Abstract
The expression of cellular proteins that play central roles in the regulation of cell growth and differentiation is frequently tightly controlled at the level of translation initiation. In this article, we provide evidence that the ETS domain transcription factor ELK-1 forms part of this class of genes. Its mRNA 5′ UTR is composed of a complexed mosaic of elements, including uAUGs, uORFs and RNA structure, that interplay to modulate ribosomal access to the ELK-1 AUG start codon. Superimposed upon this is the generation of two different 5′ UTRs via alternative splicing. The two spliced isoforms show altered cellular and tissue distributions and behave differently in polysomal recruitment assays in the presence of the drug rapamycin. We propose that repression is therefore the sum of a series of interplaying negative elements within the 5′ UTRs, a situation which may reflect the need for tight translational control of ELK-1 in different tissues and under changing physiological conditions.
INTRODUCTION
ELK-1 belongs to the ternary complex (TCF) subfamily of the ETS-domain transcription factors. It is a 428 amino acid (aa) protein (52 kDa) containing the four functional domains characteristic of all members of the TCF family. The TCFs are major nuclear targets for the RAS–MAPKs (mitogen-activated protein kinases) of the extracellular-signal regulated kinase (ERK) subfamily, and the closely related SAPK/JNK and p38MAPK stress-activated protein kinases. They therefore appear to act as an integration point for both growth and stress signals. In cultured cells, ELK-1 functions as a transcriptional activator via its association with serum response factor (SRF) in a ternary complex on the serum response element (SRE) of many immediate early genes (IEGs: e.g. c-fos. egr1, egr2 and pip92) (1).
De-regulation of ELK-1 expression has been associated with cancers of the prostrate (2) and breast (3), as well as certain T-cell malignancies (4). Mitogenic regulation of cell proliferation is accompanied by up-regulation of immediate early genes such as c-fos that play a critical role in activation of downstream AP1-dependent genes required for growth. With regards to breast cancer, recent studies on the MCF-7 cancer cell line have shown that 17β-Estradiol (E2) activates both the RAS–MAPK and the PI3K/AKT pathways. The former leads to phosphorylation of ELK-1 and the latter phosphorylation of the SRF, both of which activate the SRE in the c-fos proto-oncogene promoter (5,6). Both these pathways also impact directly on the translational read-out of the cell. However, the extent to which the E2-mediated altered growth phenotype was related to changes in the translation of the elk-1 mRNA (or other mRNAs encoding components of these signalling pathways) was not investigated.
Our lab is interested in the regulation of ELK-1 expression at the level of translation initiation, a step which is normally rate-limiting in protein synthesis. In the majority of eukaryotic mRNAs, translation begins with the binding of the eukaryotic initiation factor 4F (eIF4F) to the capped 5′ end, an interaction that is mediated via its eIF4E subunit. The eIF4F cap-binding complex is also composed of eIF4A, a member of the ‘DEAD-box’ family of RNA helicases, and eIF4G. Binding of eIF4F leads to recruitment of the ribosomal 43S preinitiation complex (containing the eIF2-GTP-tRNAMET ternary complex). The ribosome, and associated factors, is then thought to linearly scan the RNA unwinding intermolecular structure in an ATP-dependent fashion until an AUG codon is encountered and a 48S initiation complex forms. Codon–anticodon base pairing in the ribosomal P site is then thought to trigger hydrolysis of the eIF2 bound GTP, and joining of the 60S ribosomal subunit (7–9). This multi-component, coordinated series of events is strictly regulated and responds to both intra- and extra-cellular signals (10–12).
Very few studies have been performed on the translational regulation of the elk-1 gene. However, it was reported that AKT negatively regulated ELK-1 expression at the level of translation and that a region within the first 279 nt of the ELK-1 open reading frame (ORF) was necessary and sufficient for this control (13,14). These latter studies were performed using GST–ELK-1 fusion constructs; i.e. the 5′ untranslated region (UTR) of the elk-1 mRNA was removed and GST was fused N-terminally. However, the elk-1 5′ UTR exhibits many of the features associated with cellular mRNAs whose expression is tightly controlled at the level of translation initiation, including upstream open reading frames (uORFs) and predicted thermodynamically stable RNA structures positioned close to the cap. In addition, an alternatively spliced transcript with a 106-nt deletion within the 5′ UTR has been reported, opening the possibility that these alternatively spliced isoforms of the elk-1 mRNA could be linked to the translational read-out. Therefore, in this manuscript we have gone back and examined the role of these 5′ UTRs in the regulation of ELK-1 expression. The results provide intriguing insights into how the different elements identified, namely uORFs and RNA structure, impact on ribosomal access to the ELK-1 AUG initiation codon.
MATERIALS AND METHODS
Cell culture
HEK293T were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal calf serum (Brunschwig), 1% penicillin/streptomycin, in a humidified atmosphere containing 5% CO2. For the polysome analysis, cells were taken in the growing phase. They were hypertonically shocked by shifting to medium containing 300 mM NaCl for 50 min. The cells were then placed in normal isotonic medium for 30 min. When rapamycin (Rap) was used, 100 nM Rap (LC laboratories) or 0.01% DMSO (the negative control) was added during the hypertonic shock, 20 min before the transfer back to isotonic conditions. Rap and DMSO were kept on the cells throughout the recovery period.
Calcium phosphate-mediated DNA transfections were performed essentially as described in (15). All transfections were performed in triplicate. The activity of firefly (FLuc) and renilla luciferases (RLuc) in the lysates prepared from transfected cells were measured using the Dual-luciferase reporter assay system (Promega) and light emission was measured over 10 s using a TD-20/20 luminometer (Turner Designs).
Polysome gradient/RNA extraction
After treatment, cells were scraped into the culture medium and pelleted for 4 min at 800 r.p.m. The pellets were lysed for 15 min on ice in 100 mM KCl, 50 mM Tris–Cl pH 7.4, 1.5 mM MgCl2, 1 mM DTT, 1 mg/ml heparin, 1.5% NP-40, 100 μM cycloheximide, 1% aprotinin, 1 mM AEBSF and 100 U/ml of RNasin. Nuclei were pelleted by centrifugation, 10 min at 12 000 r.p.m. The supernatant was loaded on a 15–50% sucrose gradient (in 100 mM KCL, 5 mM MgCl2, 20 mM HEPES pH 7.4 and 2 mM DTT). Extracts were fractionated for 3 h 30 min at 35 000 r.p.m. at 4°C in a Beckman SW41 rotor. The gradient was analysed and the fractions collected with the ISCO UA-6.
RNA was isolated by adding the same volume of TriZol (Invitrogen) to the fractions. Samples were mixed and incubated for 15 min on ice. 0.3 volumes of chloroform was added. After centrifugation, the upper phase was collected and the RNA precipitated with 0.7 vol of isopropanol. The pellet of RNA was re-suspended in water.
RT–PCR
The RT–PCR was performed using the One Step RT-PCR kit (Qiagen) according to the manufacturer's instructions. The number of amplification cycles was first determined for each set of primers, and corresponded to the exponential phase of the different products.
DNA constructions
The bicistronics constructs were made by cloning three inserts into the pBS–KS+ multiple cloning site. The 5′ insert corresponding to renilla was generated by PCR from the pRL-SV40 Vector (Promega) with the primers (5′-GAGCTCGCTAGCCACCATGACTT-3′) and (5′-TCTCGAGCCCCTAGAATTATTGTTC-3′), and digested with SacI/XhoI. The central insert contained the 5′ UTRs of Elk-1 generated by PCR with the primers (5′-TGGGTCCATGGCTGGGGGAGTGC-3′) and (5′-GCTCTAGAGCTCGAGACATTGGGCTCCTCCTCCTCGGGCCCACGTGAGCTGTAGGGAAACGC-3′) with XhoI as the 5′ site and NcoI as the 3′ site. The 3′ FLuc insert was excised from pGL3-Basic (Promega) as an NcoI/XbaI fragment. The monocistronics constructs were generated by removing renilla. The ΔSL clones were made by removing a SmaI/BstUI fragment in the monocistronic constructs. All clones were transferred from pBS–KS to pEBS–PL (16) as SacI/Acc65I fragments. The ΔSL constructs expressed from the pEBS-PL vector retain 59 nt upstream of the first AUG codon (uAUG1) whose Kozak context also was unaltered. This expression vector was selected because its intron is positioned downstream of the cistrons. Consequently this eliminates the risk of generating a FLuc monocistronic transcript by alternative splicing (17).
The plasmid expressing the siRNA against RLuc (pBS/U6-RLi) and the empty vector control (pBS/U6ApaI) were kindly supplied by Dr Richard Llyod.
RNA structural probing within the 5′ UTR
The pBS 5′ UTRS clone was linearized with Acc65I (position 208 on the mRNA) and a T7 polymerase RNA run-off transcript generated. This was 3′ end labelled using 32P-pCp (Amersham) and RNA ligase (Amersham). The probe was purified from a 5% acrylamide–urea gel by passive diffusion. Structural probing studies were performed by titrating the RNase V1 (Ambion) and RNase T1 (Roche) precisely as outlined in the Ambion kit. Products were resolved on 5% acrylamide–urea gels and visualized by autoradiography.
RESULTS
Alternative spliced forms within the 5′ UTR
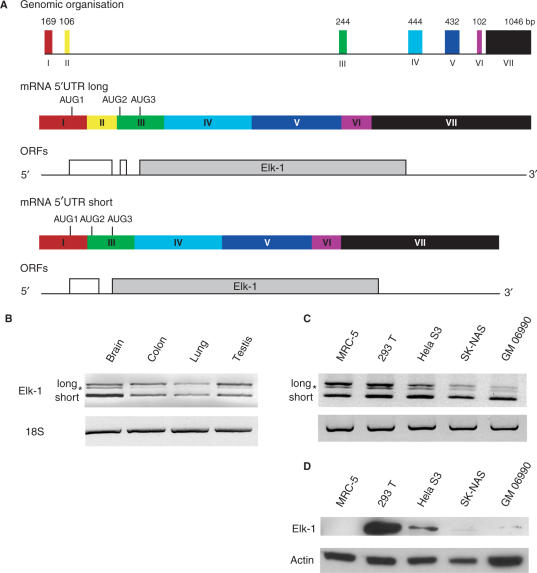
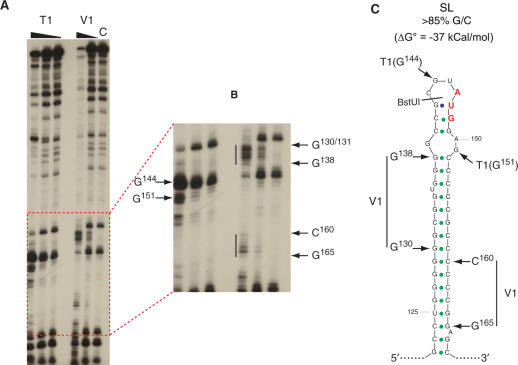
The 15.2-kb elk-1 gene spans seven exons (I to VII) and six introns, with the coding region encompassing exons III to VII (Figure 1A). However, an alternatively spliced form which removes the non-coding exon II was detected in a human hippocampus cDNA library (18). The longer form has a 5′ UTR of 346 nt [based upon the +1 position determined by (19); hereafter referred to as 5′ UTRL] and spans exons I to III. It contains two uORFs (Figure 1A, middle panel), a feature characteristic of many cellular messengers whose expression is tightly controlled at the level of translation initiation (20–22). RNA-folding programs predict considerable secondary structure. Indeed, within the first 168 nt (which are 75% G/C) there is a highly stable stem-loop (referred to hereafter as SL), with a ΔG° approaching −40 kcal/mol (Figure 2C). Earlier in vitro results indicated that such structural elements could serve as effective barriers to scanning ribosomes (23,24) and this was recently confirmed in live cell studies (20). The AUG1 is located within the loop of SL and its ORF (an ORF collinear with that of ELK-1) terminates after 54 codons. It is then followed by a second short uORF of only two codons (+1 relative to ELK-1), which terminates 17 nt before the ELK-1 AUG. The shorter transcript (5′ UTRS) is 240-nt long and is generated by the removal of the 106-bp exon II (Figure 1A, lower panel) (18). Like 5′ UTRL, it is G/C rich, and has conserved the SL and the two uAUGs. However, both are now in the same frame (i.e. there is now only a single uORF of 26 codons). RNA structural probing performed on the 5′ 208 nt (a region common to both spliced forms) clearly demonstrated V1 sensitive regions corresponding to the major part of the stem of SL and T1 sensitive sites within the loop regions (Figure 2). Therefore, the major features of the predicted fold exist in solution.
Figure 1.
Alternatively spliced isoforms of the human elk-1 mRNA. (A) The upper panel is a schematic representation of the elk-1 gene. The coloured rectangles depict the seven exons with their respective sizes (in bp) indicated above. Below is depicted the exon organization of the mature mRNAs carrying the 5′ UTRL and 5′ UTRS superimposed onto which is indicated the first three AUG codons. Below each mature mRNA is shown the ORF organization. (B) RT–PCR was performed on 50 ng of total RNA isolated from human tissues (Stratagene) using oligonucleotides that flanked the spliced exon II. As an internal control, a region of the 18S rRNA was also amplified. Products were analysed on agarose gels. (C) Similar RT–PCR analysis was performed on total RNA isolated from human cell cultures. The star indicates a DNA heterodimer composed of one long and one short chain generated during the amplification. (D) Immunoblot of total cell extracts analysed with ELK-1 specific (upper panel) and actin (lower panel) antibodies.
Figure 2.
RNA structural probing confirms the presence of SL. (A) RNA corresponding to the first 208 nts from the 5′ UTRS was transcribed in vitro and 3′ end labelled. Structure was probed using the RNase T1 and RNase V1 as outlined in Materials and Methods. The triangles above the gel indicate decreasing amounts of RNase (1U, 0.1U, 0.01U for T1, and 0.1U, 0.01U for V1). Products were resolved on a 5% acrylamide urea gel. The lane c refers to the non-treated RNA control. (B) The lower end of the gel has been expanded. The position of the bands indicated by arrows was determined using a combination of sequencing and alkaline hydrolysis ladders. The vertical lines indicate sequence regions sensitive to RNase V1. (C) The V1 and T1 sensitive sites have been superimposed on a minimum free energy prediction of the thermodynamically stable stem-loop (SL) conserved in both spliced variants. This was generated using the Vienna RNA-1.5 software (64). The position of the first uAUG (in red) and the BstUI restriction site used to generate the ΔSL mutants are also indicated.
We initially asked if we could detect these alternatively spliced transcripts in a range of human cell lines and human tissues. The elk-1 mRNA is generally expressed at very low levels and is difficult to detect in most cells even by RNase protection (data not shown). Therefore, we used semi-quantitative RT–PCR performed with oligos flanking exon II. To ensure the quantitative nature of this approach, we went to considerable effort to analyse the read-out during the linear phase of amplification (Supplementary Figure 1). Both long and short forms were readily detected (the intermediate band indicated by a star is actually a DNA heteromer composed of one light strand and one heavy strand generated during the PCR) (Figure 1B and C). In the tissue samples, the relative amounts of both isoforms were similar in colon, lung and testis (Figure 1B). However, the short form was major in the brain. Both forms were present at similar levels in the MRC-5 (lung fetal primary cells) and 293T (kidney) cell lines; however, the short transcript was dominant in HeLa S3 (cervical carcinoma), SK–NAS (neuroblastoid) and GM 06990 (lymphoblast) (Figure 1C). Attempts to perform real-time quantification proved frustrating, probably due to the extensive secondary structure around exon II. In addition, such an approach required the utilization of a primer specific for 5′ UTRS that traversed the exon I/III boundary, and one specific for 5′ UTRL (within exon II). The former in particular gave very non-reproducible read-outs. We next examined if the levels of elk-1 transcript correlated with intracellular protein levels. Immunoblots performed on extracts prepared from the five cell lines revealed no clear transcript:protein correlation (Figure 1D). Although ELK-1 was abundant in 293T cells and readily detectable in HeLa S3, it was barely visible in the SK–NAS and GM 06990 cells and totally undetectable in the MRC-5 cells (even when the blot was over-exposed). We also observed no difference in the ELK-1 protein half-life in 293T and HeLa cells (data not shown). These results indicate that ELK-1 protein levels can be regulated in a cell-specific manner at the level of translation.
The functional relevance of alternative 5′ UTRs
The 5′ UTR represents a key element in translational regulation. Therefore, despite the fact that both 5′ UTRL and 5′ UTRS retain the two uAUGs and the major structural elements, it is not inconceivable that the role of alternative splicing is to modulate the translational read-out. Such a scenario would be consistent with the variability of the two forms both in human tissue and cell lines. We decided to examine the translational behaviour of the endogenous transcripts. Translation initiation on messengers with long structured 5′ UTRs is considered to be highly sensitive to the activity of eIF4E (22,25). Therefore, we examined the recruitment of the elk-1 mRNAs onto polysomes in the presence or absence of the drug rapamycin (Rap). Rap is a lipophilic macrolide that binds mTOR preventing its signalling downstream to S6K1 and 4E-BP (26–28). This leads to the sequestration of eIF4E into an inactive complex with hypophosphorylated 4E-BP.
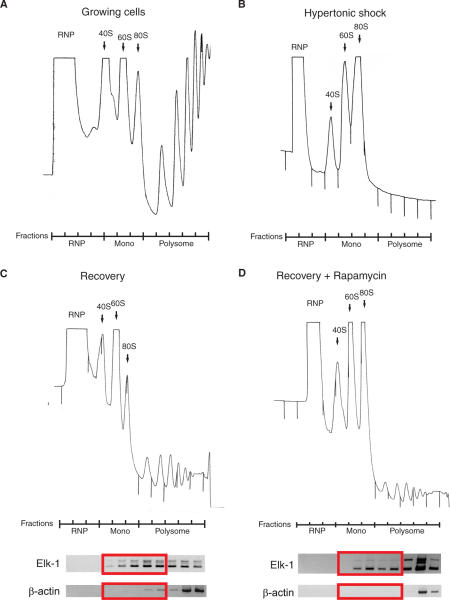
High salt (300 mM Na+) provokes a rapid inhibition of protein synthesis, disaggregation of polysomes (compare Figure 3A and B), dephosphorylation of eIF4E, 4E-BP1, rpS6 and an increased association of eIF4E and 4E-BP1 (29,30). Upon restoration of isotonic conditions these effects are reversed and the polysomal fraction reconstituted (Figure 3C). The technique is in effect an ex vivo competition experiment in which one creates a pool of free ribosomes stripped from their mRNAs by hypertonic shock then relieves this block and follows the ability of the endogenous mRNAs to compete. Using this approach, it is possible to examine what effect Rap has on the re-recruitment of RNA populations onto polysomes. In particular, one can directly examine the ability of the two endogenous elk-1 mRNA transcripts to compete for normal levels of eIF4E (Rap−), or limiting amounts (Rap+). In addition, by analysing the polysomal distribution rapidly after the restoration of isotonic conditions (i.e. 30 min) one can effectively eliminate the secondary effects of Rap on the cellular transcriptional program (31).
Figure 3.
Polysomal re-recruitment in the presence of rapamycin. Polysomal profiles were prepared from dividing 293T cells before (A) and after (B) hypertonic shock. Cells were then allowed to recover in the absence (C) or presence (D) of Rap. The gradients were fractionated and total RNA isolated. The position of the RNP, monosomal and polysomal regions of each gradient is indicated. RT–PCR was performed on the post-treatment samples using 50 ng of RNA per fraction with oligonucleotides specific for elk-1 (5′ UTR) and β-actin. The products were analysed by agarose gel electrophoresis (panels below each polysomal profile). The red rectangles indicate the monosome-light polysome fractions.
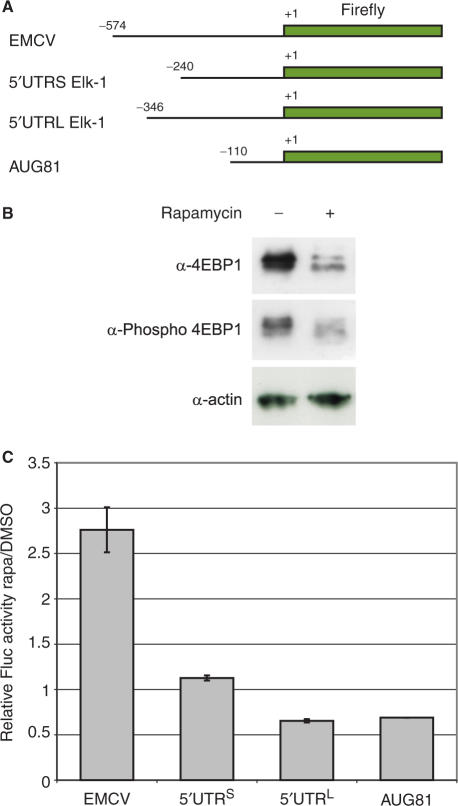
In 293T cells, 30 min after the restoration of isotonic conditions the polysomes had started to reform (Figure 3C). This process was clearly delayed in the Rap-treated cells (Figure 3D). We used RT–PCR to investigate how the two elk-1 transcripts had behaved on these gradients. Reactions were performed with 50 ng of total RNA per fraction. As a control, we followed the distribution of the β-actin mRNA (a house-keeping gene with a short 73-nt poorly structured 5′ UTR) (Figure 3C and D, lower panels). In cells treated with Rap, we observed that the 5′ UTRS was enriched in the ‘monosomal-light polysomal’ fraction of the gradient compared not only to the 5′ UTRL but also actin (Figure 3D, lower panel). This suggests that the 5′ UTRS elk-1 transcript is less sensitive to Rap and therefore preferentially re-recruited (and therefore proportionally enriched within the 50 ng of RNA). The fact that we do not see these effects in the heavy polysomal region of the gradient may reflect the short recovery time that was tested (i.e. the pattern in this region of the gradient essentially reflects the small fraction of mRNAs that remained heavy polysomal after the initial hypertonic shock). Nonetheless, these results showed a different behaviour of the 5′ UTRL and the 5′ UTRS. We decided to confirm this using an alternative approach. The two elk-1 5′ UTRs were fused to a FLuc reporter in the PolII expression vector pEBS-PL. As controls, we also fused the EMCV IRES and the 5′ UTR from the SeV P/C mRNA (AUG81: it is 110-nt long, has no detectable IRES activity, no uAUGs and little predicted structure) (Figure 4A) (32). These constructs were co-transfected into 293T cells with a vector expressing LacZ (pHR′CMV-LacZ), which served as an internal control against which FLuc activity was normalized. Cells were then treated or non-treated with Rap between 4 and 24 h post-transfection, and the effect of the drug confirmed with immunoblots against 4E-BP1 (Figure 4B). As expected, the EMCV-FLuc construct was resistant to the drug, consistent with its IRES activity. The fact that the AUG81-FLuc normalized values were <1 indicates that its 5′ UTR is more sensitive to Rap than that of the internal control (this may be a consequence of the controls shorter 5′ UTR). However, the 5′ UTRL gave normalized values nearly 2-fold lower than the 5′ UTRS, a result consistent with its increased sensitivity to the drug (Figure 4C).
Figure 4.
The 5′ UTRS confers rapamycin resistance. (A) Monocistronic FLuc constructs were constructed in the plasmid pEBS-PL with the 5′ UTRs indicated. These constructs were transfected in triplicate into 293T cells along with a reporter plasmid expressing LacZ that served as an internal control. At 4 h post-transfection, triplicate transfections were either treated or not treated with Rap. (B) The effect of Rap on downstream mTOR signalling was confirmed by immunoblots on cell extracts using 4E-BPI and phospho-4E-BPI antibodies. The blot for actin served as a loading control. (C) Cytoplasmic extracts were prepared at 24 h post-transfection. FLuc activity was measured and normalized to the internal LacZ control. The values obtained in the Rap-treated cells were then subsequently normalized to the values obtained in the non-treated control transfections. The graph depicts these FLuc activities (a value of ⩾1 indicates Rap insensitivity). The SEMs for each construct are also indicated.
No IRES, but cryptic promoter activity is associated with the 5′ UTRs
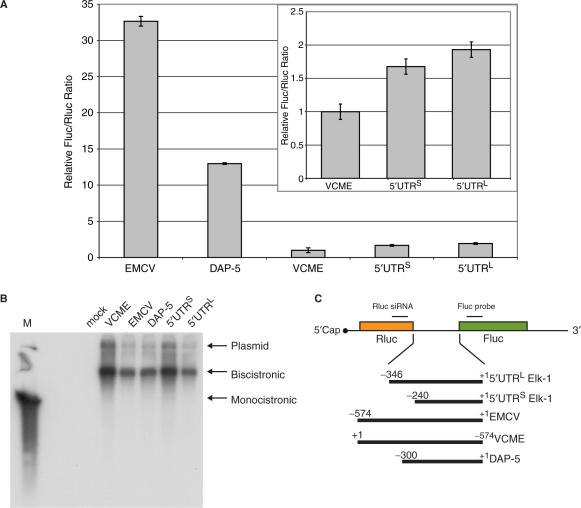
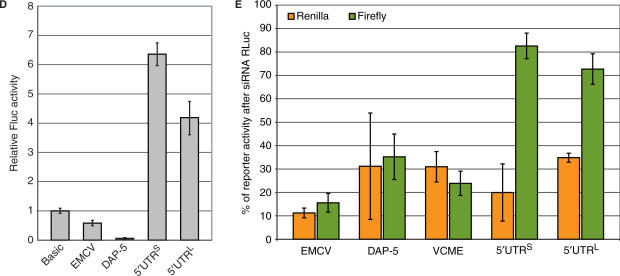
One possible source of eIF4E independent translational control mediated via the 5′ UTR (particularly those that are long, structured and contain multiple uAUGs) is an internal ribosomal entry site (IRES). IRES activity has been reported in the 5′ UTRs of cellular mRNAs that play key roles in growth control (33). We therefore tested for this activity using the now classical Rluc–FLuc bicistronic assay. The 5′ UTRL and 5′ UTRS were inserted into the intercistronic region of a bicistronic construct generated in pEBS-PL. As positive controls, we used the viral IRES of encephalomyocarditis virus (EMCV) and the 5′ UTR of Dap-5 (a reported cellular IRES). A negative control was provided by an inverted form of the EMCV IRES [referred to as RLuc-(VCME)-FLuc] (Figure 5C). Plasmids were transfected into 293T cells and reporter assays were performed at 24 h post-transfection (Figure 5A). Both EMCV and Dap-5 gave significant second cistron FLuc activity above the VCME negative control. Likewise, small but significant FLuc activity was detected with the elk-1 constructs. However, the bicistronic assay is in itself an insufficient test to unambiguously assign IRES activity (34–38). In addition, when these constructs were transfected into cells as bicistronic RNAs or translated in RRLs only the EMCV construct screened positive (data not shown). This opened the possibility that second cistron activity was derived from a shorter monocistronic construct. Northern blot analysis of total RNA from the transfected cells using either a FLuc or RLuc probe confirmed the presence of a single bicistronic transcript in all transfections (Figure 5B). However, this technique is considered relatively insensitive. We therefore decided to test for cryptic promoter activity by subcloning the (5′ UTRS)-FLuc, (5′ UTRL)-FLuc and (Dap-5)-FLuc regions into the promoter-less vector pGL3. Upon transfection into 293T cells both elk-1 5′ UTRs gave FLuc activity that was significantly higher than that observed in the negative control (pGL3-Basic FLuc) consistent with a promoter activity (Figure 5D). Reporter activity from the Dap-5 construct was consistently lower than the negative control. These results were confirmed by combining the pEBS-PL bicistronic constructs with a siRNA generated against RLuc as outlined in (34). The increased resistance of FLuc activity to a siRNA directed against RLuc in the bicistronic constructs carrying the 5′ UTRL and 5′ UTRS is consistent with the expression of a shorter monocistronic FLuc transcript (Figure 5E). The cryptic reporter activity derived from both 5′ UTRs is weak (we estimate 5% of the activity of an SV40 minimal promoter) explaining why a short monocistronic FLuc transcript was not detected by northern blot (Figure 5B). However, cellular IRESes are frequently reported to have activities significantly weaker than those of viral IRESes such as EMCV. In this context, weak promoter activity could have a major impact on the interpretation. Therefore, experiments failed to provide conclusive evidence for an IRES within either the 5′ UTRS or 5′ UTRL.
Figure 5.
No evidence for IRES activity within the 5′ UTRs. The elk-1 5′ UTRL, the elk-1 5′ UTRS, the EMCV IRES, VCME (the viral sequence inverted) and the 5′ UTR of Dap-5 were inserted between the RLuc and FLuc reporter genes in the plasmid vector pEBS-PL as depicted in (C). 293T cells were transfected in triplicate with each construct, and reporter activity was measured in cytoplasmic extracts 24 h post-transfection (A). The SEM is indicated for each plasmid clone tested. The insert magnifies the values for VCME (the negative control) and the two elk-1 5′ UTRs. Total RNA isolated from transfected cells was also analysed by northern blot (B). Ten micrograms of total RNA was resolved on a 1.2% agarose-HCHO gel, transferred to nitrocellulose and probed with a 32P-labelled DNA against FLuc. The lane M contains a monocistronic FLuc transcript generated by run-off transcription in vitro. Cryptic promoter activity was assayed by transferring the intercistronic regions into the promoterless vector pGL3 basic. 293T cells were transfected in triplicate with each construct and FLuc reporter activity assayed 24 h post-transfection (D) Each panel is representative of three independent experiments. The SEMs for each construct are also indicated. (E) The pEBS-PL bicistronic constructs were transfected in triplicate into 293T cells with either pBS/U6-RLi (a vector expressing an siRNA against RLuc) or an empty vector control (BS/U6ApaI). Cytoplasmic extracts were prepared at 48 h post-transfection and the FLuc (green bars) and RLuc (orange bars) activities in the presence of the siRNA were plotted as a percentage of the empty vector control. The SEMs for each construct are also indicated.
The role of uAUGs and uORFs
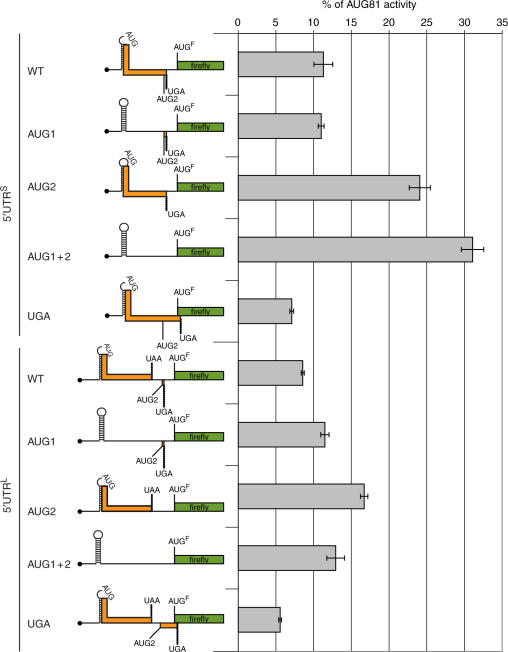
Since no IRES activity could be demonstrated within the 5′ UTRs we next analysed the role of the uAUGs as translational modulators. Although both UTRs retain two uAUGs (referred to as AUG1 and AUG2), the 5′ UTRL contains two uORFs whereas both AUGs are in frame in the short form (i.e. there is a single uORF that shares a common UGA stop codon with the second uORF in the 5′ UTRL: Figure 6). It has previously been reported that the two uORFs in the human oncogene mdm2 mRNA act synergistically to repress oncoprotein expression (39). Additionally, since ribosomes enter the elk-1 mRNA via the 5′ cap, the recognition of these uAUGs by the scanning pre-initiation complex and the fate of the ribosomes after termination of these uORFs (i.e. are they released from the mRNA or does a fraction continue to scan to subsequently reinitiate downstream?) become central elements in the regulation of ELK-1 expression (40). The question we therefore posed was ‘Does these alternative uORF organizations have a consequence for initiation at the downstream ELK-1 AUG codon?’ Starting with the (5′ UTRL)-FLuc and (5′ UTRS)-FLuc monocistronic constructs, we mutated AUG1/AUG2 both individually and together, as well as the UGA stop codon (Figure 6). Both 5′ UTRs functionally repressed reporter gene expression (∼12-fold for the 5′ UTRL and ∼11-fold for the 5′ UTRS compared with the AUG81 control). Within the context 5′ UTRS, the AUG1 change was largely neutral whereas AUG2 and AUG1 + 2 produced a marked and progressive increase in the reporter read-out which reached levels >30% of AUG81-FLuc. Curiously, the AUG1 + 2 double mutation in the context 5′ UTRL had a much more modest effect suggesting that uAUG-mediated repression may be less important in this background. Changing the UGA stop codon reduced reporter activity by ca. 2-fold for both 5′ UTRL and 5′ UTRS. This mutation extends the uORF so that it overlaps with that of FLuc by six codons (Figure 6). This drop in reporter activity suggests that a fraction of the initiation events at the ELK-1 start codon arise by re-initiation. Since it occurs in both 5′ UTR contexts, indicates that it derives from ribosomes that initiate at AUG2. Small uORFs (in this case only two codons) are thought to be favourable for re-initiation probably because the 40S subunit post-termination remains associated with the mRNA and retains many of the initiation factors that are normally lost progressively during the elongation phase (39,40).
Figure 6.
Mutational studies on the role of the uAUGs and uORFs in translational repression. Left-hand panel: A schematic representation of the 5′ UTRL-FLuc and 5′ UTRS-FLuc monocistronic constructs. The position of the uAUGs (AUG1 and 2) and the UGA stop codon that were targeted for the mutation studies are indicated. AUGF refers to the elk-1 AUG start codon that now initiates FLuc, and the presence of structure is indicated by the stem-loop. The ORFs are shown as coloured rectangles. Mutation of the UGA extends the uORF so that it now overlaps with that of FLuc. These clones were constructed in the PolII expression vector pEBS-PL. Right-hand panel: These plasmids plus an AUG81-FLuc control construct were transfected in triplicate into 293T. As an internal transfection control, a second LacZ plasmid (pHR′CMV-LacZ) was also added. Cytoplasmic extracts were prepared at 24 h post-transfection and the FLuc reporter activity was normalized to that of the internal LacZ control. All values are plotted as a percentage of the AUG81-FLuc control and SEMs for each construct are indicated. Each panel is representative of three independent experiments.
RNA structure within the 5′ UTR and shunting
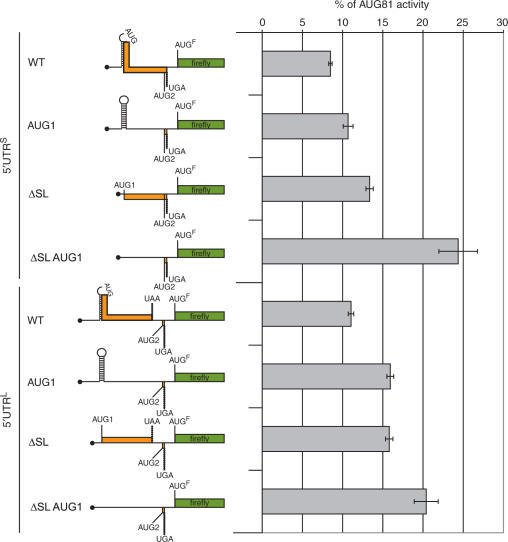
We have frequently evoked RNA structure as an element in the translational repression mediated by the 5′ UTRs. Indeed, the relatively modest effect of the AUG1 + 2 mutations within the 5′ UTRL led us to postulate that the structure was playing a central role in modulating the translational read-out. We decided to examine this directly by deleting all the structural elements in the FLuc monocistronic constructs described earlier (referred to as ΔSL). This leaves AUG1 in place with the same Kozak consensus and does not perturb the organization of the uORFs in both 5′ UTRL and 5′ UTRS (Figure 2C). When total RNA was isolated from cells transfected with these constructs and expressed in a rabbit reticulocyte lysate (RRL), the removal of the SL region had a marked positive effect on the reporter read-out (Supplementary Figure 2). In vitro systems are known to be highly sensitive to RNA structure close to the cap (23,24), and this result confirms that such elements exist in the two 5′ UTRs of elk-1. However, this change had only a very modest effect when the same constructs were assayed ex vivo (Figure 7). Why should the disruption of structural elements within the 5′ UTR have such a small effect? One possible answer arises from an examination of the structure. The SL fold positions the AUG1 in the loop region (Figure 2C), and the results from the ex vivo AUG mutation studies demonstrated that it is largely silent. We postulated that AUG1 was not always seen by the ribosome because SL promoted ribosomal shunting (or discontinuous scanning) thereby rendering it invisible (41–43). Thus in the ΔSL mutation the reporter read-out is the sum of the positive effect due to the removal of structural elements and the negative effect of the now accessible uAUG1. Therefore, mutation of AUG1 in the ΔSL background should now have a more marked positive effect on the reporter read-out. The ΔSL AUG1 double mutation was therefore introduced into the pEBS-PL 5′ UTRS-FLuc and 5′ UTRL-FLuc constructs and these were analysed by transient expression in 293T cells (Figure 7). The effect of the AUG1 mutation was clearly enhanced in the ΔSL background, particularly within the 5′ UTRS (ca. 2.5-fold). These results would suggest that at least a fraction of the scanning ribosomes bypass AUG1 by shunting, an event promoted by the stable SL (42). However, should SL be unfolded, the continued repression of ELK-1 protein levels would be assured by the uAUG1. A dual mode of pre-initiation complex displacement (i.e. linear scanning versus shunting) that responds to physiological signals within the cell has already been reported on an mRNA (43).
Figure 7.
Structural elements modulate the repressional activity of the uAUG1. Left- hand panel: A schematic representation of the uORF organization in the 5′ UTRS/5′UTRL-FLuc WT constructs and the AUG1, ΔSL and ΔSLAUG1 double mutants generated in the plasmid pEBS-PL. The presence of structure is indicated by the stem-loop as is the position of the uAUG1 in the loop of SL. Right-hand panel: Clones were transfected in triplicate into 293T cells along with an internal plasmid control expressing LacZ (pHR′CMV-LacZ). FLuc activity was normalized to β-galactosidase and values were plotted as a percentage of the AUG81-FLuc control. SEMs for each construct are indicated. WT refers to the non-mutated 5′ UTRs. Each panel is representative of three independent experiments.
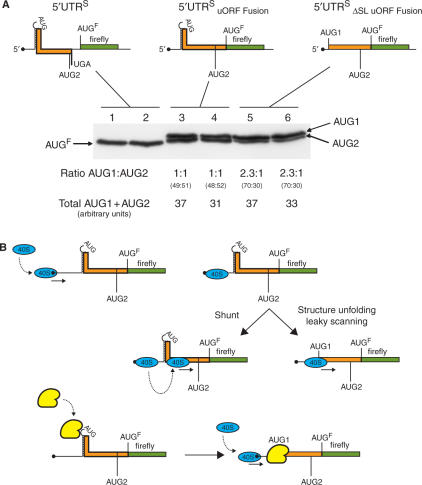
To independently confirm that ribosomal access to the uAUG1 is modulated by structure and to examine directly initiation events at the start codon, the following experiment was performed. Starting with the pEBS-PL constructs 5′ UTRS-FLuc and ΔSL 5′ UTRS-FLuc, the uORF was fused to that of the reporter (Figure 8A upper panels). These were transfected into 293T cells alongside a 5′ UTRS-FLuc control plasmid. Immunoblots performed on the 5′ UTRS-FLuc fusion transfected cell extracts produced two bands migrating slightly slower than the FLuc protein control, consistent with initiation events at the uAUG1 and uAUG2. These N-terminally extended FLuc products were expressed at equal amounts (lanes 3 and 4). However, upon deletion of the SL the protein product derived from uAUG1 became nearly 2.5-fold more abundant than that derived from uAUG2 (lanes 5 and 6). This confirms that structural elements around the uAUG1 render it less accessible to the scanning ribosome. Note also that the total amount of FLuc translation products expressed in both fusion constructs was essentially identical; indicating that at least in the 5′ UTRS background, ribosomes are very efficiently shunted past the upstream structural elements. Furthermore, the failure to detect a third band corresponding to initiation at the authentic AUG for FLuc in the 5′ UTRS fusions suggests that the FLuc protein detected in the non-mutated wild-type background (lanes 1 and 2) is the product of a re-initiation event after translation of the uORF.
Figure 8.
Ribosomal shunting renders the uAUG1 less accessible to the scanning ribosome. (A) The uORF in the pEBS-PL 5′ UTRS-FLuc and ΔSL 5′ UTRS-FLuc constructs was fused to the FLuc ORF as depicted schematically in the upper panel. These constructs, along with a pEBS-PL vector expressing the non-mutated 5′ UTRS-FLuc, were transfected in duplicate into 293T cells. Protein products containing the FLuc ORF were immunoselected with a rabbit polyclonal antibody and then resolved on a 12.5% SDS–PAGE gel. Proteins were revealed by immunoblotting using a FLuc mouse monoclonal antibody. The image was recorded on a Bio-Rad Fluor-S multi-imager and quantitated using the Bio-Rad Quantity One package. Values are indicated in arbitrary units below the gel. (B) Schematic representation of the events occurring during the scanning of the 5′ UTR. When ribosomes arrive at the RNA structural elements around the uAUG1 they can either melt the structure due to the eIF4A/4B helicase activity associated with the pre-initiation complex rendering the AUG1 visible, or bypass it by shunting. In the former scenario, the weak Kozak context of uAUG1 permits leaky scanning to downstream start sites. The lower panel depicts a situation in which the RNA structural elements are destabilized by a trans-acting cellular factor (yellow). Such factors could exhibit altered cell type distributions.
DISCUSSION
Generally, the rate-limiting step in translation initiation is the recruitment of the eIF4F complex to the 5′ cap because the eIF4E component is a limiting initiation factor in many cell types and is itself subject to regulation. Not all mRNAs compete equally for eIF4E, and this appears to be related in large part to the extent of RNA secondary structure within the 5′ UTR. Around 10% of mRNAs contain atypically long 5′ UTRs which frequently have the potential to form stable structures. These regions appear to serve as translational control elements in the expression of a number of proteins that are key players in the regulation of cell growth and differentiation (25,44,45). They function as thermodynamic barriers to the scanning ribosome and are most effective when positioned close to the 5′ cap (23–25). When 5′-proximal they probably also impede recruitment of the eIF4F cap-binding complex and hence the 40S ribosome. De-regulated expression of such mRNAs, due either to mutations within the 5′ UTR sequence itself or due to alterations in signalling pathways that impinge on the activity of the pre-initiation complex, can play a role in neoplastic transformation (44).
ELK-1 is a key player in the integration of mitogenic and stress-mediated signalling pathways, and its mRNA contains many features that would indicate tight translational regulation. An additional layer of complexity arises due to the presence of a shorter, alternatively spliced form of the 5′ UTR. The relative ratio of the two forms exhibits both cell type and tissue variability. Alternative splicing occurs in ∼35% of human genes, but is most frequently observed in the 5′ UTRs (46). Since such changes do not affect the coding potential they must play a role in regulating the read-out, an interpretation consistent with the key role of the 5′ UTR in ribosomal recruitment. Indeed, alternative splicing was reported to introduce a translational control element (a putative stem-loop) within the 5′ UTR of the human neuronal nitric-oxide sythase (nNOS) mRNA (47). Likewise, the coupling of tissue-specific alternative splicing with the translational read-out was reported for the human dicer gene (48), and the bovine growth hormone receptor (GHR) gene (49).
One attractive function for alternative splicing within the 5′ UTR is the generation, or the regulation of, an IRES. IRESes permit recruitment of the pre-initiation complex independent of the 5′ cap and therefore require a more limited set of initiation factors (in particular eIF4E). They have been reported on mRNAs whose expression continues when the activity of the eIF4F complex is compromised (e.g. during conditions of cellular stress or stages of the cell cycle). Alternative splicing coupled with the regulation of IRES activity has been reported for the mRNA encoding the neurotrophin receptor TrkB (50). However, although both the elk-1 mRNA 5′ UTRL and 5′ UTRS gave weak but positive read-outs in the bicistronic assay, this was readily attributable to cryptic promoter activity from the transfected DNA plasmids. It also explained the inability to detect ‘IRES-like’ activity when bicistronic RNAs were transfected into cells or translated in RRLs. Once again our results highlight the importance of using stringent controls when interpreting this type of assay, an observation already cited by others (34,38,51).
Having eliminated IRES elements within the 5′ UTRs we turned our attention to the role of RNA structure and the organization of the uORFs as elements modulating expression. The major secondary structural feature, namely SL, is conserved in both 5′ UTRs. This is highly G/C rich (85%), and with a predicted stability of −37 kcal/mol could be a significant barrier to the 40S scanning ribosome. Indeed, live cell studies have recently demonstrated that the steepest drop in translation efficiency occurred when stem-loop stability increased from −25 to −35 kcal/mol (20). The observation that the uAUG1 + 2 double mutant was much less effective at relieving repression in the 5′ UTRL (Figure 6) suggested that structure was a more important feature in global translational down-regulation in this background. Such a scenario would also explain the delayed re-recruitment of the endogenous 5′ UTRL elk-1 transcript onto polysomes in the presence of Rap (and hence limiting eIF4E). Nonetheless, the differences we observed between the two 5′ UTRs of the elk-1 mRNA was considerably more subtle than the >80-fold variation reported between the 5′ UTR spliced forms of the bovine ghr gene (49). This may arise because the uAUG1 is rendered invisible to the scanning ribosome due to shunting.
Both splice variants have retained the two uAUGs, and alignment shows that they are also conserved in the mouse elk-1 5′ UTR (data not shown). Not surprisingly, AUG codons are generally underrepresented in the 5′ UTR and their conservation across species is generally an indication of function (21). However, a noticeable difference between the two elk-1 transcripts is that although both have retained the two uAUGs, only the 5′ UTRL has two uORFs. Although multiple uORFs have been reported to synergistically repress translation (39), our studies failed to demonstrate any major differences in the two elk-1 5′ UTRs. However, mutation of both AUG1 and AUG2 had a more important effect in the 5′ UTRS background (reporter activities in the AUG1 + 2 double mutant ex vivo were more than 2-fold higher in 5′ UTRS than in 5′ UTRL), and mutation of AUG2 had generally a more important effect than AUG1. This may simply reflect the better ‘Kozak context’ of AUG2 (..GGGAUGG…...) relative to AUG1 (..CGUAUGG..), which will consequently sequester more of the scanning ribosomes. However, in vitro structural probing studies confirm that AUG1 is positioned in the loop of the highly stable SL, a configuration that has been reported to facilitate ribosomal shunting, a mechanism that would render AUG1 less accessible to the pre-initiation complex (20), and thereby reduce its activity as a repressor of downstream initiation (Figure 8B). Consistent with such a model was our demonstration that AUG1 served as a much more efficient initiation codon in the ΔSL background, and that its repressive effect was also more marked when the structural elements were disrupted.
The observation of an altered distribution of the two elk-1 mRNA isoforms in different tissues and cell lines, coupled with differences in sensitivity to Rap, suggests a 5′ UTR-mediated regulation of protein expression. However, in our ex vivo assays both 5′ UTRs were equally effective translational repressors. What then can be the basis of this regulation? Repression arises both as a consequence of RNA structure and uAUGs, recurrent features in the mRNAs that encode growth regulatory proteins (20–22,40). However, the relevant contribution of each of these elements to the repression phenotype may not be the same in the 5′ UTRL and 5′ UTRS. The uAUGs appeared to be more effective negative elements in the latter since their removal had a more marked effect on the reporter read-out (3-fold increase ex vivo as compared with ∼1.5-fold in 5′ UTRL; Figure 6). This led us to postulate that RNA structure may be a more important feature in the 5′ UTRL (assuming that repression is the sum total of the effect of uAUGs and global RNA structure). However, there is an important caveat in the interpretation of our ex vivo assays in that they were performed under optimal growth conditions. This does not reflect the physiological context, in which alterations in initiation factor activity occur. These changes have important consequences for the translational read-out. For example, the activity of eIF2 which forms part of the eIF2-GTP-tRNAMet ternary complex associated with the scanning ribosome is tightly regulated by cell growth and stress signals. Alterations in the eIF2
which forms part of the eIF2-GTP-tRNAMet ternary complex associated with the scanning ribosome is tightly regulated by cell growth and stress signals. Alterations in the eIF2 activity are known to modulate translational re-initiation, an event that appears to play a role in ELK-1 expression based upon the UGA mutation (Figure 6) (40). Re-initiation-mediated ELK-1 expression is probably coupled with ribosomal starts at the upstream AUG2. This small ORF, comprising only two codons, is conserved in many mammalian elk-1 mRNAs (e.g. human, mouse, rat, cow and dog). Additionally, the relevant amounts and activity of different key components of the initiation apparatus, in particular the eIF4 family (eIF4E, eIF4G, eIF4A, eIF4B), vary in a tissue-specific manner (52). Variations in the amount/activity of eIF4E (the cap-binding protein) and eIF4A/4B (the ATP-dependent helicase and its cofactor) will impact directly on the expression of mRNAs with structured 5′ UTRs, and will therefore have consequences for the translational expression of key cellular proteins. This in turn may impose variations in the 5′ UTR of genes whose expression must be tightly controlled in all environments, a scenario that could explain the variation in the ratio of the two spliced forms observed. For example, when eIF4E activity/amount is low, ELK-1 expression would arise mainly from the 5′ UTRS transcript since in this context the uAUGs rather than RNA structure are the major regulatory elements. Such a scenario would explain the preferential re-recruitment of the 5′ UTRS transcript onto polysomes in the presence of Rap, and hence reduced eIF4E activity. It is therefore not inconceivable that under certain physiological conditions, the differences in the activity associated with the two spliced variants may be more apparent than observed in our ex vivo assays. An additional order of complexity arises from the recent observation that other members of the DEAD-box helicase family can play a role in the selective translation of cellular mRNAs (53) (Figure 8B, lower panel). Many of these helicases have RNA chaperone activities and are key components of the splicing and nuclear export machinery. They provide a tantalizing explanation for how nuclear events, such as alternative splicing, can impact profoundly on the translational read-out (54–56), and may also impact on the altered polysomal recruitment of the two elk-1 transcripts. Finally, the cell lines we employed, namely 293T, are tumoural. However, most (if not all) tumoural cell lines have major perturbations in the pathways that regulate protein synthesis (a characteristic that contributes to the tumoural phenotype) (57–59), a background that may also serve to mask the functional differences between the two spliced variants.
activity are known to modulate translational re-initiation, an event that appears to play a role in ELK-1 expression based upon the UGA mutation (Figure 6) (40). Re-initiation-mediated ELK-1 expression is probably coupled with ribosomal starts at the upstream AUG2. This small ORF, comprising only two codons, is conserved in many mammalian elk-1 mRNAs (e.g. human, mouse, rat, cow and dog). Additionally, the relevant amounts and activity of different key components of the initiation apparatus, in particular the eIF4 family (eIF4E, eIF4G, eIF4A, eIF4B), vary in a tissue-specific manner (52). Variations in the amount/activity of eIF4E (the cap-binding protein) and eIF4A/4B (the ATP-dependent helicase and its cofactor) will impact directly on the expression of mRNAs with structured 5′ UTRs, and will therefore have consequences for the translational expression of key cellular proteins. This in turn may impose variations in the 5′ UTR of genes whose expression must be tightly controlled in all environments, a scenario that could explain the variation in the ratio of the two spliced forms observed. For example, when eIF4E activity/amount is low, ELK-1 expression would arise mainly from the 5′ UTRS transcript since in this context the uAUGs rather than RNA structure are the major regulatory elements. Such a scenario would explain the preferential re-recruitment of the 5′ UTRS transcript onto polysomes in the presence of Rap, and hence reduced eIF4E activity. It is therefore not inconceivable that under certain physiological conditions, the differences in the activity associated with the two spliced variants may be more apparent than observed in our ex vivo assays. An additional order of complexity arises from the recent observation that other members of the DEAD-box helicase family can play a role in the selective translation of cellular mRNAs (53) (Figure 8B, lower panel). Many of these helicases have RNA chaperone activities and are key components of the splicing and nuclear export machinery. They provide a tantalizing explanation for how nuclear events, such as alternative splicing, can impact profoundly on the translational read-out (54–56), and may also impact on the altered polysomal recruitment of the two elk-1 transcripts. Finally, the cell lines we employed, namely 293T, are tumoural. However, most (if not all) tumoural cell lines have major perturbations in the pathways that regulate protein synthesis (a characteristic that contributes to the tumoural phenotype) (57–59), a background that may also serve to mask the functional differences between the two spliced variants.
An earlier study on ELK-1 expression performed without the 5′ UTRs identified a region within the first 279 nt of the ORF that was responsible for translational down-regulation upon AKT activation (13). This region was not included in our studies. Since the AKT pathway directly impinges on eIF4E activity, via mTOR, it remains possible that this element and the 5′ UTRs may interplay in regulating ELK-1 expression (60–62). Additionally, a shorter form of ELK-1 generated from an internal AUG initiation codon was reported (sELK-1) (63). How the ribosome accesses this site (it is the seventh AUG codon on the mRNA), and the origin of its tissue specificity (it is found mainly in neuronal cells), remains unclear. The internal 279 nt AKT-responsive site did not appear to play a role in its expression. Nonetheless, we are currently investigating if this ORF element in the context of the different 5′ UTRs plays a role in modulating both ELK-1 and sELK-1 expression in response to growth/differentiation signals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The work was supported by grants from the Swiss National Science Foundation (No. 310000-116170) and the Ligue Genevoise Contre le Cancer. We would also like to acknowledge Dr Bob Hipskind and Dr Jocylene Caboche for supplying us with the ELK-1 starting material, Dr Richard Lloyd for the RLuc siRNA vector, Dr Jean-Dominique Vassalli and Dr Beatrice Conne for use of the gradient apparatus and the input of Dr Sylvain de Breyne and Dr Bernadino Conrad. Funding to pay the Open Access publication charges for this article was provided by the Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sharrocks AD. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 2.Brandlin I, Eiseler T, Salowsky R, Johannes FJ. Protein kinase C(mu) regulation of the JNK pathway is triggered via phosphoinositide-dependent kinase 1 and protein kinase C(epsilon) J. Biol. Chem. 2002;277:45451–45457. doi: 10.1074/jbc.M205299200. [DOI] [PubMed] [Google Scholar]
- 3.Xiao D, Qu X, Weber HC. GRP receptor-mediated immediate early gene expression and transcription factor Elk-1 activation in prostate cancer cells. Regul. Pept. 2002;109:141–148. doi: 10.1016/s0167-0115(02)00197-0. [DOI] [PubMed] [Google Scholar]
- 4.Rao VN, Huebner K, Isobe M, ar-Rushdi A, Croce CM, Reddy ES. elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science. 1989;244:66–70. doi: 10.1126/science.2539641. [DOI] [PubMed] [Google Scholar]
- 5.Duan R, Xie W, Burghardt RC, Safe S. Estrogen receptor-mediated activation of the serum response element in MCF-7 cells through MAPK-dependent phosphorylation of Elk-1. J. Biol. Chem. 2001;276:11590–11598. doi: 10.1074/jbc.M005492200. [DOI] [PubMed] [Google Scholar]
- 6.Duan R, Xie W, Li X, McDougal A, Safe S. Estrogen regulation of c-fos gene expression through phosphatidylinositol-3-kinase-dependent activation of serum response factor in MCF-7 breast cancer cells. Biochem. Biophys. Res. Commun. 2002;294:384–394. doi: 10.1016/S0006-291X(02)00499-0. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti A, Maitra U. Function of eukaryotic initiation factor 5 in the formation of an 80 S ribosomal polypeptide chain initiation complex. J. Biol. Chem. 1991;266:14039–14045. [PubMed] [Google Scholar]
- 8.Huang HK, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 10.Dever TE. Translation initiation: adept at adapting. Trends Biochem. Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- 11.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 12.Sonenberg N, Gingras AC. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell. Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 13.Figueroa C, Vojtek AB. Akt negatively regulates translation of the ternary complex factor Elk-1. Oncogene. 2003;22:5554–5561. doi: 10.1038/sj.onc.1206496. [DOI] [PubMed] [Google Scholar]
- 14.Galetic I, Maira SM, Andjelkovic M, Hemmings BA. Negative regulation of ERK and Elk by protein kinase B modulates c-fos transcription. J. Biol. Chem. 2003;278:4416–4423. doi: 10.1074/jbc.M210578200. [DOI] [PubMed] [Google Scholar]
- 15.Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamauchi T, Toko M, Suga M, Hatakeyama T, Isobe M. Structural organization of the human Elk1 gene and its processed pseudogene Elk2. DNA Res. 1999;6:21–27. doi: 10.1093/dnares/6.1.21. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann U, Brocke P, Dittmer J, Nordheim A. Characterization of the Human elk-1 Promoter. Potential role of a downstream intronic sequence for elk-1 gene expression in monocytes. J. Biol. Chem. 1999;274:1736–1744. doi: 10.1074/jbc.274.3.1736. [DOI] [PubMed] [Google Scholar]
- 20.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5′ untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 22.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis AE. Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell. Biol. 1999;31:73–86. doi: 10.1016/s1357-2725(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 26.Abraham RT. Identification of TOR signaling complexes: more TORC for the cell growth engine. Cell. 2002;111:9–12. doi: 10.1016/s0092-8674(02)01009-7. [DOI] [PubMed] [Google Scholar]
- 27.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 28.Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell. 2003;12:271–280. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Morley SJ, Naegele S. Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J. Biol. Chem. 2002;277:32855–32859. doi: 10.1074/jbc.C200376200. [DOI] [PubMed] [Google Scholar]
- 30.Naegele S, Morley SJ. Molecular cross-talk between MEK1/2 and mTOR signaling during recovery of 293 cells from hypertonic stress. J. Biol. Chem. 2004;279:46023–46034. doi: 10.1074/jbc.M404945200. [DOI] [PubMed] [Google Scholar]
- 31.Peng T, Golub TR, Sabatini DM. The immunosuppressant Rapamycin mimics a starvation-like signal distinct from amino acid and glucose eprivation. Mol. Cell. Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curran J, Kolakofsky D. Sendai virus P gene produces multiple proteins from overlapping open reading frames. Enzyme. 1990;44:244–249. doi: 10.1159/000468762. [DOI] [PubMed] [Google Scholar]
- 33.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 34.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene. 2003;318:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- 37.Bert AG, Grepin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1{alpha} and other cellular 5′ UTRs. RNA. 2006;12:1074–1083. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Dong Z, Han B, Yang Y, Liu Y, Zhang JT. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 2005;33:3763–3771. doi: 10.1093/nar/gki680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown CY, Mize GJ, Pineda M, George DL, Morris DR. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]
- 40.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curran J, Kolakofsky D. Scanning independent ribosomal initiation of the Sendai virus X protein. EMBO J. 1988;7:2869–2874. doi: 10.1002/j.1460-2075.1988.tb03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Futterer J, Kiss-Laszlo Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 43.Yueh A, Schneider RJ. Selective translation initiation by ribosome jumping in adenovirus- infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 44.van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell. Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 45.Stoneley M, Willis AE. Aberrant regulation of translation initiation in tumorigenesis. Curr. Mol. Med. 2003;3:597–603. doi: 10.2174/1566524033479474. [DOI] [PubMed] [Google Scholar]
- 46.Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newton DC, Bevan SC, Choi S, Robb GB, Millar A, Wang Y, Marsden PA. Translational regulation of human neuronal nitric-oxide synthase by an alternatively spliced 5′-untranslated region leader exon. J. Biol. Chem. 2003;278:636–644. doi: 10.1074/jbc.M209988200. [DOI] [PubMed] [Google Scholar]
- 48.Irvin-Wilson CV, Chaudhuri G. Alternative initiation and splicing in dicer gene expression in human breast cells. Breast Cancer Res. 2005;7:R563–R569. doi: 10.1186/bcr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang H, Lucy MC. Variants of the 5′-untranslated region of the bovine growth hormone receptor mRNA: isolation, expression and effects on translational efficiency. Gene. 2001;265:45–53. doi: 10.1016/s0378-1119(01)00356-0. [DOI] [PubMed] [Google Scholar]
- 50.Dobson T, Minic A, Nielsen K, Amiott E, Krushel L. Internal initiation of translation of the TrkB mRNA is mediated by multiple regions within the 5′ leader. Nucleic Acids Res. 2005;33:2929–2941. doi: 10.1093/nar/gki605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bert AG, Grepin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1{alpha} and other cellular 5′ UTRs. RNA. 2006;12:1074–1083. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 2005;122:865–876. doi: 10.1016/j.mod.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 54.Le HH, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 55.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holland EC. Mouse models of human cancer as tools in drug development. Cancer Cell. 2004;6:197–198. doi: 10.1016/j.ccr.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Holland EC. Regulation of translation and cancer. Cell Cycle. 2004;3:452–455. [PubMed] [Google Scholar]
- 59.Holland EC, Sonenberg N, Pandolfi PP, Thomas G. Signaling control of mRNA translation in cancer pathogenesis. Oncogene. 2004;23:3138–3144. doi: 10.1038/sj.onc.1207590. [DOI] [PubMed] [Google Scholar]
- 60.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khaleghpour K, Pyronnet S, Gingras AC, Sonenberg N. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol. Cell. Biol. 1999;19:4302–4310. doi: 10.1128/mcb.19.6.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanhoutte P, Nissen JL, Brugg B, Gaspera BD, Besson MJ, Hipskind RA, Caboche J. Opposing roles of Elk-1 and its brain-specific isoform, short Elk-1, in nerve growth factor-induced PC12 differentiation. J. Biol. Chem. 2001;276:5189–5196. doi: 10.1074/jbc.M006678200. [DOI] [PubMed] [Google Scholar]
- 64.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.