunits (Figure 3C, lanes 3–7). The A subunit alone did not show nicking or ds DNA cleavage activity (Figure 3C, lane 8). To determine the nicking strand and sequence specificity of the BtsI large subunit (B subunit), the nicked-circular product was purified and sequenced. The run-off sequencing results show sequencing peaks drop off after the sequence 5′ GCAGTG(a) 3′, indicating that BtsI large subunit (B subunit) is a bottom-strand NEase (Nb.BtsI) that specifically nicks the 5′ ↓CACTGC 3′ site. DNA sequence from the top-strand template is continuous, indicating no nicks are introduced on the top strand (data not shown). We also found that Nb.BtsI nicks a miscognate site (star site) 5′ ↓CACTGG 3′ that is one base (underlined G) different from the cognate site when 10-fold over-digestion was carried out. BtsI endonuclease also displays strong star activity in over-digestion. However, the entire spectrum of BtsI star sites has not been fully characterized (data not shown).
Figure 3.
Characterization of BtsI A/B subunits. (A) Lane 1, protein size marker; purified BtsI B subunit (lane 2) and A subunit (lane 3). The predicted molecular masses of A and B subunits are 18.6 and 37.6 kDa, respectively. (B) DNA nicking activity of the BtsI large subunit (Nb.BtsI). Lane 1, 1-kb marker; lane 2, linearized DNA by XhoI; lane 3, ϕX174 DNA; lanes 4–12, ϕX174 DNA digested with BtsI B subunit. (C) Recombinant BtsI double-strand cleavage activity reconstituted by combining purified BtsI A and B subunits. Lane 1, DNA marker; lane 2, ϕX174 DNA digested with BtsI B subunit; lanes 3–7, ϕX174 digested with BtsI A and B subunits; lane 8, ϕX174 treated with A subunit; lane 9, ϕX174 DNA; lane 10, Nt.SapI nicked ϕX174; lane 11, linearized ϕX174 by XhoI.
Amino acid sequence alignment of catalytic sites of related restriction enzymes
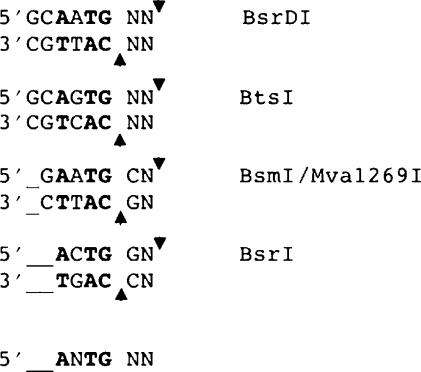
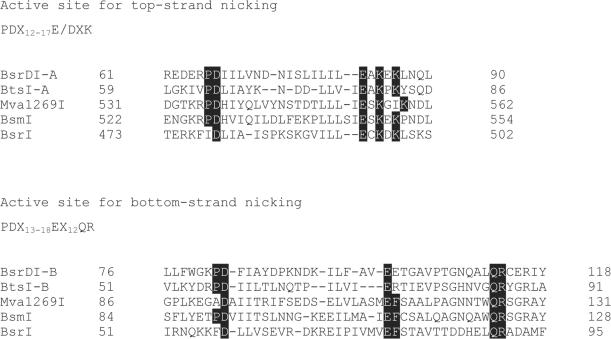
BsrDI GCAATG (2/0) and BtsI GCAGTG (2/0) share limited amino acid sequence similarity with several other Type IIS REases that cleave DNA in the same general manner, that is, close to their recognition sequences with a 2-base 3′-cohesive end. These related enzymes include BsmI/Mva1269I (GAATGC 1/-1), and BsrI (ACTGG 1/-1). Aligning the recognition sequences of these enzymes reveals that each of them cleaves DNA two bases downstream from an invariant ANTG tetranucleotide in the top strand of the recognition sequence and immediately before the complementary CANT tetranucleotide in the bottom strand (Figure 4). Interestingly, while BsrDI and BtsI are heterodimeric, the other three enzymes appear to be monomeric. These latter might have arisen by gene fusion between ancestral subunits that remain separated in BsrDI and BtsI.
Figure 4.
Recognition sequences and cleavage sites of REases that share aa sequence similarity with BsrDI and BtsI. The recognition sequences are aligned with respect to the invariant RNTG/CANY sequence (shown in bold) and the consistent 2-base 3′-overhang. Positions of cleavage are indicated by arrows; unspecified nucleotides are shown as N.
Amino acid sequence analysis and experimental observations suggest that all of these enzymes harbor two catalytic sites. The first, N-terminal, catalytic site (Cb) in each enzyme is somewhat non-canonical in sequence: PD-Xn-E-Xn-QR, whereas the second catalytic site (Ct) in each is canonical: PD-Xn-EXK (Figure 5) (16). One site (Cb, catalytic site for bottom strand) is located in the large subunit of BsrDI and BtsI, or near the N-terminus of the monomeric proteins (BsmI/Mva1269I, BsrI) and catalyzes the cleavage of the bottom strand of the recognition sequence. The other site (Ct, catalytic site for top strand), is located near the C-terminus in the monomeric enzymes, and within the small subunit in the heterodimeric ones, and probably catalyzes hydrolysis of the top-strand (see mutagenesis results section). The region between the catalytic sites is variable and perhaps comprises the DNA target-recognition domain (TRD) of each protein. To a first approximation, the domain/subunit architecture of the heterodimers BsrDI and BtsI can be described as Cb∼TRD:Ct, where ‘∼’ represents covalent polypeptide linkage and ‘:’ represents non-covalent association. Likewise, the architecture of monomers BsmI/Mva1269I, and BsrI can be described as Cb∼TRD∼Ct. As described above, the isolated large subunits of BsrDI and BtsI (composition Cb∼TRD), in the absence of their Ct-containing small subunit, nick only the bottom strand of the target sequence.
Figure 5.
Amino acid sequence alignments of two catalytic sites. Top amino acid sequences: top-strand catalytic site (Ct) residues of BsrDI and BtsI A subunits, C-terminal regions of BsmI/Mva1269I, and BsrI. Bottom amino acid sequences: bottom-strand catalytic site (Cb) residues of BsrDI and BtsI B subunits, N-terminal regions of BsmI/Mva1269I, and BsrI. BsrDI and BtsI are heterodimeric while BsmI, Mva1269I and BsrI are monomeric. The Genbank accession numbers for BsrDI, BtsI, BsmI and Mva1269I are DQ367879, DQ355163, AY079085 and DQ074451, respectively.
Site-directed mutagenesis of two catalytic sites in A/B subunits of BsrDI and BtsI endonucleases
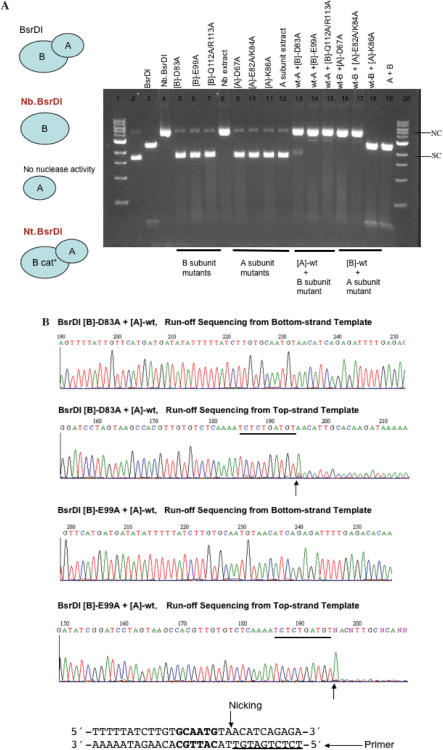
The Asp, Glu, Gln and Arg in Cb the putative catalytic site for bottom-strand nicking in BsrDI B subunit (Nb.BsrDI) were mutated to Ala. The mutant cell extracts were assayed for DNA nicking or ds cleavage activity. Figure 6A shows that [B]-D83A, [B]-E99A and the double mutant [B]-Q112A/R113A lack DNA nicking activity as anticipated. Similarly, the Asp, Glu and Lys in Ct, the putative catalytic site for top-strand nicking in the A subunit were changed to Ala by site-directed mutagenesis. BsrDI A subunit mutants [A]-D67A, [A]-E82A/K84A, [A]-K86A, as well as the wt A subunit do not show DNA nicking or restriction activity. Surprisingly, when the catalytic-deficient B subunit mutants were combined with wt A subunit, DNA nicking activity was reconstituted. To determine the nicking strand specificity of the newly restored nicking activity, the nicked circular DNA was gel-purified and subjected to run-off sequencing. Figure 6B shows that [wt]-A/[B]-D83A or [wt-A]/[B]-E99A are top-strand nicking enzymes with substrate specificity of GCAATGNN↓. [wt-A]/[B]-Q112A/R113A gave rise to the same result (data not shown). Combining BsrDI wt B subunit with A subunit mutants [A]-D67A or [A]-E82A/K84A resulted in bottom-strand nicking activity only, further confirming that [A]-Asp67, [A]-Glu82 or [A]-Lys84 are involved in top-strand catalysis. Mixing BsrDI wt B subunit with [A]-K86A produced BsrDI restriction activity, indicating [A]-Lys86 is not a catalytic residue although this Lys residue is conserved in four of the five endonucleases (see amino acid alignment in Figure 5).
Figure 6.
(A) DNA nicking or ds cleavage activity of BsrDI A/B mutants. B cat* = catalytic-deficient B subunit. Plasmid pUC19 (2 BsrDI sites) was used as the substrate for nicking or restriction assay. NC, nicked-circular DNA; SC, supercoiled DNA. (B) Run-off sequencing of gel-purified, nicked-circular DNA. The expected nicking specificity of Nt.BsrDI is GCAATGNN↓ (BsrDI site and the adjacent sequence are shown at the bottom of the figure). The extra A peak at the end of run-off sequence was added by the template independent terminal transferase activity of the Taq DNA pol.
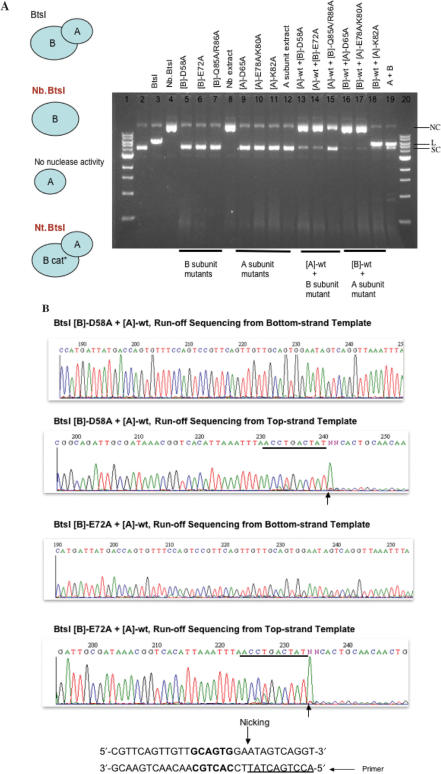
Alanine scanning was also applied to BtsI A/B subunits. The Asp, Glu, Gln and Arg in Cb, the putative catalytic site for bottom-strand nicking in BtsI B subunit (Nb.BtsI) were changed to Ala by site-directed mutagenesis. The mutant cell extracts were assayed for DNA nicking or restriction activity. Figure 7A shows that [B]-D58A, [B]-E72A and the double mutant [B]-Q85A/R86A lost DNA nicking activity as expected. Similarly, the Asp, Glu and Lys in Ct, the putative catalytic site for top-strand nicking in the A subunit were substituted by Ala. BtsI A subunit mutants [A]-D65A, [A]-E78A/K80A, [A]-K82A as well as the wt A subunit do not show nicking or restriction activity. However, when the catalytic-deficient B subunit mutants were combined with wt A subunit, DNA nicking activity was restored. Run-off sequencing of the gel-purified nicked-circular DNA products showed that [wt]-A/[B]-D58A or [wt-A]/[B]-E72A are top-strand nicking enzymes with substrate specificity of GCAGTGNN↓ (Figure 7B). [wt-A]/[B]-Q85A/R86A is also a top-strand nicking enzyme (data not shown). Combining BtsI wt B subunit with A subunit mutants [A]-D65A or [A]-E78A/K80A produced only bottom-strand nicking activity, further confirming that [A]-Asp65, [A]-Glu78 or [A]-Lys80 are involved in top-strand hydrolysis. Similar to [A]-Lys86 of BsrDI, [A]-Lys82 of BtsI is not a critical residue for top-strand cleavage because mixing BtsI wt B subunit with [A]-K82A generated BtsI restriction activity (Figure 7A).
Figure 7.
(A) DNA nicking or restriction activity of BtsI A/B mutants. B cat* = catalytic-deficient B subunit. ϕX174 RF DNA (1 BtsI site) was used as the substrate for nicking or restriction assay. NC, nicked-circular DNA; L, linear DNA; SC, supercoiled DNA. (B) Run-off sequencing of gel-purified, nicked-circular DNA. The expected nicking specificity of Nt.BtsI is GCAGTGNN↓ (BtsI site and the adjacent sequence are shown at the bottom of the figure).
DISCUSSION
We describe here cloning and expression of BsrDI and BtsI R-M systems, and the discovery of two natural nicking enzymes Nb.BsrDI and Nb.BtsI. In addition, top-strand nicking variants, Nt.BsrDI and Nt.BtsI, were created by mixing catalytic-deficient B subunits with the respective wt A subunit. Two independent catalytic sites were implicated in BsrDI/BtsI large and small subunits for top-strand and bottom-strand cleavage.
Only a few naturally occurring DNA nicking endonucleases (NEases) have been described from bacterial and viral sources. Two strand-specific and sequence-specific DNA NEases were found in the lysates of Chlorella viruses NYs-1 and NY2A (17,18). These are Nt.CviPII (previously named CviNYSI nickase, ↓CCD, D = A, G or T) and Nt.CviQII (previously named CviNY2A nickase, R↓AG, R = A or G). The CviPII and CviQII N-M systems have been cloned and expressed in heterologous hosts (19,20). Nt.BstSEI and isoschizomer Nt.BstNBI (GAGTCN4↓) were found in two strains of B. stearothermophilus (21,22). Nt.BspD6I, an isoschizomer of Nt.BstNBI with identical amino acid sequence has also been cloned (23). Nt.BstNBI was thought to be a nicking enzyme that had lost the ability to dimerize because the enzyme purified from the native strain was a NEase. Recently, however, the gene encoding its small subunit partner has been identified adjacent to the gene coding for the nicking subunit (Heiter,D. and G.G.W., unpublished data). Presumably, the small subunit of the BstNBI endonuclease was dissociated from the large subunit and was separated from the large subunit during purification, resulting in a natural top-strand nicking enzyme. Similarly, a gene encoding a small subunit of 186 aa residues had been found downstream of the Nt.BspD6I gene. BspD6I restriction activity was reconstituted by mixing Nt.BspD6I with the purified small subunit (24). Based on the mutagenesis results of BsrDI and BtsI, it should be possible to engineer bottom-strand nicking variants from BstNBI/BstSEI/BspD6I REases by mixing the catalytic-deficient variants of Nt.BstNBI/Nt.BstSEI/Nt.BspD6I with their wt small subunit, assuming that two independent catalytic sites reside in two subunits.
Strand-specific NEases have been used in DNA strand displacement amplification (SDA), EXPAR DNA amplification (25), in DNA fragment assembly and cloning, and in preparation of nicked-duplex or gapped DNA for studying DNA repair and DNA base stacking (26,27). Recently, a nicking endonuclease-mediated DNA amplification (NEMDA) using Nt.CviPII and Bst DNA polymerase in the absence of input primers has also been described (19). Nt.CviPII-digested genomic DNA into small partial duplex DNA that serves as the templates and primers for primer extension. Recently, nicking enzymes have been used to nick and label DNA for single-molecule barcoding system (28,29).
The relatively small number of NEases identified in nature has prompted efforts to engineer NEases from existing Type IIA/Type IIS/Type IIT REases by domain swapping or mutagenesis (Type IIA, REases that cleave asymmetric sites; Type IIT, heterodimeric REases). Strand-specific nicking variants have been created from AlwI, Bpu10I, BbvCI, BsaI, BsmBI, BsmAI, BsmI, BspQI, MlyI, Mva1269I and SapI (REBASE) (30). For Mva1269I, mutating the critical Asp, Glu, and Lys residues of Ct produced enzymes that nicked the bottom strand of the recognition sequence. Mutating Cb of Mva1269I produced enzymes that nicked the top strand specifically, but at very low efficiency unless the bottom strand was already nicked (15). The catalytic sites of Mva1269I appear to act sequentially, with Cb cleaving the bottom strand first, and in so-doing creating the substrate upon which Ct can act to cleave the top strand (15). Comparable results were observed with BsmI: bottom-strand nicking derivatives (Nb.BsmI, E546V) have been generated when the Glu residue of Ct was substituted for Val or Ala (Z.Z., Meixsell,T. and S.-Y.X., unpublished data). Mutating the Arg residue of the PD-Xn-E-Xn-QR motif in Cb produces a variant R123D with low nicking activity. Although the native BsrDI and BtsI prefer to nick the bottom strand first and then break the top strand in restriction of ds DNA (data not shown), the isolation of Nt.BsrDI and Nt.BtsI implies that there is no stringent requirement for nicking the bottom strand first, i.e. top-strand hydrolysis can take place without a pre-nicked bottom strand.
Possible DNA cleavage mechanism
There are a few other nucleases whose catalytic sites are held on the same polypeptide chain, on separate chains or formed by dimerization. For example, homing endonuclease PI-SceI has two catalytic sites within one polypeptide chain (one for cleavage of each strand) (31). The Tn7 transposase consists of two different subunits, TnsA and B, each of which cleaves a separate strand (32). In contrast, some recombinases (Tn10 and Tn5) use a single active site to cleave both strands. BfiI requires homodimerization to form the catalytic site located on the dimer interface. A homodimer of BfiI cleaves the bottom strand first, generating a charged group that helps the enzyme attack the top strand (1).
Previously we have generated top-strand or bottom-strand specific nicking variants from BsaI, BsmBI, BsmAI, SapI (10,11) and BspQI (P.Z. and S.-Y.X., unpublished data). Based on these results and previous biochemical studies, it was proposed that some Type IIS REases operate as monomers and carry two separate catalytic sites for top and bottom strand scission (9,10). Here we provide experimental evidence that BsrDI and BtsI possess separate catalytic sites for top-strand and bottom-strand nicking. This group of Type IIS enzymes cleaves DNA close to the recognition sequence (N0–5), suggesting a mechanism that differs from the well-studied FokI where the reach is N9–13. FokI consists of a DNA-binding domain and a catalytic domain within the same polypeptide. FokI binds to a cognate DNA site as a monomer and then forms a dimer transiently with a second molecule in the presence of divalent cation. DNA cleavage takes place at the defined position downstream of the recognition site by the non-specific nuclease domain (5,8). In FokI, dimerization activates the ds cleavage domain (6,7). In BsrDI and BtsI, however, dimerization is required for top strand cleavage only.
ACKNOWLEDGEMENTS
We thank Richard Roberts for critical comments; Iain Murray for discussion, Laurie Mazzola, Beth Ann Catin and Barton Slatko for run-off sequencing; Organic Synthesis Division of NEB for the synthesis of oligonucleotides; Donald Comb for his support and encouragement. This work is partially supported by NIH Phase II STTR grant (5 R42 HG003976-03) to S.-Y.X. Funding to pay the Open Access publication charges for this article was provided by New England Biolabs, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sasnauskas G, Halford SE, Siksnys V. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl Acad. Sci. USA. 2003;100:6410–6415. doi: 10.1073/pnas.1131003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Q, Roberts R, Guo H. Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure. Protein Sci. 2005;14:2590–2600. doi: 10.1110/ps.051565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Horton J, Maunus R, Wilson G, Roberts R, Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–1901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heiter DF, Lunnen KD, Wilson GG. Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 2005;348:631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 5.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 6.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 9.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Z, Samuelson JC, Zhou J, Dore A, Xu SY. Engineering strand-specific DNA nicking enzymes from the type IIS restriction endonucleases BsaI, BsmBI, and BsmAI. J. Mol. Biol. 2004;337:573–583. doi: 10.1016/j.jmb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Samuelson JC, Zhu Z, Xu SY. The isolation of strand-specific nicking endonucleases from a randomized SapI expression library. Nucleic Acids Res. 2004;32:3661–3671. doi: 10.1093/nar/gkh674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoker NG, Fairweather NF, Spratt BG. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 13.Jack WE, Greenough L, Dorner LF, Xu SY, Strzelecka T, Aggarwal AK, Schildkraut I. Overexpression, purification and crystallization of BamHI endonuclease. Nucleic Acids Res. 1991;19:1825–1829. doi: 10.1093/nar/19.8.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss A, Baldauf F. Molecular cloning and expression in Escherichia coli of two modification methylase genes of Bacillus subtilis. Gene. 1983;21:111–119. doi: 10.1016/0378-1119(83)90153-1. [DOI] [PubMed] [Google Scholar]
- 15.Armalyte E, Bujnicki JM, Giedriene J, Gasiunas G, Kosinski J, Lubys A. Mva1269I: a monomeric type IIS restriction endonuclease from micrococcus varians with two EcoRI- and FokI-like catalytic domains. J. Biol. Chem. 2005;280:41584–41594. doi: 10.1074/jbc.M506775200. [DOI] [PubMed] [Google Scholar]
- 16.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Y, Morgan R, Schildkraut I, Van Etten JL. A site-specific single strand endonuclease activity induced by NYs-1 virus infection of a Chlorella-like green alga. Nucleic Acids Res. 1988;16:9477–9487. doi: 10.1093/nar/16.20.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YNM, Nietfeldt J, Xia Y, Burbank D, Ropp S, Van Etten JL. Chlorella virus NY-2A encodes at least 12 DNA endonuclease/methyltransferase genes. Virology. 1998;240:366–375. doi: 10.1006/viro.1997.8936. [DOI] [PubMed] [Google Scholar]
- 19.Chan SH, Zhu Z, Van Etten JL, Xu SY. Cloning of CviPII nicking and modification system from chlorella virus NYs-1 and application of Nt.CviPII in random DNA amplification. Nucleic Acids Res. 2004;32:6187–6199. doi: 10.1093/nar/gkh958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan SH, Zhu Z, Dunigan DD, Van Etten JL, Xu SY. Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: M.CviQII methylates AG sequences. Protein Expr. Purif. 2006;49:138–150. doi: 10.1016/j.pep.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Abdurashitov MA, Belichenko OA, Shevchenko AV, Degtyarev SK. N.BstSE – site-specific nuclease from Bacillus stearothermophilus SE-589 – restriction endonuclease production. Mol. Biol. 1996;30:1261–1267. [PubMed] [Google Scholar]
- 22.Morgan RD, Calvet C, Demeter M, Agra R, Kong H. Characterization of the specific DNA nicking activity of restriction endonuclease N.BstNBI. Biol. Chem. 2000;381:1123–1125. doi: 10.1515/BC.2000.137. [DOI] [PubMed] [Google Scholar]
- 23.Perevyazova TA, Rogulin EA, Zheleznaya LA, Matvienko NI. Cloning and sequencing of the gene of site-specific nickase N.BspD6I. Biochemistry (Mosc.) 2003;68:984–987. doi: 10.1023/a:1026008528310. [DOI] [PubMed] [Google Scholar]
- 24.Yunusova A, Rogulin E, Artyukh R, Zheleznaya L, Matvienko N. Nickase and a protein encoded by an open reading frame downstream from the nickase BspD6I gene form a restriction endonuclease complex. Biochemistry (Mosc.) 2007;71:815–820. doi: 10.1134/s0006297906070157. [DOI] [PubMed] [Google Scholar]
- 25.Van Ness J, Van Ness LK, Galas DJ. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Hays JB. Mismatch repair in human nuclear extracts. Quantitative analyses of excision of nicked circular mismatched DNA substrates, constructed by a new technique employing synthetic oligonucleotides. J. Biol. Chem. 2002;277:26136–26142. doi: 10.1074/jbc.M200357200. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn H, Protozanova E, Demidov VV. Monitoring of single nicks in duplex DNA by gel electrophoretic mobility-shift assay. Electrophoresis. 2002;23:2384–2387. doi: 10.1002/1522-2683(200208)23:15<2384::AID-ELPS2384>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 28.Xiao M, Phong A, Ha C, Chan TF, Cai D, Leung L, Wan E, Kistler AL, DeRisi JL, et al. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 2007;35:e16. doi: 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jo K, Dhingra DM, Odijk T, de Pablo JJ, Graham MD, Runnheim R, Forrest D, Schwartz DC. A single-molecule barcoding system using nanoslits for DNA analysis. Proc. Natl Acad. Sci. USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts R, Vincze T, Posfai J, Macelis D. REBASE – enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:269–270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christ F, Schoettler S, Wende W, Steuer S, Pingoud A, Pingoud V. The monomeric homing endonuclease PI-SceI has two catalytic centres for cleavage of the two strands of its DNA substrate. EMBO J. 1999;18:6908–6916. doi: 10.1093/emboj/18.24.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarnovsky RJ, May EW, Craig NL. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]