Abstract
We report the results of a genetic screen to identify molecules important for synapse formation and/or maintenance. siRNAs were used to decrease the expression of candidate genes in neurons, and synapse development was assessed. We surveyed 22 cadherin family members and demonstrated distinct roles for cadherin-11 and cadherin-13 in synapse development. Our screen also revealed roles for the class 4 Semaphorins Sema4B and Sema4D in the development of glutamatergic and/or GABAergic synapses. We found that Sema4D affects the formation of GABAergic but not glutamatergic synapses. Our screen also identified the activity-regulated small GTPase Rem2 as a regulator of synapse development. A known calcium channel modulator, Rem2 may function as part of a homeostatic mechanism that controls synapse number. These experiments establish the feasibility of RNAi screens to characterize the mechanisms that control mammalian neuronal development and to identify components of the genetic program that regulate synapse formation and/or maintenance.
Introduction
Synapses are specialized sites of cell-cell contact that mediate the transmission and storage of information in the brain. The majority of synapses in the central nervous system are chemical synapses where communication occurs by the release of neurotransmitter from the presynaptic terminal which then acts on neurotransmitter receptors localized at the postsynaptic terminal (Li and Sheng, 2003). Multiple, overlapping steps are required for the development of glutamatergic synapses in the mammalian hippocampus, beginning with initial contact between the axon and the dendrite (Ziv and Garner, 2004). After initial contact, pre-assembled packets of proteins localize to the presynaptic compartment to form the active zone and the synaptic vesicle release machinery (Ziv and Garner, 2004). Simultaneously, neurotransmitter receptors, signaling molecules, and scaffolding and structural proteins cluster at the postsynaptic site (Ziv and Garner, 2004). Finally, synapses undergo a process of refinement wherein a subset of synapses are weakened and eliminated while the remainder are strengthened and maintained (Katz and Shatz, 1996).
Given the complex nature of synapse development, it is not surprising that multiple proteins are involved in regulating this process. Trans-synaptic ligand-receptor complexes and cell adhesion molecules are attractive candidates to regulate contact initiation and stabilization as these proteins have both adhesive and intracellular signaling capabilities. Indeed, the receptor tyrosine kinase EphB and its ligand ephrinB, the neurexin/neuroligin complex, and SynCAM (Biederer et al., 2002; Chih et al., 2005; Dalva et al., 2000) have been shown to play a role in glutamatergic synapse development. Moreover, the coordination of synapse development is a global cellular process that requires gene transcription in the nucleus (Flavell et al., 2006; Shalizi et al., 2006) and homeostatic regulation to insure that excitability remains within the appropriate physiological range (Turrigiano and Nelson, 2004). Although intensive study has begun to identify molecules that regulate synapse development, it is still not known at which step in synapse development most of these molecules function. An identification of the full complement of molecules that are required for synapse development would represent a major advance towards understanding the requirement for individual molecules at specific steps in the process of synapse development.
In comparison to glutamatergic synapse development, less is known about the development of inhibitory, GABAergic synapses. The relatively low abundance of GABAergic synapses on hippocampal neurons has precluded the use of biochemistry to identify proteins present at such synapses (Fritschy and Brunig, 2003). Thus far, only a few proteins such as gephyrin, a 93kD peripheral membrane protein, and dystrophin, the Duchenne muscular dystrophy gene product, have been shown to regulate GABAergic synapse development (Fritschy and Brunig, 2003).
A remaining question in the field of synapse development is how a neuron matches the correct postsynaptic neurotransmitter receptors to the appropriate glutamatergic or GABAergic presynaptic terminal. Interestingly, several proteins can regulate both glutamatergic and GABAergic synapse development and/or function (Chih et al., 2005; Elmariah et al., 2004; Graf et al., 2004; Varoqueaux et al., 2006; Weiner et al., 2005), and considerable mismatching of components of glutamatergic and GABAergic synapses occurs both in vivo and in vitro (e.g. the localization of GABAA receptors to glutamatergic presynaptic terminals) (Anderson et al., 2004; Nusser et al., 1998; Rao et al., 2000). Moreover, the fidelity of matching increases as the neurons mature (Anderson et al., 2004), suggesting that neurons may execute a general program of synapse development that is sufficient to initiate the assembly of synaptic structures and that the final specification of the postsynaptic structure as either glutamatergic or GABAergic occurs at a later step in synapse development. These observations imply that proteins likely exist in the postsynaptic cell that regulate the sorting and/or matching of the proper neurotransmitter receptors with their appropriate presynaptic terminals.
To gain insight into the molecular mechanisms that control synapse development, we developed a new strategy for discovering molecules required for the various steps in glutamatergic and GABAergic synapse development. Specifically, we established an RNA interference (RNAi)-based, forward genetic screen to identify molecules that regulate synapse development. Thus far, we have identified five genes that play a role in glutamatergic and/or GABAergic synapse development. These include: two cadherin family members, cadherin-11 and cadherin-13; two class 4 semaphorins, Sema4B and Sema4D; and the activity-regulated small GTPase Rem2.
Results
Establishment of an RNAi Screen for Synapse Development in Mammalian Neurons
We took a two-step approach to identify molecules that are important for the development of glutamatergic synapses in hippocampal neurons. First, we used transcriptional profiling to identify transcripts that are up- or down-regulated during the time period of synapse development in the postnatal rodent hippocampus. We also identified genes regulated by activity in cultured neurons, as neuronal activity has been shown to be required for synapse stabilization and maintenance (Katz and Shatz, 1996). From an analysis of the regulation of over 30,000 genes, we compiled a list of approximately 600 genes that we considered good candidates to regulate synapse development. From this list, we prioritized the genes to be targeted in the RNAi screen by choosing those that were highly induced by activity or those with relevant functional motifs such as cadherin repeats or Ig or ring finger domains (see Supplemental Table S1). We also supplemented our list with genes that we hypothesized might be involved in synapse development based on work in other organisms or on their role in related biological processes.
Next, to determine whether these candidate genes play a role in synapse development, we established an RNAi-based screen in cultured hippocampal neurons (Fig. 1). RNAi is an innate mechanism in eukaryotes in which double-stranded 21- to 23-nucleotide small interfering RNAs (siRNAs) target the corresponding mRNA for degradation in a sequence-specific manner (Novina and Sharp, 2004). We used the RNase III-family member enzyme Dicer to construct a library of siRNAs targeting the candidate gene products. The mixture of siRNAs produced by Dicer digestion has been shown to effectively target specific genes (Myers et al., 2003) (Supplemental Figs. S1, S2), and in pilot experiments, we demonstrated efficient and specific knockdown of over 15 gene products in heterologous cells using diced siRNAs (Supplemental Fig. S2 and A. E. West, A. Brunet, S.P. and M.E.G, unpublished observations).
Figure 1.
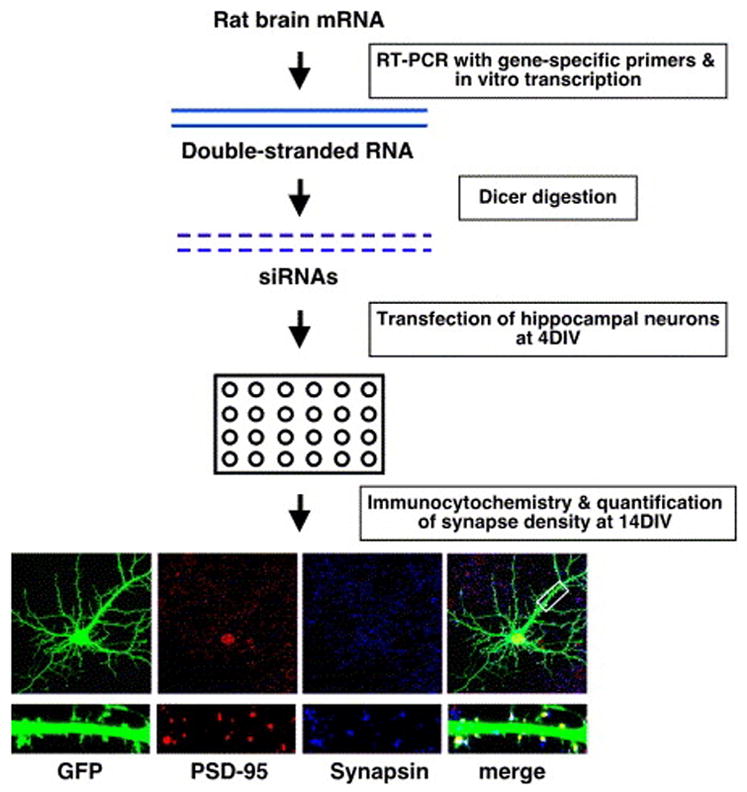
Overview of RNAi screen. Cells were stained for PSD-95 (red) and synapsin I (blue), and glutamatergic synapse density was quantified as the overlap of PSD-95/synapsin I with GFP (neuronal cell bodies were excluded from all analyses of cultured neurons; see Methods). The white box indicates the region of dendrite magnified at the bottom.
Before commencing the screen, diced siRNAs were evaluated for their ability to knockdown neuronally-expressed proteins over the time period of synapse development in hippocampal culture. As synapse development in culture occurs over a period of approximately two weeks (Rao et al., 1998), the time frame of gene knockdown that is required in a screen for genes important for synapse development is significantly longer than that required for previously reported screens. We transfected hippocampal neurons with diced siRNAs targeting the PSD-95 or GluR2 mRNA at 4DIV and assessed protein knockdown by quantitative immunocytochemistry at 14DIV. Diced siRNAs targeting the PSD-95 or GluR2 mRNA significantly reduce the expression of the targeted gene (Supplemental Fig. S1). In addition, we sought to increase the throughput of the screen by transfecting diced siRNAs into neurons as pools targeting multiple genes. We found that transfecting pools of siRNAs targeting more than one gene results in knockdown comparable to that observed with transfection of siRNAs targeting each gene individually (Supplemental Fig. S3), and thus the majority of the screen was performed using pools of diced siRNAs targeting multiple genes.
The assay that we established to assess the effect of knockdown of the targeted genes on the density of glutamatergic synapses is as follows. We co-transfected neurons with GFP and diced siRNAs at 4DIV and then fixed and stained the transfected cells at 14DIV with antibodies that recognize synapsin I, a synaptic vesicle-associated protein, and PSD-95, a scaffolding protein that is highly enriched at the postsynaptic side of glutamatergic synapses. Images of transfected neurons were acquired by confocal microscopy, and synapse density was quantified as the number of opposing synapsin I/PSD-95 puncta present along the dendrites of GFP-expressing neurons (see Methods). As we scored the number of synapses formed onto the transfected neuron, we only assessed the effect of the knockdown in the postsynaptic neuron.
Our screen was controlled for nonspecific or “off-target” effects of the RNAi methodology. For one, the low hit rate of the screen (16% of pools were initially positive, 4% were reproducibly positive) speaks to the overall immutability of glutamatergic synapse density after transfection with siRNAs (Fig. 2A). Moreover, we compared neurons transfected with GFP to neurons transfected with GFP and siRNAs targeting DsRed and found that synapse density did not differ between the two conditions (data not shown). We also used Sholl analysis to assess the complexity of the dendritic arbors in the diced siRNA-treated cells and found no change as a result of treating neurons with diced siRNAs targeting DsRed or the genes comprising the positive pools (Supplemental Fig. S4). This suggests that transfection with siRNAs did not a priori cause a deficit in cell health or dendritic complexity and that the reductions in glutamatergic synapse density observed for our positive pools are likely not a result of a perturbation of cell health or dendritic complexity.
Figure 2.
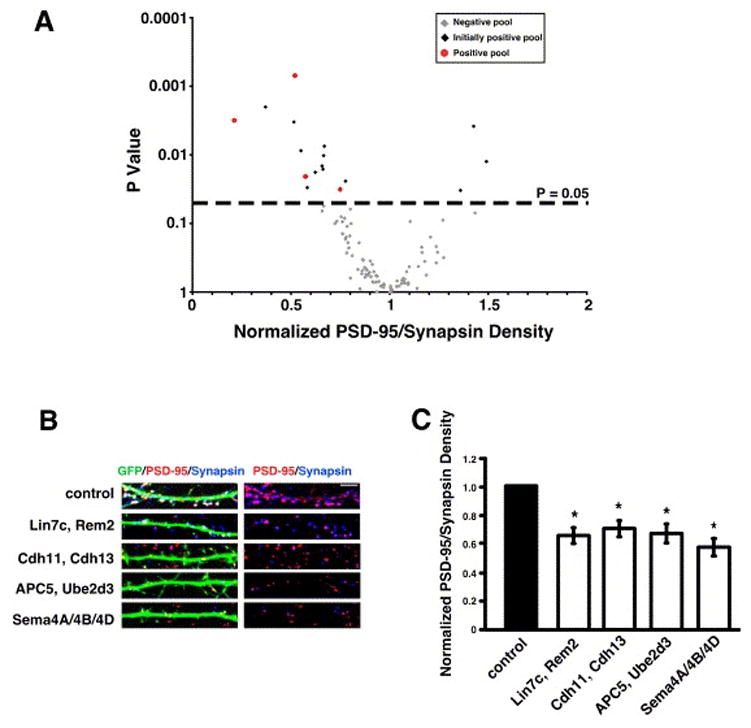
RNAi screen results. A) Scatter plot of normalized glutamatergic synapse density for neurons transfected with diced siRNAs targeting 160 different genes comprising 105 unique pools versus P value (two sample t-test). Gray diamonds indicate pools not positive by t-test, black diamonds indicate pools initially positive by t-test that did not repeat as positive, and red circles indicate pools initially positive by t-test that repeated as positive. The dashed line at P=0.05 indicates cutoff for statistical significance. B) Immunostaining for PSD-95 (red) and synapsin I (blue) in the dendrites of cells co-transfected with GFP and diced siRNAs targeting the gene products indicated on the left. Glutamatergic synapses were defined as the overlap of red and blue puncta on a green neuron (white puncta in left panels). The right panels display PSD-95 and synapsin I staining in the absence of the GFP signal; overlapping puncta appear magenta. C) Quantification of the density of PSD-95/synapsin I puncta for neurons transfected with diced siRNAs targeting the indicated gene products. n=72 for Lin7c/Rem2, n=43 for Sema4A/Sema4B/Sema4D, n=77 for APC5/Ube2d3, n=69 for cadherin-11/cadherin-13. Significance of p<0.002 by multi-factorial ANOVA is indicated by asterisk. For this and all subsequent figures, the normalized values shown are the average of at least three independent experiments and “n” refers to number of cells analyzed. Error bars are ± standard error of the average ratio. The error for the control condition is contained within the error bar for each experimental condition (see Methods). Scale bar is 5 μm in this and all subsequent figures unless otherwise indicated.
Thus far, we have analyzed 160 genes comprising 105 unique pools of diced siRNAs targeting 1–4 genes (Fig. 2A and Supplemental Table S1). A pool was considered “positive” if the pool of diced siRNAs caused a change in the density of glutamatergic synapses as compared to control (significance determined by two sample t-test using a cutoff of p<0.05; black diamonds in Fig. 2A). All positive pools were re-tested and analyzed further if the pool was positive in the second experiment (red circles in Fig. 2A). Of the 105 pools analyzed to date, four pools of diced siRNAs yielded a reproducible change in synapse density: Lin7c/Rem2, cadherin-11/cadherin-13, APC5/Ube2d3, and Sema4A/Sema4B/Sema4D (Fig. 2B, C).
Deconvolution of Positive Pools
We further characterized the hits in our screen by transfecting neurons with short-hairpin RNAs (shRNAs) targeting each of the genes from the positive pools. We thus sought to determine which gene(s) was responsible for the observed decrease in synapse density. We designed three shRNAs against each of the genes in the positive pools using current algorithms for the rational design of shRNAs (see Methods). Each shRNA matches a single 21 base pair (bp) sequence within the targeted gene. This is in contrast to the diced siRNAs where multiple 21 bp siRNAs target the gene of interest. To insure maximum efficiency of knockdown by shRNAs, we simultaneously transfected three shRNAs targeting a specific gene into neurons and assessed synapse density. We also confirmed the specificity of the diced siRNAs and the shRNAs for knockdown of individual gene products in heterologous cells (Supplemental Fig. S2).
By targeting each gene product from our positive pools using shRNAs, we found that knockdown of the Rem2, cadherin-11, cadherin-13, or Sema4B gene product causes a decrease in the density of glutamatergic synapses. The transfection of shRNAs targeting APC5 or Sema4D has no effect on glutamatergic synapse density (Fig. 3A, B), and shRNA constructs targeting the Sema4A and Ube2d3 gene products cause cell lethality.
Figure 3.
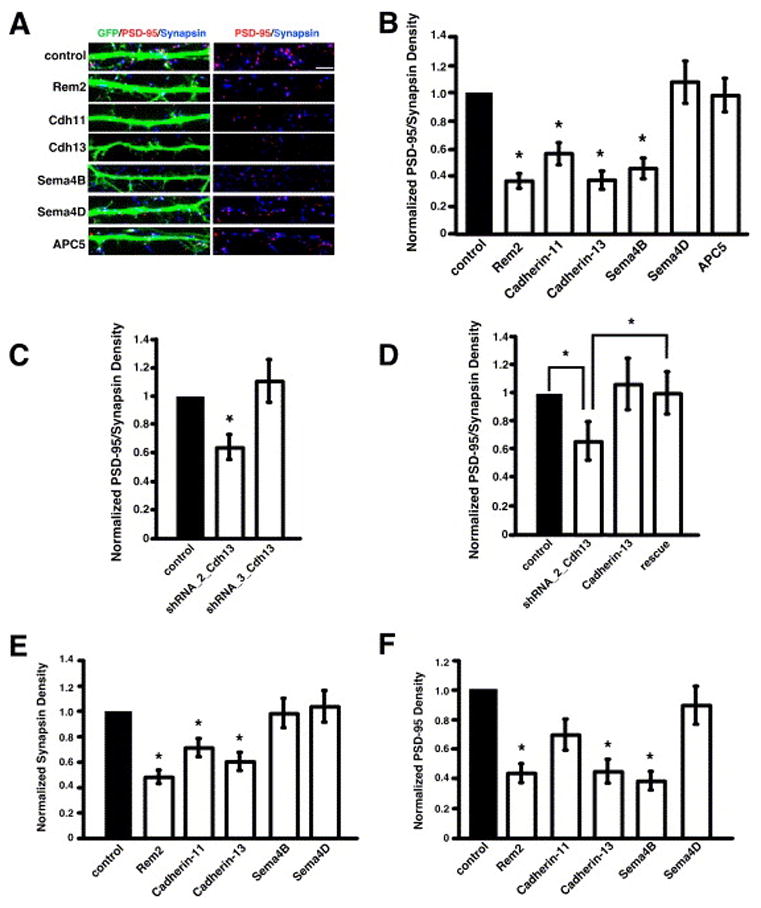
Deconvolution of positive pools demonstrates that Rem2, cadherin-11, cadherin-13, and Sema4B are required for glutamatergic synapse development. A) Immunostaining for PSD-95 (red) and synapsin I (blue) in the dendrites of cells co-transfected with GFP and shRNAs targeting the indicated gene products. Glutamatergic synapses were defined as the overlap of red and blue puncta on a green neuron (white puncta in left panels). The right panels display PSD-95 and synapsin I staining in the absence of the GFP signal; overlapping puncta appear magenta. B) Quantification of the density of PSD-95/synapsin I puncta for neurons transfected with shRNA constructs targeting the indicated gene products. n=38 for Rem2, n=44 for cadherin-11, n=43 for cadherin-13, n=46 for Sema4B, n=44 for Sema4D, n=39 for APC5. Significance of p<0.0001 by multi-factorial ANOVA is indicated by asterisk. C) Quantification of the density of PSD-95/synapsin I puncta for neurons transfected with shRNA constructs targeting the cadherin-13 gene product. n=92 for shRNA_2_Cdh13, n=56 for shRNA_3_Cdh13. Significance of p<0.0001 by multi-factorial ANOVA is indicated by asterisk. D) Quantification of the density of PSD-95/synapsin I puncta for neurons transfected with a shRNA construct targeting the cadherin-13 gene product (shRNA_2_Cdh13), a cadherin-13 cDNA construct resistant to RNAi by shRNA_2_Cdh13 (Cadherin-13), or shRNA_2_Cdh13 and the RNAi-resistant cadherin-13 construct (rescue). n=54 for shRNA_2_Cdh13, n=41 for Cadherin-13, n=60 for rescue. Significance of p<0.05 by multi-factorial ANOVA is indicated by asterisk; p=0.6 for cadherin-13 vs. rescue. E) Quantification of the density of synapsin I puncta on neurons transfected with shRNA constructs targeting the indicated gene products. n=38 for Rem2, n=44 for cadherin-11, n=43 for cadherin-13, n=46 for Sema4B, n=44 for Sema4D. Significance of p<0.03 by multi-factorial ANOVA is indicated by asterisk. F) Quantification of the density of PSD-95 puncta in neurons transfected with shRNA constructs targeting the indicated gene products. n=38 for Rem2, n=44 for cadherin-11, n=43 for cadherin-13, n=46 for Sema4B, n=44 for Sema4D. Significance of p<0.0001 by multi-factorial ANOVA is indicated by asterisk; p=0.08 for cadherin-11.
Our screen identified regulators of synapse development from multiple categories on our original list of candidate genes: activity-regulated genes, cell adhesion molecules, and putative pathfinding molecules, underscoring the complex nature of synapse development. We have investigated the importance of these genes for glutamatergic synapse development as follows: 1) analysis of the specificity of the RNAi phenotype (Figs. 3C, D, 7C, D), 2) assessment of glutamatergic synapse development by whole cell voltage clamp recording of AMPA receptor-mediated mEPSCs (Fig. 4) and immunostaining with antibodies recognizing the GluR2 subunit of the AMPAR (Fig. 5), and 3) assessment of the development of pre- and postsynaptic specializations independent of a requirement for co-localization (Fig. 3E, F). In addition, we sought to determine if the genes identified in the screen are general regulators of synapse development by testing whether they are also involved in GABAergic synapse development (Fig. 6).
Figure 7.
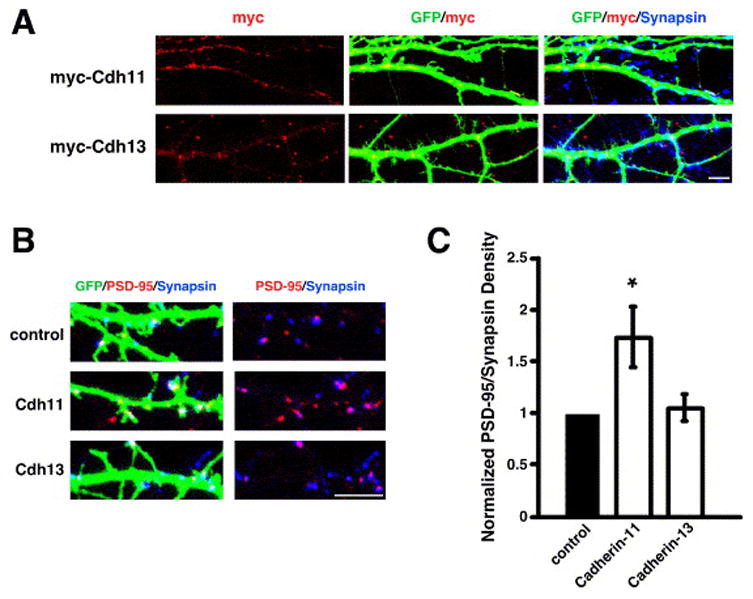
Cadherin-11 and cadherin-13 have distinct subcellular localizations in neurons and cadherin-11 over-expression promotes glutamatergic synapse development. A) Immunostaining at 14DIV for myc (red) in the dendrites of cells co-transfected with GFP and myc-tagged cDNA constructs as indicated on the left. B) Immunostaining for PSD-95 (red) and synapsin I (blue) in the dendrites of cells co-transfected with GFP and myc-tagged cDNA constructs as indicated on the left. Glutamatergic synapses were defined as the overlap of red and blue puncta on a green neuron (white puncta in left panels). The right panels display PSD-95 and synapsin I staining in the absence of the GFP signal; overlapping puncta appear magenta. C) Quantification of the density of PSD-95/synapsin I puncta for neurons transfected with a cDNA construct encoding myc-tagged Cadherin-11 or myc-tagged Cadherin-13. n=55 for Cadherin-11, n=76 for Cadherin-13. Significance of p<0.01 by multi-factorial ANOVA is indicated by asterisk.
Figure 4.
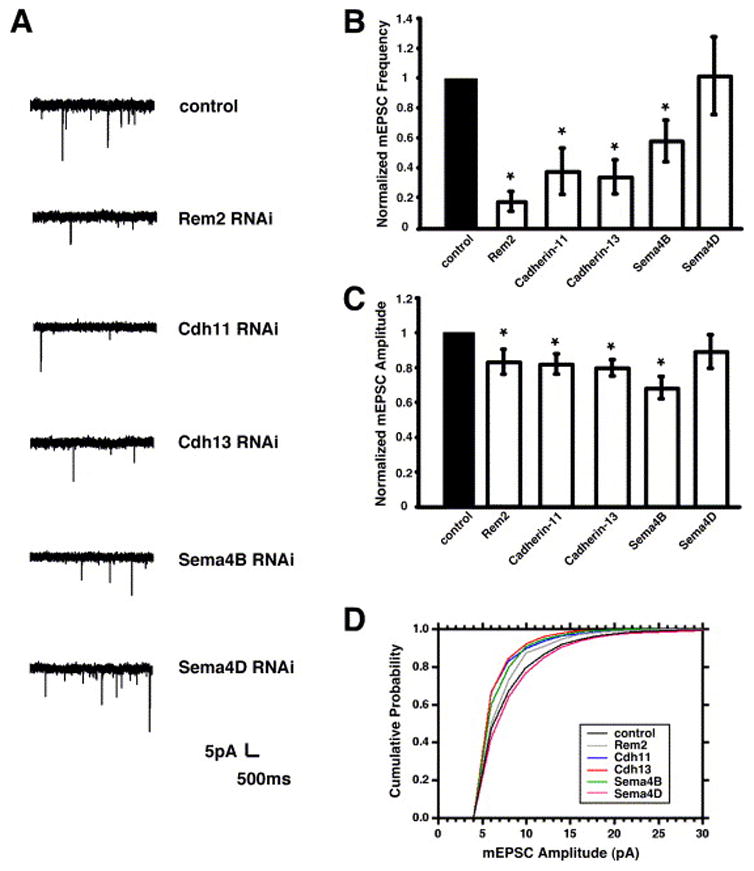
Rem2, cadherin-11, cadherin-13, and Sema4B are required for the development of functional glutamatergic synapses. A) Sample traces of mEPSCs from neurons transfected with shRNAs targeting the indicated gene products. B) Quantification of average mEPSC frequency for neurons transfected with shRNA constructs targeting the indicated gene products. n=15 for Rem2, n=15 for cadherin-11, n=14 for cadherin-13, n=15 for Sema4B, n=13 for Sema4D. Significance of p<0.05 by two sample t-test indicated by asterisk. C) Quantification of average mEPSC amplitude for neurons transfected with shRNA constructs targeting the indicated gene products. n=15 for Rem2, n=15 for cadherin-11, n=14 for cadherin-13, n=15 for Sema4B, n=13 for Sema4D. Significance of p<0.05 by two sample t-test indicated by asterisk. D) Cumulative probability distributions of mEPSC amplitudes for neurons transfected with shRNA constructs targeting the indicated gene products. n=16 for control, n=15 for Rem2, n=15 for cadherin-11, n=14 for cadherin-13, n=14 for Sema4B, n=12 for Sema4D. The cadherin-11 and cadherin-13 mEPSC amplitude distributions are different from control by Kolmogorov-Smirnov test, p<0.05.
Figure 5.
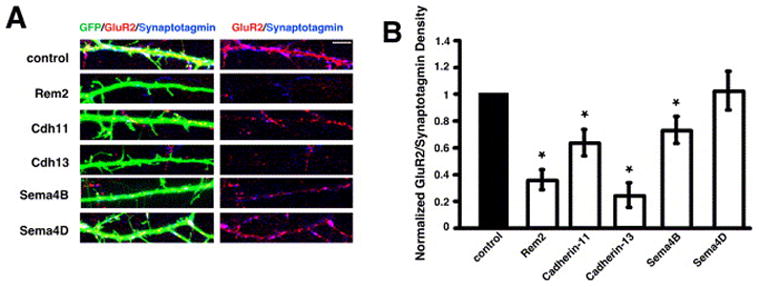
Rem2, cadherin-11, cadherin-13, and Sema4B are required for the development of AMPA receptor-containing synapses. A) Immunostaining for GluR2 (red) and Synaptotagmin I (blue) in the dendrites of cells co-transfected with GFP and shRNAs targeting the indicated gene products. Glutamatergic synapses were defined as the overlap of red and blue puncta on a green neuron (white puncta in left panels). The right panels display GluR2 and synaptotagmin I staining in the absence of the GFP signal; overlapping puncta appear magenta. B) Quantification of the density of GluR2/synaptotagmin I puncta for neurons transfected with shRNA constructs targeting the indicated gene products. n=45 for Rem2, n=36 for cadherin-11, n=54 for cadherin-13, n=50 for Sema4B, n=50 for Sema4D. Significance of p<0.04 by multi-factorial ANOVA is indicated by asterisk.
Figure 6.
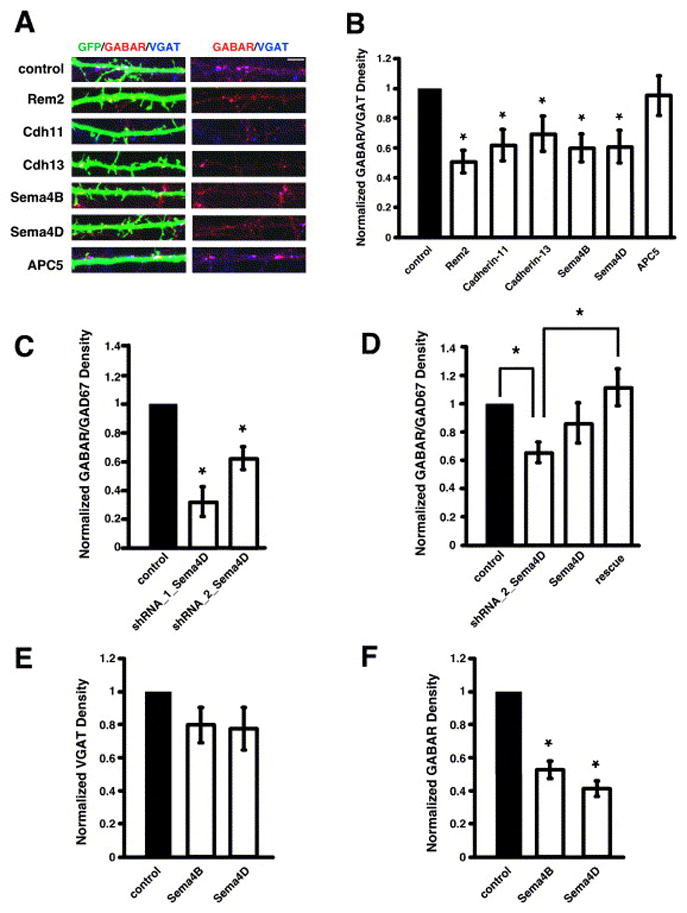
Rem2, cadherin-11, cadherin-13, Sema4B, and Sema4D are required for GABAergic synapse development. A) Immunostaining for GABAAβ2/3 receptor (red) and VGAT (blue) in the dendrites of cells co-transfected with GFP and shRNAs targeting the indicated gene products. GABAergic synapses were defined as the overlap of red and blue puncta on a green neuron (white puncta in left panels). The right panels display GABAAβ2/3 receptor and VGAT staining in the absence of the GFP signal; overlapping puncta appear magenta. B) Quantification of the density of GABAAβ2/3 receptor/VGAT puncta for neurons transfected with shRNA constructs targeting the indicated gene products. n=56 for Rem2, n=70 for cadherin-11, n=63 for cadherin-13, n=62 for Sema4B, n=60 for Sema4D, n=64 for APC5. Significance of p<0.03 by multi-factorial ANOVA is indicated by asterisk. C) Quantification of the density of GABAAγ2 receptor/GAD67 puncta for neurons transfected with shRNA constructs targeting the Sema4D gene product. n=58 for shRNA_1_Sema4D, n=103 for shRNA_2_Sema4D. Significance of p<0.005 by multi-factorial ANOVA is indicated by asterisk. D) Quantification of the density of GABAAγ2 receptor/GAD67 puncta for neurons transfected with a shRNA construct targeting the Sema4D gene product (shRNA_2_Sema4D), a Sema4D cDNA resistant to RNAi by shRNA_2_Sema4D (Sema4D), or shRNA_2_Sema4D and the RNAi-resistant Sema4D construct (rescue). n=67 for shRNA_2_Sema4D, n=44 for Sema4D, n=54 for rescue. Significance of p<0.02 by multi-factorial ANOVA is indicated by asterisk; p=0.15 for Sema4D vs. rescue. E) Quantification of the density of VGAT puncta on neurons transfected with shRNA constructs targeting the indicated gene products. n=62 for Sema4B, n=60 for Sema4D. Significance of p<0.0001 by multi-factorial ANOVA is indicated by asterisk. F) Quantification of the density of GABAAβ2/3 receptor puncta on neurons transfected with shRNA constructs targeting the indicated gene products. n=62 for Sema4B, n=60 for Sema4D. Significance of p<0.0001 by multi-factorial ANOVA is indicated by asterisk.
Cadherin-13 and cadherin-11 act via distinct mechanisms to regulate synapse development
Our screen revealed a role for the cadherin-13 and cadherin-11 gene products as positive regulators of glutamatergic synapse development. The large size of the cadherin family, the restricted expression of cadherin family members, and the localization of cadherin family members to synapses has suggested a role for the cadherin family in determining synapse specificity (Fannon and Colman, 1996). It is not known which cadherin family members are important for the development of connections between specific subpopulations of neurons. However, cadherin-11 is known to be expressed in the hippocampus during the postnatal period when robust synaptogenesis occurs (Manabe et al., 2000), and the expression of cadherin-13 is up-regulated between 2 and 7 days in culture, a time when synapse development is beginning (data not shown). We further characterized the role of cadherin-13 in synapse development and compared the function of cadherin-13 to that of cadherin-11.
We began by verifying the specificity of the cadherin-13 RNAi phenotype (Fig. 3C, D). We demonstrated that transfection of a single shRNA construct targeting cadherin-13, shRNA_2_cdh13 (Fig. 3C), produces a decrease in synapse density comparable to that observed in neurons transfected with pools of diced siRNAs (Fig. 2C) or with three shRNA constructs (Fig. 3B). The various cadherin-13 RNAi constructs target distinct regions of the cadherin-13 mRNA and cause a decrease in glutamatergic synapse density, thus the effect of the RNAi is likely due to specific knockdown of the cadherin-13 mRNA and not to an off-target effect of the RNAi.
We created a cadherin-13 expression plasmid resistant to knockdown by shRNA_2_cdh-13 (Supplemental Fig. S2B), and tested its ability to rescue the decrease in synapse density observed with shRNA_2_cdh13. The expression of this plasmid by itself does not affect synapse density (Fig. 3D). When we co-transfected the RNAi-resistant cadherin-13 expression plasmid with shRNA_2_cdh13 we found that expression of RNAi-resistant cadherin-13 was sufficient to rescue the decrease in synapse density observed with transfection of shRNA_2_cdh13 (Fig. 3D). These results suggest that the decrease in synapse density in cadherin-13 RNAi-treated neurons is due to specific knockdown of the cadherin-13 gene product by the RNAi constructs. Validation of a role for cadherin-11 in glutamatergic synapse development by rescue was complicated by the fact that over-expression of cadherin-11 causes an increase in the density of glutamatergic synapses (Fig. 6B, C). Nevertheless, the observation that the over-expression of cadherin-11 increases glutamatergic synapse density provides additional evidence of a role for cadherin-11 in synapse development (see below).
Our initial analyses utilized synapsin I/PSD-95 co-localization as a measure of synapse density. The observed decreases in synapse density could reflect a bona fide decrease in glutamatergic synapse number or alternatively, a down-regulation or mis-localization of specific synaptic proteins such as synapsin I or PSD-95. To distinguish between these possibilities, we examined the number of AMPA receptor-containing synapses in neurons transfected with shRNAs targeting the cadherin-13 or cadherin-11 gene product. We used whole-cell voltage clamp recordings in the presence of tetrodotoxin and bicuculline to measure the frequency and amplitude of AMPA receptor-mediated mEPSC events. We transfected cells with shRNA constructs targeting cadherin-13 or cadherin-11 at 4DIV and performed whole-cell voltage clamp recordings at 14–15DIV. We found a significant decrease in the frequency of mEPSCs in neurons transfected with shRNAs targeting the cadherin-13 or cadherin-11 gene product (Fig. 4A, B). In addition, we detected a small but significant decrease in the amplitude of mEPSCs in the cadherin-13 and cadherin-11 shRNA-transfected cells (Fig. 4A, C, D). By contrast, transfection of shRNAs targeting another gene product, Sema4D, had no effect on mEPSC frequency or amplitude (Fig. 4). We also found that neurons transfected with shRNA constructs targeting cadherin-11, but not cadherin-13, had increased input resistance (Supplemental Table S2), possibly due to a decrease in cell soma size (data not shown). However, this data does not confound our analysis of mEPSC frequency as an increased input resistance should enhance our ability to detect mEPSC events.
We also determined the density of AMPA receptor-containing synapses by immunocytochemistry. Transfected neurons were stained with antibodies that recognize the synaptic vesicle associated protein Synaptotagmin I and the GluR2 subunit of the AMPA receptor, and synapse density was quantified as the overlap of Synaptotagmin I and GluR2 puncta on the dendrites of transfected neurons. Neurons transfected with shRNA constructs targeting cadherin-13 or cadherin-11, but not Sema4D, show a significant decrease in the density of AMPA receptor-containing synapses (Fig. 5), suggesting that the observed decrease in mEPSC frequency in cadherin-13 or cadherin-11 shRNA-transfected neurons is due, at least in part, to a decrease in the number of functional, AMPA receptor-containing synapses.
We sought to better understand the mechanism of action of cadherin-13 and cadherin-11 by determining whether they are required for the development of the pre-and/or postsynaptic specialization. For example, if cadherin-13 or cadherin-11 regulates synapse development via a homophilic adhesion event that stabilizes contact between the axon and the dendrite, cadherin knock down might be predicted to block multiple steps of synapse development, leading to a decrease in the number of both pre- and postsynaptic structures. Alternatively, if cadherin-13 or cadherin-11 functions exclusively to regulate the localization of molecules to the postsynaptic density, cadherin RNAi might be predicted to cause a reduction in the number of post- but not presynaptic structures.
To address this question, we quantified the density of synapsin I or PSD-95 puncta using the same confocal images that were used to assess synapse density as the overlap of synapsin I/PSD-95. We found that knockdown of cadherin-13 results in a significant decrease in the density of both synapsin I and PSD-95 puncta (Fig. 3E, F), suggesting that cadherin-13 acts at an early step in synapse development required for the formation or stabilization of both the pre- and postsynaptic specialization. Knockdown of cadherin-11 causes a small but significant decrease in the density of synapsin I puncta and a trend toward a decrease in the density of PSD-95 puncta (p=0.08; Fig. 3E, F), suggesting that cadherin-11 may be required for a trans-synaptic signaling event that preferentially initiates presynaptic assembly.
We next asked if cadherin-13 or cadherin-11 is also required for GABAergic synapse development. We transfected neurons with GFP and shRNAs targeting the cadherin-13 or cadherin-11 gene products at 4DIV, and at 14DIV, we stained the cells with antibodies that recognize the presynaptic vesicular GABA transporter VGAT and the β2/3 subunit of the GABAA receptor. We quantified the number of overlapping VGAT and GABAA receptor puncta on the transfected neurons and found that RNAi of cadherin-13 or cadherin-11 causes a decrease in the density of GABAergic synapses (Fig. 6A, B). In contrast, shRNA constructs targeting another gene product, APC5, had no effect on GABAergic synapse density (Fig. 6A, B). We also assessed GABAergic synapse density using antibodies recognizing the presynaptic GABA-synthesizing enzyme GAD67 and the γ2 subunit of the GABAA receptor and found that GABAergic synapse density was significantly decreased in neurons transfected with shRNAs targeting cadherin-13 or cadherin-11 (data not shown). These results indicate that cadherin-13 and cadherin-11 regulate both glutamatergic and GABAergic synapse development, possibly at a step common to the development of both types of synapses.
To gain further insight into the function of cadherin-13 and cadherin-11, we examined the subcellular localization of N-terminal myc-tagged cadherin-13 and cadherin-11 in cultured hippocampal neurons (Fig. 7A). Cadherin-13 is unique among the cadherin family in that it is linked to the cell membrane via a glycosylphosphatidylinositol (GPI) moiety, whereas cadherin-11 is a Type II transmembrane domain-containing cadherin (Nollet et al., 2000). We observed myc-cadherin-13 immunoreactivity in discrete puncta within and next to the transfected dendrite, while myc-cadherin-11 seemed to be contained within the dendritic shaft (Fig. 7A). Although the localization of the over-expressed protein may not precisely reflect the localization of the endogenous protein, these results suggest that cadherin-13 and cadherin-11 may have different mechanisms of action. For example, cadherin-13, in contrast to cadherin-11, may be cleaved from the cell surface, and its release from the membrane may have functional consequences for its role in synapse development (Baron and Caughey, 2003).
To better understand the role of cadherin-13 and cadherin-11 in synapse development, we also asked if either protein is able to promote synapse development when over-expressed in neurons. We analyzed the density of synapsin I/PSD-95 co-localized puncta along the dendrites of neurons transfected with plasmids expressing the myc-cadherin-13 or myc-cadherin-11 proteins. We found that ectopic expression of cadherin-11, but not cadherin-13, was sufficient to increase synapse number (Fig. 7B, C). This is in contrast to previous work that demonstrated that the over-expression of N-cadherin, a related cadherin family member, is not sufficient to increase synapse number (Graf et al., 2004; Scheiffele et al., 2000; Togashi et al., 2002).
Class 4 semaphorin family members regulate both glutamatergic and GABAergic synapse development
In addition to cadherin family members, our screen identified a role for Sema4B in glutamatergic synapse development (Figs. 2, 3). Having established that RNAi knockdown of Sema4B causes a decrease in the density of synapsin I/PSD-95 co-localized puncta (Figs. 2, 3), we asked if RNAi of Sema4B also affects the frequency and amplitude of AMPA receptor-mediated mEPSCs and/or the density of AMPA receptor-containing synapses. We found that neurons transfected with RNAi constructs targeting Sema4B exhibit a decrease in the frequency and amplitude of AMPA receptor-mediated mEPSCs (Fig. 4). In addition, we found that the density of AMPA receptor-containing synapses was decreased in neurons transfected with RNAi constructs targeting Sema4B and stained for synaptotagmin I and GluR2 (Fig. 5). Thus, by three independent assays, we have identified a role for Sema4B in glutamatergic synapse development.
We asked whether Sema4B is required for the development of the pre- and/or postsynaptic specialization. We found that knockdown of Sema4B causes a decrease in the density of PSD-95, but not synapsin I, puncta (Fig. 3E, F), suggesting that Sema4B is preferentially required for development of the postsynaptic specialization. This result is intriguing as Sema4B has been shown to bind to the postsynaptic scaffolding protein PSD-95 via a C-terminal PDZ binding motif and to localize to PSD-95-containing synapses in cultured hippocampal neurons (Burkhardt et al., 2005). Indeed, we find that an N-termimal myc-tagged Sema4B protein is widely expressed throughout the neuron and is present at glutamatergic synapses (Supplemental Fig. S5), consistent with a role for Sema4B in regulating the maturation of the postsynaptic density once synapse formation has begun.
In addition, we asked if Sema4B functions specifically to regulate glutamatergic synapse development or if it also regulates GABAergic synapse development. We assayed the density of GABAergic synapses on neurons transfected with shRNAs targeting Sema4B and found that knockdown of Sema4B causes a decrease in GABAergic synapse density (Fig. 6A, B). We found that RNAi of Sema4B significantly affects the density of GABAA receptor puncta while having a less dramatic effect on the density of VGAT puncta (Fig. 6E, F). This is similar to the effect of knockdown of Sema4B on glutamatergic synapse development, where the postsynaptic specialization is principally disrupted (Fig. 3E, F). Taken together, these findings suggest that Sema4B may play a role in assembling the postsynaptic specialization at both glutamatergic and GABAergic synapses.
During our analysis of Sema4B, we used RNAi constructs targeting the related Semaphorin Sema4D as a negative control. Knockdown of Sema4D had no effect on glutamatergic synapses as assessed by the co-localization of synapsin I/PSD-95 and synaptotagmin I/GluR2 or by the recording of AMPA-mediated mEPSCs (Figs. 3, 4, 5). Quite unexpectedly, we found that knockdown of Sema4D leads to a significant decrease in GABAergic synapse density as assessed by the co-localization of GAD65/GABAARβ2/3 (Fig. 6A, B) and GAD67/GABAARγ2 (data not shown and Fig. 6C, D). We verified the specificity of the effect of Sema4D knockdown on GABAergic synapse density (Fig. 6C, D) first by demonstrating that non-overlapping shRNA constructs targeting Sema4D, shRNA_1_Sema4D or shRNA_2_Sema4D, cause a decrease in synapse density (Fig. 6C, D). Second, we rescued the decrease in synapse density caused by Sema4D knockdown by co-expressing shRNA_2_Sema4D and a full-length Sema4D construct resistant to knockdown by shRNA_2_Sema4D (Supplemental Figure S2D). Expression of RNAi-resistant Sema4D by itself did not affect GABAergic synapse density (Fig. 6D); however, the expression of RNAi-resistant Sema4D was sufficient to rescue the decrease in GABAergic synapse density observed with transfection of shRNA_2_Sema4D (Fig. 6D). Taken together, these results indicate that the observed decrease in GABAergic synapse density is due to specific knockdown of Sema4D by the RNAi constructs.
We next asked whether Sema4D regulates the development of the GABAergic pre- and/or postsynaptic specialization. We found that RNAi of Sema4D significantly affects the density of GABAA receptor puncta while having a less dramatic effect on the density of VGAT puncta (Fig. 6E, F). This suggests that Sema4D exerts its effect on GABAergic synapse development primarily via the assembly of the postsynaptic specialization.
To gain further insight into the function of Sema4D in GABAergic synapse development, we examined its expression pattern and subcellular localization. Results from transcriptional profiling experiments revealed that Sema4D transcripts are expressed in our hippocampal cultures at the time that synapses are forming, and preliminary analysis demonstrated that Sema4D mRNA is present in the principal cell layers of the hippocampus at postnatal day 7 (P7) (data not shown), a finding corroborated by both the Brain Gene Expression Map and Allen Brain Atlas (Magdaleno et al., 2006; http://www.brain-map.org/). We also used antibody staining to examine the subcellular localization of a C-terminal myc-tagged Sema4D construct and found that myc-tagged Sema4D is expressed throughout the neuron (Supplemental Fig. S5B). Taken together, these data demonstrate that Sema4D is expressed in the principal cells of the hippocampus at the time that GABAergic synapses are forming.
We next asked if Sema4D regulates GABAergic synapse development in vivo by analyzing synapse development in a mouse carrying a targeted deletion of the Sema4D gene (Shi et al., 2000). We immunostained coronal sections from P20–23 Sema4D−/− and wild-type littermates with an antibody recognizing GAD67 (Fig. 8). We imaged eight distinct regions of the hippocampus in matched wild-type and Sema4D−/− sections (see Methods) and found a modest but reproducible decrease in the average intensity of GAD67 immunoreactivity in the Sema4D−/− animals compared to wild-type animals in seven of the eight regions examined (Fig. 8 and Supplemental Table S3). We confirmed that the total number of cells in the hippocampus was unchanged between Sema4D−/− and wild-type animals by counting nuclei stained with the DNA-binding dye Hoechst 33342 (Supplemental Table S3). This result suggests that the decrease in GAD67 immunoreactivity does not reflect an overall decrease in the number of cells in the hippocampus of the Sema4D−/− mice. This initial analysis of the Sema4D−/− mice is consistent with the possibility that Sema4D functions to regulate GABAergic synapse development in the intact nervous system.
Figure 8.
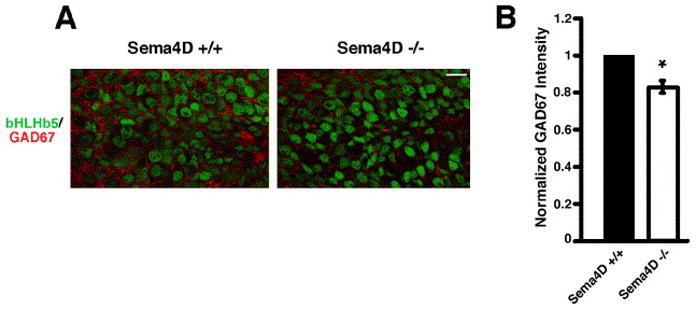
Sema4D −/− mice have reduced GAD67 immunoreactivity in the hippocampus. A) Immunostaining for bHLHb5 (green) and GAD67 (red) in the granule cell layer of the dentate gyrus for sections from Sema4D+/+ and Sema4D−/− littermates. Scale bar is 10 μm. B) Quantification of GAD67 staining intensity (normalized to bHLHb5 staining intensity) in the granule cell layer of the dentate gyrus for sections from Sema4D+/+ and Sema4D−/− littermates. n=42 for both conditions. Significance of p<0.0001 by multi-factorial ANOVA is indicated by asterisk.
The intracellular signaling molecule Rem2 is required for both glutamatergic and GABAergic synapse development
In addition to the identification of four cell surface proteins that regulate the development of glutamatergic and/or GABAergic synapses, our RNAi screen revealed a role for the cytoplasmic GTPase Rem2 in synapse development. Rem2 is a member of the RGK (Rem, Rad, and Gem/Kir) family of Ras-related small GTPases and has been shown to regulate calcium channel function and cytoskeletal rearrangements (Beguin et al., 2005; Chen et al., 2005; Finlin et al., 2005).
We found that transfection of diced siRNAs or a pool of three shRNAs targeting Rem2 causes a decrease in the density of synapsin I/PSD-95 puncta (Figs. 2B, C, 3A, B). Cells transfected with shRNA constructs targeting Rem2 also exhibit decreases in the frequency and amplitude of mEPSCs and in the density of AMPA receptor-containing synapses (Figs. 4, 5), suggesting that Rem2 plays a role in regulating glutamatergic synapse development. We sought further insight into the mechanism of action of Rem2 by asking whether it regulates the development of the pre- and/or postsynaptic specialization. Knockdown of Rem2 results in a significant decrease in the density of both synapsin I and PSD-95 puncta (Fig. 3E, F), suggesting that Rem2 acts at an early step in synapse development before the formation or stabilization of the pre- and postynaptic specialization. We also asked if Rem2 regulates GABAergic synapse development by assaying the density of GABAergic synapses on neurons transfected with RNAi constructs targeting Rem2. Knockdown of Rem2 led to a significant decrease in the density of GABAergic synapses (Fig. 6A, B), suggesting that Rem2 is a general synaptogenic factor that regulates both glutamatergic and GABAergic synapse development and/or maturation.
We next sought to gain insight into the mechanism by which Rem2 regulates synapse development. It has been shown the transcription of Rem2 is induced by extracellular stimuli (Finlin et al., 2005), and indeed, we found that the expression of Rem2 is induced several fold in response to the membrane depolarization of 5DIV cultured neurons (data not shown; confirmed by semi-quantitative RT-PCR; Supplemental Fig. S6A). This raises the possibility that Rem2 regulates synapse density as part of a feedback loop that controls calcium influx into the neuron. Finally, we examined the subcellular localization of an N-terminal myc-tagged Rem2 construct and found that myc-tagged Rem2 is expressed throughout the axon, cell body, and dendrites of hippocampal neurons (Supplemental Fig. S6B). The observation that Rem2 is localized throughout the neuron, taken together with the finding that Rem2 transcription is induced by membrane depolarization, suggests that Rem2 may act globally in response to neuronal activity to affect the ability of a neuron to develop synapses.
Discussion
In this study, we demonstrate the utility of an RNAi-based screen to identify molecules required for the development and/or maturation of synapses in the mammalian hippocampus. Interestingly, the molecules identified by our screen as mediators of glutamatergic synapse development also appear to play a role in GABAergic synapse development, suggesting that common mechanisms regulate the development of these distinct synapse subtypes. In contrast, we found that Sema4D regulates GABAergic, but not glutamatergic, synapse development, making Sema4D one of only a few proteins identified to date that regulates the development of GABAergic, but not glutamatergic, synapses. This finding also indicates that glutamatergic synapse development can occur in the absence of a normal density of GABAergic inputs and suggests that glutamatergic and GABAergic synapse development are, in part, controlled by distinct mechanisms.
Surprisingly, our screen did not identify proteins that control glutamatergic synapse development without also affecting GABAergic synapse development. A number of molecules have been shown to be required for both glutamatergic and GABAergic synapse development and/or function (Chih et al., 2005; Elmariah et al., 2004; Graf et al., 2004; Togashi et al., 2002; Varoqueaux et al., 2006; Weiner et al., 2005), suggesting that a core group of proteins may mediate cellular processes that are critical for the development of both types of synapses. For example, synapses may form promiscuously between neurons through the function of adhesion molecules such as cadherin-11, while the final identity of the postsynaptic specialization as either glutamatergic or GABAergic might be specified only after contact between the axon and the dendrite has been established. Alternatively, neurons lacking glutamatergic synapses might be less likely to form GABAergic synapses as a result of homeostatic mechanisms which maintain excitability within an appropriate physiological range (Turrigiano and Nelson, 2004). In this case, the molecules we have identified as regulators of glutamatergic synapse development might be indirectly involved in GABAergic synapse development.
Cadherin family members as mediators of synapse development
In addition to suggesting general conclusions about the mechanisms of glutamatergic and GABAergic synapse development, our RNAi screen has provided new evidence concerning the role of specific proteins in this process. Of the twenty-two cadherin family members assayed in our screen, only cadherin-13 and cadherin-11 were identified as playing a role in synapse development. This may be due to variability in the effectiveness of the RNAi-mediated knockdown of the various cadherin family members. Alternatively, a role for additional cadherin family members in synapse development may have been obscured by functional redundancy among related family members. This possibility is intriguing in that it suggests that cadherin-13 and cadherin-11 play non-redundant roles in synapse development.
It has been postulated that the expression of cadherins by distinct subsets of neurons constitutes a “code” which allows for the matching of the correct pre- and postsynaptic partners during synapse formation (Salinas and Price, 2005). However, studies to date have failed to identify which members of this family are important for the development of specific subsets of synapses. We have demonstrated that the loss of cadherin-11 causes a decrease in synapse density, while the over-expression of cadherin-11 promotes the development of glutamatergic synapses, suggesting that cadherin-11 may initiate contact between the axon and the dendrite. Cadherin-13 appears to function by a distinct mechanism, as it appears to be cleaved from the cell surface and, thus far, we have not been able to demonstrate that over-expression causes an increase in synapse density. The data presented here advances our understanding of the role of cadherin family members in synapse development by demonstrating that two family members, cadherin-13 and cadherin-11, are important regulators of synapse development that appear to function via distinct mechanisms.
Differing requirements for Sema4B and Sema4D in glutamatergic and GABAergic synapse formation
Based on the role of the EphB receptor tyrosine kinase in axonal pathfinding and synapse development, we hypothesized that other proteins implicated in neuronal pathfinding might also regulate synapse development. We assayed 41 putative membrane-bound pathfinding ligands and receptors in our screen and revealed a novel role for Sema4B and Sema4D in glutamatergic and/or GABAergic synapse development. The semaphorin family comprises over 30 members grouped into eight classes on the basis of sequence similarity and domain organization (He et al., 2002). Of the vertebrate semaphorins, the secreted class 3 semaphorins have been shown to serve as repulsive guidance cues in the nervous system; however, the role of the membrane-bound class 4–7 semaphorins in nervous system function is poorly understood.
Our data indicates that Sema4B preferentially regulates the development of the postsynaptic specialization at glutamatergic synapses. Sema4B contains extracellular Sema and Ig domains and a short intracellular C-terminal domain that includes a PDZ-binding motif (He et al., 2002). PDZ domain-containing proteins function as scaffolds to localize receptors and signaling molecules to glutamatergic synapses (McGee and Bredt, 2003). As such, Sema4B may play a role in assembling the postsynaptic specialization by recruiting scaffolding proteins to the postsynaptic density. In support of this model, Sema4B interacts with PSD-95 and is present at glutamatergic synapses (Burkhardt et al., 2005). In addition to our discovery of a role for Sema4B in synapse development, a class 3 semaphorin has recently been shown to modulate synaptic transmission in the mammalian hippocampus (Sahay et al., 2005), and a Drosophila semaphorin homolog has been shown to regulate synapse formation at the Giant Fiber synapse (Godenschwege et al., 2002). Taken together, these findings suggest that semaphorins may play a general role in regulating glutamatergic synapse development.
One of the most striking findings reported here is that Sema4D plays a role in GABAergic synapse development. It is possible that Sema4D functions to convert a stable but not yet specified synaptic contact into a GABAergic synapse, for example by recruitment of GABA receptors to the postsynaptic specialization. Although Sema4D is anchored to the cell surface via a transmembrane domain, it has been demonstrated in the immune system that the extracellular domain of Sema4D can be cleaved by a protease and released from the cell surface (Elhabazi et al., 2001). Thus, Sema4D may mediate its effects on GABAergic synapse development either as a membrane-bound molecule and/or in a non-cell autonomous manner as a cleaved protein. It is also possible that Sema4D affects GABAergic synapse development by engaging a receptor such as PlexinB1 (Tamagnone et al., 1999). Identifying the receptor that acts in concert with Sema4D has the potential to provide further insight into GABAergic synapse development.
The role of activity-regulated genes in synapse development
The importance of neuronal activity for synaptic refinement and maturation is well documented (Katz and Shatz, 1996), and a number of activity-regulated genes have been shown to play a role in synapse formation, function, and/or elimination (Flavell et al., 2006; Pak and Sheng, 2003; Sala et al., 2003; Shalizi et al., 2006; Steward and Worley, 2001; West et al., 2001). To better understand how activity-regulated genes function to control synapse development, we asked whether the activity-regulated genes we identified by transcriptional profiling were also required for synapse development.
Our screen revealed a novel role for the activity-regulated small GTPase Rem2 in the development of glutamatergic and GABAergic synapses. Previous work demonstrated a role for Rem2 in the regulation of calcium currents in neurons (Chen et al., 2005). This observation, taken together with our finding that Rem2 regulates synapse development, suggests that a connection may exist between the role of Rem2 in calcium homeostasis and synapse development. In support of this hypothesis, manipulation of the activity level of the postsynaptic neuron is known to affect the ability of the neuron to form and/or maintain synapses (Burrone et al., 2002; Pratt et al., 2003). Our success in identifying a role for the activity-regulated gene Rem2 in synapse development suggests that further study of activity-regulated genes and of Rem2 will provide new insight into how a neuron transforms membrane depolarization into a change in gene expression to regulate synapse formation and/or function.
A new approach for gene discovery in mammalian neurons
To date, technical challenges have hindered the large-scale identification of molecules that are required for synapse development in the mammalian CNS. Thus, a large-scale, unbiased strategy to identify new molecules that are important for synapse formation and/or maintenance has the potential to greatly advance our understanding of this process. Our screen demonstrates the feasibility of using RNAi in mammalian neurons to identify new molecules that function in CNS development. The success of our screen suggests that further screening would yield new insight into the genetic program that regulates both glutamatergic and GABAergic synapse development. We believe that the current screen, with modifications to the phenotypic assay, could also be used for the identification of genes required for processes such as dendritic and axonal outgrowth and activity-regulated gene transcription. The challenge for future approaches to RNAi screening in mammalian neurons will be to increase throughput using technologies that enable automated assessment of complex neuronal phenotypes.
Experimental Procedures
Screen Design
Activity- or developmentally-regulated genes were identified using Affymetrix arrays as described in the Supplemental Data. The screen was conducted such that the average density of co-localized synapsin I/PSD-95 puncta was compared between experimental and control conditions. A pool was considered “positive” if it was significantly different (p<0.05) from control by a two-sample t-test. Positive pools were re-screened, and those that were positive by the above criteria in two or more independent experiments were analyzed further. See the Supplemental Data for further details.
RNAi constructs
Recombinant Dicer enzyme was used to generate diced siRNAs as described in the Supplemental Data. shRNAs expressed from the pSuper vector targeting specific genes were generated as described in the Supplemental Data.
Cell culture and transfection
E18 hippocampal neurons were grown at low density on a glial monolayer and transfected at 4DIV by the calcium phosphate method. For a detailed description of neuronal cell culture and transfection conditions, see the Supplemental Data. HEK 293T cells were transfected by the calcium phosphate method and protein knockdown was assessed by Western blotting as described in the Supplemental Data.
Electrophysiology
mEPSC amplitude and frequency were measured by whole-cell voltage clamp recording as described in the Supplemental Data.
Immunocytochemistry
Cultured hippocampal neurons were fixed and stained for the myc antigen and various synaptic markers as described in the Supplemental Data. Images were acquired on a Zeiss LSM510 or Zeiss LSM5 Pascal confocal microscope and analyzed using OpenLab (Improvision) or Metamorph (Molecular Devices) image analysis software. For a detailed description of image acquisition and analysis, see the Supplemental Data.
Analysis of GAD67 staining in hippocampus
Hippocampal sections from P20–P23 wild-type and Sema4D−/− littermates were stained with anti-GAD67 antibody and imaged on a Zeiss LSM5 Pascal confocal microscope. The intensity of immunoreactivity was analyzed using Metamorph image analysis software. For a detailed description of the immunohistochemistry and image acquisition and analysis, see the Supplemental Data.
Data normalization and statistics
For a detailed description of the statistical treatment of the data, see the Supplemental Data.
Supplementary Material
Acknowledgments
We thank J. Myers for recombinant Dicer virus, M. Sheng and A. Dunah for NR2B antibodies, A. Kumanogoh and H. Kikutani for Sema4D reagents, S. Ross for bHLHb5 antibodies, H. Feldman and the Clinical Research Program at Children’s Hospital for help with statistical analysis, and J. Kustan, M. Salanga and the MRRC imaging core for assistance with confocal microscopy. We thank J. Kaplan, Z. Gitai and members of the Greenberg lab for help with screen design and S. Cohen, E. Hong, L. Jackson-Grusby, Z. Wills, and S. Ross for critical reading of the manuscript. This work was supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship (S.P.), The Charles H. Hood Foundation, Inc. Boston, MA (S.P.), a Helen Hay Whitney Foundation Postdoctoral Fellowship (E.C.G.), NIH HL81012 (L.F.B.), a Mental Retardation Developmental Disabilities Research Center grant HD18655 (M.E.G.) and National Institutes of Health grant NS45500 (M.E.G.). M.E.G. acknowledges the generous support of the F.M. Kirby Foundation to the Children’s Hospital Neurobiology Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson TR, Shah PA, Benson DL. Maturation of glutamatergic and GABAergic synapse composition in hippocampal neurons. Neuropharmacology. 2004;47:694–705. doi: 10.1016/j.neuropharm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Baron GS, Caughey B. Effect of glycosylphosphatidylinositol anchor-dependent and -independent prion protein association with model raft membranes on conversion to the protease-resistant isoform. J Biol Chem. 2003;278:14883–14892. doi: 10.1074/jbc.M210840200. [DOI] [PubMed] [Google Scholar]
- Beguin P, Mahalakshmi RN, Nagashima K, Cher DH, Kuwamura N, Yamada Y, Seino Y, Hunziker W. Roles of 14-3-3 and calmodulin binding in subcellular localization and function of the small G-protein Rem2. Biochem J. 2005;390:67–75. doi: 10.1042/BJ20050414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Burkhardt C, Muller M, Badde A, Garner CC, Gundelfinger ED, Puschel AW. Semaphorin 4B interacts with the post-synaptic density protein PSD-95/SAP90 and is recruited to synapses through a C-terminal PDZ-binding motif. FEBS Lett. 2005;579:3821–3828. doi: 10.1016/j.febslet.2005.05.079. [DOI] [PubMed] [Google Scholar]
- Burrone J, O’Byrne M, Murthy VN. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature. 2002;420:414–418. doi: 10.1038/nature01242. [DOI] [PubMed] [Google Scholar]
- Chen H, Puhl HL, 3rd, Niu Mitchell SLDC, Ikeda SR. Expression of Rem2, an RGK family small GTPase, reduces N-type calcium current without affecting channel surface density. J Neurosci. 2005;25:9762–9772. doi: 10.1523/JNEUROSCI.3111-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Elhabazi A, Delaire S, Bensussan A, Boumsell L, Bismuth G. Biological activity of soluble CD100. I. The extracellular region of CD100 is released from the surface of T lymphocytes by regulated proteolysis. J Immunol. 2001;166:4341–4347. doi: 10.4049/jimmunol.166.7.4341. [DOI] [PubMed] [Google Scholar]
- Elmariah SB, Crumling MA, Parsons TD, Balice-Gordon RJ. Postsynaptic TrkB-mediated signaling modulates excitatory and inhibitory neurotransmitter receptor clustering at hippocampal synapses. J Neurosci. 2004;24:2380–2393. doi: 10.1523/JNEUROSCI.4112-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Finlin BS, Mosley AL, Crump SM, Correll RN, Ozcan S, Satin J, Andres DA. Regulation of L-type Ca2+ channel activity and insulin secretion by the Rem2 GTPase. J Biol Chem. 2005;280:41864–41871. doi: 10.1074/jbc.M414261200. [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Brunig I. Formation and plasticity of GABAergic synapses: physiological mechanisms and pathophysiological implications. Pharmacol Ther. 2003;98:299–323. doi: 10.1016/s0163-7258(03)00037-8. [DOI] [PubMed] [Google Scholar]
- Godenschwege TA, Hu H, Shan-Crofts X, Goodman CS, Murphey RK. Bi-directional signaling by Semaphorin 1a during central synapse formation in Drosophila. Nat Neurosci. 2002;5:1294–1301. doi: 10.1038/nn976. [DOI] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang KC, Koprivica V, Ming G, Song HJ. Knowing how to navigate: mechanisms of semaphorin signaling in the nervous system. Sci STKE. 2002;119:RE1. doi: 10.1126/stke.2002.119.re1. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Li Z, Sheng M. Some assembly required: the development of neuronal synapses. Nat Rev Mol Cell Biol. 2003;4:833–841. doi: 10.1038/nrm1242. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe T, Togashi H, Uchida N, Suzuki SC, Hayakawa Y, Yamamoto M, Yoda H, Miyakawa T, Takeichi M, Chisaka O. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol Cell Neurosci. 2000;15:534–546. doi: 10.1006/mcne.2000.0849. [DOI] [PubMed] [Google Scholar]
- McGee AW, Bredt DS. Assembly and plasticity of the glutamatergic postsynaptic specialization. Curr Opin Neurobiol. 2003;13:111–118. doi: 10.1016/s0959-4388(03)00008-4. [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- Nusser Z, Sieghart W, Somogyi P. Segregation of different GABAA receptors to synaptic and extrasynaptic membranes of cerebellar granule cells. J Neurosci. 1998;18:1693–1703. doi: 10.1523/JNEUROSCI.18-05-01693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Pratt KG, Watt AJ, Griffith LC, Nelson SB, Turrigiano GG. Activity-dependent remodeling of presynaptic inputs by postsynaptic expression of activated CaMKII. Neuron. 2003;39:269–281. doi: 10.1016/s0896-6273(03)00422-7. [DOI] [PubMed] [Google Scholar]
- Rao A, Cha EM, Craig AM. Mismatched appositions of presynaptic and postsynaptic components in isolated hippocampal neurons. J Neurosci. 2000;20:8344–8353. doi: 10.1523/JNEUROSCI.20-22-08344.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Kim E, Sheng M, Craig AM. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J Neurosci. 1998;18:1217–1229. doi: 10.1523/JNEUROSCI.18-04-01217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay A, Kim CH, Sepkuty JP, Cho E, Huganir RL, Ginty DD, Kolodkin AL. Secreted semaphorins modulate synaptic transmission in the adult hippocampus. J Neurosci. 2005;25:3613–3620. doi: 10.1523/JNEUROSCI.5255-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci. 2003;23:6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas PC, Price SR. Cadherins and catenins in synapse development. Curr Opin Neurobiol. 2005;15:73–80. doi: 10.1016/j.conb.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shi W, Kumanogoh A, Watanabe C, Uchida J, Wang X, Yasui T, Yukawa K, Ikawa M, Okabe M, Parnes JR, et al. The class IV semaphorin CD100 plays nonredundant roles in the immune system: defective B and T cell activation in CD100-deficient mice. Immunity. 2000;13:633–642. doi: 10.1016/s1074-7613(00)00063-7. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc Natl Acad Sci U S A. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Garner CC. Cellular and molecular mechanisms of presynaptic assembly. Nat Rev Neurosci. 2004;5:385–399. doi: 10.1038/nrn1370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


