Abstract
The genome of influenza A virus consists of eight single-stranded RNA molecules of negative polarity. Their replication and transcription take place in the nucleus of infected cells using ribonucleoprotein complexes (RNPs) as templates. Two of the viral transcripts, those generated by RNPs 7 and 8, can be spliced and lead to two alternative protein products (M1 and M2, NS1 and NEP/NS2, respectively). Previous studies have shown that when expressed from cDNA, NS1 protein alters the splicing and transport of RNA polymerase II-driven transcripts. Here we used a transient replication/transcription system, in which RNP 8 is replicated and transcribed by recombinant RNA and proteins, to study the splicing and nucleo-cytoplasmic transport of true viral transcripts. Our results show that the encoded NS1 protein inhibits the splicing of the collinear transcript. This regulation is mediated by the N-terminal region of the protein but does not involve its RNA-binding activity. We also show that NS1 protein preferentially blocks the nucleo-cytoplasmic transport of the collinear RNP 8 transcript in an RNA-binding dependent manner. These results rule out previous models to explain the regulation of mRNA processing and transport by NS1 and underlines the relevance of NS1 protein in the control of virus gene expression.
INTRODUCTION
The influenza A viruses are members of the family Orthomyxoviridae with a genome consisting of eight single-stranded RNA segments of negative polarity. Each one is encapsidated by binding to the polymerase complex and to a number of nucleoprotein (NP) monomers (1), forming a ribonucleoprotein (RNP) complex (2). Transcription and replication of each virus RNP take place in the nucleus of the infected cell [reviewed in (3)] and hence viral mRNAs are potential substrates for the cellular splicing machinery and need to be exported from the nucleus to express the viral proteins. Indeed, the collinear transcripts from segments 7 and 8 can be directly expressed or can become spliced to generate M1 and M2 or NS1 and NEP(NS2) proteins, respectively (4–7). In productively infected cells, these splicing events are regulated in such a way that the steady-state ratios of spliced versus non-spliced mRNAs is only a few percent [reviewed in (8)]. These relative levels can be modified in non-productive cells (9) or by virus mutation (10) and are not constant along virus infection (11), suggesting the involvement of virus and cellular factors in the regulation of virus mRNA splicing.
The NS1 protein is a crucial regulatory factor during virus infection [reviewed in (12–14)], affecting cellular and viral gene expression and virus counteraction of the interferon response. It accumulates in the nucleus early during virus infection (15) and when expressed from cloned DNA (16–18), but it can be found in association with polysomes later in the infection (19,20). NS1 is a RNA-binding protein that interacts in vitro with dsRNA (21,22), vRNA (23,24), poly-A-containing RNAs (25) and U6 snRNA (26). The RNA-binding domain is located within the N-terminal half of the protein (24,27), and the rest of the protein appears to contain an effector domain (28). NS1 protein interacts with many viral and cellular factors. These include the virus RNP and/or polymerase (29), cellular proteins involved in translation, like hStaufen, PABPI and eIF4G (20,30–32) and cellular factors involved in post-transcriptional processing of RNA, like CPSF (33), PABPII (34) and NS1–BP, a potential splicing-related factor (35).
The splicing of NS1 mRNA has been studied in vitro. Early work indicated that it is a non-spliceable transcript under normal in vitro conditions (36), due the block after formation of a 55S pre-splicing complex (37). Subsequent mapping experiments led to the identification of RNA sequences within NS1 mRNA that may down-regulate its splicing efficiency (38). In vivo studies have also been carried out to analyze the splicing of NS1 mRNA. When expressed from a RNA PolII-driven cDNA it can be spliced (16,39) and its splicing can be blocked in trans by the expression of the encoded NS1 protein (40,41).
In addition of the splicing inhibition, PolII-driven expression of NS1 from a cDNA led to a general retention of mRNAs in the nucleus (25,28,41). The interaction of NS1 with the CPSF polyadenylation factor (33) suggested a mechanism for this transport block, as NS1–CPSF interaction would inhibit the 3′-processing of cellular transcripts. Furthermore, it was proposed that CPSF interaction would indirectly down-regulate the splicing of cellular, but not viral, single-intron containing transcripts (42).
The main limitation of these in vivo studies is that NS1 expression was carried out by transfection and the ‘viral’ mRNAs analysed are indeed PolII-driven transcripts. To avoid these problems we have established a transient virus replication-transcription system in vivo in which a recombinant NS RNP is amplified and transcribed by the viral polymerase. Using this more physiological system we show here that NS1 protein down-regulates the splicing and block the nuclear export of its own viral mRNA. Furthermore, we report a genetic analysis of the roles of the various NS1 domains for such regulation.
METHODS
Biological materials
The origin of the HEK293T cells (43) has been described previously (44). The cells were cultivated as described (45). The pHH plasmid containing the NS cDNA of A/Victoria/3/75 strain RNA under control of the polI promoter has been described (44). The antisera specific for NS1 and NEP(NS2) proteins were generated by hiperimmunization of rabbits or rats with purified proteins.
In vitro mutagenesis and plasmid constructions
To generate mutant plasmid pHHΔNS1, a site-directed mutagenesis was carried out on plasmid pHHVicNS (46) using the QuickChangeTM kit from Stratagene and the following mutagenic oligonucleotides: NS1mut1 (5′GCTTCCTTTGGCATGTCTGATAACAAATTGTAGACCAAGAACTAGG3′) and NS1mut2 (5′CCTAGTTCTTGGTCTACAATTTGTTATCAGACATGCCAAAGGAAGC 3′). In this way, two successive stop codons were introduced in the NS1 reading frame within the intron sequence. Likewise, a NS1 mutant plasmid was generated in which RNA binding is abolished (pHHVicNSR38A/K41A) (22,47), using the following mutagenic oligonucleotides: NS1mut3 (5′TCGGCTTCGCGCAGATCAGGCGTCCCTAAGG3′) and NS1mut4 (5′CCTTAGGGACGCCTGATCTGCGCGAAGCCGA3′).
Protein analyses
For protein labelling in vivo, cultures were washed, incubated for 30 min in a DMEM medium lacking met-cys and labelled for 1 h at 36 hours post-transfection with 35S-met-cys to a final concentration of 200 μCi/ml (48). Total extracts were prepared in Laemmli sample buffer and processed by polyacrylamide gel electrophoresis and autoradiography.
Immunoprecipitation was carried out as described (44). Protein-A-Sepharose immune matrices were prepared with rabbit anti-NS2 IgG and were incubated with soluble cell extracts prepared in 100 mM NaCl-1 mM EDTA-50 mM Tris–HCl-1% NP40, pH 7.5. After washing with the same buffer, the bound material was eluted in Laemmli sample buffer and processed by polyacrylamide-gel electrophoresis and autoradiography.
For immunofluorescence, transfected or mock-transfected HEK293T cells were washed with PBS, fixed for 20 min with 3% paraformaldehyde at room temperature and permeabilized with 0.5% Triton X100 for 5 min. The cells were blocked with PBS-3%BSA and incubated with rat anti-NS1 serum diluted in PBS-0.1% BSA, for 1 h at room temperature. After washing with PBS, the bound antibodies were revealed with FITC-conjugated goat anti-rat antibodies, by incubation for 1 h at room temperature. Finally, the cells were washed with PBS and the preparations were mounted with Mowiol. Images were obtained using a Leica fluorescence microscope.
For western blot analysis, cell extracts were separated by polyacrylamide–SDS gel electrophoresis and transferred to Immobilon filters. The filters were blocked for 1 h at room temperature with PBS-3% BSA and incubated with the primary antibodies diluted in 0.1% BSA-0.05% Tween-20 in PBS, for 1 h at room temperature. After washing four times for 15 min with 0.25% Tween-20 in PBS, the filters were incubated with a dilution of goat anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase in 0.1% BSA-0.05% Tween-20 in PBS. After washing in PBS, the filters were developed by enhanced chemiluminiscence.
RNA analyses
The transfected or mock-transfected cells were washed with PBS and collected by centrifugation for 5 min at 2500 r.p.m. The isolation of total RNA was carried out using ULTRASPEC RNA Isolation Reagent (Biotecx, Houston, TX, USA) according to the manufacturer's instructions. For cell fractionation, the cultures were resuspended in isotonic buffer (150 mM NaCl-1.5 mM MgCl2-10 mM Tris–HCl-0.5% NP40-2mM VRC-1 mM DTT, pH 8.5) and the nuclei were separated by centrifugation for 5 min at 3000 r.p.m. and 4°C. After an additional wash in the same buffer, the supernatants were pooled as cytoplasmic fraction and the pellet was used as nuclear fraction. The extraction of RNA from the nuclear fraction was carried out as indicated above for total RNA. For RNA extraction from the cytoplasmic fraction it was incubated with proteinase K (1 μg/ml) in 100 mM NaCl-50 mM Tris–HCl-5 mM EDTA-0.5% SDS, pH 7.5) for 1 h at 37°C, extracted with phenol–chloroform–isoamylalcohol and recovered by ethanol precipitation. The nucleic acids were digested with RNAse-free DNAse (1 U/μg RNA) and further extracted with phenol-chloroform-isoamylalcohol and precipitated with ethanol as above. Quantitation of ribosomal RNAs and ribosomal RNA precursors in the subcellular fractions indicated that around 80% of total cell rRNAs were present and no rRNA precursors could be detected in the cytoplasmic fraction.
Purification of polyA-containing RNA was carried out by oligo-dT cellulose chromatography. RNA samples were resuspended in 0.5% sarkosyl in DEPC-treated H2O and boiled for 3 min. After dilution in 0.5 M NaCl-10 mM Tris–HCl-0.5% sarkosyl, pH 7.5, the samples were applied to the column and incubated for 1 min. The flow-through was re-applied again and the column was washed with the same buffer. The polyA-containing RNA was eluted in two steps by washing first with 0.5% sarkosyl and then with DEPC-treated H2O. Finally, the polyA-containing RNA obtained was boiled again and used as input for a second step of oligo-dT cellulose chromatography as above, to ensure the removal of viral genomic RNAs complementary to the virus mRNAs. To control the recovery of the oligo-dT cellulose chromatography a known amount of a polyA-containing labelled riboprobe was added and the radiactivity present in the various fractions was determined. An average recovery of 75% was obtained in the experiments carried out.
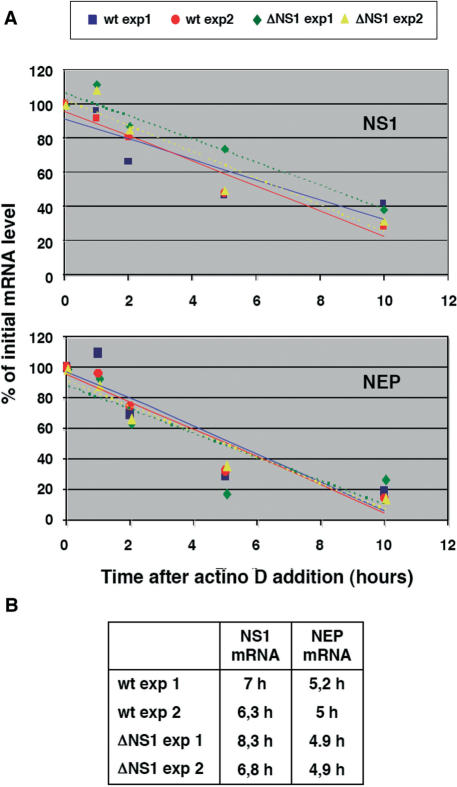
To determine the stability of NS1 and NEP(NS2) mRNAs, actinomycin D (50 μg/ml) was added to the transfected cells at 36 h post-transfection and total RNA was isolated at various times thereafter. PolyA-containing RNA was purified and the accumulation of NS1 and NEP(NS2) mRNAs was determined by RNAse-protection assay.
For RNAse-protection assay, the polyA-containing RNA samples were co-precipitated with a molar excess of labelled riboprobe and resuspended in 400 mM NaCl-40 mM Pipes-5 mM EDTA-80% deionised formamide, pH 6.4. After denaturation for 5 min at 85°C, hybridization was carried out overnight at 50°C. The mixture was diluted in a RNAse solution containing 300 mM NaCl-5 mM EDTA-10 mM Tris–HCl, pH 7.5, 50 μg/ml of RNAse A and 1 u/μl of RNAse T1 and incubated for 2 h at 37°C. After proteinase K/SdS treatment as indicated above, phenol extraction and ethanol precipitation, the samples were resuspended in formamide loading buffer and analysed by polyacrylamide-urea gel electrophoresis and autoradiography. Quantitation was performed in a phosphorimager.
RESULTS
A transient replication–transcription system to analyse virus mRNA processing and transport
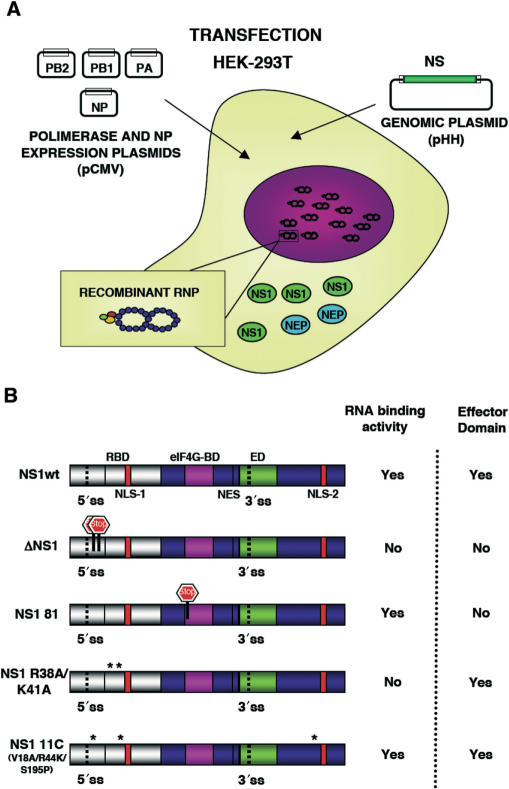
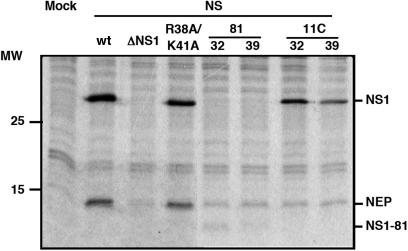
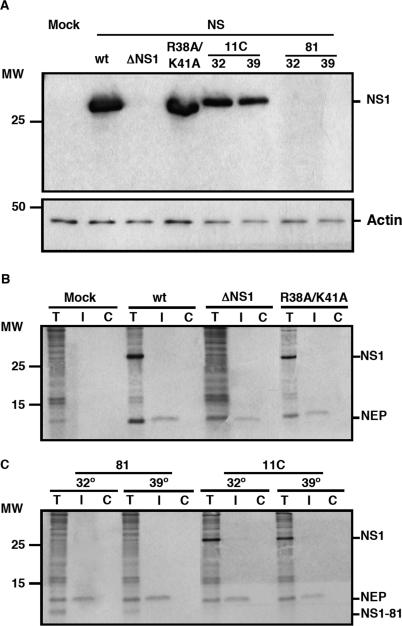
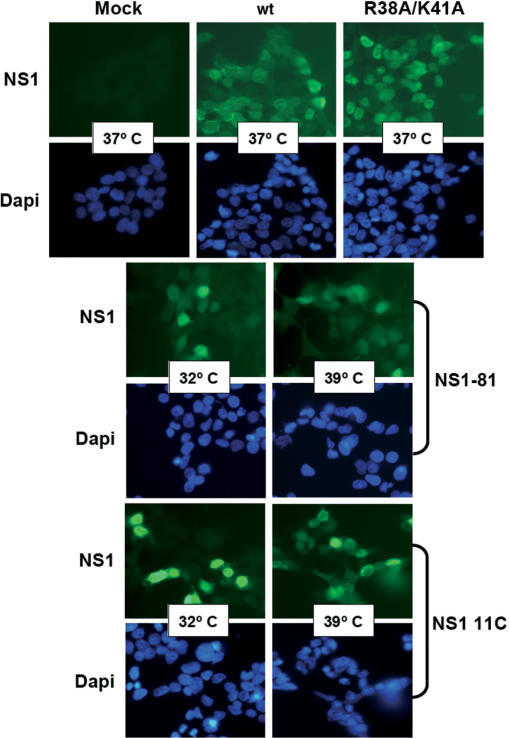
The aim of this work was the analysis of the splicing and nucleo-cytoplasmic transport of influenza virus mRNA, independent of other virus processes but nevertheless in a virus-like environment. To this aim we set up a transient viral replication–transcription system in which the generation of the viral transcripts would be driven by the normal virus machinery. As previous work of several groups has implicated NS1 protein in the control of mRNA maturation and transport, we expressed the polymerase and NP by RNA PolII-driven transcription and a segment 8 genomic RNA under control of an RNA PolI promoter (Figure 1A). In this way, the consequences of NS1 expression on the splicing and nucleo-cytoplasmic transport of segment 8-specific mRNAs could be analysed under conditions more similar to a virus infection. Indeed, when the synthesis of NS proteins was determined in cultures transiently replicating and transcribing a recombinant NS replicon a clear signal for both NS1 and NEP(NS2) proteins was observed (Figure 2; wt) and the accumulation of NS1 and NEP(NS2) proteins were verified by western-blot or immunoprecipitation using specific antibodies (Figure 3A, B; wt). Both the synthesis and accumulation levels were similar to those obtained in a normal infected cell (data not shown). This experimental setting allowed the genetic analysis of the implication of NS1 protein in viral mRNA maturation and transport by simply mutating the replicon included in the system. Thus, a number of altered replicons in which NS1 had been mutated or replicons derived from NS1 mutant viruses (44,46) were prepared and their genotype is described schematically in Figure 1B. Mutant ΔNS1 contained two stop codons close to the N-terminus of the protein but 22 nt away from the splicing 5′-donor site, leading to the synthesis of a 18 amino acids long peptide. Accordingly, neither synthesis nor accumulation of NS1 protein was observed in the system with this replicon (Figures 2 and 3A), while the synthesis of NEP(NS2) protein was still detectable (Figures 2 and 3B). Likewise, NS1 mutants lacking the effector domain (NS1 81) or devoid of RNA-binding activity (NS1 R38A/K41A) and mutants with ts phenotype (NS1 81 and NS1 11C) could be expressed in the system and showed expression levels according to the described phenotypes (Figures 2, 3A–C) (44,46). Furthermore, immunofluorescence studies indicated that a large fraction of the cultures in which the NS replicons had been transiently established showed NS1 expression (Figure 4), allowing biochemical analysis of the transcripts generated. In these cultures, the levels of NS1 expression were similar to those observed during infection and most of the signal was found in the nuclei of the cells (Figure 4).
Figure 1.
A transient replication-transcription system with a recombinant NS replicon. (A) Diagram showing the process for reconstitution of a recombinant NS replicon in HEK293T cells. Transfection of the polymerase expression plasmids (PB1, PB2 and PA) and NP expression plasmid, as well as the NS segment genomic plasmid (pHHNS) leads to the replication and expression of NS RNPs. The process results in the expression of NS1 and NEP(NS2) proteins. (B) Structure and properties of the NS1 proteins expressed from the NS replicons used in this study. The location of the RNA-binding domain (RBD), effector domain (ED), eIF4G-binding domain (eIF4G-BD), the nuclear localization signals (NLS-1 and NLS-2), the nuclear export signal (NES) and the splicing sites (5′-ss and 3′-ss) are indicated. The position of translation stops introduced by mutation in ΔNS1 and NS1 81 mutants are indicated. The point mutations present in mutants R38AK41A and 11C are indicated by asterisks.
Figure 2.
Synthesis of NS1 and NEP(NS2) proteins from the recombinant replicons. Cultures of HEK293T cells were transfected with the appropriate plasmids to establish the indicated wt or mutant NS replicons or mock transfected (Mock). At 36 h post-transfection, the cultures were pulse labelled with 35S methionine-cysteine and whole cell extracts were analysed by polyacrylamide gel electrophoresis and autoradiography. Cells containing the NS1 81 or the 11C replicons were incubated at either permissive (32) or restrictive (39) temperature. The position of the relevant proteins is indicated to the right. The mobility of molecular weight markers in shown to the left.
Figure 3.
Expression of viral genes encoded in the recombinant replicons. Cultures of HEK293T cells were transfected with the appropriate plasmids to establish the indicated wt or mutant NS replicons or mock transfected (Mock). (A) At 36 h post-transfection, total cell extracts were prepared and analysed by western blot using NS1-specific antibodies. To control the loading, a western-blot against actin was used. The position of NS1 and actin proteins is indicated to the right. The NS1 81 mutant protein was not detectable with the antibody used. The mobility of molecular weight markers is shown to the left. (B, C) At 36 h post-transfection, the cultures were pulse-labelled with 35S-met-cys as indicated in the legend to Figure 2 and soluble cell extracts (T) were prepared. The extracts were immunoprecipitated with anti-NEP(NS2) (I) or with control antibodies (C) and the products were analysed by polyacrylamide gel electrophoresis and autoradiography. The position of NEP(NS2) and NS1 proteins is indicated to the right. The mobility of molecular weight markers is shown to the left.
Figure 4.
Intracellular localization of NS1 protein expressed from recombinant NS replicons. Cultures of HEK293T cells were transfected with the appropriate plasmids to establish the indicated wt or mutant NS replicons or mock transfected (Mock). At 36 h post-transfection, the cultures were fixed and processed for immunofluorescence using anti-NS1 serum (NS1). The cultures were counterstained with Dapi to reveal the nuclei (Dapi). Cells containing the NS1 81 or the 11C replicons were incubated at either permissive (32) or restrictive (39) temperature.
The stability of NS segment mRNAs is independent of NS1 protein expression
To study the potential role of NS1 protein in the maturation and transport of NS segment transcripts we first determined whether the expression of NS1 could alter the stability of the collinear or spliced mRNAs, encoding NS1 and NEP(NS2) proteins, respectively. Total cell RNA was isolated from cultures in which wt or ΔNS1 NS replicons had been established, prior to and at various times after actinomycin D chase. The polyA-containing fraction of these preparations was analysed by RNAse-protection assay using a probe that could distinguish collinear, unspliced mRNA from the spliced transcript. The results are shown in Figure 5 and indicate that the presence or absence of NS1 did not alter the half-lives of NS1 or NEP(NS2) mRNAs. Therefore, we can use the relative accumulation of NS1 versus NEP(NS2) transcripts as a measure of the extent of NS collinear mRNA splicing.
Figure 5.
Stability of NS-encoded viral mRNAs. (A) Cultures of HEK293T cells were transfected with the appropriate plasmids to establish the wt or ΔNS1 NS replicons. At 36 h post-transfection, actinomycin D was added to the cultures and total RNA was extracted at the times thereafter indicated in the Figure. The poly A-containing RNA was selected by oligo dT-cellulose chromatography and the amounts of NS1 and NEP mRNA were determined by RNAse-protection assay. The results from two independent experiments are shown and the half-lives obtained from each experiment are presented in the part B of the Figure.
The N-terminal region of NS1 protein is sufficient for the down-regulation of NS collinear mRNA splicing
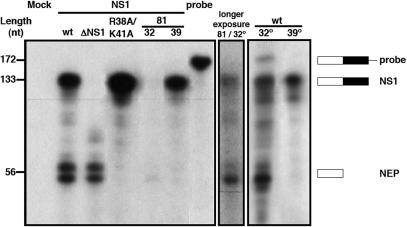
To determine the extent of splicing of NS collinear transcript we establish the various NS replicons by transfection of cultures of HEK293T cells as described above. Total cell RNA was isolated and the polyA-containing RNA fraction was purified and analysed by quantitative RNAse-protection assay. The conditions of the assay were set to ensure an excess of probe for all samples. The results of a representative experiment are presented in Figure 6 and Table 1 summarizes the data obtained from three independent experiments. When wt NS replicon was used, both NS1 collinear and NEP(NS2) spliced mRNAs were detected and an extent of splicing of around 45% was obtained (Figure 6, wt; Table 1). However, deletion of NS1 led to dramatic change in the splicing efficiency, as almost no collinear transcript could be detected (Figure 6, ΔNS1; Table 1). These results indicate that NS1 protein down-regulates the splicing of its own mRNA.
Figure 6.
Effect of NS1 mutations on the accumulation of NS-encoded mRNAs. Cultures of HEK293T cells were transfected with the appropriate plasmids to establish the indicated wt or mutant NS replicons. Cells containing the NS1 81 replicon were incubated at either permissive (32) or restrictive (39) temperature. At 36 h post-transfection total RNA was extracted and the poly A-containing RNA was selected by oligo dT-cellulose chromatography. The amounts of NS1 and NEP mRNA were determined by RNAse-protection assay using a probe overlapping the first exon and the intron sequences (probe). The protected fragments were separated by polyacrylamide-urea gel electrophoresis, revealed by autoradiography and quantitated in a phosphorimager. The results obtained for mutant NS1 81 at permissive temperature are shown to the right after longer exposure. The mobility of the probe and the protected fragments is indicated to the right and the position of molecular weight markers is shown to the left.
Table 1.
Ratio of NS1 and NEP mRNAs
| NS Replication | NS1 mRNA | NEP mRNA | SDb |
|---|---|---|---|
| wt | 53a | 47a | 4.2 |
| wt (32°C) | 30 | 70 | 8.9 |
| wt (39°C) | 93 | 7 | 3.7 |
| ΔNS1 | 2 | 98 | 1.2 |
| 81 (32°C) | 26 | 74 | 5 |
| 81 (39°C) | 85 | 15 | 8.7 |
| R38A/K41A | >98 | < 2 | 1.2 |
aPercent of total NS segment mRNA.
bStandard deviation.
To test whether the N-terminal RNA-binding or the C-terminal effector domains in NS1 are involved in this down-regulation we determined the splicing phenotypes of mutants affected in either one. Mutant NS1 81 (44) lacks the sequences 81-238 in NS1 protein and hence does not contain the effector domain. The NS1 81 mutant virus is temperature-sensitive and shows a major defect in late gene expression and minor reduction in vRNA replication. Therefore, we examined the splicing of transcrips from a NS1 81 replicon at both permissive (32°C) and restrictive (39°C) temperatures. The extent of splicing at permissive temperature was similar to wt (Figure 6, NS1 81; Table 1), and therefore we can conclude that the N-terminal region of NS1 protein is sufficient for down-regulation of the splicing of its own mRNA and the C-terminal effector domain is not required for such function. Although splicing efficiency was reduced at restrictive temperature (Figure 6, NS1 81; Table 1), the wt replicon showed the same decrease (Figure 6, wt; Table 1), indicating that this effect is not due to the mutation. Such a reduction for the wt replicon could be the consequence of a low-splicing efficiency of the cell machinery at high temperature, as the corresponding wt recombinant virus showed reduced replication at 39°C (46), or may be connected to the temperature-sensitivity of vRNA replication reported earlier (49).
In view of these results we tested whether the NS1 RNA-binding activity, which has been mapped to the N-terminal half of the protein (22,24,50), is required to down-regulate NS1 mRNA splicing. Mutations R38A and K41A, known to abolish NS1–RNA binding (22,47) were introduced in the NS replicons and the ratio of NS1 versus NEP(NS2) mRNAs was determined in cells in which the mutated replicon had been established. The results are presented in Figure 6 and Table 1 and show that essentially no splicing of the NS collinear transcript is observed. Thus, we can conclude that RNA-binding by NS1 protein is not necessary for it to down-regulate the splicing of its own mRNA and a mutant lacking RNA-binding activity behaves as an highly efficient blocker of NS1 mRNA splicing. The splicing efficiency of NS1 ts mutant 11C (V18A, R44K, S195P) was similar to that observed for mutant NS1 81 (data not shown).
The NS1 protein preferentially blocks nucleo-cytoplasmic transport of NS collinear mRNA
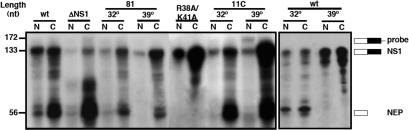
The transcription, maturation and nucleo-cytoplasmic transport of mammalian mRNAs are linked processes and influenza virus transcription depends functionally and structurally on the cellular transcription apparatus (51–53). In view of the alterations induced by NS1 in the splicing of NS transcripts we seeked for potential changes in the nucleo-cytoplasmic transport of these mRNAs. The various NS replicons described above were established in HEK293T cell cultures and nuclear and cytoplasmic fractions were prepared. The polyA-containing RNAs from these fractions were isolated and the recovery of NS1 and NEP(NS2) mRNAs was determined in each fraction by quantitative RNAse-protection assay. The results of a representative experiment are presented in Figure 7 and the data obtained from three independent experiments is summarized in Table 2. The distribution of these mRNAs in both cellular fractions indicated that nucleo-cytoplasmic export was more efficient for the spliced NEP(NS2) than for the collinear NS1 mRNA (Figure 7, wt). When NS1 expression was abolished, most of the transcript was spliced and efficiently exported to the cytoplasm (Figure 7, ΔNS1). However, the collinear NS1 mRNA generated by the R38A/K41A mutant replicon, which constitutes most of the NS transcript produced (see Figure 6) was exported very efficiently (Figure 7, R38A/K41A). Therefore, we can conclude that NS1 protein down-regulates the export of its own mRNA and that this regulation requires the RNA-binding activity of NS1. A temperature sensitive phenotype was found for transport of NS1 mRNA with mutants NS1 81 and 11C. Thus, while the phenotype obtained at 32°C was indistinguishable from wt, the export of NS1 mRNA was very efficient at restrictive temperature (Figure 7, NS1 81; 11C). Contrary to the results for NS1 mRNA splicing (Table 1), the transport of NS mRNAs generated from a wt NS replicon was not affected at high temperature (Figure 7; Table 2), thereby verifying that mutants NS1 81 and 11C are temperature-sensitive for the block of NS1 mRNA export.
Figure 7.
Effect of NS1 mutations on the nucleo-cytoplasmic transport of NS-encoded mRNAs. Cultures of HEK293T cells were transfected with the appropriate plasmids to establish the indicated wt or mutant NS replicons. Cells containing the NS1 81 or the 11C replicons were incubated at either permissive (32) or restrictive (39) temperature. At 36 h post-transfection the cells were fractionated into nuclear and cytoplasmic fractions. Total RNA was extracted from each fraction and the poly A-containing RNA was selected by oligo dT-cellulose chromatography. The amounts of NS1 and NEP mRNA were determined by RNAse-protection assay using a probe overlapping the first exon and the intron sequences (probe). The protected fragments were separated by polyacrylamide-urea gel electrophoresis, revealed by autoradiography and quantitated in a phosphorimager. The mobility of the probe and the protected fragments is indicated to the right and the position of molecular weight markers is shown to the left.
Table 2.
Nucleo-cytoplasmic distribution of NS1 and NEP mRNAs
| NS Replicon | NS1 mRNA | NEP mRNA | ||||
|---|---|---|---|---|---|---|
| Nucleus | Cytoplasm | SDb | Nucleus | Cytoplasm | SDb | |
| wt | 43a | 57a | 5.6 | 16a | 84a | 3.1 |
| wt (32°C) | 42 | 58 | 4.8 | 28 | 72 | 4.3 |
| wt (39°C) | 43 | 57 | 4.9 | n.d | n.d | – |
| ΔNS1 | 18 | 82 | 4.2 | 6 | 94 | 2.5 |
| 81 (32°C) | 33 | 67 | 2.5 | 26 | 74 | 4.1 |
| 81 (39°C) | 13 | 87 | 4.8 | <4 | >96 | 1.5 |
| R38A/K41A | 16 | 84 | 3.7 | n.d | n.d | – |
| 11C (32°C) | 28 | 72 | 4.1 | <4 | >96 | 2.1 |
| 11C (39°C) | 6 | 94 | 1.5 | <2 | >98 | 1.2 |
aPercent of total NS1 or NEP cellular mRNA.
bStandard deviation.
n.d. not applicable.
DISCUSSION
In this report, we have analysed the consequences of NS1 protein expression in the splicing and nucleo-cytoplasmic transport of NS transcripts using a transient system for the replication and transcription of a recombinant influenza virus NS replicon. This is a convenient system that can be used as a model of the situation of an infected cell. Indeed, the experimental approach used is a development of the amplification in vivo of transcriptionally active viral mini-RNPs employed in the past for structural studies (2,54,55). Furthermore, full-length RNPs generated in a similar way have been rescued into infectious virus (44,46). Here we show that NS replicons can be transiently amplified and express both NS1 and NEP(NS2) proteins in proportions and to levels similar to those produced in the virus infection (Figures 2–4). We felt that this should be a convenient system to carry out a genetic analysis of the role of NS1 in the maturation and export of viral transcripts as the studies could be made independent of other possible phenotypes described for NS1 mutants. Thus, mutants NS1 81 and 11C show predominant alterations in late virus gene expression and particle formation (44,46), aspects not relevant in the experimental setting used here. On the other hand, a ΔNS1 virus mutant is difficult to handle in a cell line with a normal interferon response (56) but can be studied normally in a transient situation as the one described here. Although it has been reported that the presence of NEP(NS2) protein inhibits viral RNA replication in a similar recombinant system (57), we consider this effect non-relevant to our studies, that deal with the post-transcriptional processing and export of viral mRNAs.
The results presented here indicate that NS1 protein inhibits the splicing of the NS collinear transcript in a RNA-independent manner, as mutant NS1 R38A/K41A behaves as wt (Figure 6; Table 1), and preferentially blocks the nucleo-cytoplasmic export of NS1 mRNA by a mechanism dependent on NS1–RNA interaction, since mutant NS1 R38A/K41A permits an efficient export of this mRNA (Figure 7; Table 2). Although cells in which a mutant NS R38A/K41A has been established accumulate large amounts of NS1 mRNA and very little NEP(NS2) mRNA in their cytoplasms (Figures 6 and 7), quasi-normal synthesis of NEP(NS2) protein is produced (Figures 2 and 3B). This apparent contradiction can be explained by the fact that, at early times in the replication and transcription of the NS R38A/K41A replicon, no NS1 protein is present in the system. Until NS1 protein is accumulated to the required intracellular levels, efficient splicing of the collinear transcript is to be expected, as this is the situation with a NS ΔNS1 replicon (Figure 6). Nevertheless, to account for the observed levels of NEP(NS2) synthesis it would be necessary to invoke a preferential translation of NEP(NS2) mRNA. It is conceivable that NEP(NS2) mRNA is efficiently incorporated into the cell translation machinery because it is generated at the early phase of the replicon gene expression. On the contrary, NS1 mRNA is produced at later times, when NS1 protein has accumulated. At that point a large fraction of the cell-derived mRNA is being used for virus-induced cap-snatching and incorporation of new mRNAs into the cell translation machinery might be affected. In addition, the level of NS1 protein present in each setting may influence the translation ability of the NEP mRNA present (19,30,31). Thus, when NS1 is not expressed (ΔNS1 replicon), translation of NEP mRNA might be much less efficient than in those cases in which NS1 protein is abundantly expressed, as for instance in cells harbouring the wt NS replicon.
The effects of NS1 in the splicing and nucleo-cytoplasmic transport of mRNAs have been studied previously by several groups, including ours, but the analyses were carried out in vitro or using as targets cellular mRNAs or viral ORFs expressed as RNA polymerase II transcripts, but not true virus transcripts. On the basis of these studies, several models have been put forward along the years for the inhibition of pre-mRNA splicing by NS1: (i) From the results of in vitro experiments it was first proposed that the binding of NS1 to a specific motif in U6 snRNA would impede proper formation of U6-U2 and U6-U4 snRNP interactions, leading to the block of the splicing reaction (26,40). Such inhibitory interaction was postulated to occur in splicing reactions on cellular pre-mRNAs but not on viral mRNAs (40). This model would not explain the alterations induced by NS1 in the use of alternative splice sites (41) and is not supported by the results presented in this report, as we show that NS1 protein indeed inhibits the splicing of a true viral pre-mRNA (the NS collinear transcript) and the RNA-binding activity of NS1 is not required for such inhibition (Figure 6). (ii) After reporting that the effector domain of NS1 protein binds the 30 kDa subunit of CPSF and blocks cellular mRNA cleavage and polyadenylation (33), it was proposed that the splicing inhibition by NS1 would be an indirect consequence of the failure in 3′-end processing, as the definition of the exon in a single-exon containing mRNA depends on the 3′-end polyadenylation (42). This model would explain the inhibition of cellular mRNA splicing and the lack of such inhibition for intron-containing viral mRNAs (40), since the 3′-polyadenylation of the latter is carried out by polymerase stuttering (58). Again, our results do not support the proposed model, as NS1 protein indeed blocks the splicing of the NS collinear transcript and a NS1 mutant lacking the effector domain fully inhibits the splicing of its own mRNA (Figure 6).
We cannot rule out the possibility that NS1 inhibition of cellular mRNA 3′-end formation would indirectly affect the splicing of cellular pre-mRNAs, but the results presented here suggest that NS1 protein would inhibit splicing by a more general mechanism. The interaction with NS1–BP, a human protein involved in splicing (35) is one possibility, while a de-regulation of the phosphorylation state of SR proteins is an alternative that would explain the modification in alternative splice site usage by NS1 expression (41). A general inhibition of the splicing machinery would be in line with the changes in the localization of snRNPs induced by NS1 expression or virus infection (59) and could be considered as part of the viral response to inhibit the expression of cellular antiviral factors (60). Whatever the mechanism involved, our data indicate that inhibition would be mediated by the N-terminal region of NS1 but would not require NS1–RNA interactions (Figure 6).
As indicated earlier, the nucleo-cytoplasmic transport of mRNAs is inhibited by expression of NS1 from cloned DNA (25,28,41) and a model has been proposed for such inhibition based upon the interaction of NS1 with the 30 kDa subunit of the CPSF and the subsequent inhibition of 3′-end mRNA processing (33). In addition, the interaction of NS1 with nuclear PABPII would block the export of mRNAs that could partially escape from the inhibition of 3′-end formation (34). This model suggested that the export of true viral mRNAs would not be affected by this transport block because their poly A-tails are produced by the viral polymerase in a way independent of normal cell mRNA polyadenylation (58). In addition, the interaction of NS1 with the TAP/p15 pathway for mRNA export has been recently described (61), leading to the block of transport of cellular mRNAs that have undergone splicing. However, the results shown in this report indicate that a preferential block of NS1 mRNA export is produced when NS1 is expressed from a NS replicon and further suggest that the RNA-binding activity of NS1 is required for the inhibition of viral mRNA export (Figure 7). These results suggest that, in addition to the block of cellular, spliced mRNA export, mediated by the inhibition of cellular mRNA 3′-end processing (33) or by targetting the TAP/p15 mRNA export pathway (61), NS1 knocks down the export of its own virus mRNA, a non-spliced mRNA, by RNA-binding. The mechanism for this nuclear retention is not clear at this point in time but a recent publication showing that nuclear export of some virus mRNAs, including NS1 mRNA, requires their association to the cellular transcription machinery might shed some light (53). Thus, NS1 might act by avoiding the coupling of the viral transcription events to the RNA polymerase II complex after virus RNP-mediated cap-snatching and hence interfering with the association of shuttling hnRNP proteins to the virus transcripts. This interference would not take place on the spliced NEP(NS2) mRNA because it would associate to the standard EJC/TAP-p15-dependent mRNA export pathway (62).
The nuclear retention of NS1 mRNA could be a way to improve the possibilities for this collinear transcript to form a spliceosomal complex in the context of general splicing inhibition described above. Thus, complete inhibition of the splicing of the NS collinear transcript would be deletereous for virus infection, as no NEP(NS2) protein would be formed, and the virus might have evolved an escape mechanism to avoid a complete block of viral mRNA splicing. Such scenario is compatible with the phenotypes observed in the genetic analysis presented here. When NS1 protein is not expressed (ΔNS1), no inhibition of splicing and export is produced, the collinear NS1 transcript is almost fully spliced and NEP(NS2) mRNA is efficiently exported. If a RNA-binding mutant of NS1 is expressed, splicing inhibition persists but the export block is eliminated because no RNA recognition can take place, leading to an efficient export of collinear NS transcript.
In summary, the results presented in this report demonstrate that NS1 regulates the splicing and nucleo-cytoplasmic export of its own collinear mRNA, in addition to the previously reported inhibitions of cellular mRNA processing and export. The simplest interpretation of these results implies that the mechanisms for splicing and export inhibition are distinct and at least in part different from the way NS1 appears to alter cellular mRNA metabolism.
ACKNOWLEDGEMENTS
The authors would like to thank J.C. de la Torre and J. Ávila for providing biological materials. The technical assistance of Y. Fernández and N. Zamarreño is gratefully acknowledged. U.G. was a fellow from Programa de Formación de Personal Investigador. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grant BFU2004-00491) and the European Network of Excellence ViRgil. Funding to pay the Open Access publication charges for this article was provided by Grant BFU2004-00491.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martín-Benito J, Area E, Ortega J, Llorca O, Valpuesta JM, Carrascosa JL, Ortín J. Three dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elton D, Digard P, Tiley L, Ortín J. In: Current Topics in Influenza Virology. Kawaoka Y, editor. Norfolk: Horizon Scientific Press; 2005. pp. 1–92. Structure and function of the influenza virus RNP. [Google Scholar]
- 4.Inglis SC, Barret T, Brown CM, Almond JW. The smallest genome RNA segment of influenza virus contains two genes that may overlap. Proc. Natl Acad. Sci. USA. 1979;76:3790–3794. doi: 10.1073/pnas.76.8.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis SC, Brown CM. Spliced and unspliced RNAs encoded by virion RNA segment 7 of influenza virus. Nucleic Acid Res. 1981;9:2727–2740. doi: 10.1093/nar/9.12.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb RA, Lai CJ. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980;21:475–485. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- 7.Lamb RA, Lai CJ, Choppin PW. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl Acad. Sci. USA. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palese P, Shaw M. In: Fields Virology. 5th. Knipe DM, Howley PM, editors. Vol. 1. Philadelphia, PA: Lippincott Williams & Wilkins; 2006. pp. 1647–1689. Orthomyxoviridae: The viruses and their replication. [Google Scholar]
- 9.Bradshaw GL, Schwartz CD, Schlesinger RW. Replication of H1N1 influenza viruses in cultured mouse embryo brain cells: virus strain and cell differentiation affect synthesis of proteins encoded in RNA segments 7 and 8 and efficiency of mRNA splicing. Virology. 1990;176:390–402. doi: 10.1016/0042-6822(90)90009-g. [DOI] [PubMed] [Google Scholar]
- 10.Smith DB, Inglis SC. Regulated production of an influenza virus spliced mRNA mediated by virus-specific products. EMBO J. 1985;4:2313–2319. doi: 10.1002/j.1460-2075.1985.tb03932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valcárcel J, Portela A, Ortín J. Regulated M1 mRNA splicing in influenza virus-infected cells. J. Gen. Virol. 1991;72:1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Sastre A. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- 13.Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 14.Ortín J. Multiple levels of post-transcriptional regulation of Influenza virus gene expression. Semin. Virol. 1998;3:335–342. [Google Scholar]
- 15.Briedis DJ, Conti G, Munn EA, Mahy BWJ. Migration of influenza virus-specific polypeptides from cytoplasm to nucleus of infected cells. Virology. 1981;111:154–164. doi: 10.1016/0042-6822(81)90661-9. [DOI] [PubMed] [Google Scholar]
- 16.Lamb RA, Lai CJ. Expression of unspliced NS1 mRNA, spliced NS2 mRNA, and a spliced chimera mRNA from cloned influenza virus NS DNA in an SV40 vector. Virology. 1984;135:139–147. doi: 10.1016/0042-6822(84)90124-7. [DOI] [PubMed] [Google Scholar]
- 17.Portela A, Melero JA, Martinez C, Domingo E, Ortin J. A primer vector system that allows temperature dependent gene amplification and expression in mammalian cells: regulation of the influenza virus NS1 gene expression. Nucleic Acids Res. 1985;13:7959–7977. doi: 10.1093/nar/13.22.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenspan D, Palese P, Krystal M. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Luna S, Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falcón AM, Fortes P, Marión RM, Beloso A, Ortín J. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 1999;27:2241–2247. doi: 10.1093/nar/27.11.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatada E, Saito S, Okishio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J. Gen. Virol. 1997;78:1059–1063. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- 24.Marión RM, Aragón T, Beloso A, Nieto A, Ortín J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4277. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu Y, Krug RM. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J. Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu Y, Nemeroff M, Krug RM. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 27.Chien CY, Xu Y, Xiao R, Aramini JM, Sahasrabudhe PV, Krug RM, Montelione GT. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- 28.Qian X-Y, Alonso-Caplen F, Krug RM. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 1994;68:2433–2441. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marión RM, Zürcher T, de la Luna S, Ortín J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 30.Aragón T, de la Luna S, Novoa I, Carrasco L, Ortín J, Nieto A. Translation factor eIF4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cel. Biol. 2000;20:6259–6268. doi: 10.1128/mcb.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgui I, Aragón T, Ortín J, Nieto A. PABP1 and eIF4GI associate to influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 2003;84:3263–3274. doi: 10.1099/vir.0.19487-0. [DOI] [PubMed] [Google Scholar]
- 32.Marión RM, Fortes P, Beloso A, Dotti C, Ortín J. A human sequence homologue of staufen is an RNA-binding protein that localizes to the polysomes of the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Li Y, Krug RM. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. Embo J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff T, O’Neill RE, Palese P. NS1-Binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plotch SJ, Krug RM. In vitro splicing of influenza viral NS1 mRNA and NS1-beta-globin chimeras: possible mechanisms for the control of viral mRNA splicing. Proc. Natl Acad. Sci. USA. 1986;83:5444–5448. doi: 10.1073/pnas.83.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agris CH, Nemeroff ME, Krug RM. A block in mammalian splicing occurring after formation of large complexes containing U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins. Mol. Cell. Biol. 1989;9:259–267. doi: 10.1128/mcb.9.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nemeroff ME, Utans U, Kramer A, Krug RM. Identification of cis-acting intron and exon sequences in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol. Cell. Biol. 1992;12:962–970. doi: 10.1128/mcb.12.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portela A, Melero JA, de la Luna S, Ortin J. Construction of cell lines that regulate by temperature the amplification and expression of influenza virus non-structural protein genes. EMBO J. 1986;5:2387–2392. doi: 10.1002/j.1460-2075.1986.tb04508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Qian XY, Krug RM. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 41.Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks RNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Chen ZY, Wang W, Baker CC, Krug RM. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA. 2001;7:920–931. doi: 10.1017/s1355838201010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falcón AM, Marión RM, Zürcher T, Gómez P, Portela A, Nieto A, Ortín J. Defective RNA replication and late gene expression in temperature-sensitive (A/Victoria/3/75) influenza viruses expressing deleted forms of NS1 protein. J. Virol. 2004;78:3880–3888. doi: 10.1128/JVI.78.8.3880-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortín J, Nájera R, López C, Dávila M, Domingo E. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene. 1980;11:319–331. doi: 10.1016/0378-1119(80)90072-4. [DOI] [PubMed] [Google Scholar]
- 46.Garaigorta U, Falcon AM, Ortin J. Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. J Virol. 2005;79:15246–15257. doi: 10.1128/JVI.79.24.15246-15257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zürcher T, Marión RM, Ortín J. The protein synthesis shut-off induced by influenza virus infection is independent of PKR activity. J. Virol. 2000;74:8781–8784. doi: 10.1128/jvi.74.18.8781-8784.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalton RM, Mullin AE, Amorim MJ, Medcalf E, Tiley LS, Digard P. Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes. Virol. J. 2006;3:58. doi: 10.1186/1743-422X-3-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein [letter] Nat. Struct. Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 51.Chan AY, Vreede FT, Smith M, Engelhardt OG, Fodor E. Influenza virus inhibits RNA polymerase II elongation. Virology. 2006;351:210–217. doi: 10.1016/j.virol.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Engelhardt OG, Smith M, Fodor E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 2005;79:5812–5818. doi: 10.1128/JVI.79.9.5812-5818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amorim MJ, Read EK, Dalton RM, Medcalf L, Digard P. Nuclear export of influenza A virus mRNAs requires ongoing RNA Polymerase II Activity. Traffic. 2007;8:1–11. doi: 10.1111/j.1600-0854.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 54.Ortega J, Martín-Benito J, Zürcher T, Valpuesta JM, Carrascosa JL, Ortín J. Ultrastructural and functional analyses of of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 2000;74:156–163. doi: 10.1128/jvi.74.1.156-163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Area E, Martín-Benito J, Gastaminza P, Torreira E, Valpuesta JM, Carrascosa JL, Ortín J. Three-dimensional structure of the influenza virus RNA polymerase: localization of subunit domains. Proc. Natl Acad. Sci. USA. 2004;101:308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 57.Bullido R, Gomez-Puertas P, Saiz MJ, Portela A. Influenza A virus NEP (NS2 protein) downregulates RNA synthesis of model template RNAs. J. Virol. 2001;75:4912–4917. doi: 10.1128/JVI.75.10.4912-4917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poon LLM, Pritlove DC, Fodor E, Brownlee GG. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fortes P, Lamond AI, Ortín J. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 1995;76:1001–1007. doi: 10.1099/0022-1317-76-4-1001. [DOI] [PubMed] [Google Scholar]
- 60.Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 61.Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl Acad. Sci. USA. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stutz F, Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]