Abstract
GW bodies (or P-bodies) are cytoplasmic granules containing proteins involved in both mRNA degradation and storage, including the RNA interference machinery. Their mechanism of assembly and function are still poorly known although their number depends upon the flux of mRNA to be stored or degraded. We show here that silencing of the translational regulator CPEB1 leads to their disappearance, as reported for other GW body components. Surprisingly, the same results were obtained with several siRNAs targeting genes encoding proteins unrelated to mRNA metabolism. The disappearance of GW bodies did not correlate with the silencing activity of the siRNA and did not inhibit further silencing by siRNA. Importantly, in most cases, GW bodies were rapidly reinduced by arsenite, indicating that their assembly was not prevented by the inhibition of the targeted or off-target genes. We therefore propose that some siRNA sequences affect mRNA metabolism so as to diminish the amount of mRNA directed to the GW bodies. As an exception, GW bodies were not reinduced following Rck/p54 depletion by interference, indicating that this component is truly required for the GW body assembly. Noteworthy, Rck/p54 was dispensable for the assembly of stress granules, in spite of their close relationship with the GW bodies.
INTRODUCTION
GW bodies, also called dcp bodies, or P-bodies in yeast, are recently described cytoplasmic structures involved in mRNA metabolism. They were first implicated in mRNA degradation. In eukaryotes, mRNA degradation is initiated by the removal of the polyA tail followed by either 3' to 5' degradation by the exosome or decapping of the 5' extremity and 5' to 3' degradation by Xrn1. GW bodies contain all the proteins of the 5' to 3' mRNA degradation machinery, including the decapping complex Dcp1/2, its cofactors LSm1-7 and Rck/p54 (also known as Dhh1 in yeast, Me31 in drosophila and Cgh1 in Caenorhabditis) and the exonuclease Xrn1 (1). In yeast, experiments designed to slow down the final steps of mRNA degradation enhance P-bodies in size and number. This has been observed following mutations of Dcp1 or Xrn1 (2). Moreover, when a poly(G) tract is introduced into a reporter mRNA to block exonucleolysis, it accumulates in P-bodies, indicating that they are active sites of mRNA degradation (2). Similarly, in mammals, polyadenylated RNA are detected in GW bodies following the stable depletion of Xrn1 by RNA interference (3). Furthermore, inhibiting translation with a drug which releases free mRNA, such as puromycin, leads to an increase of the GW body number (4). Conversely, when mRNAs are frozen on polysomes by a translation inhibitor such as cycloheximide, GW bodies disappear (2,3). Taken together, these data indicate that GW bodies are formed from a pool of untranslated mRNAs available for degradation.
GW bodies also contain the post-transcriptional gene silencing machinery, including both proteins of the RNA-induced silencing complex (RISC), such as Argonaute (Ago), and short RNAs, whether short interfering RNAs (siRNAs) or micro-RNAs (miRNAs) (5–8). One of the GW body markers, GW182, which was initially identified as a human autoantigen, turned out to be a direct Ago partner (5). These observations have led to the proposal that GW bodies are the sites of RNA interference activity. This issue is controversial, as some studies report the inhibition of both mi-RNA- and si-RNA-mediated interference in the absence of GW bodies (8), while others report a clear inhibition of mi-RNA-mediated interference and a slight inhibition of si-RNA-mediated interference (5,9); a few even indicate no inhibition of the si-RNA-mediated interference (10,11). The presence of RISC in the GW bodies is consistent with the need to degrade the fragments generated by the initial siRNA-mediated mRNA cleavage (12). At first glance, the presence of miRNAs was more puzzling, as they guide translation inhibition and not mRNA cleavage. Recently, it has become clear that miRNA-mediated silencing can result in RNA decay, initiated by deadenylation and decapping rather than endonucleolytic cleavage (1). In addition, GW bodies can also play a role in mRNA storage. In Huh7 hepatoma cells, the CAT1 mRNA is repressed by miR122 and localized in GW bodies. Following amino acid deprivation, its translation resumes and it disappears from the GW bodies (13).
A more general role for GW bodies in mRNA storage is still uncertain in mammals but has been established for P-bodies in yeast. When glucose deprivation leads to translation arrest and accumulation of mRNAs in P-bodies, glucose re-addition leads to mRNAs leaving the P-bodies and resuming translation (14). Therefore, P-bodies have a dual role in mRNA degradation and storage, albeit it is still unclear how these two opposite functions are coordinated. In addition, mammalian cells harbor distinct cytoplasmic structures involved in mRNA storage, the stress granules, which are induced by stress. They contain their own set of proteins, including translation initiation factors, such as eIF3, and translation repressors, such as TIA1/TIAR, as well as some of the GW body proteins, such as Xrn1 (15). Stress granules are frequently in contact with GW bodies. In some cases, they recruit GW bodies and seem to fuse with them. We have proposed that this intermingling between stress granules and GW bodies could trigger the transition from mRNA storage to mRNA degradation during stress (4).
The number of GW bodies per cell is variable, even within a clonal cell population. How this number could impact on their presumed function is unknown. First, it is unclear whether a GW body needs to be macroscopic in order to fulfill its function. Second, the GW body assembly pathway is still unknown. One of the reported parameters is the position of the cells in the cell cycle. Most GW bodies disappear before mitosis, reassemble in the G1 phase and enlarge in late S and G2 phases (16). It has been proposed, as a consequence, that the efficiency of RNA interference may vary during the cell cycle (17). In addition, several proteins of the GW bodies have been reported to be essential for the GW body assembly, because their depletion by RNA interference leads to the disappearance of GW bodies. This has been observed for Ccr4, Rck/p54, Lsm1 and eIF4ET (18); GW182 (16), Ge1 (19) and Rap55 (20). One interpretation of this data is that some of these proteins have a scaffolding role in GW body assembly. Alternatively, all of them may be necessary to ensure a certain level of repressed mRNA required for the GW body assembly (1).
We have previously shown that the translational regulator CPEB1 is enriched in GW bodies. This raised the possibility that it could play an active role in the degradation of the mRNA to which it is bound. Alternatively, it could be passively attracted by its target mRNA as a consequence of the translation repression (4). While investigating CPEB1 function using RNA interference, we found that siRNAs targeting CPEB1 also led to GW body disassembly. Surprisingly, this property was shared with several other siRNAs and appeared to be independent of the gene targeted. The siRNA-guided RNA interference had the same efficiency in the absence of GW bodies, demonstrating that these bodies were not the sites of action of the RISC machinery. Finally, except in the case of si-p54, arsenite could counteract the effect of these siRNAs, despite the fact that this drug prevents protein synthesis, thus confirming that CPEB1 protein is not involved in GW body assembly whereas p54 is truly required.
MATERIALS AND METHODS
Cell culture
Epithelioid carcinoma HeLa cells and retinal pigment epithelial RPE-1 cells (BD Biosciences Clontech, France) were routinely maintained in DMEM and DMEM/F12, respectively, supplemented with 10% fetal calf serum. IFN-α2b was a kind gift of Pierre Eid (Institut André Lwoff, Villejuif, France). For stress induction, cells were treated with 0.5 mM arsenite (Sigma Aldrich, France) for 30 min.
Transfections were performed with 3 μg si-RNA or polyI/C per 35-mm diameter dish using a standard calcium phosphate procedure, as described previously (4). siRNAs and polyI/C were purchased from MWG (MWG Biotech, France) and Sigma (Sigma Aldrich, France), respectively. The siRNA concentration was checked both by spectrophotometry and on gel (data not shown). In Figure 1, control cells were transfected with 1.3 µg of a human CPEB1-long expression vector (IMAGE 6047179). Si-Eg5 was transfected as previously described (21).
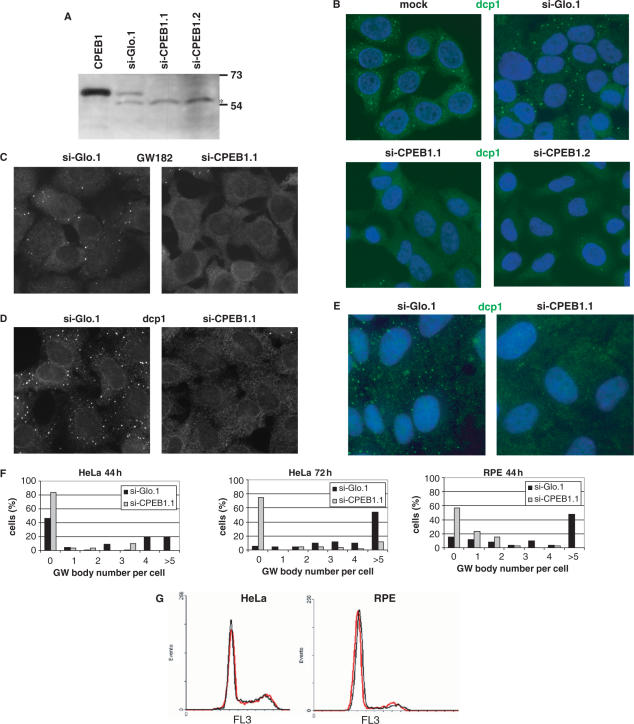
Figure 1.
CPEB1 silencing leads to the disappearance of GW bodies. (A) Silencing of CPEB1. HeLa cells were transfected with indicated siRNAs. After 44 h, proteins were extracted and 50 µg of this was analyzed by western blot with anti-CPEB1 antibody. Ten micrograms of proteins from HeLa cells transfected with a human CPEB1-long expression vector were used as a size control. Note that the antibody recognizes a non-CPEB1 protein (*) that attests an equal protein loading. The migration of molecular weight markers is indicated on the right. (B–E) Disappearance of GW bodies following si-CPEB1 transfection. HeLa (B– D) or RPE (E) cells were transfected with indicated siRNAs. Mock is a control transfection with siRNA buffer alone. After 44 h (B, C, E) or 72 h (D), cells were fixed and stained with anti-Dcp1 (B, D) or anti-GW182 (C) antibodies and observed by fluorescence microscopy. For (B) and (E), nuclei were stained with DAPI (blue). (F) Quantification of the disappearance of GW bodies. The number of GW bodies per cell, counted in 100 cells, is presented for the experiments illustrated in (B–E). (G) Cell cycle analysis. HeLa and RPE cells were transfected with si-Glo.1 (red) or si-CPEB1.1 (black) and cell cycle was analyzed by cytofluorimetry 48 h later.
IFN dosage was performed as previously described (22). Each sample was tested in duplicate and the dosage was repeated twice. Briefly, UPIL reporter cells were plated in 100 µl in a 96-well plate and cultured overnight following the addition of 25 µl of each culture medium sampled from siRNA-transfected cells. The standard curve was obtained by culturing the cells in the presence of serial dilutions of IFN-α2 at 100 000 units/ml. Luciferase activity was then measured using a Steady-Glo luciferase assay system (Promega, France).
Antibodies
Monoclonal anti-CPEB1 and anti-Kif15 antibodies were raised in our laboratory (4,23). Rabbit polyclonal anti-p54 and mouse monoclonal DM1 anti-α-tubulin were purchased from Bethyl Laboratories Inc. (Texas, USA) and Sigma Aldrich (France), respectively. The anti-GW182 human index serum was a kind gift from Theophany Eystathoy (University of Calgary, Alberta, Canada), the anti-hDcp1 rabbit antibody from Bertrand Séraphin (Centre de Génétique Moléculaire, Gif, France) and the anti-eIF3 goat antibody from John Hershey (University of California, Davis, CA). Secondary antibodies conjugated to rhodamine, FITC and horseradish peroxidase were purchased from Jackson Immunoresearch Laboratories (Immunotech, France). Monoclonal TS9 anti-CD9 and TS81 anti-CD81 antibodies (24) were obtained from Diaclone (Besançon, France).
Western blot Analysis
Cells were scraped in PBS, resuspended in a lysis buffer (50 mM Tris-HCl pH8, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40) supplemented with Complete Protease Inhibitor cocktail (Roche Diagnostics, France) and incubated on ice for 30 min. Soluble proteins were recovered after centrifugation at 15 000 g and 4°C for 10 min and quantitated by the Bradford method (Bio-Rad, France). Proteins were separated on a 7.5% polyacrylamide SDS-PAGE gel along with the Prestained Protein Ladder 10–180 kDa (MBI Fermentas, France) and transferred to a nitrocellulose Hybond-C-Extra membrane (Amersham Pharmacia, France). All incubations were then performed at room temperature. Non-specific protein-binding sites were blocked by incubation in PBS-T (PBS, 0.1% Tween-20) containing 5% (wt:vol) non-fat dry milk for 1 h. The membrane was successively incubated with primary antibody diluted in PBS-T containing 5% non-fat dry milk for 1 h, washed in PBS-T, incubated with horseradish-peroxidase-conjugated secondary antibody for 45 min and washed in PBS-T. Immune complexes were detected using the SuperSignal® Chemiluminescent Substrate detection reagent (Perbio Science, France), and quantified by densitometry of the X-Ray film. The membrane was dehybridized for 30 min by incubation in 50 mM Tris-HCl pH 6.7, 10 mM β-mercaptoethanol, 0.5% SDS at 52°C before a second hybridization.
Immunofluorescence
Cells were grown on glass coverslips and fixed in −20°C methanol for 3 min. Cells were incubated with the primary antibody for 1 h, rinsed with phosphate buffered saline (PBS), incubated with the secondary antibody for 30 min, rinsed with PBS and stained with 0.12 µg/ml DAPI for 1 min, all steps being performed at room temperature. Slides were mounted in Citifluor (Citifluor, UK) and observed on a Leica DMR microscope (Leica, Heidelberg, Germany) using a 63X1.32 oil immersion objective. Photographs were taken using a Micromax CCD camera (Princeton Instruments).
Flow Cytometry
For the quantification of CD9 and CD81 expression, each cell sample was divided in two. All steps were then performed on ice. Cells were rinsed twice in PBS containing 0.1% BSA and incubated in the presence or absence of the corresponding primary antibody for 20 min. Cells were rinsed twice in PBS 0.1% BSA and incubated in the presence of TRITC-conjugated anti-mouse antibodies for 20 min. After rinsing three times, samples were analyzed with a FACSscan (Becton Dickinson, USA) and populations were defined using the WinMDI software. The percentage of silencing was calculated using the position of the fluorescence peak in silenced cells with respect to unsilenced ones.
Cell cycle analysis following si-Eg5 transfection was performed after propidium iodide staining of the DNA. After precipitation overnight in 70% ethanol at −20°C, cells were resuspended in PBS containing 50 µg/ml propidium iodide and 20 µg/ml RNase A and incubated at 37°C for 30 min. Samples were analyzed with a FACSscan (Becton Dickinson, USA) and cell cycle partition was calculated using the WinMDI software.
RT-PCR
Total RNA was extracted using the SV Total RNA Isolation kit (Promega, France) and quantified by spectrophotometry. Reverse transcription reactions were performed with 1 µg RNA using random primers and Mu-MLV reverse transcriptase (Invitrogen, France). One-tenth of the reaction was used for PCR amplification with OAS2 primers [(GCTTTGATGTGCTTCCTGCCTT) and (ACCCCTTTGGCTTCAGTTTCCTT)], which amplified a 249-bp product or β-actin primers [(AGAGCTACGAGCTGCCTGAC) and (AGTACTTGCGCTCAGGAGGA)], which amplified a 300- bp product. PCR conditions were 35 cycles (94°C for 45 s, 60°C for 60 s and 72°C for 45 s) and 17 cycles (94°C for 30 s, 55°C for 30 s and 72°C for 30 s) for OAS2 and β-actin, respectively. The amplification products were separated on a 1.2% agarose gel, stained with ethidium bromide and quantified using a Fluor-S-Max imaging system (Bio-Rad, France). Each primer set targeted two separate exons in order to distinguish reverse transcribed mRNAs from residual genomic DNA.
RESULTS
Silencing of CPEB1 leads to the disappearance of GW bodies
We have previously shown the presence of the translational inhibitor CPEB1 in GW bodies. In order to gain insight into its function at that location, we used RNA interference to repress endogeneous CPEB1. HeLa cells were transfected with two siRNAs directed against human CPEB1 (si-CPEB1.1 and si-CPEB1.2), and the efficiency of the depletion was checked after 44 h by western blot using 4A2 monoclonal anti-CPEB1 antibody (Figure 1A). Because this antibody detects two bands in the 50–70 kD region, cells transfected with a human CPEB1 expression vector were used as a control of CPEB1 migration. A siRNA targeting rabbit beta-globin (si-Glo.1) was used as an irrelevant siRNA. Both si-CPEB1s led to the disappearance of endogeneous CPEB1 protein. In a parallel culture dish, cells were fixed and GW bodies were immunostained with anti-Dcp1 antibodies. The number of GW bodies was reduced after si-CPEB1.1 and si-CPEB1.2 transfection, compared to mock or si-Glo.1 transfected cells (Figure 1B). The same result was obtained using an antibody directed against GW182 as a second marker of the GW bodies (Figure 1C). This effect persisted 72 h after transfection (Figure 1D). We repeated the experiment in a second human cell line, RPE, and observed a similar effect (Figure 1E). The phenomenon was quantified by measuring the distribution of GW bodies per cell (Figure 1F). Compared to si-Glo.1, si-CPEB1.1 led to a similar decrease in HeLa and RPE cells, which intensified between 44 and 72 h. The effect was general rather than concerning a subpopulation of cells.
Because several targets of the CPEB1 regulatory pathway play a role in cell division during early development (Cyclin B1, Histone H3, kinesin Eg5, etc.) the depletion of CPEB1 protein could have an impact on cell proliferation. As the GW body number was reported to vary during the cell cycle, HeLa and RPE cells were transfected with si-CPEB1.1 or si-Glo.1 and stained 44 h later with propidium iodide in order to analyze the cell cycle by cytofluorimetry (Figure 1G). No difference was observed, indicating that the disappearance of GW bodies was not due to the disturbance of the cell cycle by CPEB1 repression. Taken together, these observations suggested that CPEB1 is required for GW body maintenance.
The disappearance of GW bodies is induced by a variety of siRNAs
Regulation by CPEB1 is only one among several translation regulation pathways. CPEB1 is an mRNA-binding protein known to be involved in the mRNA storage of a subset of specific mRNAs and, so far, not in mRNA degradation. It was therefore surprising that it could play a general role in GW body assembly. In order to clarify the significance of our observation, we tested two other siRNAs targeting CPEB1 and a set of 10 additional siRNAs targeting mRNA encoding proteins of various functions: (i) Rck/p54 DEAD-box helicase, which is a CPEB1 partner localizing to the GW bodies, (ii) the kinesin Kif15 and the kinesin-related protein HSET as examples of cytoplasmic proteins absent from GW bodies, (iii) the tetraspanins CD9 and CD81 as examples of transmembrane proteins and (iv) the rabbit β-Globin and the mouse Lymphotoxin-α (LT-α) as irrelevant controls, since the orthologous genes are not expressed in these cells. As a non-siRNA control, we also tested poly I/C, which is a double-stranded homopolymer of RNA supplied as a mixture of 100–600-bp-long molecules (data not shown).
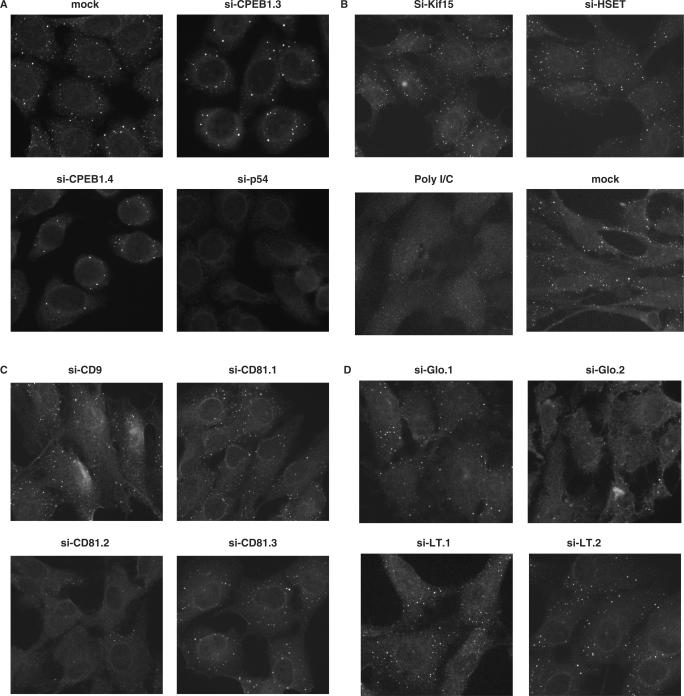
HeLa cells and RPE cells were transfected with equal amounts of these siRNAs and analyzed 48 h later. The pattern was identical in the two cell lines, with the number of GW bodies being either similar to what is seen in untransfected cells, or strongly decreased, depending on the siRNA. Si-CPEB1.3 and si-CPEB1.4 had little effect whereas si-p54 led to the complete disappearance of the GW bodies, as previously described (18) (Figure 2A). PolyI/C partially decreased the number of GW bodies (Figure 2B). In addition, one of the three siRNAs targeting CD81, si-CD81.2, and one of the irrelevant siRNAs, si-Glo.2, led to the disappearance of GW bodies (Figure 2C and D). Taken together, poly I/C showed an intermediate effect, while 5 of the 15 siRNAs tested led to a strong diminution of the GW bodies, with no obvious link with the function of their expected target: si-CPEB1.1, si-CPEB1.2, si-p54, si-Glo.2 and si-CD81.2.
Figure 2.
Various siRNAs lead to the disappearance of GW bodies. HeLa and RPE cells were transfected with indicated siRNAs. Cells were fixed after 48 h, stained with anti-Dcp1 antibodies and observed by fluorescence microscopy. Representative photographs from either HeLa (A) or RPE (B–D) cells are presented.
siRNA effect on GW bodies does not depend on their interference activity
Because some siRNAs are more potent in RNA interference than others, we investigated whether the effect on GW bodies correlated with the efficiency of silencing. In fact, si-CPEB1.1 and si-CPEB1.2 are active both in decreasing the GW body number and in depleting CPEB1 (Figure 1A), whereas si-CPEB1.3 and si-CPEB1.4 are inactive on both (Figures 2A and 3A). We therefore systematically measured the activity of the siRNAs, except for the irrelevant β-globin and LT-α siRNAs, which have no predictable mRNA targets expressed in HeLa or RPE cells.
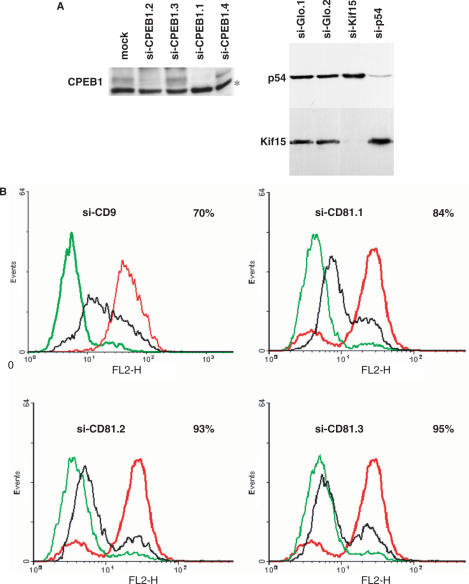
Figure 3.
si-RNA-mediated interference is efficient in the absence of GW bodies. (A) si-CPEB1, si-p54 and si-Kif15 silencing efficiency. HeLa cells were transfected with indicated siRNAs. After 48 h, proteins were extracted and 70 µg of this was analyzed by western blotting with anti-CPEB1 antibody (left panel) or 30 µg was analyzed with anti-p54 and anti-Kif15 antibodies, successively (right panel). (B) si-CD9 and si-CD81 silencing efficiency. HeLa cells were transfected with indicated siRNAs. After 48 h, cells were stained with anti-CD9 or anti-CD81 antibodies and analyzed by cytofluorimetry (in black). The background fluorescence (in green) was measured in the absence of primary antibody and the initial expression (in red) was measured after a transfection procedure with buffer alone. The percentage of silencing is indicated at the right hand corner of each panel.
The activity of si-Kif15 and si-p54 was measured by western blot 44 h after transfection (Figure 3A). Both siRNAs had similar silencing activity, whereas only si-p54 led to the disappearance of GW bodies. The activity of the siRNA targeting the tetraspanins was measured by cytofluorimetry using antibodies directed against CD9 and CD81 (Figure 3B). The four siRNAs were active on their respective targets, with 70% silencing efficiency for si-CD9 and 84–95% efficiency for CD81 siRNAs. Strikingly, the only siRNA which led to the disappearance of GW bodies, si-CD81.2, had the same silencing activity as the two other si-CD81s. This data is summarized in Table 1. There is clearly no correlation between the silencing activity of the siRNAs and their effect on the GW body number.
Table 1.
Silencing efficiency of the siRNAs and GW body number
| siRNA | Sequence | Silencing efficiency | GW body number |
|---|---|---|---|
| si-CPEB1.1 | CACCUUCCGUGUUUUUGGCUCTT | + | − |
| si-CPEB1.2 | CUCUCAGAUUUGAUUUCAATT | + | − |
| si-CPEB1.3 | GAAGGUUCAGAUUGACCCCTT | − | + |
| si-CPEB1.4 | CACCCUCAGUUAGAGGAUCTT | − | + |
| si-p54 | CCAAAGGAUCUAAGAAUCATT | + | − |
| si-Kif15 | GCACAACUCCUGCAAAUUCTT | + | + |
| si-HSET | CCAGCAGCTTCAGGACCAATT | + | + |
| si-CD81.1 | GCCCAACACCUUCUAUGUATT | + | + |
| si-CD81.2 | CACGUCGCCUUCAACUGUATT | + | − |
| si-CD81.3 | GCACCAAGUGCAUCAAGUATT | + | + |
| si-CD9 | GAGCAUCUUCGAGCAAGAATT | + | + |
| si-Glo.1 | GGUGAAUGUGGAAGAAGUUTT | n.a.* | + |
| si-Glo.2 | GGCUCAUGGCAAGAAGGUGTT | n.a.* | − |
| si-LT.1 | GCAGAACUCACUGCUCUGGUU | n.a.* | + |
| si-LT.2 | UAACCUGGAGCUCUCACGGUU | n.a.* | + |
| polyI/C | poly (I/C)100–600 | n.a.* | +/− |
*n.a. not applicable.
siRNA effect on GW bodies does not involve the interferon pathway
Having found that the effect of siRNAs on the GW bodies is related to their sequence, as not all of them decrease the GW body number, but neither to their expected target nor to their interference activity, as illustrated by the three si-CD81s, we hypothesized that they could activate a pathway distinct from the silencing machinery. Although siRNAs are short double-stranded RNA molecules 21 nt long, it has been reported that some of them do activate the interferon (IFN) pathway in certain cell lines in a sequence-dependent manner (25,26). We therefore considered this pathway as a candidate mechanism for the reduction of GW body number.
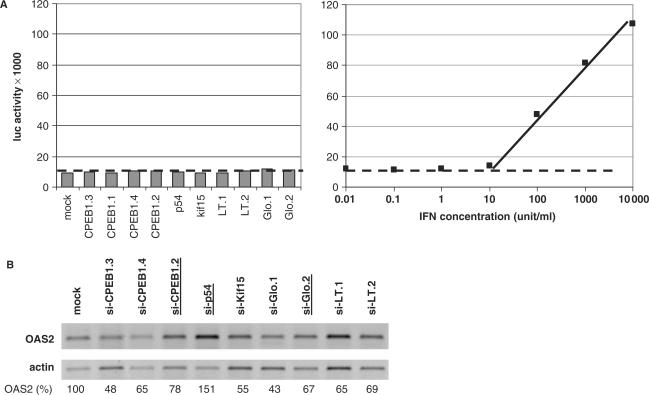
We first investigated whether IFN was produced in response to the transfection of the various siRNAs. RPE cells were transfected as described previously and the culture medium was sampled 48 h later. The presence of IFN was assayed using the UPIL5 reporter cell line which expresses luciferase under the control of an IFN responsive promoter (22). Despite the sensitivity of this assay, no IFN could be detected, indicating that its production, if any, is less than 50 units per ml (Figure 4A). In parallel, we analyzed the expression of OAS2 which has been shown to be one of the IFN-responsive genes that is the most induced by siRNAs (15-fold at 48 h in RCC1 cells, as reported in Sledz et al. (27)). RNA was extracted and the OAS2 expression was analyzed by RT-PCR along with a β-actin control. Overall, siRNA transfection rather led to a slight decrease of OAS2 expression with respect to β-actin (Figure 4B), including si-CPEB1.2 and si-Glo.2 and both of these decrease the GW body number. This suggested that the IFN pathway is not induced in these transfection conditions.
Figure 4.
IFN pathway is not activated following siRNA transfection. (A) Absence of detectable IFN in the culture medium. The cell culture medium was taken at 48 h and assayed on UPIL cells (left panel), along with an IFN standard (right panel). The luciferase activity is presented as a function of IFN concentration. The dotted line indicates the limit of sensitivity of the assay. (B) Absence of OAS2 induction. RPE cells were transfected with indicated siRNAs. After 48 h, OAS2 and actin mRNA were amplified by RT-PCR. The OAS2 to actin ratio, expressed as the percentage of the value in mock transfected cells, is indicated underneath each lane. siRNAs decreasing the GW body number are underlined.
Finally we analyzed directly the effect of IFN on GW bodies. HeLa cells were cultured in the presence of 103 u/ml IFN-α for up to 48 h and GW bodies were analyzed by immunofluorescence. At this time, the IFN treatment had no effect on the GW body number (data not shown). The full activation of the IFN pathway involves the presence of double-stranded RNA which, in our siRNA transfection experiments, could be provided by the siRNA itself. We used polyI/C as a conventional co-activator of the IFN response. Cells were cultured in the presence of 103 u/ml IFN-α for 40 h and then transfected with polyI/C in the presence of IFN-α. GW bodies were analyzed by immunofluorescence 8 h later, at a time where polyI/C transfection alone does not affect the GW body number. Even under these conditions, IFN had no effect on GW bodies (data not shown). In conclusion, we find no evidence for an involvement of the IFN pathway in the disappearance of GW bodies following siRNA transfection.
The disappearance of GW bodies does not affect RNA interference
The silencing machinery accumulates in GW bodies, including proteins of the RISC complex and siRNAs. Some of the GW body components are then required for the degradation of the cleaved RNA products. We therefore investigated whether the disappearance of GW bodies decreases the efficiency of RNA interference by siRNAs.
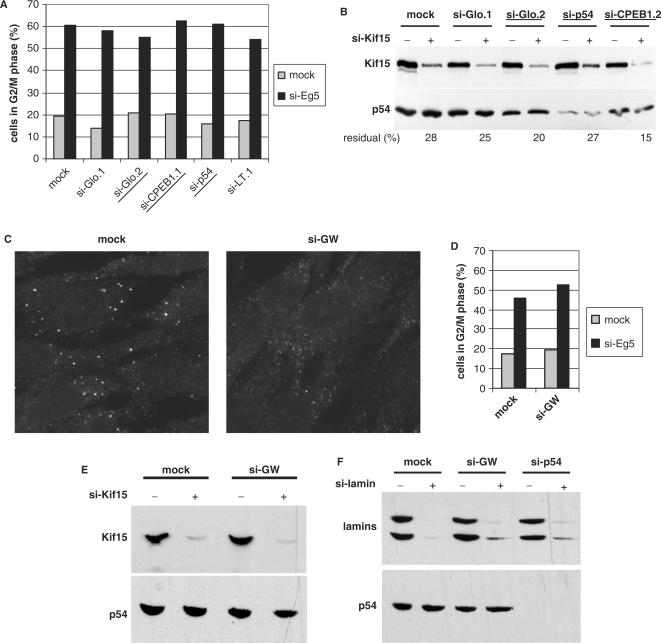
We first used si-Eg5 as a reporter of siRNA activity. The Eg5 kinesin is required for mitosis and its depletion leads to a phenotype of prometaphase arrest which can be quantified, as previously described (21). HeLa cells were mock transfected, transfected with si-Glo.1 or si-LT.1, which do not change the number of GW bodies, or transfected with si-Glo.2, si-CPEB1.1 or si-p54, which do decrease their number. After 40 h, cells were divided in two and transfected 8 h later with either buffer alone or si-Eg5. The cell cycle was analyzed 40 h later by cytofluorimetry after propidium iodide staining (Figure 5A). A G2/M blockage of about 60% was observed in all cultures, indicating that the si-Eg5 is as efficient in the absence as in the presence of GW bodies.
Figure 5.
Silencing is efficient in the absence of GW bodies. (A) Silencing of Eg5. HeLa cells were transfected with indicated siRNAs for 48 h and transfected again with buffer alone or si-Eg5. After 24 h the DNA content of the cells was analyzed by cytometry following propidium iodide staining. The percentage of 4n cells is plotted. siRNAs decreasing the GW body number are underlined. (B) Silencing of Kif15. RPE cells were transfected with the indicated siRNA for 31 h and transfected again with buffer alone or si-Kif15. Proteins were analyzed by western blot with anti-Kif15 and anti-p54 antibodies after 24 h. The percentage of Kif15 residual expression is indicated at the bottom. siRNAs decreasing the GW body number are underlined. (C) Disappearance of GW bodies following si-GW182 transfection. RPE cells were transfected with si-GW182 and analyzed as in Figure 2. (D, E) Silencing of Eg5 and Kif15 following si-GW182 transfection. Silencing was assayed as in (A) and (B). (F) Silencing of lamins. RPE cells were transfected with the indicated siRNA for 31 h and transfected again with buffer alone or si-lamin. Proteins were analyzed by western blot with anti-lamin antibodies after 24 h.
In case this phenotype was dependent on an Eg5 protein threshold and therefore not sensitive to small variations in silencing efficiencies, we performed a similar experiment in RPE cells using si-Kif15 as a reporter of siRNA activity and testing its efficiency directly by western blot analysis. RPE cells were mock transfected, transfected with si-Glo.1, which does not change the number of GW bodies, or transfected with si-Glo.2, si-p54 or si-CPEB1.1, which do decrease their number. After 24 h, cells were divided in two and transfected 7 h later with buffer alone or si-Kif15. Proteins were analyzed 24 h later by western blot (Figure 5B). At this time, Kif15 depletion was incomplete and similar in all samples, confirming that the silencing efficiency does not depend on the presence of GW bodies.
Similar experiments were previously reported using a siRNA targeting GW182 to decrease the GW body number, and subsequently testing interference of lamin A/C (8). The authors observed a strong inhibition of lamin silencing, suggesting the requirement of either intact GW bodies or GW182 for an efficient interference (8). To enable the comparison with these results, we investigated the effect of the same si-GW182 on Eg5 and Kif15 silencing. Despite a clear reduction of GW bodies (Figure 5C), we did not observe any significant inhibition of further silencing (Figure 5D and E). Neither did si-GW182 or si-p54 have any significant effect on the subsequent silencing of lamin A/C (Figure 5F), confirming that si-RNA-driven interference does not require intact GW bodies.
GW body disassembly is reversed by arsenite in a Rck/p54-dependent manner
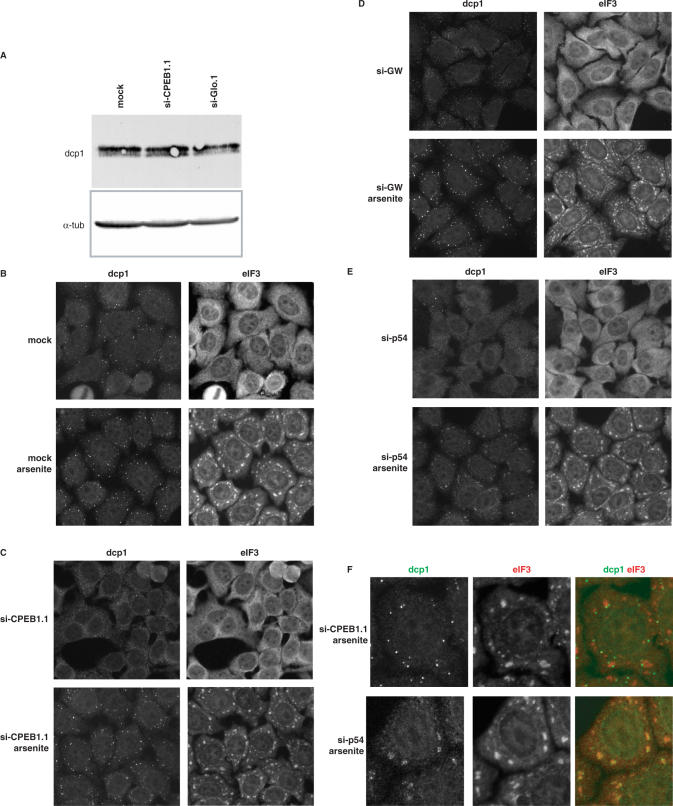
The disappearance of GW bodies could be due either to the degradation of its components or to their dispersal. To address this issue, Dcp1 protein was analyzed in HeLa cells 48 h after siRNA transfection (Figure 6A). The protein had the same abundance in cells transfected with si-CPEB1.1, which strongly decreases the GW body number, as in mock or si-Glo.1 transfected cells. This indicated a redistribution of GW body components toward the cytoplasm rather than a degradation.
Figure 6.
Disappearance of GW bodies is reversible. (A) Persistance of Dcp1 protein. RPE cells were transfected with the indicated siRNA for 48 h. Proteins were extracted and 60 µg of this was analyzed by western blotting with anti-Dcp1 and anti-α−tubulin antibodies, successively. (B–F) Reinduction of GW bodies by arsenite. HeLa cells were mock transfected (B) or transfected with si-CPEB1.1 (C), si-GW182 (D) or si-p54 (E) for 48 h, and treated or not with 0.5 mM arsenite for 30 min, as indicated. After fixation, cells were stained with a combination of anti-Dcp1 (left panel) and anti-eIF3 (right panel) antibodies and observed by fluorescence microscopy. A single cell from (C) and (E) is enlarged in (F) upper panel and lower panel, respectively, with Dcp1 and eIF3 staining in green and red, respectively.
In order to determine if the disassembly of GW bodies by siRNA is reversible, we investigated the effect of arsenite after siRNA transfection. Indeed, beside its role as an inducer of stress granules, arsenite has been shown to promote the assembly of GW bodies (4). HeLa cells were mock transfected or transfected with si-CPEB1.1, si-Glo.2, si-p54 or si-GW182 for 48 h and then treated or not with arsenite for 30 min. GW bodies and stress granules were analyzed by immunofluorescence using anti-Dcp1 and anti-eIF3 antibodies, respectively. Arsenite fully induced stress granule assembly in the five samples (Figure 6B–E and data not shown). The number of GW bodies strongly increased in mock transfected cells, as previously described, with 100% of the cells having numerous GW bodies (Figure 6B). The same increase was observed in si-CPEB1.1, si-Glo.2 and si-GW182 transfected cells (Figure 6C and D and data not shown). By contrast, only a few cells recovered GW bodies after si-p54 transfection (Figure 6D). Instead, the Dcp1 protein relocalized in the newly assembled stress granules (Figure 6E). The same results were obtained in RPE cells. This indicated that, in most cases, arsenite is sufficient to counteract the siRNA action on GW bodies. Interestingly, at the dose used, arsenite is a strong translation inhibitor (28). Therefore, the GW body reinduction occurs in the absence of neo-synthesized protein, confirming that neither the CPEB1 nor the GW182 protein are necessary for GW body assembly. However, the p54 protein is truly required for the GW body assembly, at least in response to arsenite.
DISCUSSION
GW bodies contain proteins involved in mRNA degradation, in RNA interference as well as in mRNA storage. CPEB1 is known as a specific translation regulator acting within a large multiprotein complex bound to the 3' untranslated region of its target mRNAs. We have used RNA interference to investigate its function in human cells and found that the depletion of the CPEB1 protein is accompanied by the disappearance of GW bodies. While we had previously shown that the CPEB1 protein accumulates in GW bodies, it seemed unlikely that the depletion of a specific regulator present at low abundance could induce GW body disassembly. We therefore tested a larger set of siRNAs, including a siRNA targeting Rck/p54, which is a CPEB1 partner also found in GW bodies, siRNAs targeting mRNAs encoding proteins that should not be involved in mRNA metabolism, such as kinesins Kif15 and HSET, and tetraspanins CD9 and CD81, which are transmembrane proteins expressed at the cell surface, as well as irrelevant siRNAs designed against mRNAs that are not expressed in the cells under study, such as β-Globin and LT-α. Out of 15 siRNAs, five shared this property of making GW bodies disappear: two targeting CPEB1 and three targeting Rck/p54, CD81 and β-Globin, respectively. The effect on GW bodies was therefore not restricted to the siRNAs depleting proteins obviously involved in mRNA metabolism.
The main parameter described so far to affect GW body number in mammals is the cell cycle, and the CPEB protein is clearly involved in mitosis (16). In Xenopus oocytes, CPEB-regulated genes include cyclins A and B, Mos and Aurora-A kinases, Eg5 kinesin, histone H4, all of which are involved in the resumption of meiosis (29). In mammals, cyclin B1 translation is subject to a similar regulation in the MCF7 cell line (30). The depletion of CPEB1 in HeLa and RPE cells could therefore lead to cyclin B1 misregulation and cell cycle abnormalities. However, in spite of a drastic depletion of the CPEB1 protein, we did not observe any change in the cell cycle, indicating either that CPEB1 is not responsible for cyclin B1 regulation in these cells or that it can be replaced by a functionally identical protein. Consequently, the disappearance of the GW bodies was not related to a particular accumulation in M or early G1 phase, which are the two cell cycle phases where GW bodies are sparse.
It has also been reported that the number of GW bodies decreases in 3T3 quiescent cells (16). RPE cells, which have been immortalized by expression of the telomerase, are highly sensitive to contact inhibition and their proliferation is down-regulated at confluence, yet the number of GW bodies increased in confluent cells compared to that in exponentially growing cells (data not shown). Similarly, the number of GW bodies increased at confluence in HeLa cells (compare 44- and 72-h panels in Figure 1F). Thus, confluence and quiescence enhance GW bodies in these cell lines, which is in agreement with the situation in yeast, where P-bodies increase in number and brightness with cell density (31). However, we observed no marked difference of cell density 44 h after transfection of the various siRNAs (data not shown).
The fact that an siRNA designed against β-globin led to the disappearance of GW bodies raised the possibility that it was due to off-target silencing. First, as there is no sequence similarity between the various siRNAs, they would be expected to silence different off-target genes, yet lead to a common phenotype, which seems very unlikely. Second, in the case of β-Globin and CPEB1 siRNAs, GW bodies could be efficiently reinduced in 30 min by arsenite, despite the fact that arsenite fully inhibits protein synthesis. This indicated that whatever proteins might be involuntarily depleted, they were not required for the GW body assembly. This strongly suggests that the disappearance of the GW bodies in these cases is not due to the depletion of a particular protein but rather to some deregulation of GW body genesis.
One potential consequence of introducing double-stranded RNA in mammalian cells is the activation of the IFN pathway. Although short molecules are less potent than long ones, siRNAs can nevertheless induce part of the IFN response. This has been reported following transfection of in vitro synthetized siRNAs (27), plasmid-encoded shRNAs (32) and chemically synthetized siRNAs (25,26). In the last case, some siRNAs were found to be more potent than others, depending on their sequence. Although these data were obtained in cells from the immune system, it could have provided an explanation for our observations. We tested this hypothesis by measuring both IFN in the culture medium of the transfected cells and OAS2 expression. Indeed OAS2 was reported to be highly induced 48 h after siRNA transfection in RCC1 cells (27). Production of IFN was undetectable and OAS2 mRNA was not induced after siRNA transfection, providing no evidence of IFN pathway activation. Moreover, IFN addition to the culture medium did not affect the GW body number.
In the literature, the role of GW bodies in RNA interference is still unclear. On the one hand, in one study, GW182 depletion using RNA interference simultaneously decreased the GW body number and the silencing by miRNAs and siRNAs (8). In other studies, GW182 depletion led to a clear inhibition of miRNA-mediated silencing but only to a slight inhibition of siRNA activity (5,9). On the other hand, two studies reported that cells lacking GW bodies could still perform efficient siRNA-mediated silencing. These cells were obtained either by stably depleting Drosha protein using shRNA (10) or by transiently silencing Lsm1 using siRNAs (11). The latter study also reported that Rck/p54 protein is associated to RISC but is not required for siRNA-mediated cleavage activity. Our data are in full agreement with these two studies, as no inhibition of siRNA activity was observed following the disappearance of GW bodies. We therefore conclude that RNA interference can take place outside of the GW bodies.
A possible explanation for our observations could be that, in certain cases, the large amount of transfected siRNA molecules saturates RISC and progressively clears it out of the GW bodies. Our functional data do not support such a saturation effect. First, the effect on GW body number is not related to the silencing efficiency of the siRNA, which might be expected to be related to its affinity for RISC. Second, the silencing of Eg5 or Kif15 remained intact after the disappearance of GW bodies, indicating no saturation of the silencing machinery. Alternatively, siRNAs could sequester, outside of the GW bodies, RNA-binding proteins that are not involved in silencing activity but that are required for GW body assembly. The arsenite experiment argues against this possibility, as a large number of GW bodies reappear in 30 min while protein synthesis is inhibited. Finally, a third hypothesis would be that some siRNAs alter mRNA metabolism through a mechanism still to be identified, leading to a decrease of mRNAs directed to GW bodies. These would be restored when translation is inhibited by arsenite.
Whatever be the explanation, it remains clear that the disappearance of GW bodies following siRNA transfection does not obligatorily indicate that the targeted gene is important for GW body assembly. RNA interference is now a straightforward strategy to investigate protein function in mammalian cells, and it has led to the conclusion that many proteins of the GW bodies are essential for their assembly: Ccr4, Rck/p54, Lsm1, eIF4ET (18), GW182 (16), Ge1 (19) and Rap55 (20). How so many proteins could all be required has not been much discussed, and our results open up the possibility that some of these proteins are not really essential for GW body assembly. Our data indicate that GW182 is not the GW body scaffold protein, as previously hypothesized (16). It can be noted that Lsm1 depletion by a genetic approach in yeast leads to more numerous P-bodies rather than to their disappearance (2,33); it would be interesting to use arsenite treatment to confirm its importance for GW body assembly. We found that depletion of Rck/p54 protein not only leads to the disappearance of GW bodies but also prevents the induction of GW bodies by arsenite. This protein is therefore strictly required for the assembly of GW bodies, at least in response to arsenite. This is reminiscent of the situation in yeast, where Dhh1 works redundantly with Pat1, and the double Dhh1/Pat1 mutation prevents P-body formation in response to glucose deprivation (34). Rck/p54 is an RNA-binding protein of the DEAD box helicase family, with multiple properties: it associates to Dcp1 and enhances decapping in yeast (35), it associates to RISC and is required for miRNA-mediated silencing in mammals (11) and it belongs to a translation repressing complex along with CPEB in Xenopus oocytes (36). It is an abundant protein which could play a scaffolding or remodeling role in GW bodies due to its helicase activity (37). Alternatively, it could be part of the machinery which drives quiescent mRNA to the GW bodies for storage and/or degradation.
Although GW bodies and stress granules are related mRNP in terms of both structure and composition, Rck/p54 silencing had absolutely no effect on the stress granule assembly. Interestingly, we found that Dcp1, which is normally localized in the GW bodies independently of arsenite treatment, relocated to stress granules in the absence of Rck/p54. We have previously reported a similar behavior when stress granules are induced by CPEB1 expression (4). These two data suggest that, when GW bodies are missing, stress granules can be a second default localization of Dcp1 protein. Our data, obtained in the same cellular context during arsenite treatment, establish that, despite strong structural and functional similarities between GW bodies and stress granules, their assembly follows distinct pathways.
ACKNOWLEDGEMENTS
We thank Christophe Lallemand for his help with IFN measurement, Eric Rubinstein and Claude Boucheix for their advice with tetraspanin interference and Michèle Ernoult-Lange and Linda Pritchard for critical reading of the manuscript. Florence Le Roy was supported by a fellowship from the Fondation pour la Recherche Médicale. This work was supported by the Ligue contre le Cancer, the Association pour la Recherche sur le Cancer and the Agence Nationale pour la Recherche. Funding to pay the Open Access publication charge was provided by Ligue contre le Cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 2.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 7.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 9.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 14.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 17.Lian S, Jakymiw A, Eystathioy T, Hamel JC, Fritzler MJ, Chan EK. GW bodies, microRNAs and the cell cycle. Cell Cycle. 2006;5:242–245. doi: 10.4161/cc.5.3.2410. [DOI] [PubMed] [Google Scholar]
- 18.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weil D, Garcon L, Harper M, Dumenil D, Dautry F, Kress M. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques. 2002;33:1244–1248. doi: 10.2144/02336st01. [DOI] [PubMed] [Google Scholar]
- 22.Lallemand C, Bayat-Sarmadi M, Blanchard B, Tovey MG. Identification of a novel transcriptional regulatory element common to the p53 and interferon regulatory factor 1 genes. J. Biol. Chem. 1997;272:29801–29809. doi: 10.1074/jbc.272.47.29801. [DOI] [PubMed] [Google Scholar]
- 23.Buster DW, Baird DH, Yu W, Solowska JM, Chauviere M, Mazurek A, Kress M, Baas PW. Expression of the mitotic kinesin Kif15 in postmitotic neurons: implications for neuronal migration and development. J. Neurocytol. 2003;32:79–96. doi: 10.1023/a:1027332432740. [DOI] [PubMed] [Google Scholar]
- 24.Charrin S, Le Naour F, Oualid M, Billard M, Faure G, Hanash SM, Boucheix C, Rubinstein E. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 2001;276:14329–14337. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- 25.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 26.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 28.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 30.Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 31.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 33.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase,Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]